Abstract
Mitotic cyclin-dependent kinases (CDKs) control entry into mitosis, but their role during mitotic progression is less well understood. Here we characterize the functions of CDK activity associated with the mitotic cyclins Clb1, Clb2, and Clb3. We show that Clb–CDKs are important for the activation of the ubiquitin ligase Anaphase-Promoting Complex/Cyclosome (APC/C)–Cdc20 that triggers the metaphase–anaphase transition. Furthermore, we define an essential role for Clb–CDK activity in anaphase spindle elongation. Thus, mitotic CDKs serve not only to initiate M phase, but are also needed continuously throughout mitosis to trigger key mitotic events such as APC/C activation and anaphase spindle elongation.
Keywords: APC/C, CDK, Cdc20, spindle elongation, mitosis
The purpose of mitosis is the faithful partitioning of the duplicated genome between two daughter cells. Chromosome segregation occurs during anaphase and is triggered by the dissolution of the linkages that hold sister chromatids together (Nasmyth 2002). These linkages are mediated by cohesin complexes, which in budding yeast are composed of Scc1/Mcd1, Scc3, Smc1, and Smc3 (Nasmyth 2002). At the onset of anaphase, the cohesin subunit Scc1/Mcd1 is cleaved by the protease Separase (Esp1 in budding yeast), leading to the loss of cohesion between sister chromatids and subsequent chromosome segregation (Nasmyth 2002). Prior to anaphase, Separase/Esp1 is held inactive by the binding of Securin (Pds1 in budding yeast) (Nasmyth 2002). At the metaphase–anaphase transition, Securin/Pds1 is targeted for proteosomal degradation by an ubiquitin ligase known as the Anaphase-Promoting Complex/Cyclosome (APC/C) and its specificity factor Cdc20 (APC/C–Cdc20) (Peters 2006).
APC/C–Cdc20 is regulated at multiple levels. Surveillance mechanisms such as the DNA damage and spindle assembly checkpoints negatively regulate APC/C–Cdc20 activity. In budding yeast, the DNA damage checkpoint arrests the cell cycle in metaphase by stabilizing Securin/Pds1 (Cohen-Fix and Koshland 1997; Sanchez et al. 1999; Tinker-Kulberg and Morgan 1999; Wang et al. 2001; Agarwal et al. 2003; Searle et al. 2004). In response to unattached kinetochores, the spindle assembly checkpoint prevents the metaphase–anaphase transition by inhibiting the interaction between Cdc20 and APC/C (Musacchio and Salmon 2007). However, these two checkpoints are not the only mechanisms that restrict APC/C–Cdc20 activity to the metaphase–anaphase transition because their combined inactivation does not cause precocious anaphase entry (Lee et al. 2002). APC/C–Cdc20 phosphorylation by cyclin-dependent kinases (CDKs) (and other kinases) is believed to activate the ubiquitin ligase at the metaphase–anaphase transition. However, the data supporting this conclusion are largely derived from in vitro studies (Peters 2006). The only in vivo evidence implicating CDKs in the regulation of the APC/C comes from studies in the budding yeast. Compromising mitotic CDK activity led to defects in Securin/Pds1 degradation, and mutation of the CDK phosphorylation sites within the budding yeast APC/C subunits Cdc16, Cdc23, and Cdc27 to alanines causes a 15-min delay in metaphase (Rudner and Murray 2000). Whether the subtlety of this phenotype is due to additional CDK phosphorylation sites on other APC/C components or whether CDK’s contribution to APC/C–Cdc20 activation is minor is unclear.
Mitotic CDKs are best known for their role in triggering entry into mitosis (Miele 2004). They are composed of a catalytic CDK subunit and a regulatory B-type cyclin subunit. Mitotic CDKs promote nuclear envelope breakdown, spindle assembly and organization, chromosome condensation, and Golgi fragmentation, and contribute to APC/C regulation (Nigg 2001; Miele 2004). Budding yeast contains a single CDK subunit, CDC28, and six B-type cyclins, CLB1–6, that have overlapping functions in S phase and mitosis (Andrews and Measday 1998). Clb1, Clb2, Clb3, and Clb4 together are required for entry into mitosis (Fitch et al. 1992; Richardson et al. 1992; Amon et al. 1993), which in yeast is defined by the severing of a bridge connecting the two duplicated spindle pole bodies (SPBs) (yeast centrosomes) and the formation of a bipolar spindle. Clb1 and Clb2 are also required for progression through mitosis subsequent to entry into mitosis (Surana et al. 1991; Fitch et al. 1992; Richardson et al. 1992). The combined inactivation of CLB1 and CLB2 arrests cells with a single undivided nucleus and a short bipolar spindle (Surana et al. 1991; Fitch et al. 1992; Richardson et al. 1992). The molecular basis for this arrest, however, was unknown.
Here we characterize the consequences of losing Clb1/2–CDK or Clb2/3–CDK activity on progression through mitosis. We find that Clb1/2–CDK activity is required for the timely degradation of Securin/Pds1 at the metaphase–anaphase transition, providing evidence for a requirement for CDK activity in APC/C–Cdc20 activation in vivo. In addition, our studies reveal a new role for Clb1/2–CDK activity in promoting the metaphase–anaphase transition after cohesin cleavage. The observation that cleavage of cohesin is sufficient to trigger anaphase spindle elongation (Uhlmann et al. 2000) led to the notion that anaphase spindle elongation was merely a consequence of loss of sister chromatid cohesion. We find that this is not the case, but that Clb1/2–CDK activity is required for anaphase spindle elongation to occur. Finally, we show that the combined inactivation of Clb2 and Clb3–CDK activity also leads to defects in Securin/Pds1 degradation and spindle elongation, suggesting that overall levels of Clb–CDK activity are important for these two mitotic events to occur. Our work indicates that mitotic CDKs not only initiate entry into M phase, they are needed at multiple steps throughout mitosis for successful completion of the key events in this cell cycle stage.
Results
Nuclear division is impaired in cells lacking Clb1/2–CDK activity
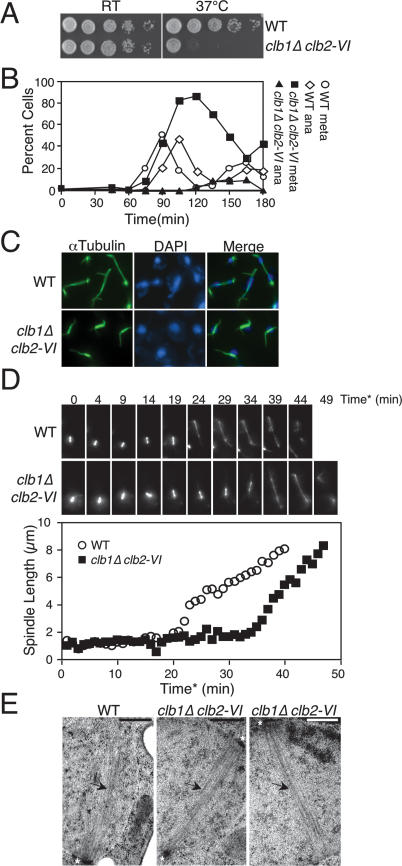
To investigate the role of Clb1–CDK and Clb2–CDK complexes in mitotic progression, we examined the phenotype of cells devoid of these cyclins by combining a temperature-sensitive allele of CLB2, clb2-VI (Amon et al. 1993), with a deletion of CLB1, henceforth clb1Δ clb2-VI. This allele combination is recessive (Supplemental Fig. 1A) and, at the permissive temperature (25°C), clb1Δ clb2-VI cells are viable and progress through the cell cycle with wild-type kinetics (Fig. 1A; Supplemental Fig. 1B,C). At temperatures between 36.5°C and 37.5°C, however, the viability of clb1Δ clb2-VI cells is greatly reduced (Fig. 1A). The clb2-VI allele, which contains four amino acid substitutions (D232G, L286S, K353R, and D485G), is not protein-null at the restrictive temperature (Supplemental Fig. 1D), indicating that Clb2–CDK kinase complexes are inactive in these cells.
Figure 1.
CLB1 and CLB2 are required for progression through mitosis. (A) Tenfold serial dilution of wild-type (A1411) and clb1Δ clb2-VI cells (A3000), each carrying a Cdc14-3HA fusion on YEPD plates. Plates were incubated at room temperature (RT) or 37°C for 3 d. (B) Wild-type cells carrying a Cdc14-3HA fusion (A1411) and clb1Δ clb2-VI cells carrying Pds1-13Myc and Cdc14-3HA fusions (A12159) were arrested in G1 in YEPD with α-factor (5 μg/mL) for 2.5 h at room temperature. Cells were washed with 10 vol YEP and released into pheromone-free YEPD media prewarmed to 37°C. The percentage of cells in metaphase and anaphase was determined at the indicated time points. At least 100 cells were counted at each time point. (C) Spindle morphology of wild-type (A1411) and clb1Δ clb2-VI (A3000) cells, each carrying a Cdc14-3HA fusion 120 min after release from a pheromone-induced G1 arrest. Microtubules are shown in green, and DNA is shown in blue. (D) Time-lapse series of wild-type (A16772) and clb1Δ clb2-VI (A17122) cells carrying a Tub1-GFP fusion. Cells were prepared as described in the Materials and Methods. For time-lapse series, eight 0.5-μm Z-stacks were collected every minute and the Z-stacks were then projected in the XY plane for spindle length measurements. Spindle length measurements were performed using softWoRx software. Time* corresponds to time after SPB separation, with the 0 time point defined as the first time point at which two separated SPBs are clearly resolved. (Top panel) A time-lapse series of a wild-type cell undergoing anaphase (shown in Supplemental Movie 1). This cell was chosen because it initiated anaphase at a time similar to the wild-type population’s mean anaphase onset time. (Bottom panel) A time-lapse series of a clb1Δ clb2-VI cell undergoing anaphase (shown in Supplemental Movie 2). This cell was chosen because it initiated anaphase at a time similar to the clb1Δ clb2-VI population’s mean anaphase onset time. (Graph) The distance between the two separated SPBs in each cell was measured at every time point in which the spindle was in focus. (E) Electron micrographs of a section of yeast cells prepared by high-pressure freezing and freeze substitution (see Material and Methods). The SPBs are marked by asterisks and microtubules are indicated by arrows. The bar in each image represents 500 nm. (Left panel) Section from a wild-type cell undergoing nuclear division at 37°C. Only one SPB was visible in this section and one-half spindle was clearly resolved. (Middle panel) Spindle from a clb1Δ clb2-VI cell at 37°C. The top SPB was clearly captured in this section whereas segments of the bottom SPB are just visible. The microtubules nucleated by both SPBs are clearly visible and a bipolar spindle is evident. (Right panel) A second representative spindle from a clb1Δ clb2-VI cell at 37°C. The bipolar spindle emanating from two separated SPBs is clearly visible (although only one SPB was contained in this section).
