Abstract
MtHAP2-1 is a CCAAT-binding transcription factor from the model legume Medicago truncatula. We previously showed that MtHAP2-1 expression is regulated both spatially and temporally by microRNA169. Here we present a novel regulatory mechanism controlling MtHAP2-1 expression. Alternative splicing of an intron in the MtHAP2-1 5′leader sequence (LS) becomes predominant during the development of root nodules, leading to the production of a small peptide, uORF1p. Our results indicate that binding of uORF1p to MtHAP2-1 5′LS mRNA leads to reduced accumulation of the MtHAP2-1 transcript and may contribute to spatial restriction of MtHAP2-1 expression within the nodule. We propose that miR169 and uORF1p play essential, sequential, and nonredundant roles in regulating MtHAP2-1 expression. Importantly, in contrast to previously described cis-acting uORFs, uORF1p is able to act in trans to down-regulate gene expression. Our work thus contributes to a better understanding of the action of upstream ORFs (uORFs) in the regulation of gene expression.
Keywords: HAP2, Medicago truncatula, alternative splicing, symbiosis, transcription factor, upstream ORF
Alternative splicing (AS) is an important post-transcriptional regulatory mechanism contributing to the functional complexity of higher eukaryotes. It can participate in increasing protein diversity by generating multiple transcripts from a single gene (Graveley 2001; Kazan 2003). In addition, AS can reduce gene expression by yielding isoforms that are degraded by nonsense-mediated mRNA decay (NMD) or other mechanisms (Hillman et al. 2004). In some cases, AS occurs within the 5′ leader sequence (LS) of the gene, leading to post-transcriptional regulation or inhibition of translation, as well as to the production of proteins with different properties (Procissi et al. 2002; Halterman et al. 2003; Hu et al. 2005; Roberts et al. 2005).
Overall organism complexity does not necessarily correlate with the complexity of the respective genome (∼25,000 genes in humans and ∼19,000 genes in nematodes, for example). Indeed, the human genome contains significantly fewer genes than was initially predicted at the outset of the Human Genome Project, with only ∼1.5% of the total length serving as protein-coding exons (International Human Genome Sequencing Consortium 2001). However, human genes are more complex, with AS generating a larger number of protein products. In certain cases, one gene can generate thousands of spliced variants (Soller 2006). Traditional gene-by-gene investigations of AS mechanisms are now being complemented by global approaches (Matlin et al. 2005; Blencowe 2006; Cuperlovic-Culf et al. 2006) and the publication of AS databases (http://www.ebi.ac.uk/asd). Recent genome-wide studies have indicated that up to 70% of human genes undergo AS (Baek et al. 2005). A big proportion of these genes encode proteins involved in signaling and gene regulation (Modrek and Lee 2002).
Although our knowledge of the extent of this phenomenon in higher plants is still poor, recent computational and experimental studies suggest that AS probably plays a far more significant role in the generation of plant proteome diversity than was previously thought. The proposal that 21% of all predicted Arabidopsis genes are alternatively spliced (Wang and Brendel 2006) is probably an underestimate since (1) the number of publicly available expressed sequenced tags (ESTs) from Arabidopsis (∼1.5 million) is far below that for other relatively well-studied genomes (nearly 8 million ESTs for the human genome; http://www.ncbi.nlm.nih.gov/dbEST/dbEST_summary.html), (2) full-length cDNAs are available for only approximately half of all predicted genes (Chung et al. 2006), and (3) ESTs from genes with low expression levels or with expression at a particular developmental stage or physiological condition, which are more likely to be alternatively spliced, are likely to be poorly represented in the databases.
Among the various cis elements in mRNAs that participate in regulating translation are AUG codons within transcript leaders (upstream AUGs) and, in some cases, associated upstream ORFs (uORFs). Up to 50% of eukaryotic mRNAs contain AUG codons within their transcript leader regions (Suzuki et al. 2000). uAUGs are conspicuously common in genes involved in the control of cellular growth and differentiation, including oncogenes and transcription factor genes (Kozak 1987, 1991, 2002). The best documented mode of action of uORFs is the inhibition of translation through a mechanism called ribosome stalling. In this case, the ribosome stalls during either the elongation or termination phase of uORF translation, creating a block in subsequent ribosome scanning.
The symbiotic interaction between leguminous plant species and bacteria collectively known as rhizobia leads to the de novo development of differentiated organs, known as nodules, on the roots of host plants. A fine-tuned molecular dialogue is established between plant and bacteria during nodulation, for which key players are the bacterial-produced lipooligosaccharides called Nod factors. Four main differentiated zones can be identified in a mature nodule: the meristematic apical zone (a plant stem cell compartment), the infection zone, the fixation zone, and the senescent zone (Vasse et al. 1990; Patriarca et al. 2004). Although early nodulation events, including Nod factor signaling and infection, have been extensively investigated, later stages of nodulation such as meristem differentiation and nodule organogenesis remain poorly understood (Foucher and Kondorosi 2000; Patriarca et al. 2004).
We previously identified MtHAP2-1, a HAP2-type transcription factor of the CCAAT-box-binding family (CBF), called NF-Y in humans and HAP in plants and yeast (El Yahyaoui et al. 2004). HAP proteins form heterotrimeric complexes that bind to the CCAAT consensus motif that is present in 30% of eukaryotic promoters (Mantovani 1998). HAP protein complexes appear to play central roles during development by regulating cell division and/or differentiation in eukaryotes (Bhattacharya et al. 2003; Di Agostino et al. 2006). In plants, HAP genes have been shown to be key regulators of embryogenesis, chloroplast biogenesis, flowering-time control, or blue light and abscisic acid responses, for example (Lotan et al. 1998; Kwong et al. 2003; Lee et al. 2003; Miyoshi et al. 2003; Ben-Naim et al. 2006; Warpeha et al. 2007). MtHAP2-1 plays a key role during nodule development, possibly by regulating nodule meristem function, and its expression is regulated by a microRNA, miR169 (Combier et al. 2006). The complementary expression pattern of MtHAP2-1 and miR169 indicates that the function of miR169 may be to restrict MtHAP2-1 expression to the meristematic zone as part of a process that is essential for nodule differentiation (Combier et al. 2006).
Here we describe a previously unknown post-transcriptional mechanism that regulates MtHAP2-1 expression during nodulation. AS of an intron in the MtHAP2-1 5′LS increases during nodule development, leading to the production of a small polypeptide, uORF1p. Interestingly, in contrast to previously described cis-acting uORFs, uORF1p is able to act in trans to down-regulate gene expression. We present a model in which both miR169 and uORF1p contribute to spatial and temporal regulation of MtHAP2-1 expression during nodule development.
Results
AS of the MtHAP2-1 intron 1, located in the 5′LS
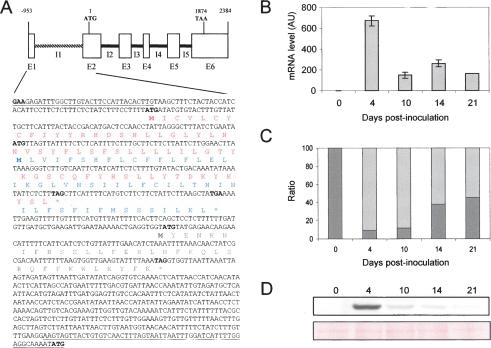
The 5′LS of MtHAP2-1 comprises two short exon sequences, E1 (33 base pairs [bp]) and E2 (55 bp), flanking a large intron, I1 (865 bp) (Fig. 1A). Analysis of the available Medicago truncatula EST databases (http://medicago.toulouse.inra.fr/Mt/EST) identified a set of eight ESTs corresponding to MtHAP2-1. Interestingly, one of these (MtBB39H03) contains part of the MtHAP2-1 5′leader intron sequence, suggesting that AS may occur in the MtHAP2-1 5′LS.
Figure 1.
