Abstract
Although simian/human immunodeficiency virus (SHIV) strain DH12 replicates to high titers and causes immunodeficiency in pig-tailed macaques, virus loads measured in SHIVDH12-infected rhesus monkeys are consistently 100-fold lower and none of 22 inoculated animals have developed disease. We previously reported that the administration of anti-human CD8 mAb to rhesus macaques at the time of primary SHIVDH12 infection resulted in marked elevations of virus loads. One of the treated animals experienced rapid and profound depletions of circulating CD4+ T lymphocytes. Although the CD4+ T cell number partially recovered, this monkey subsequently suffered significant weight loss and was euthanized. A tissue culture virus stock derived from this animal, designated SHIVDH12R, induced marked and rapid CD4+ cell loss after i.v. inoculation of rhesus monkeys. Retrospective analyses of clinical specimens, collected during the emergence of SHIVDH12R indicated: (i) the input cloned SHIV remained the predominant virus during the first 5–7 months of infection; (ii) variants bearing only a few of the SHIVDH12R consensus changes first appeared 7 months after the administration of anti-CD8 mAb; (iii) high titers of neutralizing antibody directed against the input SHIV were detected by week 10 and persisted throughout the infection; and (iv) no neutralizing antibody against SHIVDH12R ever developed.
Because HIV-1 establishes infections only in humans and chimpanzees and induces disease only in humans, chimeric simian/human immunodeficiency viruses (SHIVs) have been constructed to examine the pathogenicity and develop vaccines pertaining to HIV-1-encoded proteins in the more tractable Asian macaque model system. We have reported that SHIVDH12, previously designated SHIVMD14YE, replicates to high titers, induces p27 antigenemia, and induces immunodeficiency in pig-tailed macaques (1). However, in SHIVDH12-infected rhesus monkeys, virus loads are consistently 100-fold lower, plasma p27 is undetectable, and none of the inoculated animals have ever developed CD4+ T lymphocyte depletion or signs of disease (2).
A large body of evidence now indicates that virus-specific cytotoxic T lymphocytes (CTL) play a major role in resolving acute primate lentiviral infections. In HIV-1 infections of humans, a CTL response, demonstrable before seroconversion, is temporally linked to declining virus loads (3–5). In this regard, we previously reported that the administration of the anti-human CD8 mAb T87PT3F9 at the time of primary infection of rhesus macaques with SHIVDH12 led to markedly elevated (50- to 100-fold) plasma and cell-associated viral loads compared with untreated animals; p27 antigenemia also became detectable for the first time in the CD8 mAb-treated SHIVDH12-infected rhesus monkeys (2). The antibody used in that study induced only partial and transient depletions of CD8+ T lymphocytes in rhesus recipients, and the responses observed varied from animal to animal. A recent study, using considerably higher doses of a humanized mouse anti-human CD8 mAb, reported more complete ablation of CD8+ T cells in naive rhesus monkeys that lasted for 17–60 days (6). Nonetheless, the two monkeys receiving the T87PT3F9 anti-CD8 mAb 3 days before and 4 days after i.v. SHIVDH12 inoculation both experienced severe CD4+ T lymphocyte declines that lasted several weeks (2).
Although the anti-CD8mAb-treated animal (565Z) with the highest virus loads eventually resolved its primary SHIVDH12 infection, it experienced a second wave of viremia and CD4+ T cell depletion, became symptomatic, and subsequently was euthanized. The SHIV recovered from macaque 565Z caused rapid CD4+ cell loss after its i.v. inoculation into two naïve rhesus monkeys and was used to prepare a tissue culture-derived virus stock with similar properties. The availability of cells and plasma collected from macaque 565Z during the course of its infection permitted a retrospective immunologic and genetic analysis of a SHIV as it evolved from a nonpathogenic to an acutely pathogenic virus during a single passage in a rhesus monkey.
Methods
Virus.
The structure of SHIVDH12 and the preparation of virus stocks in macaque peripheral blood mononuclear cells (PBMCs) for animal inoculation has been described (1). SHIVDH12 also was prepared in MT4 cells for immunoblotting.