To identify the essential function(s) of Clb1/2–CDKs, we examined the phenotype of clb1Δ clb2-VI cells undergoing a synchronous cell cycle at 37°C. Wild-type and clb1Δ clb2-VI cells were arrested in G1 with α-factor pheromone (at room temperature) and then released into the cell cycle at 37°C. Wild-type cells progressed through metaphase 90 min after release, proceeded into anaphase by 105 min, and exited mitosis 120 min after release from the G1 block (Fig. 1B). Consistent with previous studies (Surana et al. 1991; Fitch et al. 1992; Richardson et al. 1992), we find that cells in clb1Δ clb2-VI cultures accumulated with short bipolar spindles and undivided nuclei (Fig. 1B,C). Live-cell microscopy confirmed this result. During live-cell imaging, we defined anaphase onset as the time point after which elongation of metaphase spindles (at least 2 μm long) was observed in at least four consecutive time points. Using this criterion, wild-type cells entered anaphase 24.3 min after SPB separation (SD = 14.1; n = 8) (an example is shown in Fig. 1D; Supplemental Movie 1). In clb1Δ clb2-VI cells, anaphase entry did not occur until 47.4 min after SPB separation (SD = 16.4; n = 8) (an example is shown in Fig. 1D; Supplemental Movie 2). This difference was highly significant (P = 0.009; two-sided Student’s t-test).
Analysis of fixed cells using indirect in situ immunofluorescence indicated that only a few clb1Δ clb2-VI cells entered anaphase (Fig. 1B). Instead, spindle disassembly occurred at later time points, indicating that complete CDK inactivation and hence exit from mitosis eventually occur in clb1Δ clb2-VI cells. It is however important to note that exit from mitosis is substantially delayed in clb1Δ clb2-VI cells because they undergo spindle disassembly at least 45 min after wild-type cells (Fig. 1B). In the live-cell analysis, clb1Δ clb2-VI cells behaved slightly differently. After a delay, most cells elongated their spindles and completed anaphase (Fig. 1D; Supplemental Movie 2). The basis for this difference in behavior is at present unknown. It is nevertheless clear that, under both analysis conditions, the onset of anaphase spindle elongation was impaired in clb1Δ clb2-VI cells.
Ultrastructural analysis of the short bipolar spindles observed in clb1Δ clb2-VI cells revealed that they resembled those of wild-type cells (Fig. 1E; see Winey et al. 1995, for a picture of a short wild-type bipolar spindle in which both SPBs are visible). We conclude that the bipolar spindles formed by clb1Δ clb2-VI cells are normal in appearance, at least at the electron microscopic level, and that CLB1 and CLB2 are required for nuclear division.
The mitotic progression defect of clb1Δ clb2-VI cells is independent of inhibitory Tyr19 phosphorylation on Cdc28
In budding yeast, spindle formation occurs during S phase. It is thus not possible to know whether a cell cycle arrest/delay characterized by short bipolar spindles reflects a G2 or metaphase defect. G2 delays can be caused by phosphorylation of Cdc28 on Tyr19, which is catalyzed by the Saccharomyces cerevisiae ortholog of the Wee1 kinase, Swe1 (Mendenhall and Hodge 1998). Swe1 has been shown to block cells in G2 in response to activation of the morphogenesis checkpoint (for review, see Keaton and Lew 2006). We inactivated SWE1 or utilized a CDC28 allele resistant to Tyr19 phosphorylation, cdc28Y19F, to test whether phosphorylation of Tyr19 could account for the cell cycle delay of clb1Δ clb2-VI cells. Inactivation of SWE1 did not affect cell cycle progression of clb1Δ clb2-VI cells (Fig. 2A). Identical results were obtained with the cdc28Y19F allele (Supplemental Fig. 2). Therefore, the accumulation of cells with short bipolar spindles in clb1Δ clb2-VI cultures cannot be attributed to persistent Y19 phosphorylation.
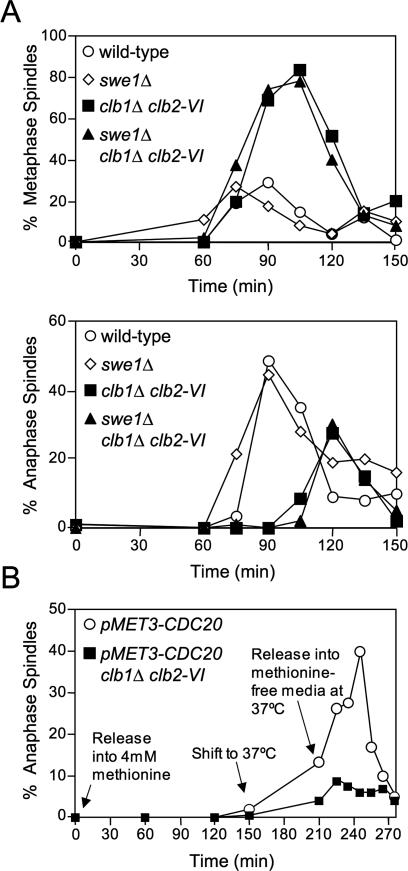
Figure 2.
Clb1/2–CDK activity is required at the metaphase–anaphase transition to bring about anaphase entry. (A) Wild-type (A1411), swe1Δ (A7507), clb1Δ clb2-VI (A3000), and swe1Δ clb1Δ clb2-VI (A15936) cells, each carrying Cdc14-3HA, were grown and analyzed as described in Figure 1B. (Top panel) Percentage of cells with metaphase spindles. (Bottom panel) Percentage of cells with anaphase spindles. (B) pMET3-CDC20 (A7334) and pMET3-CDC20 clb1Δ clb2-VI Pds1-13Myc (A15112) cells were grown in methionine-free complete synthetic media (CSM) and arrested in G1 with α-factor (5 μg/mL) for 2.5 h at room temperature. The cells were washed with 10 vol CSM and released at room temperature into α-factor-free CSM containing 4 mM methionine to arrest cells in metaphase. After 2.5 h in methionine, cells were shifted to 37°C for 1 h to inactivate Clb2-VI–CDK activity in metaphase. Cells were then released into methionine-free CSM at 37°C to restore CDC20 expression while maintaining the inactivation of Clb2-VI–CDK. Fifteen percent of cells enter anaphase in the presence of methionine upon temperature shift to 37°C. We believe this is due to a temperature-induced transient derepression of the MET3 promoter.
Clb1/2–CDK activity is required during metaphase to promote anaphase
Our experiments suggest that Clb1/2–CDK activity is needed at a point between the G1/S transition and metaphase for anaphase spindle elongation. To determine if Clb1/2–CDK complexes were required during metaphase to bring about anaphase entry, we examined the effect of inactivating these CDK complexes during metaphase. Cells can be reversibly arrested in metaphase by placing CDC20 under the control of the methionine-repressible MET3 promoter (Uhlmann et al. 2000). We arrested pMET3-CDC20 and pMET3-CDC20 clb1Δ clb2-VI cells in metaphase by the addition of methionine and then inactivated Clb2-VI–CDKs by shifting cells to 37°C. When >80% of cells were arrested in metaphase, they were released into methionine-free medium at 37°C to maintain Clb2-VI inactivation. Upon methionine removal, CDC20 expression was rapidly restored and pMET3-CDC20 cells progressed into anaphase (Fig. 2B). In contrast, entry into anaphase did not occur in pMET3-CDC20 clb1Δ clb2-VI cells (Fig. 2B). Thus, Clb1/2–CDK activity is needed during metaphase to bring about anaphase entry.
Clb1/2–CDK activity is needed for Securin degradation
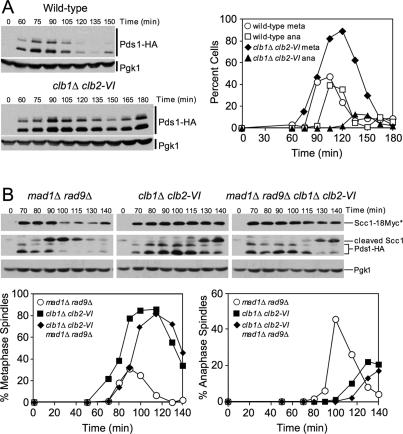
Securin/Pds1 degradation marks anaphase entry. To determine whether Clb1/2–CDK activity was required for Securin/Pds1 degradation, we examined the levels of Securin/Pds1 in clb1Δ clb2-VI cells. Following the release from a G1 block at 37°C, wild-type cells initiated Securin/Pds1 degradation and anaphase entry 105 min after release from the arrest (Fig. 3A). clb1Δ clb2-VI cells, however, were substantially delayed in Securin/Pds1 degradation (Fig. 3A), indicating that Clb1/2–CDK activity is required for Securin/Pds1 degradation.
Figure 3.
clb1Δ clb2-VI cells are defective in Securin degradation. (A) Wild-type cells carrying Pds1-HA (A1015) and clb1Δ clb2-VI cells carrying Pds1-HA and Scc1-18Myc (A15111) were grown and treated as in Figure 1B. The percentage of cells with metaphase and anaphase spindles (graph) was determined and Pds1-HA protein levels were examined by Western blot analysis. Pgk1 was used as a loading control in Western blots. (B) mad1Δ rad9Δ (A15999), clb1Δ clb2-VI (A15111), and mad1Δ rad9Δ clb1Δ clb2-VI (A16000) cells, each carrying Pds1-HA and Scc1-18Myc, were grown and treated as in Figure 1B. The percentage of cells with metaphase (left graph) and anaphase (right graph) spindles, and the amounts of full-length Scc1-18Myc (Scc1*), the C-terminal cleavage fragment of Scc1-18Myc (cleaved Scc1), and Pds1-HA were examined. Pgk1 was used as a loading control in Western blots.
Two surveillance mechanisms, the DNA damage and spindle assembly checkpoints, are known to prevent the APC/C–Cdc20-mediated degradation of Securin/Pds1. To examine whether the stabilization of Securin/Pds1 in clb1Δ clb2-VI cells was caused by the activation of either checkpoint, we examined the effects of deleting components of both surveillance mechanisms on Securin/Pds1 degradation and cell cycle progression in clb1Δ clb2-VI cells. RAD9 and MAD1 were deleted in order to inactivate the DNA damage and spindle assembly checkpoint, respectively. In mad1Δ rad9Δ cells, the degradation of Securin/Pds1 at 90 min coincided with the formation of the cleavage product of Scc1/Mcd1 and the appearance of anaphase spindles (Fig. 3B). In clb1Δ clb2-VI strains, Securin/Pds1 degradation, the formation of the Scc1/Mcd1 cleavage product, and the appearance of anaphase spindles were severely delayed (Fig. 3B). Securin/Pds1 degradation was similarly delayed in mad1Δ rad9Δ clb1Δ clb2-VI (Fig. 3B). Importantly, the delay in anaphase spindle elongation paralleled that of Scc1/Mcd1 cleavage, demonstrating that spindle morphology accurately reflects cell cycle progression of clb1Δ clb2-VI cells.