MtHAP2-1 intron 1 is alternatively spliced. (A) Schematic representation of MtHAP2-1 gene structure. Exons (E1–E6) are represented with boxes and introns (I1–I5) are shown as thick lines. MtHAP2-1 first intron (I1), shown with a discontinuous bar, is located in the 5′LS. Sequence of the 5′LS is shown at the bottom with exons E1 and E2 underlined. Deduced amino acid sequences of uORF1 (red), uORF2 (blue), and uORF3 (gray), are shown under the nucleotide sequence. (B) Expression analysis of MtHAP2-1 during nodule development. MtHAP2-1 transcript levels were determined by qRT–PCR with cDNA obtained from M. truncatula roots inoculated with S. meliloti at the indicated time points. The expression values were normalized using the expression level of MtEF1α as an internal standard (El Yahyaoui et al. 2004) and related to the value of MtHAP2-1 expression level before inoculation (0 dpi), which is set at 1. Mean expression values and SEM values were calculated from the results of three independent experiments. (C) qRT–PCR analysis of the expression levels of alternatively (dark gray) and normally spliced (light gray) MtHAP2-1 transcripts. The expression values were normalized using the expression level of MtEF1α as an internal standard and related to the value of the total MtHAP2-1 gene expression at each time point, which is set at 100%. (D) MtHAP2-1 protein expression analysis. (Top panel) Total proteins from nodules were extracted at the indicated time points, expressed in days after inoculation, and protein extracts (50 μg) were separated and analyzed by immunoblotting using anti-MtHAP2-1 polyclonal antibodies. (Bottom panel) Equal loading in each lane was confirmed by Ponceau S staining.
Real-time RT–PCR analysis showed that MtHAP2-1 expression is very low in noninoculated M. truncatula roots. During nodule development, MtHAP2-1 expression in nodules peaks 4 d after inoculation with Sinorhizobium meliloti, and then expression decreases during later stages of nodulation (Fig. 1B; Combier et al. 2006). Since AS of introns located in 5′LSs may have an important effect on gene expression and regulation (Lahousse et al. 2004; Roberts et al. 2005), we further investigated AS of the intron I1 in the MtHAP2-1 5′LS. Primers within either the intron1 or the exon1–exon2 junction were used to quantify the expression of the alternatively and normally spliced forms of MtHAP2-1, respectively. As a control, PCR using primers hybridizing within the fourth intron (I4) of MtHAP2-1 resulted in no amplification, allowing us to rule out the presence of contaminating genomic DNA. As shown in Figure 1C, low expression of MtHAP2-1 in roots correlates with the presence of the alternatively spliced form in noninoculated roots and in nodules 14 and 21 dpi, whereas in young nodules (4 dpi), which show the highest level of MtHAP2-1 expression, about 90% of MtHAP2-1 is normally spliced. Intriguingly, AS increases during nodule development, with the alternatively spliced form reaching about 50% of total MtHAP2-1 expression 21 d after inoculation. Furthermore, Western blot analysis using a polyclonal anti-MtHAP2-1 antibody revealed that, as for the MtHAP2-1 transcript, MtHAP2-1 protein expression in nodules peaks 4 d after inoculation and then decreases during nodule development (Fig. 1D). Interestingly, this decrease in MtHAP2-1 protein levels correlates with an increase of the MtHAP2-1 alternatively spliced form, 14 and 21 dpi. In addition, qRT–PCR analysis showed that the weak expression of MtHAP2-1 in different plant organs correlates with the presence of the alternatively spliced form (data not shown). Taken together, these results suggest that AS of the intron 1 of MtHAP2-1 is regulated during nodule development and may result in reduced MtHAP2-1 accumulation.
The alternatively spliced version of MtHAP2-1, containing intron 1, will be hereafter referred to as MtHAP2-1-AS and the normally spliced form will be called MtHAP2-1-NS.
Specific silencing of MtHAP2-1-AS leads to a nodule growth defect
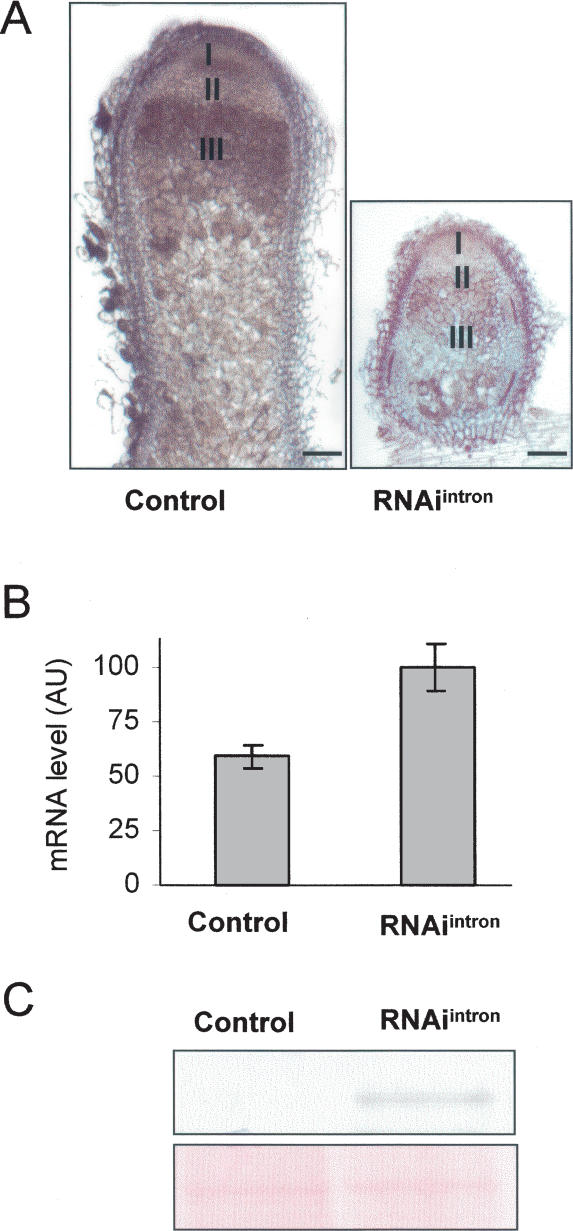
RNAi has been successfully used in cultured Drosophila cells to specifically silence the unspliced form of a given gene, allowing segregation between the unspliced and spliced variants (Celotto and Graveley 2002). To test the functional role of AS of MtHAP2-1 during nodule development, an RNAi construct was generated using a sequence located in the first intron of MtHAP2-1 (RNAiintron1). Specific silencing of MtHAP2-1-AS was confirmed by qRT–PCR analysis (Supplemental Fig. S1). We showed previously that silencing of both alternatively and normally spliced forms of MtHAP2-1, using a sequence located in the 3′UTR (RNAiMtHAP2-1), led to the formation of small, nonfunctional (non-nitrogen-fixing) nodules, arrested in growth (Combier et al. 2006). In contrast, nodules formed on transgenic roots carrying the RNAiintron1 silencing cassette are nitrogen fixing, with a normal differentiated structure and an active and persistent meristem (Fig. 2A). Nevertheless, due to the small size of these RNAiintron1 nodules compared with control nodules, it is clear that growth has been significantly modified. Importantly, MtHAP2-1 transcript and protein levels in these nodules, 21 d after inoculation, were significantly higher than for wild-type plants (Fig. 2B,C). These results imply that the MtHAP2-1-AS is required for normal nodule development, and suggest that this form negatively regulates MtHAP2-1 transcript and protein accumulation.
Figure 2.
RNAiintron1 transgenic roots show a nodule growth defect. (A) Light microscopy images of longitudinal sections (50 μm) of nodules of M. truncatula transgenic roots expressing either an RNAiintron1 construct (right) or an empty-vector control (left). Pictures were taken 21 d after inoculation with S. meliloti. Bar, 100 μm. (B) MtHAP2-1 transcript levels were determined by qRT–PCR with cDNA obtained from empty-vector control, and RNAiintron1 M. truncatula nodules 21 d after inoculation with S. meliloti. The expression values were normalized using the expression level of MtEF1α as an internal standard and related to the value of the MtHAP2-1 transcript level in the RNAiintron1 transgenic plants, which is set at 100%. Mean expression values and SEM values were calculated from the results of two independent experiments. (C, top panel) Total proteins from empty-vector control and RNAiintron1 M. truncatula nodules were extracted 21 d after inoculation with S. meliloti, and protein extracts (50 μg) were analyzed by immunoblotting using anti-MtHAP2-1 polyclonal antibodies. (Bottom panel) Equal loading in both lanes was confirmed by Ponceau S staining.