The SHIVDH12R stock was prepared by combining 2 × 106 lymph node cells, 7 × 106 splenocytes plus thymocytes, and 9 × 105 PBMCs, collected from rhesus macaque 25112 at the time of its necropsy, with 1.3 × 106 PBMCs from rhesus monkey RQ974 (obtained 3 weeks postinfection), resuspending the mixture in RPMI 1640 medium, supplemented with 20% of heat-inactivated FBS (HyClone) in a total volume of 10 ml, and incubating the suspension for 20 hr at 37°C. The cell mixture then was cocultivated with PBMCs from two uninfected rhesus macaques (40.5 × 106 and 61.5 × 106 cells, respectively in 20 ml), previously stimulated with 25 μg/ml of Con A (Amersham Pharmacia Biotech) for 20 hr. Medium was collected daily from the coculture supernatant and samples containing the highest reverse transcriptase activities (days 5–7) were pooled, filtered through a 0.45-μm membrane, aliquoted, and stored at −70°C until use.
Animal Experiments.
Rhesus macaques (Macaca mulatta) were maintained in accordance with the Guide for the Care and Use of Laboratory Animals (7) and were housed in a biosafety level 2 facility; biosafety level 3 practices were followed. Animals were anesthetized with i.m. injections of Tiletamine-HCl and Zolazepam-HCl (Telazol; Fort Dodge Laboratories, Fort Dodge, IA) during phlebotomies or SHIV inoculations. Blood samples were collected with ACD-A solution or EDTA as the anticoagulant. PBMCs or plasma samples were prepared from ACD-A anticoagulated blood and stored in liquid nitrogen or −70°C, respectively, until use. Lymphocyte immunophenotyping and complete blood counts were performed by using EDTA anticoagulated blood samples.
The SHIVDH12-infected rhesus monkey 565Z developed immunodeficiency and was euthanized 68 weeks postinfection. At the time of its necropsy, a suspension of splenocytes (1.4 × 106 cells) and lymph node cells (7.5 × 105 cells) was combined with 10 ml of its own anticoagulated blood and 6 ml of bone marrow, and the mixture was inoculated i.v. into two rhesus monkeys (macaques RQ974 and 25112). Rhesus macaques H27 and 5981 were inoculated i.v. with 4.1 × 105 TCID50 and 656 TCID50 of the SHIVDH12R tissue culture-derived stock, respectively.
Virus Load Measurements.
Plasma viral RNA levels were determined by real-time PCR (ABI Prism 7700 Sequence Detection System, Perkin–Elmer) using reverse-transcribed viral RNA from macaque plasma samples as templates. Plasma viral RNA was extracted by using a QIAamp Viral RNA Kit (Qiagen, Valencia, CA), and reverse-transcribed with MultiScribe reverse transcriptase (TaqMan Reverse Transcription Reagents Kit, Perkin–Elmer/Roche). The cDNA was amplified (45 cycles/default settings) with Ampli Taq Gold DNA polymerase (PCR Core Reagents Kit, Perkin–Elmer/Roche) by using a pair of primers derived from the vpr gene (forward, nucleotides 6220–6240, and reverse, nucleotides 6358–6338) of SHIVDH12. SHIVDH12R-infected animal plasma and SHIVDH12-infected rhesus PBMC culture supernatants, which previously were prequantified by the branched DNA method (8), served as standards for the assay.
p27 antigenemia was measured by an SIV core antigen assay (Coulter), using a murine mAb to capture the p27 SIV Gag protein in plasma prepared from EDTA anticoagulated blood samples.
Long env Gene PCR.