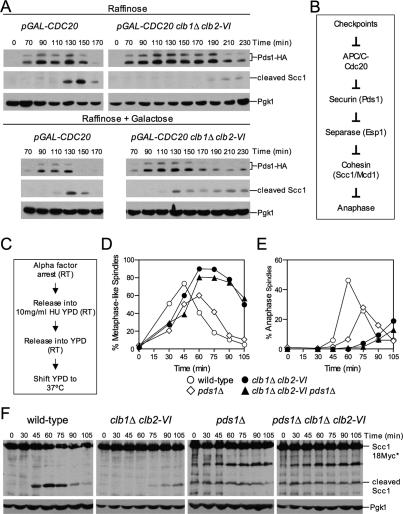
Our data indicate that cells lacking Clb1/2–CDK activity are defective in targeting Securin/Pds1 for degradation and thus are unable to fully activate Separase. This defect appears to be independent of the checkpoints known to control this event and thus may reflect a direct role for Clb1/2–CDK activity in activating APC/C–Cdc20. To test this idea, we examined the effects of high levels of Cdc20 on Securin/Pds1 degradation and cohesin cleavage in clb1Δ clb2-VI cells. Overexpression of CDC20 from the GAL1–10 promoter partially rescued the delay in Securin/Pds1 degradation and cohesin cleavage in clb1Δ clb2-VI cells. Under conditions when the GAL1–10 promoter is inactive (Raffinose), pGAL-CDC20 cells initiated cohesin cleavage at least 60 min prior to pGAL-CDC20 clb1Δ clb2-VI cells (Fig. 4A). In contrast, when Cdc20 was overproduced (Raffinose + Galactose), the delay in Securin/Pds1 degradation and cohesin cleavage was less pronounced (Fig. 4A). This observation provides further evidence that the APC/C is hypoactive in the absence of Clb1/2–CDK activity.
Figure 4.
Clb1/2–CDK activity governs anaphase entry in multiple ways. (A) Wild-type (A16651) and clb1Δ clb2-VI (A16580) strains, each carrying Pds1-HA, Scc1-18Myc, and three copies of pGAL-CDC20 at the URA3 locus, were arrested with α-factor (5 μg/mL) in YEP medium containing 2% Raffinose (YEPR) for 2.5 h at room temperature. Subsequently, cells were washed with 10 vol YEP and released into prewarmed YEPR at 37°C. The cultures were immediately split in two, and galactose (2%) was added to one of the two cultures. Protein levels of Scc1-18Myc and Pds1-HA were monitored during the cell cycle. Pgk1 was used as a loading control. The blot for Pds1-HA in Raffinose media is from a 2.5-min exposed film, while the Pds1-HA blot in galactose media is from an overnight exposure. The overnight exposure was required because Pds1-HA levels are very low in strains that overexpress CDC20. All Scc1-18Myc blots are from overnight exposures. All Pgk1 blots are from 2.5-min exposures. (B) A schematic diagram of the pathway regulating the metaphase–anaphase transition. (C–F) Wild-type (A3160), pds1Δ (A2015), clb1Δ clb2-VI Pds1-HA (A15111), and clb1Δ clb2-VI pds1Δ (A15840), each carrying a Scc1-18Myc fusion, were arrested in G1 as described in Figure 1B. Cells were then released into YEPD containing 10 mg/mL HU to arrest cells in S phase at room temperature (RT). This step is necessary because pds1Δ cells have a temperature-sensitive defect at the G1/S transition that is likely due to impaired Separase/Esp1 accumulation in the nucleus at 37°C (Yamamoto et al. 1996a, b; Agarwal and Cohen-Fix 2002). After 75 min in HU, cells were washed extensively and resuspended in HU-free YEPD at room temperature. The flasks were then placed in a 30°C water bath and the water bath was then set to 37°C. We found that this step-wise temperature shift produced the most synchronous release from the HU block into 37°C YEPD. The 0-min time point was taken before cells were released from the HU block. (D) Percentage of cells with metaphase-like spindles. Spindles were classified as metaphase-like because metaphase, as defined by 2N cells with a bipolar spindle containing Securin/Pds1 and noncleaved cohesin, does not occur in pds1Δ cells. (E) Percentage of cells with anaphase spindles. (F) Cohesin cleavage was monitored by examining the amount of full-length Scc1-18Myc (Scc1*) and the C-terminal cleavage fragment of Scc1-18Myc (cleaved Scc1). Scc1-18Myc blots from pds1Δ strains released from HU consistently show a 90-kDa protein that cross-reacts with the Myc antibody, the identity of which is unknown. Pgk1 was used as a loading control in Western blots.
clb1Δ clb2-VI cells are defective in spindle elongation in the absence of PDS1
Our data show that clb1Δ clb2-VI cells are defective in Securin/Pds1 degradation. If this defect was solely responsible for the metaphase delay of clb1Δ clb2-VI cells, deletion of PDS1 should eliminate the anaphase entry defect of clb1Δ clb2-VI cells (Fig. 4B). We used cohesin (Scc1/Mcd1) cleavage to follow anaphase entry in pds1Δ cells. Wild-type, pds1Δ, clb1Δ clb2-VI, and pds1Δ clb1Δ clb2-VI cells were arrested in early S phase with hydroxyurea (HU) at 25°C (Fig. 4C) to permit sufficient accumulation of Separase/Esp1 in the nucleus of pds1Δ cells (Yamamoto et al. 1996a, b; Agarwal and Cohen-Fix 2002). Upon release from the S phase block at 37°C, cohesin cleavage was delayed in clb1Δ clb2-VI cells (Fig. 4F). In contrast, cohesin cleavage occurred prematurely in cells lacking PDS1 (Fig. 4F). clb1Δ clb2-VI cells lacking PDS1 exhibited the same premature cohesin cleavage as the pds1Δ single mutant (Fig. 4F), indicating that the failure to degrade Pds1 was likely to be the sole reason why cohesin cleavage was delayed in clb1Δ clb2-VI cells.
We also examined the effects of deleting PDS1 on cell cycle progression. Wild-type and pds1Δ cells progressed from metaphase into anaphase with similar kinetics (Fig. 4D,E). In contrast, clb1Δ clb2-VI cells were delayed in entering anaphase (Fig. 4D,E). Surprisingly, deletion of PDS1 did not accelerate spindle elongation in clb1Δ clb2-VI cells (Fig. 4D,E). Therefore, clb1Δ clb2-VI cells have a defect in anaphase entry that is independent of Securin/Pds1 stabilization and cohesin cleavage. Consistent with this idea is the finding that overproduction of CDC20, despite partially rescuing the cohesin cleavage defect in this strain, failed to rescue the spindle elongation defect of clb1Δ clb2-VI cells (data not shown).
Inactivation of cohesin does not suppress the spindle elongation defect of clb1Δ clb2-VI cells
In budding yeast, cleavage of the Scc1/Mcd1 subunit of the cohesin complex has been shown to be necessary and sufficient for anaphase spindle elongation (Uhlmann et al. 2000). In cells carrying a mutant form of cohesin that contains a target site for the TEV protease, expression of TEV protease is sufficient to induce chromosome segregation, thus bypassing the requirement for APC/C–Cdc20 or Separase/Esp1 in triggering spindle elongation (Uhlmann et al. 2000). Similarly, temperature-sensitive alleles of SCC1/MCD1 (mcd1-1 or scc1-73) allow spindle elongation and sister chromatid separation in the absence of Separase/Esp1 activation (Fig. 4B; Guacci et al. 1997; Severin et al. 2001b). If the inability of clb1Δ clb2-VI cells to form anaphase spindles was due to a failure to trigger Scc1/Mcd1 cleavage, inactivation of cohesins should cause spindle elongation (Fig. 4B). To test this hypothesis, we employed the temperature-sensitive mcd1-1 allele, which has been shown previously to bypass the metaphase arrest of cells lacking APC/C–Cdc20 activity (Guacci et al. 1997).
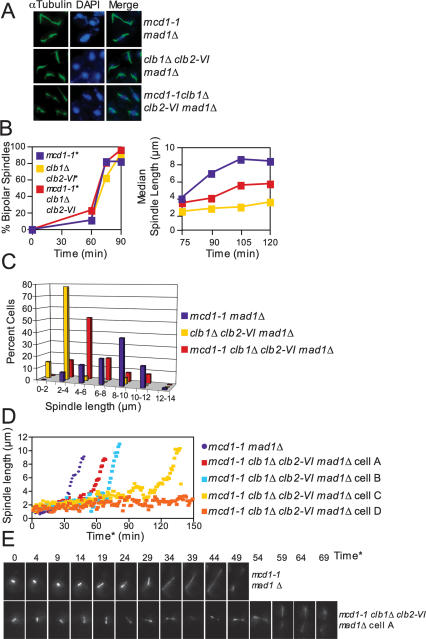
Inactivation of sister chromatid cohesion causes activation of the spindle assembly checkpoint because kinetochore–microtubule attachments that are not under tension are swiftly severed (Biggins and Murray 2001; Tanaka et al. 2002; Dewar et al. 2004). To avoid indirect effects of spindle checkpoint activation on the kinetics of spindle elongation, we deleted the spindle checkpoint component MAD1. Upon release from a G1 block at 37°C, all strains entered mitosis with similar kinetics as judged by bipolar spindle formation (Fig. 5B). Surprisingly, mcd1-1 clb1Δ clb2-VI cells failed to elongate spindles to the extent seen in mcd1-1 cells, as judged by spindle length measurements (Fig. 5B; Supplemental Fig. 3A). At 105 min after release from G1, mcd1-1 cells exhibited a broad distribution of spindle lengths, with a mode at 8–10 μm. clb1Δ clb2-VI cells exhibited a mode at 2–4 μm (Fig. 5C; Supplemental Fig. 3A). The distribution of spindle lengths in mcd1-1 clb1Δ clb2-VI cells was slightly broader than that of clb1Δ clb2-VI cells and centered at 4–6 μm (Fig. 5C; Supplemental Fig. 3A).
Figure 5.