Overexpression of MtHAP2-1 without its regulatory sequences results in small, nitrogen-fixing nodules
Our results indicate that both MtHAP2-1 5′ and 3′-terminal sequences play key roles in the regulation of nodule development. To further test this idea, MtHAP2-1, without its 5′ and 3′ regulatory regions, was overexpressed in M. truncatula roots. Overexpression of MtHAP2-1 in the transgenic roots was confirmed by both qRT–PCR and Western blot analysis using antiMtHAP2-1 polyclonal antibodies (Supplemental Fig. S2B,C). Transgenic roots developed nitrogen-fixing nodules, with a normal and differentiated structure. However, nodule size was again significantly reduced, as compared with the controls (Supplemental Fig. S2A). This result provides further evidence that altered spatial and temporal expression of MtHAP2-1 results in altered nodule organogenesis.
A GUS fusion reveals that uORF1 is expressed in the nodule infection zone
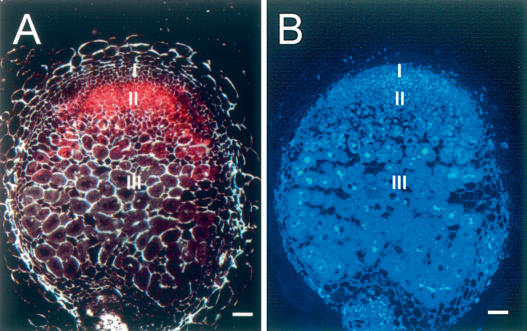
Analysis of the sequence of the first intron of MtHAP2-1 revealed 11 uORFs potentially encoding polypeptides of between 4 and 62 amino acids. We focused our analysis on the three longest ORFs, named uORF1, uORF2, and uORF3, potentially encoding polypeptides of 62, 50, and 34 amino acids, respectively (Fig. 1). If the first intron is unspliced, ribosome scanning could initiate translation at the uAUGs codons of these uORFs rather than at the AUG at the start of the main MtHAP2-1 ORF. To test this hypothesis, ∼2.3-, ∼2.4-, and ∼2.6-kb upstream MtHAP2-1 genomic promoter sequences extending as far as the uATGs of uORF1, uORF2, and uORF3, respectively, were fused to a GUS reporter gene and M. truncatula roots were transformed with these three constructs. Whereas roots expressing fusions with uORF2 and uORF3 did not show any GUS expression (data not shown), the fusion with uORF1 revealed GUS expression (red staining) located in the infection zone of mature nodules (zone II) (Fig. 3A) but not in the meristematic zone (zone I) where MtHAP2-1 is expressed. This observation strongly suggests that the AUG of uORF1 can initiate translation, and therefore may lead to the synthesis of the 62-amino-acid polypeptide in nodule tissues.
Figure 3.
uORF1 is expressed in the infection zone of mature nodules. Histochemical localization of β-glucuronidase (GUS) activity (A, red color) and nuclear staining using DAPI (B) of longitudinal sections of nodules from M. truncatula transgenic roots expressing a uORF1-GUS fusion construct. Dark-field (A) and fluorescence (B) microscopy images were taken 21 d after inoculation with S. meliloti. (B). Differentiated zones can be distinguished in the mature nodule after nuclear staining with DAPI (Vasse et al. 1990). (I) Meristematic zone. (II) Infection zone. (III) Nitrogen fixation zone. Bar, 50 μm.
Overexpression of uORF1 leads to a severe nodule developmental defect
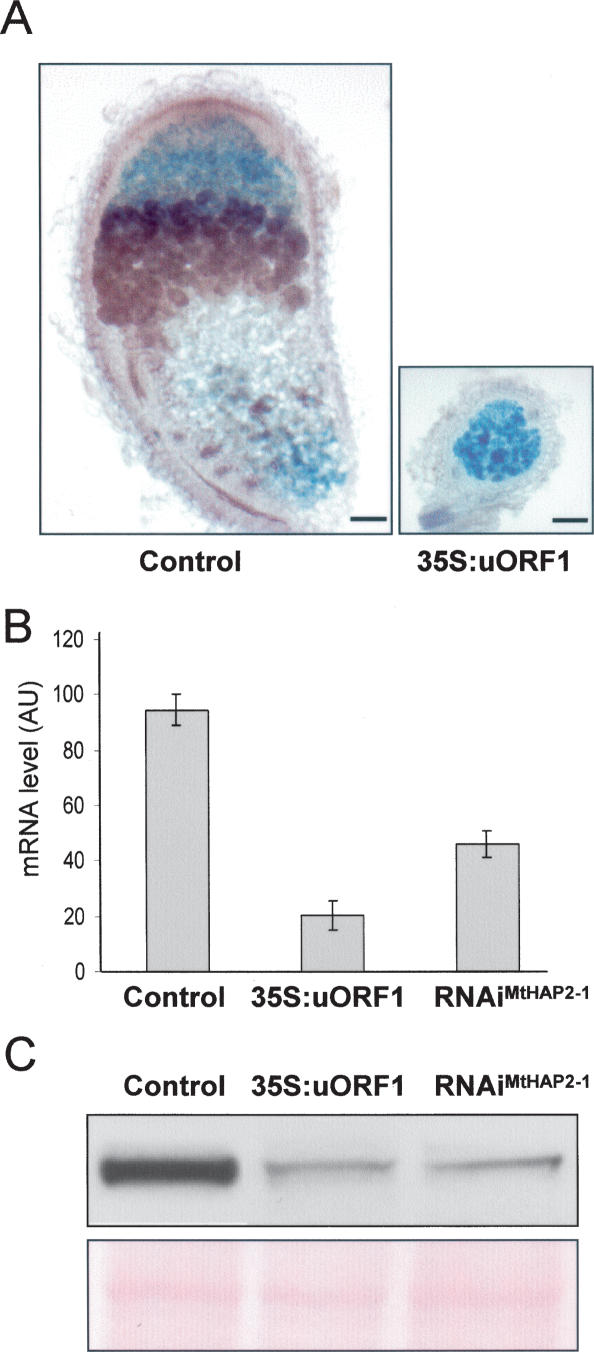
To obtain more functional information, uORF1 was overexpressed in M. truncatula roots, under the control of the constitutive 35S promoter. Very small nodules were formed on transgenic roots 21 d after inoculation, and nodule sectioning revealed only poorly differentiated zones, enclosed by the endodermis, thus indicative of early growth arrest (Fig. 4A). Importantly, MtHAP2-1 transcript and protein levels were greatly reduced in these nodules 5 d after inoculation (Fig. 4B,C). This phenotype is very similar to that observed in RNAiMtHAP2-1 nodules, for which MtHAP2-1 transcript and protein levels are also significantly reduced (Fig. 4B,C; Combier et al. 2006). These data imply that uORF1 can act in trans to negatively regulate MtHAP2-1 accumulation, and confirm that the precise regulation of MtHAP2-1 expression is required for normal nodule development.
Figure 4.
Overexpression of uORF1 in roots leads to MtHAP2-1 degradation and a defect in nodule organogenesis. (A) Light microscopy images (50 μm) of longitudinal sections of nodules of M. truncatula transgenic roots expressing either a 35S:uORF1 construct (right) or an empty-vector control (left). Pictures were taken 21 d after inoculation with S. meliloti. Bar, 100 μm. (B) MtHAP2-1 transcript levels were determined by qRT–PCR with cDNA obtained from empty-vector control, 35S:uORF1 and RNAiMtHAP2-1 M. truncatula transgenic roots 5 d after inoculation with S. meliloti. The expression values were normalized using the expression level of MtEF1α as an internal standard and related to the value of the MtHAP2-1 transcript level in the control plants, which is set at 100%. Mean expression values and SEM values were calculated from the results of two independent experiments. (C, top panel) Total proteins from empty-vector control, 35S:uORF1 and RNAiMtHAP2-1 M. truncatula transgenic roots were extracted 5 d after inoculation with S. meliloti, and protein extracts (50 μg) were analyzed by immunoblotting using anti-MtHAP2-1 polyclonal antibodies. (Bottom panel) Equal loading was confirmed by Ponceau S staining.