Five-microliter samples of Hirt fractionated (9) DNA (approximately 105 cell equivalents), prepared from previously frozen PBMC samples [cultured, after thawing, for 18 hr in RPMI 1640 containing 4 mM glutamine, penicillin (25 units/ml), streptomycin (25 μg/ml), and 10% FBS at a density of 106 cells/ml], were used as templates for PCR amplification of a 3,048-bp region (map positions 6144–9171) of SHIVDH12 proviral DNA, which encompassed the entire HIV-1DH12 Env coding region. Amplifications were performed by using the Expand Long Template PCR system (Boehringer Mannheim #1681834). Reaction mixtures consisted of 5 μl Hirt DNA, 500 μM each dNTP, PCR buffer #2 (containing 2.25 μM MgCl2), 0.3 μM each primer [6144(+), 5′-CAGTAGATCCTAGACTAGAGC-3′; 9190(−), 5′-CCACCTCTCCTAAGAGTCTC-3′], and 0.75 μl of the enzyme mixture in a final volume of 50 μl overlaid with 30 μl mineral oil. In cases where more than 30 cycles of amplification were required, a second round of 30 cycles of amplification was performed by using nested primers [6317(+), 5′-CACAGTCAGACTGATCAAGC-3′; 9137(−), 5′-CTGTCGCAGATCTCCAGAC-3′] under the same reaction conditions. Amplifications were performed as follows: one cycle of denaturation (94°C, 2 min); 10 cycles of amplification (94°C, 20 s/63°C, 30 s/68°C, 4 min); an additional 20 cycles of amplification and extension (94°C, 20 s/63°C, 30 s/68°C, 4 min + 20 s per cycle); a final extension at 68°C for 7 min. The PCR products were gel-purified on 0.6% agarose gels, and then cloned directly into pCR2.1 (Promega Biotec #K2030–01) for sequence analysis of individual molecular clones.
Lymphocyte Immunophenotyping.
EDTA-treated blood samples were stained with fluorochrome-conjugated mAbs [CD3-FITC (Serotec); CD4-allophycocyanin, CD8-peridinin chlorophyll protein, and CD20-phycoerythrin, (Becton Dickinson Immunocytometry Systems)] and analyzed by flow cytometry (FACSort, Becton Dickinson) as described (1).
Antibody Assays.
Limiting dilution, end-point neutralization assays (100% neutralization against 100 TCID50 of SHIVDH12 or SHIVDH12R) were performed as described (10). Serial 3-fold dilutions of plasma were incubated with virus, and the mixture then was used as inoculum for quadruplicate, 2-week infections of MT-4 cells. Non-neutralized culture supernatants appear as black dots on the autoradiograms of these 32P reverse transcriptase (RT) assays, reflecting the presence of particle-associated RT activity in the medium. The neutralization titers were calculated by the method of Reed and Muench (11). For immunoblotting, SHIVDH12 particles were concentrated by ultracentrifugation (100,000 × g for 80 min by using an SW28 rotor in an L8–70M Ultracentrifuge, Beckman Instruments, Fullerton, CA). The pellet was resuspended in PBS and subjected to ultracentrifugation through 20% sucrose in PBS (200,000 × g for 75 min by using an SW55 rotor in a L8–70M Ultracentrifuge, Beckman Instruments). The concentrated SHIV particles then were mixed with purified HIV-1DH12 gp120, expressed from a recombinant vaccinia virus as reported (12). After incubation with SDS/PAGE sample buffer (13), the viral proteins were resolved by SDS/PAGE, transferred to a poly(vinylidene difluoride) membrane (Immobilon-P, Millipore), and incubated with serially collected plasma samples (diluted 1:100) that had been preadsorbed with sonicated lysates of Escherichia coli and MT4 cells, followed by incubation with sheep anti-human Ig conjugated with horseradish peroxidase (Amersham Pharmacia Biotech). Protein bands were visualized on x-ray film (Kodak Biomax MR) after incubation with chemiluminescent substrate (SuperSignal West Pico Chemiluminescent Substrate, Pierce).
Results and Discussion
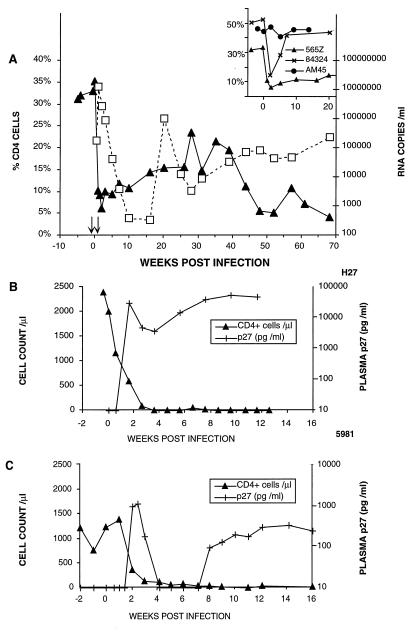
We previously reported that the two rhesus monkeys (565Z and 84324) receiving the T87PT3F9 anti-CD8 mAb before (−3 days) and immediately after (+4 days) inoculation with SHIVDH12 suffered significant CD4+ T cell losses of several weeks duration (Fig. 1A, Inset). In both of these animals, the mAb-induced CD8+ T cell decline (from 1,375 to 217 cells/μl in macaque 565Z; from 795 to 407 cells/ml in macaque 84324) was inversely related to the peak levels of plasma viremia (12 × 106 copies/ml in 565Z and 0.56 × 106 copies/ml for 84324). These virus loads were 10- to 20-fold higher than macaques not receiving the anti-CD8 mAb.