Clb1/2–CDK activity is required after cohesin cleavage to bring about anaphase spindle elongation. (A–C) mcd1-1 mad1Δ (A2784), clb1Δ clb2-VI mad1Δ Pds1-13Myc (A14770), and mcd1-1 mad1Δ clb1Δ clb2-VI Pds1-13Myc (A14771) cells, each carrying a Cdc14-3HA fusion, were arrested in G1 at room temperature and released into 37°C YEPD as described in Figure 1B. MAD1 was deleted in each strain because cohesin mutants trigger the spindle assembly checkpoint (Severin et al. 2001b). (A) Microtubule and DNA morphologies are shown 105 min after release from G1. Microtubules are shown in green, and DNA is shown in blue. Spindles appeared fragile in mcd1-1 mad1Δ and mcd1-1 mad1Δ clb1Δ clb2-VI cells. This is due to premature separation of sister chromatids, which results in extremely short kinetochore microtubules. (B) The percentage of cells with bipolar spindles (left graph) and the median spindle length of each strain (right graph) were determined at the indicated times (n = 100 spindles at 75 min and 90 min, n = 80 spindles at 105 min, and n = 70 spindles at 120 min). (*) MAD1 was deleted in each strain to prevent activation of the spindle assembly checkpoint. These time points were chosen because they had the highest percentage of cells with bipolar spindles. Box and whisker plots of spindle lengths from this experiment are shown in Supplemental Figure 3A. (C) Spindle length distributions of mcd1-1 mad1Δ, clb1Δ clb2-VI mad1Δ, and mcd1-1 clb1Δ clb2-VI mad1Δ cells 105 min after release from G1 (n = 80 spindles). The 105-min time point was chosen because it had the highest percentage of cells with bipolar spindles. (D,E) Time-lapse series of spindle elongation in mcd1-1 mad1Δ (A17974) and mcd1-1 clb1Δ clb2-VI mad1Δ (A17977) cells, each carrying Tub1-GFP and Cdc14-3HA fusions. Cells were prepared for imaging as described in Figure 1D and in the Materials and Methods. Time* refers to time after SPB separation with 0 defined as the first time point at which SPB separation is visible. (D) The distance between the two separated SPBs in each cell was measured at every time point in which the spindle was in focus. The mcd1-1 clb1Δ clb2-VI madΔ cells A (Supplemental Movie 4) and B, C, and D (Supplemental Movie 5) are shown because they represent the range of spindle elongation phenotypes observed in mcd1-1 clb1Δ clb2-VI mad1Δ cells. (E, top panel) A time-lapse series of a mcd1-1 mad1Δ cell undergoing anaphase (shown in Supplemental Movie 3). This cell was chosen because it initiated anaphase at a time similar to the mcd1-1 mad1Δ population’s mean anaphase onset time. (Bottom panel) A time-lapse series of a mcd1-1 clb1Δ clb2-VI mad1Δ cell undergoing anaphase (shown in Supplemental Movie 4). This cell was chosen because it initiated anaphase at a time similar to the mcd1-1 clb1Δ clb2-VI mad1Δ population’s mean anaphase onset time.
Live-cell analysis also showed that clb1Δ clb2-VI mcd1-1 cells failed to elongate their spindles. mcd1-1 cells reached a spindle length of 4 μm, on average 29.9 min (SD = 14.2; n = 14) after SPB separation (Fig. 5D,E; Supplemental Movie 3), and only five out of 28 cells failed to elongate their spindles (data not shown). In contrast, in mcd1-1 clb1Δ clb2-VI cells, spindle elongation either did not occur at all (six out of 16 cells) (i.e., cell D in Fig. 5D; Supplemental Movie 5) or occurred with a delay (48.6 min after SPB separation; SD = 14.3; n = 9; note that cells that failed to elongate their spindles were not included in this calculation) (Fig. 5D,E; Supplemental Movies 4, 5). The delay in anaphase onset was more variable in clb1Δ clb2-VI cells lacking sister chromatid cohesion than in clb1Δ clb2-VI cells, but the difference between mcd1-1 and mcd1-1 clb1Δ clb2-VI cells was nevertheless highly significant (P = 0.006; two-sided Student’s t-test). The spindle behavior of clb1Δ clb2-VI cells lacking cohesion was also more complex than that of clb1Δ clb2-VI cells with sister chromatid linkages. The mitotic spindles went through continuous cycles of stretching to a length between 3 and 4 μm, collapse, and reformation (Supplemental Movies 4, 5). We conclude that loss of cohesins did not fully restore spindle elongation to clb1Δ clb2-VI cells, indicating that Clb1/2–CDK activity is required for anaphase spindle elongation independently of cohesin removal. Furthermore, the defect in Securin/Pds1 degradation in clb1Δ clb2-VI cells contributes to their short spindle phenotype, because a shift to higher spindle lengths is observed upon inactivation of cohesion with the mcd1-1 allele.
Clb1/2–CDK activity is required for anaphase spindle elongation
The inability of the mcd1-1 allele to restore spindle elongation to clb1Δ clb2-VI cells indicates that Clb1/2–CDK complexes have additional functions in promoting chromosome segregation that are distinct from their role in promoting APC/C–Cdc20 complex activation. What are these additional functions? Clb1/2–CDK activity could be required either to resolve cohesin-independent linkages between sister chromatids or to promote anaphase spindle elongation.
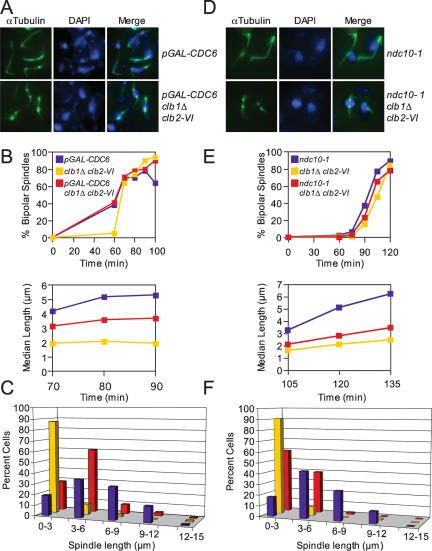
To test the possibility that the absence of Clb1/2–CDK activity triggers aberrant forms of sister chromatid entanglement that are cohesion-independent or to resolve commonly occurring forms of sister chromatid linkages, such as catenations, we examined how the complete lack of a sister chromatid affects spindle elongation in clb1Δ clb2-VI cells. Cdc6 is an essential component of prereplicative complexes (pre-RCs) (Toone et al. 1997). Strains carrying CDC6 under the control of the glucose-repressible GAL1–10 promoter fail to replicate their DNA in the presence of glucose (Piatti et al. 1995; Biggins and Murray 2001; Stern and Murray 2001; Tanaka et al. 2002). To avoid indirect effects of spindle checkpoint activation on the kinetics of spindle elongation when CDC6 is depleted, we deleted the spindle checkpoint component MAD1 (Stern and Murray 2001). Upon release from a pheromone-induced G1 arrest at 37°C, DNA replication was largely blocked in pGAL-CDC6 and pGAL-CDC6 clb1Δ clb2-VI, but not clb1Δ clb2-VI cells (Supplemental Fig. 4). pGAL-CDC6 cells progressively elongated their spindles, while those of clb1Δ clb2-VI cells remained short (Fig. 6B; Supplemental Fig. 3B). pGAL-CDC6 clb1Δ clb2-VI cells failed to elongate their spindles to lengths seen in pGAL-CDC6 cells and spindles had the characteristic fragile appearance of spindles with short kinetochore microtubules (Fig. 6A,B; Supplemental Fig. 3B). Spindle length measurements at 90 min following the release from G1 showed that 43% of cells in the pGAL-CDC6 cultures contained spindles >6 μm, while only 2% of clb1Δ clb2-VI cells and 10% of pGAL-CDC6 clb1Δ clb2-VI cells contained spindles of this length (Fig. 6C; Supplemental Fig. 3B). We conclude that unresolved sister chromatid linkages cannot account for the spindle elongation defect of clb1Δ clb2-VI cells.
Figure 6.
clb1Δ clb2-VI cells are defective in anaphase spindle elongation. (A–C) mad1Δ cdc6Δ pGAL-Ubi-R-CDC6 Pds1-18Myc (A15078), mad1Δ clb1Δ clb2-VI Pds1-13Myc Cdc14-3HA (A14770), and mad1Δ cdc6Δ pGAL-Ubi-R-CDC6 clb1Δ clb2-VI Pds1-18Myc (A15334) cells were grown in YEP Raffinose + Galactose (YEPRG) at room temperature and arrested in G1 with α-factor (5 μg/mL) for 3 h. Cells were released into α-factor-free YEPRG at room temperature and, after 20 min, glucose was added to repress pGAL-CDC6. Glucose was added after 20 min to allow cells enough time to accumulate sufficient Cdc6 for one cell cycle. α-Factor was added again 75 min after release to arrest cells in G1 of the next cell cycle while pGAL-CDC6 was repressed. Two-hundred-twenty-five minutes after α-factor addition (3 h total), cells were released from their second G1 arrest into YEPD at 37°C. (A) Microtubule and DNA morphologies are shown 90 min after release from G1. Microtubules are shown in green, and DNA is shown in blue. (B) The percentage of cells with bipolar spindles (top graph) and the median spindle length of each strain (bottom graph) were determined at the indicated times (at least 100 cells were examined per time point). These time points were chosen because they had the highest percentage of cells with bipolar spindles. Box and whisker plots of spindle lengths from this experiment are shown in Supplemental Figure 3B. (C) The distribution of spindle lengths 90 min after release from G1 is shown (at least 100 spindles were measured). The 90 min time point was chosen because it had the highest percentage of cells with bipolar spindles. (D–F) ndc10-1 (A2733), clb1Δ clb2-VI (A3000), and ncd10-1 clb1Δ clb2-VI (A15260), each carrying a Cdc14-3HA fusion, were grown in YEPD at room temperature and treated as described in Figure 1B. (D) Microtubule and DNA morphologies are shown 120 min after release from the G1 arrest. Microtubules are shown in green, and DNA is shown in blue. (E) The percentage of cells with bipolar spindles (top graph) and the median spindle length of each strain (bottom graph) were determined at the indicated times (at least 100 cells were examined per time point). These time points were chosen because they had the highest percentage of cells with bipolar spindles. Box and whisker plots of spindle lengths from this experiment are shown in Supplemental Figure 3C. (F) The distribution of spindle lengths 120 min after release from G1 is shown (n = 100 spindles). The 120 min time point was chosen because it had the highest percentage of cells with bipolar spindles.