Expression of the uORF1 protein in trans leads to reduced accumulation of a MtHAP2-1 5′LS-AS containing transcript
To gain further insight into the control of MtHAP2-1 expression by uORF1, a 35S:GUS fusion construct, with or without the alternatively spliced, intron1-containing, MtHAP2-1 5′LS (Fig. 5A), was coexpressed with either 35S:uORF1, 35S:uORF2, or 35S:uORF3 in Nicotiana benthamiana leaves. Both MtHAP2-1 5′LS-AS and MtHAP2-1 5′LS-NS transcripts are detected after transient delivery of a MtHAP2-1 5′LS-AS-GUS fusion (Supplemental Fig. S3), confirming that AS of MtHAP2-1 5′LS occurs in N. benthamiana and validating our strategy to use N. benthamiana as a heterologous system. Importantly, overexpression of uORF1 led to decreased GUS expression and reduced GUS transcript and protein levels only when coexpressed with the 35S:5′LS-GUS construct (Fig. 5B–D, lanes 1,2). The higher GUS expression level observed with the 35S:5′LS-AS-GUS compared with the 35S:GUS construct is probably due to intron-mediated enhancement, a phenomenon that has been reported to confer up to 100-fold enhancement of gene expression (Kim et al. 2006). No effect on GUS expression was detected when uORF1 was coexpressed with 35S:GUS-3′UTR (data not shown) or when uORF2 or uORF3 were used for coinfiltration, thus indicating a clear specificity for uORF1 action on the MtHAP2-1 5′LS (Supplemental Fig. S4). To confirm that uORF1 is indeed translated in N. benthamiana, an HA-tagged version of uORF1 was generated. As for the untagged version of uORF1, expression of 35S:uORF1-HA led to decreased GUS expression, and reduced GUS transcript and protein levels, only when coexpressed with the GUS construct containing the MtHAP2-1 5′LS (Fig. 5B–D, lanes 1,3). Expression of the uORF1-HA fusion protein was confirmed by Western blotting using anti-HA antibodies. No effect on the expression of 35S:5′LS-GUS was observed when using a mutated form of uORF1, in which the ATG start codon had been mutated to AGG to prevent protein translation initiation (Supplemental Fig. S4). Finally, we found that uORF1p is able to down-regulate expression of both MtHAP2-1 5′LS-AS and MtHAP2-1 5′LS-NS-containing transcripts (Supplemental Fig. S3).
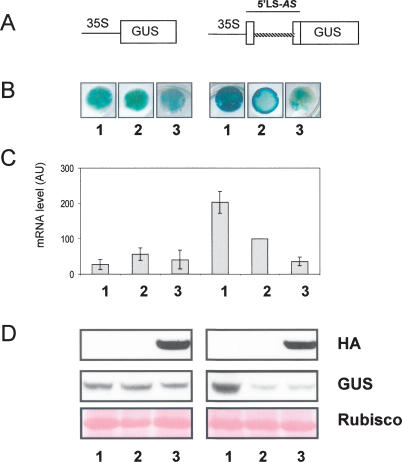
Figure 5.
uORF1p expression in N. benthamiana leaves leads to reduced levels of GUS fused to MtHAP2-1 5′LS-AS. Expression analysis of GUS (β-glucuronidase) 2 d after Agrobacterium-mediated transient expression of the GUS fusions shown in A together with either an empty vector control (1), untagged 35S:uORF1 (2), or 35S:uORF1-HA (3). (A) Schematic representation of the GUS fusions used in this experiment. MtHAP2-1 5′LS, containing the first intron (discontinuous line), was fused to the β-glucuronidase (GUS) gene under the control of the 35S promoter. (B) GUS expression determined by GUS histochemical staining of N. benthamiana leaf discs. (C) GUS transcript levels determined by qRT–PCR. The expression values were normalized using the expression level of NbEF1α as an internal standard and related to the expression value of 35S:5′LS-GUS + untagged 35S:uORF1, which is set at 100%. Mean expression values and SEM values were calculated from the results of seven independent experiments. (D) Western blot analysis showing GUS protein (middle panel) and uORF1-HA (top panel) expression levels. Ponceau S staining of ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) for confirmation of equal loading in each lane is shown at the bottom.
Taken together, these data provide strong evidence that the uORF1p peptide acts in trans to down-regulate MtHAP2-1 expression by targeting the 5′LS.
uORF1p binds to both MtHAP2-1-AS and MtHAP2-1-NS-containing mRNA
To investigate whether uORF1p can interact directly with the MtHAP2-1 5′LS-containing mRNA, we made use of an RNA pull-down assay. uORF1p, transiently expressed in N. benthamiana leaves, was found to bind both MtHAP2-1 5′LS-AS and MtHAP2-1-NS biotinylated RNA (Fig. 6A, lanes 2,3). The specificity of this binding was demonstrated by the inability of uORF1p-HA to bind to the streptavidin resin alone (data not shown) or after incubation with a biotinylated control RNA of the same length corresponding to the unrelated Arabidopsis transcription factor AtMYB30 (Fig. 6A, lane 4). Interestingly, no uORF1p binding was detected after incubation with a biotinylated RNA corresponding to MtHAP2-1 deleted from its 5′LS (MtHAP2-1 Δ5′LS) (Fig. 6A, lane 5), strongly suggesting that uORF1p physically interacts with the E1 and/or E2 exon sequence(s) present in MtHAP2-1 5′LS (Fig. 1).
Figure 6.
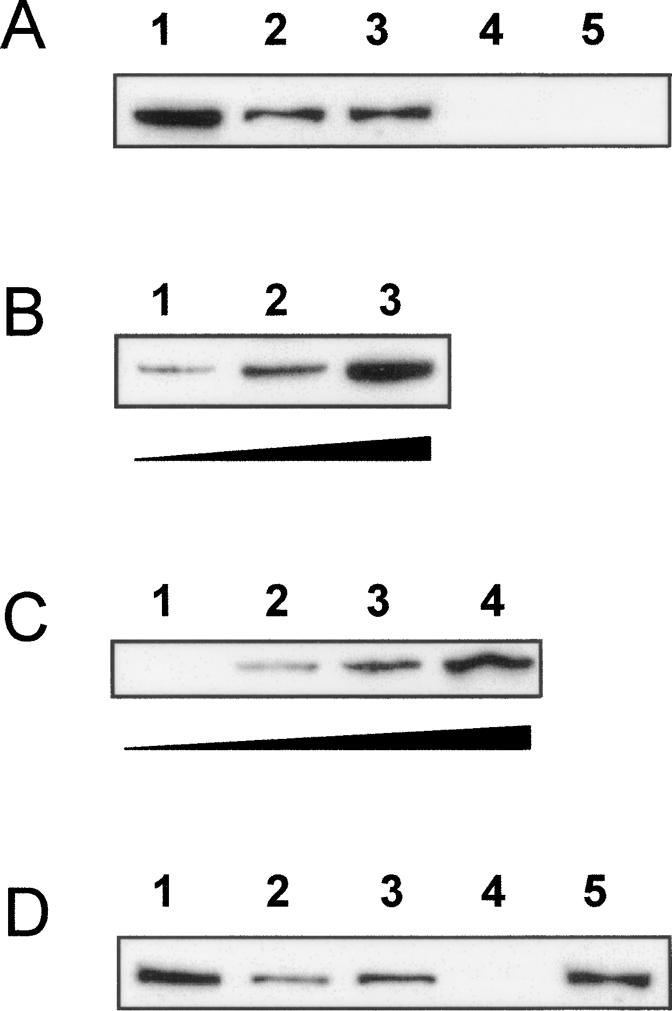
uORF1p binds specifically to MtHAP2-1 5′LS RNA. Western Blots with an anti-HA antibody showing uORF1p-HA recovered after the RNA pull-down assays. (A, lane 1) Input of uORF1p-HA corresponds to 10% of the total N. benthamiana protein extract used for each binding assay. uORF1p-HA was eluted from the streptavidin beads after incubation with biotinylated MtHAP2-1 5′LS-AS-corresponding (lane 2) or MtHAP2-1-NS-corresponding (lane 3) RNA. uORF1p-HA was not eluted from the streptavidin beads after incubation with the biotinylated AtMYB30- (lane 4) or MtHAP2-1 Δ5′LS-corresponding RNA (lane 5). (B) Increasing amounts of total N. benthamiana protein extracts expressing uORF1p (1 mg, lane 1; 5 mg, lane 2; and 25 mg, lane 3) were used for the binding assay. uORF1p-HA was eluted from the streptavidin beads after incubation with biotinylated MtHAP2-1 5′LS-AS-corresponding RNA. (C) Increasing ratios of biotinylated to nonbiotinylated MtHAP2-1 5′LS-AS-corresponding RNA (0:1, lane 1; 1:3, lane 2; 1:1, lane 3; and 1:0, lane 4) were used for incubation with the streptavidin beads prior to binding of uORF1p-HA. (D) Competition experiment in which uORF1p was first bound to streptavidin beads containing a 1:1 mix of biotinylated to nonbiotinylated MtHAP2-1 5′LS-AS RNA (input of uORF1p, lane 1) prior to elution using MtHAP2-1 5′LS-AS (eluted uORF1p, lane 2) or AtMYB30 nonbiotinylated RNAs (eluted uORF1p, lane 4). The amount of uORF1p remaining on the beads after treatment with MtHAP2-1 5′LS-AS (lane 3) or AtMYB30 nonbiotinylated RNAs (lane 5) is also shown.