Figure 1.
(A) The dynamics of viral RNA levels and CD4+ T lymphocyte counts during the SHIVDH12 infection of rhesus macaque 565Z. The arrows indicate the times when anti-human CD8 mAb was administered. (B and C) SHIVDH12R induces p27 antigenemia and causes rapid CD4+ T cell depletion in rhesus macaques. Blood collected at the indicated times from two infected rhesus macaques were fractionated into plasma and cells and levels of p27 and CD4+ T lymphocytes, respectively, were determined as described in Methods.
In monkey 565Z, the CD4+ cell number in the peripheral blood fell to 92 cells/ml 5 weeks postinfection and, as reported (2), the % CD4+ T lymphocytes in lymph node biopsies declined from 49%, before infection, to 12% thereafter. The circulating CD4+ cell levels in animal 565Z remained low for 2–3 months before gradually rising to approximately 60% of the preinfection level at week 35 (Fig. 1A). This partial recovery of CD4+ cells accompanied the resolution of the initial plasma viremia, which fell to approximately 300 RNA copies/μl at week 16. However, after week 30, the virus load again increased and was accompanied by a second CD4+ T lymphocyte decline, which reached 44 cells/μl at week 48. Monkey 565Z suffered significant weight loss by week 66, became moribund on week 68, and was euthanized at that time.
To determine whether a pathogenic variant of SHIVDH12 had emerged in rhesus 565Z, a cell suspension (lymph nodes, spleen, and bone marrow), prepared from tissues collected at the time of its necropsy, was mixed with anticoagulated whole blood and inoculated into two naïve rhesus monkeys (RQ974 and 25112). Both animals experienced marked and rapid CD4+ T cell declines by day 10 postinfection (to 200 and 72 cells/μl, respectively), indicating that during a single passage in macaque 565Z, SHIVDH12 had evolved into a highly pathogenic virus. Because we wanted to obtain a tissue culture-derived stock of a potentially highly pathogenic SHIV, a suspension of splenocytes, thymocytes, lymph node cells and PBMCs from macaque 25112 was combined with PBMCs from animal RQ974 and cocultivated with a 10-fold excess of activated PBMCs from naive rhesus monkeys. The resulting culture supernatant, designated SHIVDH12R (4.1 × 105 TCID50/ml), exhibited comparable in vitro infection kinetics in rhesus and human PBMCs as its SHIVDH12 parent and directed the production of similar amounts of infectious virus progeny (approximately 1 to 4 × 105 TCID50/ml). SHIVDH12R, however, induced more extensive cell killing and generated more numerous and extremely large syncytia in both PBMC types. SHIVDH12R also replicated to high levels in the continuous human MT4 T cell line.
Intravenous inoculation of two rhesus macaques (H27 and 5981) with 4.1 × 105 TCID50 or 650 TCID50 of the SHIVDH12R tissue culture-derived stock, respectively, resulted in rapid and irreversible CD4+ T cell depletion and high virus loads in both animals (Fig. 1 B and C). Animal H27 subsequently suffered marked weight loss (>30%), experienced respiratory distress, and was euthanized 12.6 weeks postvirus inoculation. An autopsy revealed multifocal interstitial pulmonary fibrosis associated with giant multinucleated cells. Rhesus monkey 5981 developed leukocytosis (>15,000 cells/μl) at week 9, chronic diarrhea, and weight loss and was euthanized at week 23 with a CD4+ cell count of 2 cells/ml. A titration (4.1 × 105 TCID50 to 1 TCID50) of SHIVDH12R in rhesus monkeys by the i.v. route consistently induced rapid and irreversible depletion of CD4+ T lymphocytes in all of the inoculated animals (Y.E., T.I., and M.M., unpublished work). Several macaques, which received 25 TCID50 or less, remain alive 1 year after SHIVDH12R infection with CD4+ T cell levels in the 100 to 200 cells/μl range.