Our data thus far show that Clb1/2–CDK activity is required for anaphase spindle formation independently of sister chromatid cohesion and suggest that Clb1/2–CDK activity is required for spindle elongation. To test this hypothesis more directly, we asked if Clb1/2–CDK activity was required for the precocious spindle elongation that occurs in some kinetochore mutants. Kinetochore mutants were examined because spindle attachment to kinetochores normally restrains spindle elongation prior to anaphase (due to sister chromatid cohesion). In cells carrying a temperature-sensitive allele of the NDC10 gene (ndc10-1), which encodes an integral kinetochore component, spindle elongation is uncoupled from cell cycle progression (Goh and Kilmartin 1993). At the restrictive temperature, ndc10-1 cells prematurely elongate their spindles without chromosome segregation (since chromosomes are not attached to microtubules of the mitotic spindle) (Fig. 6D; Goh and Kilmartin 1993; Ciosk et al. 1998; Tavormina and Burke 1998; Fraschini et al. 2001). Upon release from a G1 arrest at 37°C, ndc10-1, clb1Δ clb2-VI, and ndc10-1 clb1Δ clb2-VI cells assembled bipolar spindles with similar kinetics (Fig. 6E). The ndc10-1 mutant cells elongated their spindles in the absence of nuclear division as judged by DAPI staining, indicating that the ndc10-1 allele was inactivated (Fig. 6D). Surprisingly, the median spindle length of ndc10-1 clb1Δ clb2-VI cells was substantially shorter than that of ndc10-1 cells at all time points examined (Fig. 6E; Supplemental Fig. 3C). One-hundred-twenty minutes after release from the G1 block, spindle lengths were broadly distributed between 3 and 12 μm in ndc10-1 mutants (Fig. 6F; Supplemental Fig. 3C). In contrast, spindle lengths were between 0 and 6 μm in clb1Δ clb2-VI and ndc10-1 clb1Δ clb2-VI cells (Fig. 6F; Supplemental Fig. 3C). We conclude that Clb1/2–CDK activity is required for anaphase spindle elongation. As ndc10-1 mutants lack microtubules linking kinetochores to the mitotic spindle, we speculate that Clb1/2–CDK activity is required for anaphase B, which is the elongation of pole–pole microtubules.
Clb2/3–CDK activity is required for Securin/Pds1 degradation and anaphase spindle elongation
Is the anaphase entry defect observed in clb1Δ clb2-VI cells specific to this particular CLB cyclin mutant combination, or does the combined inactivation of other CLB cyclins also cause defects in anaphase entry? To address this question, we first examined the consequences of inactivating all CDKs on mitotic progression by using an ATP analog-sensitive allele of CDC28 (cdc28-as1) (Bishop et al. 2000). Cells carrying the cdc28-as1 allele as the sole source of CDC28 were released from a pheromone-induced G1 arrest and inhibitor was added prior to mitosis (65 min after release), when cells entered metaphase (95 min after release), and when cells were in anaphase (120 min after release). Consistent with previous data showing that Clb–CDK activity was required for the maintenance of a mitotic spindle (Fitch et al. 1992; Richardson et al. 1992; Amon et al. 1993; Bishop et al. 2000), spindle collapse occurred shortly after inhibitor addition, irrespective of which cell cycle stage cells were in (Supplemental Fig. 5). Thus, a minimal amount of Clb–CDK activity is necessary for maintenance of a mitotic spindle.
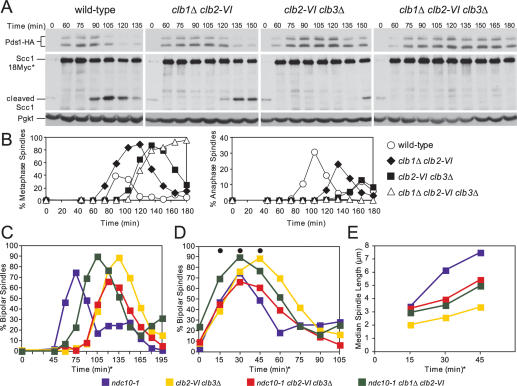
To determine whether the combined inactivation of other CLB cyclins causes anaphase entry defects, we combined a deletion of CLB3 with the temperature-sensitive clb2-VI allele. Inactivation of CLB2 and CLB3, like deletion of CLB1 and CLB2, is lethal (Fitch et al. 1992; Richardson et al. 1992), indicating an essential requirement for this cyclin pair in cell cycle progression. In addition, we examined the consequences of inactivating CLB1, CLB2, and CLB3 using a clb1Δ clb2-VI clb3Δ triple mutant. Upon release of cells from a pheromone-induced G1 arrest at 37°C, clb2-VI clb3Δ and clb1Δ clb2-VI clb3Δ cells, but not clb1Δ clb2-VI cells, exhibited a 30- and 40-min delay in bipolar spindle formation, respectively (Fig. 7B). This observation is consistent with the previous finding that Clb–CDK activity is required for bipolar spindle formation (Fitch et al. 1992; Richardson et al. 1992).
Figure 7.
clb2-VI clb3Δ cells are defective in Securin degradation and anaphase spindle elongation. (A,B) Wild-type (A1951), clb1Δ clb2-VI (A15111), clb2-VI clb3Δ (A19136), and clb1Δ clb2-VI clb3Δ (A1938) cells, each carrying Pds1-HA and Scc1-18Myc, were grown and treated as described in Figure 1B. (A) The amounts of full-length Scc1-18Myc (Scc1-18Myc*), the C-terminal cleavage fragment of Scc1-18Myc (cleaved Scc1), and Pds1-HA were also examined. Pgk1 was used as a loading control in Western blots. (B) The percentage of cells with metaphase (left graph) and anaphase (right graph) spindles was determined at the indicated times. (C–E) ndc10-1 (A2733), clb2-VI clb3Δ (A2887), ncd10-1 clb2-VI clb3Δ (A19288), and ncd10-1 clb1Δ clb2-VI (A17126), each carrying a Cdc14-3HA fusion, were grown and treated as described in Figure 1B. The percentage of cells with bipolar spindles is shown in C (n = 200). In the graph shown in D, the time points were adjusted so that onset of bipolar spindle formation occurred concomitantly. The time points chosen for analysis were marked with a black circle. For the ndc10-1 strain, the 0 time point in D corresponds to the 45-min time point in C; for the ncd10-1 clb1Δ clb2-VI strain, the 0 time point corresponds to the 75-min time point in C; for strains clb2-VI clb3Δ and ncd10-1 clb2-VI clb3Δ the 0-min time points correspond to the 90-min time point in C. The delay in bipolar spindle assembly in the ncd10-1 clb1Δ clb2-VI strain relative to ndc10-1 strain is not reproducible and not characteristic of this mutant. (E) The median spindle length of each strain. The time points correspond to the time points in D marked with a black circle. At least 140 cells were examined per time point. Box and whisker plots of spindle lengths from this experiment are shown in Supplemental Figure 3D.
We followed cell cycle progression in these strains and found that the combined function of CLB2 and CLB3 is also needed for anaphase entry. Like clb1Δ clb2-VI cells, clb2-VI clb3Δ and clb1Δ clb2-VI clb3Δ cells exhibited a delay in Securin/Pds1 degradation, cohesin cleavage, and anaphase spindle elongation. Taking into account the 30-min delay in spindle formation in clb2-VI clb3Δ cells, the delay in Securin/Pds1 degradation, cohesin cleavage, and anaphase entry appeared similar between clb1Δ clb2-VI and clb2-VI clb3Δ cells (Fig. 7A,B). The delay in the triple mutant was more pronounced (Fig. 7A,B).
To determine whether Clb2/3–CDK activity was also required for anaphase spindle elongation independently of Securin/Pds1 degradation and cohesin cleavage, we examined the kinetics of spindle elongation in clb2-VI clb3Δ cells lacking NDC10. As in clb1Δ clb2-VI cells, spindle elongation was greatly hampered in clb2-VI clb3Δ cells (Fig. 7C–E; Supplemental Fig. 3D). Compared with the ndc10-1 single mutant, clb1Δ clb2-VI ndc10-1 exhibited a 30-min delay in entry into mitosis, while both clb2-VI clb3Δ and clb2-VI clb3Δ ndc10-1 cells exhibited a 45-min delay in entry into mitosis (Fig. 7C). To compare the kinetics of mitotic spindle elongation between the strains, we therefore adjusted the graphs so that the onset of bipolar spindle formation coincided (Fig. 7D). Spindle length measurements at matched time points revealed that, whereas ndc10-1 cells reached a median spindle length of 7.4 μm, spindles in ndc10-1 clb2-VI clb3Δ and ndc10-1 clb1Δ clb2-VI cells reached median lengths of only 5.3 and 4.9 μm, respectively (Fig. 7E; Supplemental Fig. 3D). clb2-VI clb3Δ cells reached a median spindle length of 3.2 μm. Our results indicate that inactivation of CLB2 and CLB3 elicits similar metaphase defects as inactivation of CLB1 and CLB2. Furthermore, they suggest that the overall level of Clb–CDK activity is important for entry into anaphase.
Discussion
Mitotic CDKs are best known for their essential function in initiating mitosis, where they promote mitotic spindle formation, chromosome condensation, and the breakdown of the nuclear envelope (Nigg 2001; Miele 2004). The functions of these kinases in later stages of mitosis are less well understood. Here we show that Clb–CDK activity triggers two key aspects of chromosome segregation: (1) activation of APC/C–Cdc20 to facilitate Securin degradation, and (2) anaphase spindle elongation to facilitate the partitioning of the genome between the future daughter nuclei. Thus, it appears that mitotic CDK activity not only initiates mitosis, it is also a key regulator of chromosome segregation.
Clb–CDK activity is required for APC/C–Cdc20 activation in vivo
The APC/C is a large ∼1.5-MDa E3 ubiquitin ligase complex that is composed of at least 12 subunits in humans (13 in budding yeast) (Peters 2006). Many kinases including CDK, Polo-like kinases, and PKA phosphorylate APC/C–Cdc20 in vitro and most of these phosphorylations, except for those catalyzed by PKA, are thought to activate APC/C–Cdc20 (Lahav-Baratz et al. 1995; Kotani et al. 1998; Patra and Dunphy 1998; Shteinberg et al. 1999; Kramer et al. 2000; Rudner and Murray 2000; Yudkovsky et al. 2000; Golan et al. 2002; Kraft et al. 2003; Searle et al. 2004; Herzog et al. 2005; Peters 2006). In vivo, there are at least 51 phosphorylation sites in the human APC/C (Kraft et al. 2003). This large number of sites has precluded mutational analysis and therefore the functional significance of these phosphorylation events remains unknown.