uORF1p was able to bind MtHAP2-1 5′LS-AS biotinylated RNA in a dose-dependent manner (Fig. 6B, lanes 1–3). No uORF1p was eluted from the beads when a nonbiotinylated MtHAP2-1-AS RNA was used for incubation with the streptavidin resin (Fig. 6C, lane 1). Different ratios of nonbiotinylated to biotinylated MtHAP2-1 5′LS-AS RNAs were used for binding to the streptavidin beads, further demonstrating the quantitative character of uORF1p binding to MtHAP2-1 5′LS (Fig. 6C, lanes 1–4). Finally, a competition experiment was performed in which uORF1p was first bound to MtHAP2-1 5′LS-AS RNA-containing beads (Fig. 6D, lane 1) and then partially eluted from the resin using a nonbiotinylated MtHAP2-1 5′LS-AS- (Fig. 6D, lane 2) but not a nonbiotinylated AtMYB30- (Fig. 6D, lane 4) corresponding RNA. Figure 6D also shows the amount of uORFp1 protein that remained attached to the beads after competition with both RNAs (nonbiotinylated MtHAP2-1 5′LS-AS [lane 3] and AtMYB30 [lane 5]).
These results clearly show that uORF1p is able to bind in a quantitative manner to an mRNA containing either the alternatively or the normally spliced MtHAP2-1 5′LS, and suggest that this could be the mechanism by which uORF1p mediates the negative regulation of MtHAP2-1 expression in planta.
Discussion
In this study, we show that AS of intron 1, present in the 5′LS of MtHAP2-1, regulates MtHAP2-1 expression during nodule formation. Unlike the many examples described for Drosophila and Caenorhabditis elegans, there are very few cases of developmental switches being controlled by AS in plants, and more particularly, in legumes (Lorkovic et al. 2000; Macknight et al. 2002; Cheung et al. 2006). Regulation of MtHAP2-1 expression thus provides a good model to determine the molecular mechanisms that have evolved in plants to control pre-mRNA processing.
A dual regulation by miR169 and uORF1p controls MtHAP2-1 expression and nodule function
Data presented here, together with our previous work on regulation of MtHAP2-1 by miR169, suggest that a dual regulation by miR169 and uORF1p controls MtHAP2-1 expression. We previously demonstrated that miR169 is expressed in the infection zone of the nodule (Combier et al. 2006). Likewise, GUS expression in the infection zone of nodules from uORF1-GUS transgenic plants (Fig. 3) strongly suggests that ORF1p expression also localizes to this region of the nodule. Misregulation of either miR169 or uORF1p expression has a major effect on MtHAP2-1 transcript and protein expression, which correlates with a nodule growth defect: Roots expressing the RNAiMtHAP2-1 silencing cassette or overexpressing miR169 or uORF1p possess significantly reduced MtHAP2-1 levels that correlate with precocious nodule meristem abortion, whereas roots expressing the RNAiintron1 silencing cassette or a miR169-resistant form of MtHAP2-1, or overexpressing MtHAP2-1 without its regulatory regions, possess increased MtHAP2-1 levels that correlate with a nodule growth defect. Although one could argue that nodule growth deficiency, in RNAiintron1 or 35S:uORF1 transgenic plants, might lead to altered MtHAP2-1 expression, this seems unlikely, since MtHAP2-1 expression is unaltered in Medicago mutants presenting a reduced nodule growth phenotype (T. Vernie, J.-P. Combier, P. Gamas, and A. Niebel, unpubl.), such as lin (Kuppusamy et al. 2004) and nip (Veereshlingam et al. 2004). We therefore conclude that nodule growth defect in RNAiintron1 or 35S:uORF1 plants is (1) not sufficient to explain the reduced expression of MtHAP2-1 and (2) due to reduced MtHAP2-1 expression, and not vice versa.
We propose that restriction of MtHAP2-1 expression to the meristematic zone of the nodule is essential for normal nodule development, and that this is achieved by both miR169 targeting of the MtHAP2-1 3′UTR and alternative processing of the MtHAP2-1 5′LS. The pattern of both miR169 expression and AS occurrence in nodules suggests that the control of MtHAP2-1 expression is finely regulated. miR169 expression and AS appear to be complementary from a temporal point of view, since miR169 transcript abundance peaks 10 d after inoculation and decreases later during nodule formation (Supplemental Fig. S5), whereas AS increases during nodule development, reaching a maximum 21 d after inoculation (Fig. 1C; Supplemental Fig. S5).
Figure 7 proposes a model for the regulation of MtHAP2-1 expression. In this model, miR169 and uORF1 play essential, sequential, and nonredundant roles in the control of MtHAP2-1 expression, with miR169 acting during the early stages of nodule development and uORF1p taking over this function later in nodulation. Our data show that uORF1p acts on both alternatively and normally spliced MtHAP2-1 transcripts, since (1) silencing of MtHAP2-1-AS also leads to the accumulation of MtHAP2-1-NS (Supplemental Fig. S1), (2) uORF1p is able to down-regulate the expression of both the alternatively and normally spliced forms of MtHAP2-1 5′LS in N. benthamiana (Supplemental Fig. S3), and (3) uORF1p binds to both MtHAP2-1-AS and MtHAP2-1-NS-corresponding RNAs (Fig. 6A). Taken together, these results strongly suggest that E1 and/or E2 exons are the target sequences of uORF1p binding for regulation of MtHAP2-1 expression.
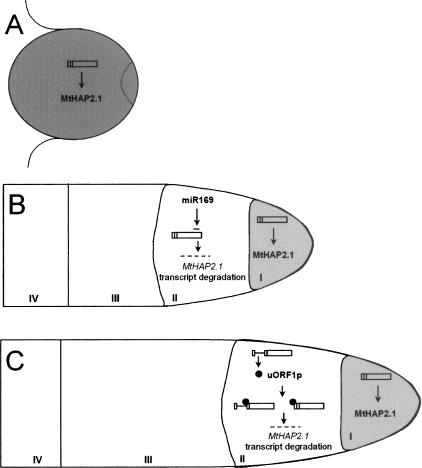
Figure 7.
Model for regulation of MtHAP2-1 expression to orchestrate nodule development. (A) Four days after inoculation, the nodule structure is not well differentiated. miR169 and uORF1p not being expressed, MtHAP2-1 presents a higher expression level, evenly distributed throughout the nodule (data not shown). (B) Ten days after inoculation, miR169 is expressed in the nodule infection zone (II), leading to MtHAP2-1 transcript down-regulation and restricting MtHAP2-1 expression to the meristematic zone (I) and normal nodule growth (Combier et al. 2006). (C) Twenty-one days after inoculation, AS of MtHAP2-1 occurs, resulting in expression of uORF1p in the infection zone. uORF1p binding to the MtHAP2-1 transcript induces MtHAP2-1 transcript degradation, thereby restricting MtHAP2-1 expression to the meristematic zone of the nodule.