The unexpected emergence of the disease-inducing SHIVDH12R during a single in vivo passage of a molecularly cloned virus, rather than after multiple animal-to-animal transfers of whole blood/bone marrow as reported by others (14, 15), provided the opportunity to retrospectively follow changes introduced into the SHIV genome and to monitor the immunologic responses to both the input SHIVDH12 and the emergent SHIVDH12R attending the acquisition of the pathogenic phenotype.
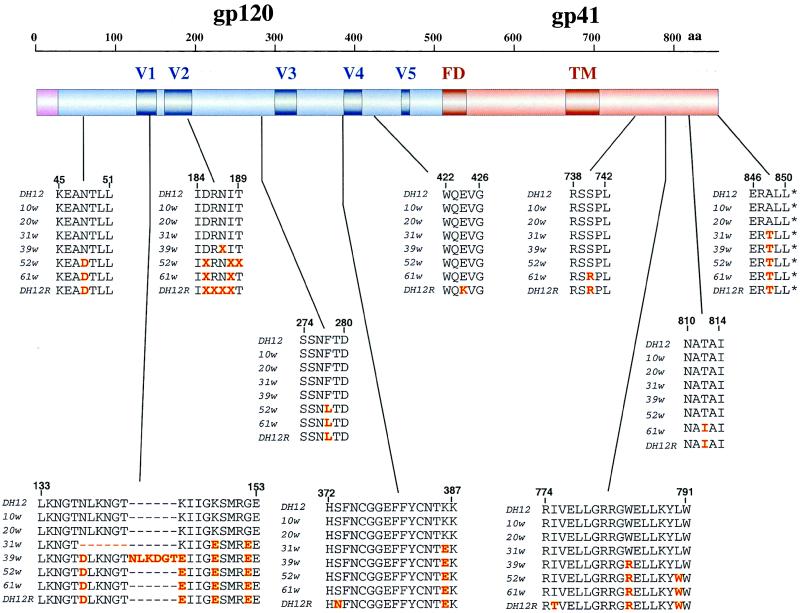
Alterations of HIV-1 env gene sequences have been previously reported to be associated with the emergence of highly pathogenic SHIVs (16, 17). Therefore, a 3,046-bp segment, which extended from the nine 3′ terminal nt vpr to the first 36 codons of nef, and encompassing the entire HIV-1 Env coding region, was PCR-amplified from Hirt-fractionated DNA, prepared from the same rhesus PBMCs used to generate the SHIVDH12R stock. Nucleotide sequence analyses of 22 independent PCR clones revealed that 13 aa in gp120 and 6 aa in gp41 of SHIVDH12R had changed compared with analogous residues in the input, molecularly cloned SHIVDH12 parent. These altered amino acids are indicated in the bottom row (DH12R) of each sequence group shown in Fig. 2. A majority of the altered gp120 amino acids mapped to the V1 and V2 regions whereas the gp41 changes clustered in the cytoplasmic tail. The changes in the V2 region of the SHIVDH12R gp120 were quite heterogeneous, consisting of two or more amino acid substitutions and/or deletions at the indicated positions (denoted by X in Fig. 2). Some of the gp120 alterations also resulted in a loss of an N-linked glycosylation site mapping to V2. No significant change was observed in the V3 region, consistent with preliminary experiments (A. Trkola and J. P. Moore, personal communication) indicating that SHIVDH12R primarily uses CCR5 and CXCR4 and is therefore indistinguishable from its dual-tropic SHIVDH12 parent (18).
Figure 2.