In our study, we observed that Clb–CDK activity is required for the timely destruction of the APC/C–Cdc20 substrate Securin/Pds1. Since the failure of clb1Δ clb2-VI cells to degrade Securin/Pds1 was independent of the checkpoints known to inhibit the APC/C–Cdc20, we conclude that a yet-to-be-identified Clb1/2–CDK-dependent process regulates APC/C–Cdc20 function or that Clb1/2–CDK activity directly controls APC/C–Cdc20 activation in vivo. We favor the latter possibility because Rudner and Murray (2000) observed that strains carrying a version of the APC/C in which all CDK phosphorylation sites within the APC/C subunits Cdc16, Cdc23, and Cdc27 were mutated to alanine (“apc/c-12A”) exhibit a 15-min delay in metaphase and require CLB2 for viability (Rudner and Murray 2000). Given that the cell cycle delay of the apc/c-12A mutant was modest compared with that exhibited by the clb1Δ clb2-VI or clb2-VI clb3Δ strains, this result suggests that additional Clb–CDK targets exist among the APC/C–Cdc20 subunits. Cdc20 is a likely candidate because human Cdc20 is phosphorylated in vivo (Kraft et al. 2003). More importantly, mutation of the four serines within the four CDK consensus sequences in budding yeast Cdc20 to alanines (cdc20-4A) does not interfere with CDC20 function in an otherwise wild-type strain, but causes lethality when combined with the apc/c-12A mutant and cell cycle arrest in metaphase (R. Rahal, unpubl.). This observation is consistent with the idea that phosphorylation of both Cdc20 and the APC/C is essential for the activation of the ubiquitin ligase at metaphase. However, because we have thus far not been able to demonstrate that the four CDK consensus sites within Cdc20 are phosphorylated in vivo, we cannot exclude the possibility that the cdc20-4A allele is hypomorphic due to structural rather than regulatory defects.
Mitotic CDK activity is required for spindle elongation during anaphase
Deletion of PDS1 in clb1Δ clb2-VI cells revealed a second essential function for Clb1/2–CDKs in the initiation of anaphase. Several lines of evidence indicate that Clb1/2–CDK activity does not mediate this function through resolving linkages between sister chromatids. mcd1-1 strains fail to maintain sister chromatid cohesion, and the mcd1-1 allele suppresses the anaphase entry defect of APC/C–Cdc20 and Separase mutants (Guacci et al. 1997; Severin et al. 2001b). In clb1Δ clb2-VI strains, however, the suppression of the anaphase entry defect by cohesin inactivation was only partial. We interpret this partial rescue to result from the fact that the mcd1-1 mutation bypasses the Securin/Pds1 degradation defect of clb1Δ clb2-VI strains, allowing some spindle elongation to occur (due to the lack of sister chromatid cohesion that normally resists microtubule pulling forces). Nevertheless, the spindles of mcd1-1 clb1Δ clb2-VI cells never fully elongated to lengths seen in mcd1-1 cells, indicating that Clb1/2–CDK activity was needed for more than cohesion removal to bring about chromosome movement. Eliminating DNA replication did not permit spindle elongation in clb1Δ clb2-VI strains either, further excluding the possibility that cohesin-independent linkages are responsible for the anaphase spindle elongation defect of cells lacking Clb1/2–CDK activity.
These findings leave us with the conclusion that the process of spindle elongation itself requires Clb1/2–CDK activity. This idea is supported by the observation that eliminating kinetochore function failed to promote anaphase B spindle elongation in clb1Δ clb2-VI strains. The same result was obtained in cells lacking Clb2/3–CDK activity, indicating that this requirement was not specific to the late mitotic cyclin pair Clb1 and Clb2. At first glance, the idea that Clb–CDK activity is required for spindle elongation may sound paradoxical because mitotic CDK activity starts to decline as cells enter anaphase. It is, however, important to note that, at least in yeast, a decrease in mitotic CDK activity is not required for anaphase spindle elongation. Overexpression of a nondegradable version of Clb2 does not interfere with spindle elongation (Surana et al. 1993). It is also worth pointing out that our data do not call into question that the decline in mitotic CDK activity commences with the activation of APC/C–Cdc20. They merely indicate that a certain amount of Clb–CDK activity is required for anaphase spindle elongation that is not met in cells lacking either Clb1/2–CDK or Clb2/3–CDK activity.
How does Clb–CDK activity promote spindle elongation during anaphase?
Our electron microscopy analysis revealed that the bipolar spindles formed by clb1Δ clb2-VI cells appear normal in overall structure, indicating that a general structural defect is not responsible for the inability of cells lacking Clb1/2–CDK activity to elongate their spindles. The protein phosphatase Cdc14 has been shown previously to be required for timely anaphase spindle elongation and the switch from metaphase microtubule dynamics to an anaphase one (Higuchi and Uhlmann 2005). The defect in anaphase spindle elongation in cdc14 mutants is, however, subtle compared with that of clb1Δ clb2-VI or clb2-VI clb3Δ cells. This finding indicates that Clb–CDK’s role in anaphase spindle elongation cannot be explained by its function in Cdc14 activation at the metaphase–anaphase transition (Azzam et al. 2004). It is also unlikely that Clb–CDKs promote spindle elongation through their role in APC–Cdc20 activation because high levels of Cdc20 suppress the Securin/Pds1 degradation defect of clb1Δ clb2-VI cells, but not the spindle elongation defect of the strain (R. Rahal, unpubl.).
Anaphase spindle elongation occurs in two steps: an initial rapid phase in which the spindle elongates to ∼3–4 μm, followed by a second phase in which the spindle slowly elongates to a final length of ∼7–8 μm (Yeh et al. 1995; Straight et al. 1997, 1998; Pearson et al. 2001). Live-cell microscopy showed that spindles in mcd1-1 clb1Δ clb2-VI cells went through cycles of rapid spindle elongation to a length between 3 and 4 μm, which was followed by spindle collapse and reformation. This finding raises the interesting possibility that the second, slow elongation phase is impaired in mcd1-1 clb1Δ clb2-VI cells. This slow elongation step could require Clb–CDK-mediated changes in microtubule motor function. We attempted to test this hypothesis in several ways, but neither overexpression of spindle elongation promoting factors such as CIN8 (Hildebrandt and Hoyt 2000) or STU2 (Severin et al. 2001a; Krishnan et al. 2004) nor deletion of spindle elongation inhibitors such as KIP3 (Hildebrandt and Hoyt 2000; Gupta et al. 2006; Varga et al. 2006) rescued the spindle elongation defect of clb1Δ clb2-VI cells and mad1Δ mcd1-1 clb1Δ clb2-VI cells (Supplemental Fig. 6; data not shown). We favor the hypothesis that Clb–CDK activity triggers spindle elongation by regulating the activity of multiple motors and/or microtubule-associated proteins, especially those at the spindle mid-zone. In this regard, it is interesting to note that mutation of the nine CDK consensus sequences in the chromosomal passenger protein Bir1 to amino acids that can no longer be phosphorylated reduces the extent of spindle elongation and prevents the localization of proteins such as Ndc10 to the spindle mid-zone during anaphase (Widlund et al. 2006). The Clb–CDK-dependent recruitment of proteins to the spindle mid-zone, we speculate, could be important for full anaphase spindle elongation.
Is the amount of Clb–CDK activity or Clb–CDK specificity (or both) important?
One key question arising from our data is whether differences in substrate specificity among the six Clb cyclins or changes in overall levels of Clb–CDK activity are needed to bring about the various mitotic transitions. Cells lacking Clb2/3–CDK activity exhibit a delay in bipolar spindle formation, a phenotype that was not shared by cells lacking Clb1/Clb2–CDK activity, raising the possibility that different Clb–CDKs execute different mitotic events. However, several other observations argue for increasing amounts of Clb–CDK activity triggering consecutive mitotic events. First, inactivation of not only CLB1 and CLB2 but also CLB2 and CLB3 causes a delay in Securin/Pds1 degradation and a defect in anaphase spindle elongation. Second, deletion of CLB2 and CLB4 also leads to a delay in mitosis (Fitch et al. 1992; Richardson et al. 1992), whereas deletion of CLB1, CLB3, and CLB4 has no discernable phenotype (Fitch et al. 1992; Richardson et al. 1992). Given that nearly 70% of mitotic CDK activity is associated with Clb2 (Surana et al. 1991), it appears that the severity of the metaphase delay correlates with the amount of CDK activity associated with a particular cyclin. Furthermore, we found that expression of CLB3-HA from the CLB2 promoter rescued the anaphase entry defect of clb1Δ clb2-VI cells to a similar degree as expression of CLB2-HA from the CLB2 promoter (Supplemental Fig. 7). Provided that the effects of the HA epitope tag on Clb2 and Clb3 are the same (both tagged versions of the proteins are not fully functional) (Supplemental Fig. 7; data not shown), this result indicates that Clb2 and Clb3 are similar in their ability to promote progression through mitosis. In this “Clb quantity model,” the defect in bipolar spindle formation that appears unique to cells lacking Clb2/3–CDK activity could be explained by the fact that CLB3 is expressed earlier during the cell cycle and thus provides the amount of Clb–CDK activity (with Clb2) necessary for the timely formation of a bipolar spindle. We suggest that changes in overall Clb–CDK levels bring about consecutive mitotic events, but will not exclude the possibility that minor differences in substrate preference exist among the different mitotic Clb–CDKs.