Expression of MtHAP2-1 is controlled by a novel post-transcriptional regulatory mechanism
uORF-mediated regulation of gene expression is well studied, especially in animals, where inhibition of translation is the best documented mode of action (Morris et al. 2000; Kozak 2002). Translation of a uORF can produce a cis-acting peptide that causes effector molecule-dependent stalling of the ribosomes at the end of the uORF. In other cases it is the length or position, or other features of the uORF, rather than the encoded peptide, that determine the efficiency with which ribosomes reinitiate downstream translation. Translation of the uORF can also control gene expression by affecting the stability of the mRNA (Gaba et al. 2005). Finally, trans-acting factors may participate in regulatory mechanisms (Vilela and Mc Carthy 2003). uORF-mediated translational regulation in cis has been reported for several plant genes including Opaque 2 and Lc in maize, and SAMDC, ATB2, and SAC51 in Arabidopsis (Imai et al. 2006, and references therein).
Here we describe a novel mode of action for a uORF. The major findings to emerge from our experiments are the following: (1) The regulatory effect of uORF1p on MtHAP2-1 expression is dependent on AS of the first intron in the 5′LS of MtHAP2-1; (2) despite the fact that the context of uORF1 start codon (CCTTTTATGA) is not very favorable (Kozak 2002), initiation of translation at uAUG1 can be detected in the nodule infection zone (Fig. 3); (3) roots overexpressing uORF1 display a nodule developmental defect that correlates with reduced MtHAP2-1 transcript and protein expression (Fig. 4); (4) transient expression of the uORF1 protein in N. benthamiana leads to a reduction on the level of a GUS fusion containing the alternatively spliced MtHAP2-1 5′LS (Fig. 5); and (5) uORF1p is able to bind both the alternatively and normally spliced MtHAP2-1 5′LS-corresponding mRNAs (Fig. 6).
This work demonstrates that uORF1p is able to act in trans to down-regulate gene expression, although our data do not allow us to exclude the possibility of additional cis regulation mediated by MtHAP2-1 uORF1. It is still unclear how binding of uORF1p to the MtHAP2-1 5′LS-corresponding mRNA leads to reduced transcript accumulation. We can exclude this to be due to transitive silencing mediated by the 5′LS in N. benthamiana, because expression of uORF2, uORF3, or a mutated version of uORF1 in which the ATG start codon had been mutated to AGG, does not result in transcript down-regulation. Since uORF1p acts in trans, we do not expect down-regulation of MtHAP2-1 expression to be due to ribosome stalling mediated by the uORF nascent peptide, in contrast to previously described cis-acting uORFs. It is possible that overexpression of uORF1p somehow results in blockage of either (1) transcription, resulting in reduced accumulation of MtHAP2-1 mRNA, or (2) translation, leading to degradation of a nontranslated transcript by NMD or other mechanisms. However, the fact that a mutant version of uORF1p also binds the MtHAP2-1 5′LS-corresponding mRNA but does not result in transcript degradation (J.P. Combier and S. Rivas, unpubl.) makes these two possibilities less likely. Alternatively, it is tempting to speculate that uORF1p may recruit additional proteins involved in nucleic acid degradation. Since uORF1p-dependent mRNA down-regulation is active in N. benthamiana, it is likely that, although BLAST analysis did not identify any uORF1-like gene in other organisms, the protein(s) required for uORF1p-dependent mRNA down-regulation are conserved among different plant species.
To our knowledge, this is the first report of a uORFp acting in trans to down-regulate gene expression following AS, and therefore it contributes to a broader understanding of the role played by uORFs in the regulation of gene expression. Further studies are now required to determine to what extent this mechanism can be generalized beyond the control of MtHAP2-1 expression during nodule development.
Materials and methods
Biological material
M. truncatula Gaertn cv. Jemalong genotype A17 was grown aeroponically as described (Journet et al. 2001) and inoculated with the S. meliloti strain RCR2001 pXLGD4 (GMI6526) (Ardourel et al. 1994) as described previously (El Yahyaoui et al. 2004). Agrobacterium-mediated transient expression in N. benthamiana leaves was performed as described (Rivas et al. 2004). Root transformations were performed as reported previously (Combier et al. 2006).
Vectors and constructs
PCR amplification was performed using Pfx polymerase (Invitrogen) and used primers are shown in Supplemental Table S1.
For M. truncatula root transformation, pPEX-derived vectors were used (Combier et al. 2006). To generate the RNAiintron1 construct, a 249-bp sequence from the intron 1 was amplified using intron1RNAi5′ and intron1RNAi3′ primers, and cloned into pPEX RNAi vector. For the uORFs-GUS fusions, 2300-, 2373-, and 2616-bp sequences upstream of uATG1, uATG2, and uATG3, respectively, were amplified with primers MtHAP2-1Pro5′ and uORF1GUS3′, uORF2GUS3′, and uORF3GUS3′, respectively, and cloned into a pPEX-GUS vector. The 35S:uORF1, 35S:uORF2, and 35S:uORF3 constructs were made after uORF1, uORF2, and uORF3 PCR amplification, with uORF1 5′ and uORF1 3′, uORF2 5′ and uORF2 3′, and uORF3 5′ and uORF3 3′ primers, respectively, and cloning into pPEX. For the MtHAP2-1 overexpression construct, the MtHAP2-1 gene was amplified with MtHAP2-1 5′ and MtHAP2-1 3′2 primers and cloned into the pPEX vector.
For the N. benthamiana transient assays, a 35S:ORF1-HA construct was generated after amplification of uORF1 using uORF1 5′ and uORF1 3′ primers and cloning into the pBIN19g vector (Rivas et al. 2004). For overexpression of HA-tagged uORF1 mutated in the start codon ATG, we used the uORF1 AGG 5′ primer. To generate the GUS fusion to the alternatively spliced MtHAP2-1 5′LS, the 5′LS was amplified using 5′LS5′ and 5′LS3′ and cloned in position 5′ of the GUS cassette in the pPEX GUS vector.
Histochemical staining and microscopy studies
Nodule sections (50 μm thick) were prepared and observed as described (Combier et al. 2007). Histology of the nodule was performed on fixed material embedded in Technovit 7100 resin (Hereaus Kulzer). Four-micron thick sections were observed in dark field or epifluorescence microscopy after nuclear staining with DAPI (diaminophenylindol). In all cases, shown figures are representative of three to five independent transformation experiments, in which about 30 transgenic roots were chosen and ∼10–20 individual nodules were observed.
RNA extraction and qRT–PCR analysis
Material for RNA analysis was ground in liquid nitrogen, and total RNA was isolated using the SV total RNA extraction kit (Promega) according to the manufacturer’s recommendations. Reverse transcription was performed using 3 μg of total RNA and SuperScript reverse transcriptase II (Invitrogen). qPCR was run on a LightCycler system (Roche Diagnostics) according to the manufacturer’s recommendations. MtEF1α or NbEF1α were used as an internal standard (El Yahyaoui et al. 2004). Primers used in the different experiments are listed in Supplemental Table S1. The specificity of the amplifications was verified by sequencing of the PCR products and by melting curve analysis at the end of each experiment. Efficiency of the amplification was verified by the analysis of standard curves.
Primers uORF1q5′ and uORF1q3′ were used to quantify the MtHAP2-1 unspliced form, and exon1–2 and MtHAP2-1TP4 primers were used for quantification of the spliced form. Amplification of either form was confirmed by sequence analysis of the obtained PCR fragments. Intron4 5′ and intron4 3′ primers, aligning at intron 4 of MtHAP2-1, were used to confirm the absence of contaminating genomic DNA.
Anti-MtHAP2-1 antibody production
Anti-MtHAP2-1 polyclonal antibodies were raised in rabbit by Eurogentec (http://www.eurogentec.be) against two peptides in the N terminus of MtHAP2-1; namely, 1MAMQPVYLKEHEGNV15 and 64APSKNLVRGVEQLFD78.