Highly pathogenic SHIVDH12R evolves in a stepwise manner during the course of infection in rhesus macaque 565Z. The deduced amino acid sequence for the cloned SHIVDH12 (DH12) and the consensus sequence from 22 independent SHIVDH12R (DH12R) PCR clones are indicated in the top and bottom lines of each grouping. Independent PCR clones also were derived from PBMC samples collected at weeks 10 (9 clones), 20 (5 clones), 31 (6 clones), 39 (14 clones), 52 (8 clones), and 61 (6 clones) postinfection as described in Methods and their nucleotide sequence was determined. The locations of amino acid changes in gp120 and gp41 at various times during the evolution of SHIVDH12 to SHIVDH12R are indicated in red. The positions of the fusion domain (FD) and transmembrane (TM) regions of gp41 are shown.
Analyses of independent PCR clones, generated from amplified PBMC-associated DNA collected and frozen at different times during the 68-week infection of animal 565Z, also were carried out to ascertain when and how SHIVDH12R arose. These 3-kb PCR clones were collected at weeks 10, 20, 31, 39, 52, and 61 postinfection. The deduced amino acid sequence of the SHIV Env region at these six times during the infection of rhesus monkey 565Z is indicated for each sequence grouping shown in Fig. 2. The sequence indicated at each sampling time represents the residues present in the majority of independent PCR clones analyzed. The env gene segments from weeks 10 and 20 were both indistinguishable from the starting SHIVDH12 env region, harboring none of the SHIVDH12R “signature” changes. In contrast, the week 31 and 39 samples contained several residues not present in the original SHIVDH12 Env glycoprotein. The V1 sequence underwent drastic changes during this period, including a 6-aa in-frame deletion present at week 31. By week 39, however, the V1 hexapeptide deletion had disappeared and was replaced with a 6-aa insertion, which included the nearly perfect triplication of the pentapeptide motif LKNGT, present, but unrecognized, in the original SHIVDH12 gp120. Week 39 also marked the time when V2 began to undergo extensive remodeling. Additional amino acid changes accumulated with time until the animal’s demise at week 68. These sequence analyses clearly indicate that SHIVDH12, not SHIVDH12R, was responsible for the CD4+ T cell decline and the associated high levels of viremia monitored during the first 5 weeks of the acute infection. The second, irreversible fall of CD4 and associated long-lasting viremia probably was caused by an evolving virus “swarm” from which SHIVDH12R ultimately emerged.
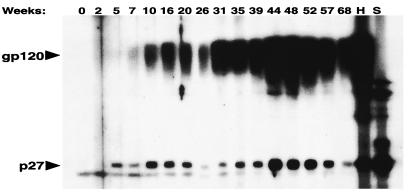
The kinetics of humoral immune responses during the evolution of SHIVDH12 in rhesus monkey 565Z initially was examined by immunoblotting (Fig. 3). As expected, antibody levels rose in response to virus production in vivo. Antibody to the p27 SIV capsid protein (CA) component of SHIVDH12 was first detected at week 5, and reactivity with gp120 was observed a few weeks later (Fig. 3). The initial peak of antibody production occurred between weeks 10 and 16 for anti-p27 and weeks 16 and 20 for anti-gp120 and subsequently fell over the ensuing 10 weeks. This decline in p27 reactivity coincided with a gradual increase in the % CD4+ T lymphocytes, which reached 23% on week 28 (Fig. 1A). However, virus loads and the levels of both antibodies gradually increased a second time, peaking between weeks 44 and 48 postinfection. This rise in plasma viremia was accompanied by a second, unremitting loss of CD4+ T cells and declining levels of both antibodies despite continuous virus production.
Figure 3.
Antiviral antibody responses during the course of the SHIVDH12 infection of rhesus monkey 565Z. Samples of blood, collected at the indicated times, were analyzed by immunoblotting as described in Methods. Lanes H and S, purified IgG (0.45 μg) from a chimpanzee infected with HIV-1DH12 (26) or plasma (1:200) from an SIVsm-infected rhesus macaque, respectively, were used as positive controls.