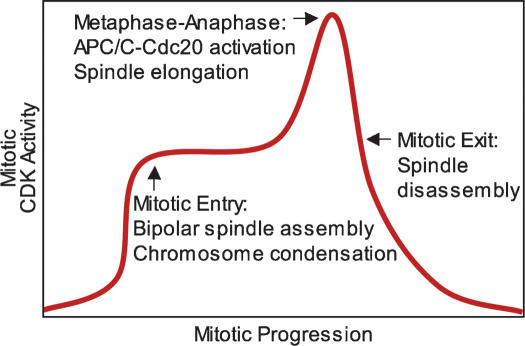
A model for how the rise in CDK activity during early mitosis establishes order to mitotic progression
It was suggested previously that increasing amounts of CDKs establish consecutive cell cycle events, with low CDK levels promoting S phase and high levels of CDKs triggering M phase (Stern and Nurse 1996). Our data indicate that increasing amounts of Clb–CDKs are also responsible for triggering consecutive mitotic events. Inactivation of CLB1 and CLB2 causes no delay in cell cycle progression prior to metaphase, whereas inactivation of CLB2 and CLB3 causes a modest 30-min delay. In contrast, inactivation of CLB1, CLB2, CLB3, and CLB4 causes cell cycle arrest in G2, with replicated DNA and unseparated SPBs (Surana et al. 1991; Fitch et al. 1992; Richardson et al. 1992; Amon et al. 1993). Furthermore, Clb–CDK activity rises as cells progress through mitosis (Surana et al. 1991; Fitch et al. 1992; Richardson et al. 1992; Surana et al. 1993). Based on these observations, we propose that higher amounts of mitotic CDK activity are needed for entry into anaphase than for entry into mitosis (Fig. 8). This steady rise in mitotic CDK activity helps establish the order of events during early mitosis, with a lower threshold of Clb–CDK activity triggering entry into mitosis and a second, higher one triggering entry into anaphase. Finally, once anaphase entry has been initiated, Clb proteolysis causes a decline in Clb–CDK activity, triggering exit from mitosis (during which thresholds may also play a role) (Wolf et al. 2007). Interestingly, increasing amounts of mitotic CDK activity may also govern progression through early stages of mitosis in mammalian cells. Mitotic CDK activity rises as cells progress from G2 into metaphase (Arion et al. 1988; Labbe et al. 1988, 1989; Gautier et al. 1989; Murray and Kirschner 1989; Lindqvist et al. 2007). Furthermore, complete inactivation of Cdk1 by RNAi-based methods prevents entry into mitosis, whereas partial Cdk1 inactivation delays entry into anaphase (Lindqvist et al. 2007). Therefore, requiring a steady rise in mitotic CDK activity for mitotic progression may be a general mechanism by which all eukaryotic cells ensure that chromosome segregation occurs only after chromosomes have condensed and a mitotic spindle has formed.
Figure 8.
A model for how increasing levels of CDK activity help establish the order of early mitotic events. See the text for details.
Materials and methods
All strains were derivatives of strain W303 (K699) and are listed in Supplemental Table 1. Growth conditions for each experiment are described in the figure legends. Western blot analysis of Pds1-HA, Scc1-18Myc, and Pgk1 was performed as described in Cohen-Fix et al. (1996), Uhlmann et al. (1999), and Monje-Casas et al. (2007), respectively. Indirect immunofluorescence for tubulin was performed as described in Monje-Casas et al. (2007). Spindle length measurements were carried out using Openlab 3.0.2 imaging program. Cdc6 depletion experiments were performed as described in Biggins and Murray (2001). Electron microscopy analysis of cells was performed as described in Winey et al. (2005). Briefly, cycling cultures of wild-type (A1411) and clb1Δ clb2-VI cells (A3000), each carrying a Cdc14-3HA fusion, were grown overnight and then shifted for 3 h to a 37°C water bath. Cell cultures were filtered into a paste and immediately high-pressure-frozen using a Leica EM PACT 2 freezer. The cells were freeze-substituted in 2% osmium tetroxide and 0.1% uranyl acetate at −80°C, then slowly warmed to room temperature and embedded in TAAB Epon. Sections were picked up on copper grids and stained with 2% uranyl acetate in acetone and lead citrate. The sections were ∼80 nm thick. Imaging was performed using a Tecnai G2 Spirit BioTWIN microscope (FEICompany) and a 2k CCD camera (AMT).
For live-cell imaging, cells were arrested in G1 in YEPD with α-factor (5 μg/mL) for 2 h at room temperature and then placed in a 37°C water bath for 30 min while in α-factor. Cells were then released as described in Figure 1B. Thirty minutes after release, cells were briefly sonicated and 5 μL of cells were placed in an imaging chamber (Lab-Tek chambered coverglass) beneath a thick 4% agar-CSM (complete synthetic media) pad to limit their movement. Three-hundred microliters of CSM prewarmed to 37°C were then added to the imaging chamber. Imaging was performed at 37°C on a DeltaVision RT (Applied Precision) microscope with a plan Apo 100×-NA 1.4 oil objective (Olympus), with FITC (488 nm excitation/528 nm emission) filters, a Photometrics CoolSNAP HQ camera (Roper Scientific), and softWoRx software (Applied Precision). The exposure time was 200 msec and images were binned two by two. Different strains were imaged on separate days because only one group of cells could be followed during each imaging time course.
Acknowledgments
We are grateful to Kerry Bloom, Andy Hoyt, Andrew Murray, Adam Rudner, and Sue Biggins for strains, reagents, and advice. We thank Eliza Vasile at the MIT CCR Microscopy and Imaging facility for her help with live-cell microscopy. We thank Maria Ericsson and Elizabeth Benecchi at the Harvard Medical School EM Facility for their help with the spindle ultrastructure analysis. We thank Andreas Hochwagen, Frank Solomon, and members of the Amon Laboratory for their critical reading of the manuscript. This work was supported by a National Institutes of Health grant GM 56800. A.A. is also an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1638308.
References
- Agarwal R., Cohen-Fix O. Phosphorylation of the mitotic regulator Pds1/securin by Cdc28 is required for efficient nuclear localization of Esp1/separase. Genes & Dev. 2002;16:1371–1382. doi: 10.1101/gad.971402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agarwal R., Tang Z., Yu H., Cohen-Fix O. Two distinct pathways for inhibiting pds1 ubiquitination in response to DNA damage. J. Biol. Chem. 2003;278:45027–45033. doi: 10.1074/jbc.M306783200. [DOI] [PubMed] [Google Scholar]
- Amon A., Tyers M., Futcher B., Nasmyth K. Mechanisms that help the yeast cell cycle clock tick: G2 cyclins transcriptionally activate G2 cyclins and repress G1 cyclins. Cell. 1993;74:993–1007. doi: 10.1016/0092-8674(93)90722-3. [DOI] [PubMed] [Google Scholar]
- Andrews B., Measday V. The cyclin family of budding yeast: Abundant use of a good idea. Trends Genet. 1998;14:66–72. doi: 10.1016/s0168-9525(97)01322-x. [DOI] [PubMed] [Google Scholar]
- Arion D., Meijer L., Brizuela L., Beach D. cdc2 is a component of the M phase-specific histone H1 kinase: Evidence for identity with MPF. Cell. 1988;55:371–378. doi: 10.1016/0092-8674(88)90060-8. [DOI] [PubMed] [Google Scholar]
- Azzam R., Chen S.L., Shou W., Mah A.S., Alexandru G., Nasmyth K., Annan R.S., Carr S.A., Deshaies R.J. Phosphorylation by cyclin B-Cdk underlies release of mitotic exit activator Cdc14 from the nucleolus. Science. 2004;305:516–519. doi: 10.1126/science.1099402. [DOI] [PubMed] [Google Scholar]
- Biggins S., Murray A.W. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes & Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop A.C., Ubersax J.A., Petsch D.T., Matheos D.P., Gray N.S., Blethrow J., Shimizu E., Tsien J.Z., Schultz P.G., Rose M.D., et al. A chemical switch for inhibitor-sensitive alleles of any protein kinase. Nature. 2000;407:395–401. doi: 10.1038/35030148. [DOI] [PubMed] [Google Scholar]
- Ciosk R., Zachariae W., Michaelis C., Shevchenko A., Mann M., Nasmyth K. An ESP1/PDS1 complex regulates loss of sister chromatid cohesion at the metaphase to anaphase transition in yeast. Cell. 1998;93:1067–1076. doi: 10.1016/s0092-8674(00)81211-8. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O., Koshland D. The anaphase inhibitor of Saccharomyces cerevisiaePds1p is a target of the DNA damage checkpoint pathway. Proc. Natl. Acad. Sci. 1997;94:14361–14366. doi: 10.1073/pnas.94.26.14361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Fix O., Peters J.M., Kirschner M.W., Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes & Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Dewar H., Tanaka K., Nasmyth K., Tanaka T.U. Tension between two kinetochores suffices for their bi-orientation on the mitotic spindle. Nature. 2004;428:93–97. doi: 10.1038/nature02328. [DOI] [PubMed] [Google Scholar]
- Fitch I., Dahmann C., Surana U., Amon A., Nasmyth K., Goetsch L., Byers B., Futcher B. Characterization of four B-type cyclin genes of the budding yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1992;3:805–818. doi: 10.1091/mbc.3.7.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraschini R., Beretta A., Lucchini G., Piatti S. Role of the kinetochore protein Ndc10 in mitotic checkpoint activation in Saccharomyces cerevisiae. Mol. Genet. Genomics. 2001;266:115–125. doi: 10.1007/s004380100533. [DOI] [PubMed] [Google Scholar]
- Gautier J., Matsukawa T., Nurse P., Maller J. Dephosphorylation and activation of Xenopus p34cdc2 protein kinase during the cell cycle. Nature. 1989;339:626–629. doi: 10.1038/339626a0. [DOI] [PubMed] [Google Scholar]
- Goh P.Y., Kilmartin J.V. NDC10: A gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 1993;121:503–512. doi: 10.1083/jcb.121.3.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golan A., Yudkovsky Y., Hershko A. The cyclin–ubiquitin ligase activity of cyclosome/APC is jointly activated by protein kinases Cdk1–cyclin B and Plk. J. Biol. Chem. 2002;277:15552–15557. doi: 10.1074/jbc.M111476200. [DOI] [PubMed] [Google Scholar]
- Guacci V., Koshland D., Strunnikov A. A direct link between sister chromatid cohesion and chromosome condensation revealed through the analysis of MCD1 in S. cerevisiae. Cell. 1997;91:47–57. doi: 10.1016/s0092-8674(01)80008-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.L., Carvalho P., Roof D.M., Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat. Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Herzog F., Mechtler K., Peters J.M. Identification of cell cycle-dependent phosphorylation sites on the anaphase-promoting complex/cyclosome by mass spectrometry. Methods Enzymol. 2005;398:231–245. doi: 10.1016/S0076-6879(05)98019-1. [DOI] [PubMed] [Google Scholar]
- Higuchi T., Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt E.R., Hoyt M.A. Mitotic motors in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 2000;1496:99–116. doi: 10.1016/s0167-4889(00)00012-4. [DOI] [PubMed] [Google Scholar]