Expression analysis
Histochemical GUS staining were performed and analyzed as described (Combier et al. 2006).
To analyze protein expression, root material or N. bentamiana leaf discs were ground in liquid nitrogen and 50–100 mg of powder were resuspended in 100 μL of extraction buffer (50 mM Tris-HCl at pH 7.4, 150 mM NaCl, 10% glycerol [v/v], 1 mM DTT, 1 mM PMSF, 1% plant protease inhibitor cocktail [Sigma]) and centrifuged at 10,000g for 10 min at 4°C. Protein concentration in the supernatant was determined with the Bradford protein assay kit (Bio-Rad), using BSA as a standard. Fifty micrograms of total protein were separated on a NuPage 4%–12% Bis-Tris gels (Invitrogen) according to the manufacturer's instructions and transferred onto Protran BA85 nitrocellulose membranes (Schleicher & Schuell) by wet electroblotting (Mini-Protean II system; Bio-Rad). For detection of MtHAP2-1 and GUS, the blots were incubated with anti-MtHAP2-1 polyclonal antibodies (1:10,000 dilution) and rabbit anti-β glucuronidase polyclonal antibodies (1:1,000 dilution; Fitzgerald Industries International), respectively. After incubation with the secondary antibody (anti-rabbit IgG-peroxidase, 1:15,000; Sigma), bands were visualized using the ECL Plus kit (Amersham Pharmacia Biotech) under standard conditions. For detection of HA-tagged uORF1p, blots were incubated with anti-HA rat monoclonal (clone EF10) antibodies (Roche), linked to horseradish, at a final dilution of 1:1,000.
RNA pull-down assays
MtHAP2-1 with no 5′LS was amplified by PCR from cDNA from M. truncatula nodules, 4 d after inoculation with S. meliloti, using primers MtHAP2-1 5′ and 3′LS3′ and cloned into a pGEM-T vector (Promega). This construct was named MtHAP2-1Δ5′LS.
Alternatively spliced MtHAP2-1 5′LS was amplified using primers 5′LS5′ and 5′LS3′ from M. truncatula genomic DNA, cloned into pGEM-T and named MtHAP2-1 5′LS-AS.
Finally, normally spliced MtHAP2-1 was amplified from M. truncatula nodules, 4 d after inoculation with S. meliloti, using primers 5′LS5′ and 3′LS3′ and cloned into pGEM-T. This construct was named MtHAP2-1-NS.
As a control, the coding sequence of the Arabidopsis transcription factor AtMYB30 (At3g28910) was also amplified and cloned into pGEM-T.
PCR fragments containing either MtHAP2-1Δ5′LS, MtHAP2-1 5′LS-AS, or MtHAP2-1-NS were amplified using the M13 Fwd and M13 Rev primers, and used for in vitro transcription using a Riboprobe Combination System SP6/T7 (Promega), in the presence of biotinylated UTP. The biotinylated RNAs (or different ratios of nonbiotinylated to biotinylated RNAs) were bound to streptavidin magnetic beads (μMACS Streptavidin Kit; Milteny Biotec) for 1 min at room temperature and incubated with various amounts of a total protein extract containing HA-tagged uORF1p from N. benthamiana agroinfiltrated leaves, for 1 h at 4°C, as described (Campalans et al. 2004). Bound proteins were eluted in protein loading buffer and analyzed by Western Blot using anti-HA rat monoclonal (clone EF10) antibodies (Roche) as described above. Nonbiotinylated MtHAP2-1 5′LS-AS-corresponding RNA was also used in competition experiments to elute the bound uORF1p.
Accession numbers
The GenBank accession number of MtHAP2-1 and uORF1p described in this study is EF488826.
Acknowledgments
We thank Martin Crespi and David Barker for critical reading of the manuscript. This work was supported by FP6 Grain Legume Integrated Project (FP6-2002-FOOD-1-506223).
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.461808.
References
- Ardourel M., Demont N., Debelle F., Maillet F., de Billy F., Prome J.C., Denarie J., Truchet G. Rhizobium meliloti lipooligosaccharide nodulation factors: Different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek D., Green P. Sequence conservation, relative isoform frequencies, and nonsense-mediated decay in evolutionarily conserved alternative splicing. Proc. Natl. Acad. Sci. 2005;102:12813–12818. doi: 10.1073/pnas.0506139102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Naim O., Eshed R., Parnis A., Teper-Bamnolker P., Shalit A., Coupland G., Samach A., Lifschitz E. The CCAAT binding factor can mediate interactions between CONSTANS-like proteins and DNA. Plant J. 2006;46:462–476. doi: 10.1111/j.1365-313X.2006.02706.x. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A., Deng J.M., Zhang Z., Behringer R., de Crombrugghe B., Maity S.N. The B subunit of the CCAAT box binding transcription factor complex (CBF/NF-Y) is essential for early mouse development and cell proliferation. Cancer Res. 2003;63:8167–8172. [PubMed] [Google Scholar]
- Blencowe B.J. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- Campalans A., Kondorosi A., Crespi M. Enod40, a short open reading frame-containing mRNA, induces cytoplasmic localization of a nuclear RNA binding protein in Medicago truncatula. Plant Cell. 2004;16:1047–1059. doi: 10.1105/tpc.019406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celotto A.M., Graveley B.R. Exon-specific RNAi: A tool for dissecting the functional relevance of alternative splicing. RNA. 2002;8:718–724. doi: 10.1017/s1355838202021064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung F., Haas B.J., Goldberg S.M., May G.D., Xiao Y., Town C.D.2006Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology BMC Genomics 7272 . 10.1186/1471-2164-7-272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung B.Y., Simons C., Firth A.E., Brown C.M., Hellens R.P. Effect of 5′UTR introns on gene expression in Arabidopsis thaliana. BMC Genomics. 2006;7:120. doi: 10.1186/1471-2164-7-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J.P., Frugier F., de Billy F., Boualem A., El-Yahyaoui F., Moreau S., Vernie T., Ott T., Gamas P., Crespi M., et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes & Dev. 2006;20:3084–3088. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Combier J.P., Vernie T., de Billy F., El Yahyaoui F., Mathis R., Gamas P. The MtMMPL1 early nodulin is a novel member of the matrix metalloendoproteinase family with a role in Medicago truncatula infection by Sinorhizobium meliloti. Plant Physiol. 2007;144:703–716. doi: 10.1104/pp.106.092585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuperlovic-Culf M., Belacel N., Culf A.S., Ouellette R.J. Microarray analysis of alternative splicing. OMICS. 2006;10:344–357. doi: 10.1089/omi.2006.10.344. [DOI] [PubMed] [Google Scholar]
- Di Agostino S., Strano S., Emiliozzi V., Blandino G., Piaggio G. Gain of function of mutant p53: The mutant p53/NF-Y protein complex reveals an aberrant transcriptional mechanism of cell cycle regulation. Cancer Cell. 2006;10:191–202. doi: 10.1016/j.ccr.2006.08.013. [DOI] [PubMed] [Google Scholar]
- El Yahyaoui F., Kuster H., Ben Amor B., Hohnjec N., Puhler A., Becker A., Gouzy J., Vernie T., Gough C., Niebel A., et al. Expression profiling in Medicago truncatula identifies more than 750 genes differentially expressed during nodulation, including many potential regulators of the symbiotic program. Plant Physiol. 2004;136:3159–3176. doi: 10.1104/pp.104.043612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucher F., Kondorosi E. Cell cycle regulation in the course of nodule organogenesis in Medicago. Plant Mol. Biol. 2000;43:773–786. doi: 10.1023/a:1006405029600. [DOI] [PubMed] [Google Scholar]
- Gaba A., Jacobson A., Sachs M. Ribosome occupancy of the yeast CPA1 uORF termination codon modulates non-sense-mediated mRNA decay. Mol. Cell. 2005;20:449–460. doi: 10.1016/j.molcel.2005.09.019. [DOI] [PubMed] [Google Scholar]
- Graveley B.R. Alternative splicing: Increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- Halterman D.A., Wei F., Wise R.P. Powdery mildew-induced Mla mRNAs are alternatively spliced and contain multiple upstream open reading frames. Plant Physiol. 2003;131:558–567. doi: 10.1104/pp.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hillman R.T., Green R.E., Brenner S.E. An unappreciated role for RNA surveillance. Genome Biol. 2004;5:R8. doi: 10.1186/gb-2004-5-2-r8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Fang C., Xu Y. The effect of isoforms of the cell polarity protein, human ASIP, on the cell cycle and Fas/FasL-mediated apoptosis in human hepatoma cells. Cell. Mol. Life Sci. 2005;62:1974–1983. doi: 10.1007/s00018-005-5134-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai A., Hanzawa Y., Komura M., Yamamoto K.T., Komeda Y., Takahashi T. The dawrf phenotype of the Arabidopsis acl5 mutant is suppressed by a mutation in an upstream ORFof a bHLH gene. Development. 2006;133:3575–3585. doi: 10.1242/dev.02535. [DOI] [PubMed] [Google Scholar]
- International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Journet E.P., El-Gachtouli N., Vernoud V., de Billy F., Pichon M., Dedieu A., Arnould C., Morandi D., Barker D.G., Gianinazzi-Pearson V. Medicago truncatula ENOD11: A novel RPRP-encoding early nodulin gene expressed during mycorrhization in arbuscule-containing cells. Mol. Plant Microbe Interact. 2001;14:737–748. doi: 10.1094/MPMI.2001.14.6.737. [DOI] [PubMed] [Google Scholar]
- Kazan K. Alternative splicing and proteome diversity in plants: The tip of the iceberg has just emerged. Trends Plant Sci. 2003;8:468–471. doi: 10.1016/j.tplants.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Kim M.J., Kim H., Shin J.S., Chung C.H., Ohlrogge J.B., Suh M.C. Seed-specific expression of sesame microsomal oleic acid desaturase is controlled by combinatorial properties between negative cis-regulatory elements in the SeFAD2 promoter and enhancers in the 5′-UTR intron. Mol. Genet. Genomics. 2006;276:351–368. doi: 10.1007/s00438-006-0148-2. [DOI] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr. 1991;1:111–115. [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Pushing the limits of the scanning mechanism for initiation of translation. Gene. 2002;299:1–34. doi: 10.1016/S0378-1119(02)01056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuppusamy K.T., Endre G., Prabhu R., Penmetsa R.V., Veereshlingam H., Cook D.R., Dickstein R., VandenBosch K.A. LIN, a Medicago truncatula gene required for nodule differentiation and persistence of rhizobial infections. Plant Physiol. 2004;136:3682–3691. doi: 10.1104/pp.104.045575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwong R.W., Bui A.Q., Lee H., Kwong L.W., Fischer R.L., Goldberg R.B., Harada J.J. LEAFY COTYLEDON1LIKE defines a class of regulators essential for embryo development. Plant Cell. 2003;15:5–18. doi: 10.1105/tpc.006973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahousse S., Smorowski A.L., Denis C., Lantoine D., Kerckaert J.P., Galiegue-Zouitina S. Structural features of hematopoiesis-specific RhoH/ARHH gene: High diversity of 5′-UTR in different hematopoietic lineages suggests a complex post-transcriptional regulation. Gene. 2004;343:55–68. doi: 10.1016/j.gene.2004.08.022. [DOI] [PubMed] [Google Scholar]
- Lee H., Fischer R.L., Goldberg R.B., Harada J.J. Arabidopsis LEAFY COTYLEDON1 represents a functionally specialized subunit of the CCAAT binding transcription factor. Proc. Natl. Acad. Sci. 2003;100:2152–2156. doi: 10.1073/pnas.0437909100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorkovic Z.J., Wieczorek Kirk D.A., Lambermon M.H., Filipowicz W. Pre-mRNA splicing in higher plants. Trends Plant Sci. 2000;5:160–167. doi: 10.1016/s1360-1385(00)01595-8. [DOI] [PubMed] [Google Scholar]
- Lotan T., Ohto M., Yee K.M., West M.A., Lo R., Kwong R.W., Yamagishi K., Fischer R.L., Goldberg R.B., Harada J.J. Arabidopsis LEAFY COTYLEDON1 is sufficient to induce embryo development in vegetative cells. Cell. 1998;93:1195–1205. doi: 10.1016/s0092-8674(00)81463-4. [DOI] [PubMed] [Google Scholar]
- Macknight R., Duroux M., Laurie R., Dijkwel P., Simpson G., Dean C. Functional significance of the alternative transcript processing of the Arabidopsis floral promoter FCA. Plant Cell. 2002;14:877–888. doi: 10.1105/tpc.010456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998;26:1135–1143. doi: 10.1093/nar/26.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlin A.J., Clark F., Smith C.W.J. Understanding alternative splicing: Towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- Miyoshi K., Ito Y., Serizawa A., Kurata N. OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J. 2003;36:532–540. doi: 10.1046/j.1365-313x.2003.01897.x. [DOI] [PubMed] [Google Scholar]
- Modrek B., Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- Morris D.R., Geballe A.P. Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patriarca E.J., Tate R., Ferraioli S., Iaccarino M. Organogenesis of legume root nodules. Int. Rev. Cytol. 2004;234:201–262. doi: 10.1016/S0074-7696(04)34005-2. [DOI] [PubMed] [Google Scholar]
- Procissi A., Piazza P., Tonelli C. A maize r1 gene is regulated post-transcriptionally by differential splicing of its leader. Plant Mol. Biol. 2002;49:239–248. doi: 10.1023/a:1014959230492. [DOI] [PubMed] [Google Scholar]
- Rivas S., Rougon-Cardoso A., Smoker M., Schauser L., Yoshioka H., Jones J.D.G. CITRX thioredoxin interacts with the tomato Cf-9 resistance protein and negatively regulates defence. EMBO J. 2004;23:2156–2165. doi: 10.1038/sj.emboj.7600224. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Roberts A.G., Redding S.J., Llewellyn D.H. An alternatively-spliced exon in the 5′-UTR of human ALAS1 mRNA inhibits translation and renders it resistant to haem-mediated decay. FEBS Lett. 2005;579:1061–1066. doi: 10.1016/j.febslet.2004.12.080. [DOI] [PubMed] [Google Scholar]
- Soller M. Pre-messenger RNA processing and its regulation: A genomic perspective. Cell. Mol. Life Sci. 2006;63:796–819. doi: 10.1007/s00018-005-5391-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y., Ishihara D., Sasaki M., Nakagawa H., Hata H., Tsunoda T., Watanabe M., Komatsu T., Ota T., Isogai T., et al. Statistical analysis of the 5′ untranslated region of human mRNA using ‘Oligo-Capped’ cDNA libraries. Genomics. 2000;64:286–297. doi: 10.1006/geno.2000.6076. [DOI] [PubMed] [Google Scholar]
- Vasse J., de Billy F., Camut S., Truchet G. Correlation between ultrastructural differentiation of bacteroids and nitrogen fixation in alfalfa nodules. J. Bacteriol. 1990;172:4295–4306. doi: 10.1128/jb.172.8.4295-4306.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veereshlingam H., Haynes J.G., Penmetsa R.V., Cook D.R., Sherrier D.J., Dickstein R. nip, a symbiotic Medicago truncatula mutant that forms root nodules with aberrant infection threads and plant defense-like response. Plant Physiol. 2004;136:3692–3702. doi: 10.1104/pp.104.049064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela C., Mc Carthy J.E. Regulation of fungal gene expression via short ORFs in the mRNA 5′untranslated region. Mol. Microbiol. 2003;49:859–867. doi: 10.1046/j.1365-2958.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- Wang B.B., Brendel V. Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. 2006;103:7175–7180. doi: 10.1073/pnas.0602039103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warpeha K.M., Upadhyay S., Yeh J., Adamiak J., Hawkins S.I., Lapik Y.R., Anderson M.B., Kaufman L.S. The GCR1, GPA1, PRN1, NF-Y signal chain mediates both blue light and abscisic acid responses in Arabidopsis. Plant Physiol. 2007;143:1590–1600. doi: 10.1104/pp.106.089904. [DOI] [PMC free article] [PubMed] [Google Scholar]