- Keaton M.A., Lew D.J. Eavesdropping on the cytoskeleton: Progress and controversy in the yeast morphogenesis checkpoint. Curr. Opin. Microbiol. 2006;9:540–546. doi: 10.1016/j.mib.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Kotani S., Tugendreich S., Fujii M., Jorgensen P.M., Watanabe N., Hoog C., Hieter P., Todokoro K. PKA and MPF-activated polo-like kinase regulate anaphase-promoting complex activity and mitosis progression. Mol. Cell. 1998;1:371–380. doi: 10.1016/s1097-2765(00)80037-4. [DOI] [PubMed] [Google Scholar]
- Kraft C., Herzog F., Gieffers C., Mechtler K., Hagting A., Pines J., Peters J.M. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer E.R., Scheuringer N., Podtelejnikov A.V., Mann M., Peters J.M. Mitotic regulation of the APC activator proteins CDC20 and CDH1. Mol. Biol. Cell. 2000;11:1555–1569. doi: 10.1091/mbc.11.5.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan V., Nirantar S., Crasta K., Cheng A.Y., Surana U. DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol. Cell. 2004;16:687–700. doi: 10.1016/j.molcel.2004.11.001. [DOI] [PubMed] [Google Scholar]
- Labbe J.C., Lee M.G., Nurse P., Picard A., Doree M. Activation at M-phase of a protein kinase encoded by a starfish homologue of the cell cycle control gene cdc2+ Nature. 1988;335:251–254. doi: 10.1038/335251a0. [DOI] [PubMed] [Google Scholar]
- Labbe J.C., Picard A., Peaucellier G., Cavadore J.C., Nurse P., Doree M. Purification of MPF from starfish: Identification as the H1 histone kinase p34cdc2 and a possible mechanism for its periodic activation. Cell. 1989;57:253–263. doi: 10.1016/0092-8674(89)90963-x. [DOI] [PubMed] [Google Scholar]
- Lahav-Baratz S., Sudakin V., Ruderman J.V., Hershko A. Reversible phosphorylation controls the activity of cyclosome-associated cyclin–ubiquitin ligase. Proc. Natl. Acad. Sci. 1995;92:9303–9307. doi: 10.1073/pnas.92.20.9303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.H., Amon A., Prinz S. Spo13 regulates cohesin cleavage. Genes & Dev. 2002;16:1672–1681. doi: 10.1101/gad.989302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindqvist A., van Zon W., Karlsson Rosenthal C., Wolthuis R.M. Cyclin B1–Cdk1 activation continues after centrosome separation to control mitotic progression. PLoS Biol. 2007;5:e123. doi: 10.1371/journal.pbio.0050123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendenhall M.D., Hodge A.E. Regulation of Cdc28 cyclin-dependent protein kinase activity during the cell cycle of the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1998;62:1191–1243. doi: 10.1128/mmbr.62.4.1191-1243.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele L. The biology of cyclins and cyclin-dependent protein kinases: An introduction. Methods Mol. Biol. 2004;285:3–21. doi: 10.1385/1-59259-822-6:003. [DOI] [PubMed] [Google Scholar]
- Monje-Casas F., Prabhu V.R., Lee B.H., Boselli M., Amon A. Kinetochore orientation during meiosis is controlled by Aurora B and the monopolin complex. Cell. 2007;128:477–490. doi: 10.1016/j.cell.2006.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray A.W., Kirschner M.W. Cyclin synthesis drives the early embryonic cell cycle. Nature. 1989;339:275–280. doi: 10.1038/339275a0. [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: The molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Nigg E.A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2001;2:21–32. doi: 10.1038/35048096. [DOI] [PubMed] [Google Scholar]
- Patra D., Dunphy W.G. Xe-p9, a Xenopus Suc1/Cks protein, is essential for the Cdc2-dependent phosphorylation of the anaphase-promoting complex at mitosis. Genes & Dev. 1998;12:2549–2559. doi: 10.1101/gad.12.16.2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C.G., Maddox P.S., Salmon E.D., Bloom K. Budding yeast chromosome structure and dynamics during mitosis. J. Cell Biol. 2001;152:1255–1266. doi: 10.1083/jcb.152.6.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters J.M. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Piatti S., Lengauer C., Nasmyth K. Cdc6 is an unstable protein whose de novo synthesis in G1 is important for the onset of S phase and for preventing a ‘reductional’ anaphase in the budding yeast Saccharomyces cerevisiae. EMBO J. 1995;14:3788–3799. doi: 10.1002/j.1460-2075.1995.tb00048.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson H., Lew D.J., Henze M., Sugimoto K., Reed S.I. Cyclin-B homologs in Saccharomyces cerevisiae function in S phase and in G2. Genes & Dev. 1992;6:2021–2034. doi: 10.1101/gad.6.11.2021. [DOI] [PubMed] [Google Scholar]
- Rudner A.D., Murray A.W. Phosphorylation by Cdc28 activates the Cdc20-dependent activity of the anaphase-promoting complex. J. Cell Biol. 2000;149:1377–1390. doi: 10.1083/jcb.149.7.1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez Y., Bachant J., Wang H., Hu F., Liu D., Tetzlaff M., Elledge S.J. Control of the DNA damage checkpoint by chk1 and rad53 protein kinases through distinct mechanisms. Science. 1999;286:1166–1171. doi: 10.1126/science.286.5442.1166. [DOI] [PubMed] [Google Scholar]
- Searle J.S., Schollaert K.L., Wilkins B.J., Sanchez Y. The DNA damage checkpoint and PKA pathways converge on APC substrates and Cdc20 to regulate mitotic progression. Nat. Cell Biol. 2004;6:138–145. doi: 10.1038/ncb1092. [DOI] [PubMed] [Google Scholar]
- Severin F., Habermann B., Huffaker T., Hyman T. Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 2001a;153:435–442. doi: 10.1083/jcb.153.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F., Hyman A.A., Piatti S. Correct spindle elongation at the metaphase/anaphase transition is an APC-dependent event in budding yeast. J. Cell Biol. 2001b;155:711–718. doi: 10.1083/jcb.200104096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shteinberg M., Protopopov Y., Listovsky T., Brandeis M., Hershko A. Phosphorylation of the cyclosome is required for its stimulation by Fizzy/cdc20. Biochem. Biophys. Res. Commun. 1999;260:193–198. doi: 10.1006/bbrc.1999.0884. [DOI] [PubMed] [Google Scholar]
- Stern B.M., Murray A.W. Lack of tension at kinetochores activates the spindle checkpoint in budding yeast. Curr. Biol. 2001;11:1462–1467. doi: 10.1016/s0960-9822(01)00451-1. [DOI] [PubMed] [Google Scholar]
- Stern B., Nurse P. A quantitative model for the cdc2 control of S phase and mitosis in fission yeast. Trends Genet. 1996;12:345–350. [PubMed] [Google Scholar]
- Straight A.F., Marshall W.F., Sedat J.W., Murray A.W. Mitosis in living budding yeast: Anaphase A but no metaphase plate. Science. 1997;277:574–578. doi: 10.1126/science.277.5325.574. [DOI] [PubMed] [Google Scholar]
- Straight A.F., Sedat J.W., Murray A.W. Time-lapse microscopy reveals unique roles for kinesins during anaphase in budding yeast. J. Cell Biol. 1998;143:687–694. doi: 10.1083/jcb.143.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surana U., Robitsch H., Price C., Schuster T., Fitch I., Futcher A.B., Nasmyth K. The role of CDC28 and cyclins during mitosis in the budding yeast S. cerevisiae. Cell. 1991;65:145–161. doi: 10.1016/0092-8674(91)90416-v. [DOI] [PubMed] [Google Scholar]
- Surana U., Amon A., Dowzer C., McGrew J., Byers B., Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993;12:1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T.U., Rachidi N., Janke C., Pereira G., Galova M., Schiebel E., Stark M.J., Nasmyth K. Evidence that the Ipl1–Sli15 (Aurora kinase–INCENP) complex promotes chromosome bi-orientation by altering kinetochore–spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Tavormina P.A., Burke D.J. Cell cycle arrest in cdc20 mutants of Saccharomyces cerevisiae is independent of Ndc10p and kinetochore function but requires a subset of spindle checkpoint genes. Genetics. 1998;148:1701–1713. doi: 10.1093/genetics/148.4.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinker-Kulberg R.L., Morgan D.O. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes & Dev. 1999;13:1936–1949. doi: 10.1101/gad.13.15.1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone W.M., Aerne B.L., Morgan B.A., Johnston L.H. Getting started: Regulating the initiation of DNA replication in yeast. Annu. Rev. Microbiol. 1997;51:125–149. doi: 10.1146/annurev.micro.51.1.125. [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Lottspeich F., Nasmyth K. Sister-chromatid separation at anaphase onset is promoted by cleavage of the cohesin subunit Scc1. Nature. 1999;400:37–42. doi: 10.1038/21831. [DOI] [PubMed] [Google Scholar]
- Uhlmann F., Wernic D., Poupart M.A., Koonin E.V., Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- Varga V., Helenius J., Tanaka K., Hyman A.A., Tanaka T.U., Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat. Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Wang H., Liu D., Wang Y., Qin J., Elledge S.J. Pds1 phosphorylation in response to DNA damage is essential for its DNA damage checkpoint function. Genes & Dev. 2001;15:1361–1372. doi: 10.1101/gad.893201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widlund P.O., Lyssand J.S., Anderson S., Niessen S., Yates J.R., Davis T.N. Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol. Biol. Cell. 2006;17:1065–1074. doi: 10.1091/mbc.E05-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Mamay C.L., O’Toole E.T., Mastronarde D.N., Giddings T.H., McDonald K.L., McIntosh J.R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J. Cell Biol. 1995;129:1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Morgan G.P., Straight P.D., Giddings T.H., Mastronarde D.N. Three-dimensional ultrastructure of Saccharomyces cerevisiae meiotic spindles. Mol. Biol. Cell. 2005;16:1178–1188. doi: 10.1091/mbc.E04-09-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf F., Sigl R., Geley S. ‘The end of the beginning’: cdk1 thresholds and exit from mitosis. Cell Cycle. 2007;6:1408–1411. [PubMed] [Google Scholar]
- Yamamoto A., Guacci V., Koshland D. Pds1p is required for faithful execution of anaphase in the yeast, Saccharomyces cerevisiae. J. Cell Biol. 1996a;133:85–97. doi: 10.1083/jcb.133.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A., Guacci V., Koshland D. Pds1p, an inhibitor of anaphase in budding yeast, plays a critical role in the APC and checkpoint pathway(s) J. Cell Biol. 1996b;133:99–110. doi: 10.1083/jcb.133.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Skibbens R.V., Cheng J.W., Salmon E.D., Bloom K. Spindle dynamics and cell cycle regulation of dynein in the budding yeast, Saccharomyces cerevisiae. J. Cell Biol. 1995;130:687–700. doi: 10.1083/jcb.130.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudkovsky Y., Shteinberg M., Listovsky T., Brandeis M., Hershko A. Phosphorylation of Cdc20/fizzy negatively regulates the mammalian cyclosome/APC in the mitotic checkpoint. Biochem. Biophys. Res. Commun. 2000;271:299–304. doi: 10.1006/bbrc.2000.2622. [DOI] [PubMed] [Google Scholar]