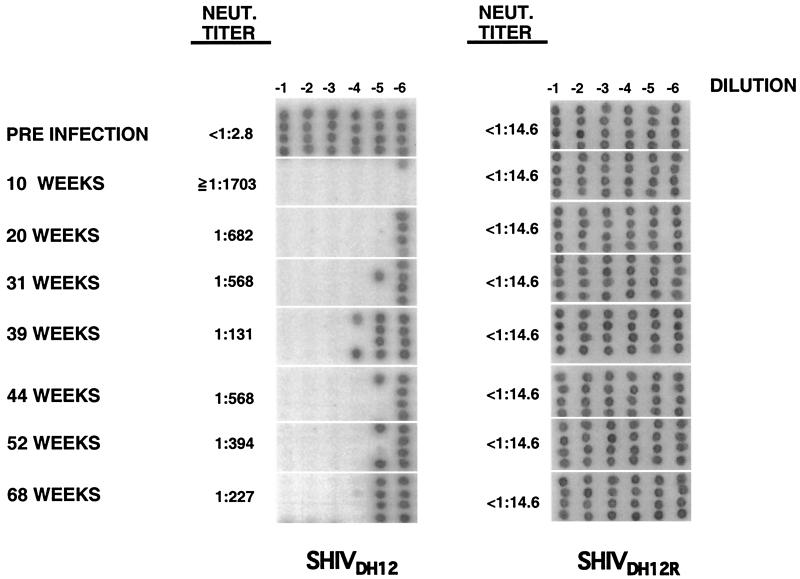
Neutralizing antibodies, directed against SHIVDH12 or SHIVDH12R, also were examined during the course of the macaque 565Z infection, using an end-point assay that measures 100% neutralization (10, 19). High titers of antibodies, capable of neutralizing the input SHIVDH12 but not SHIVDH12R, were evident 10 weeks postinfection (Fig. 4), coinciding with the emergence of binding antibody to gp120 (Fig. 3). This result indicates that the initial immune response, which peaked between weeks 10 and 20, was elicited by SHIVDH12, not SHIVDH12R. Neutralizing titers against SHIVDH12 decreased somewhat after week 10 and persisted at a lower level throughout the remainder of the infection. No demonstrable neutralizing antibodies against SHIVDH12R were present in macaque 565Z at any time during the infection (Fig. 4).
Figure 4.
Neutralizing antibody responses during the course of the SHIVDH12 infection of rhesus monkey 565Z. Plasma samples prepared at the indicated times were subjected to neutralization assay as described (10).
Several important conclusions can be drawn from this study regarding the contribution of viral and host factors to the development of lentivirus-induced immunodeficiency. First and foremost, the pathogenic derivative SHIVDH12R emerged only in rhesus 565Z, whose CD8+ T lymphocytes had been markedly depleted by mAb treatment at the time of primary viral infection. None of 16 other rhesus monkeys, inoculated with SHIVDH12 but not treated with the anti-CD8 mAb, or two other animals (2) that failed to generate high virus loads after mAb treatment, developed immunodeficiency. Our results therefore suggest the following scenario of events in animal 565Z: (i) Suppression of the initial cytotoxic T lymphocyte response, induced by the administration of the anti-CD8 mAb, interfered with the clearance of the input SHIVDH12. (ii) As a result, SHIVDH12 underwent multiple cycles of replication and became widely disseminated in the infected animal during primary infection. (iii) The primary SHIVDH12 infection finally was brought under control by week 12, although by that time, a pool of viral variants possibly emerged as a minor SHIV subpopulation. (iv) Variants bearing a few of the SHIVDH12R “signature” residues became predominant and could be detected by week 31, nearly 7 months after cessation of the anti-CD8 mAb treatment. (v) Stepwise evolution of the envelope sequences finally resulted in the appearance of the highly pathogenic SHIVDH12R. One might conclude, therefore, that the administration of the anti-CD8 mAb, if not the direct stimulus for the emergence of SHIVDH12R, was indirectly involved in its ultimate appearance.
Although the selective forces in vivo responsible for the emergence of the highly pathogenic SHIVDH12R are presently unknown, SHIVDH12R certainly can be classified as a neutralization-escape virus variant based on the assay shown in Fig. 4. Our recent study of the clearance of physical and infectious HIV particles in vivo demonstrated that neutralizing antibodies can immediately inactivate autologous virus in plasma and greatly enhance its physical clearance from the blood (20). This result suggests that neutralizing antibodies are able to exert strong selective pressure, but may not, per se, be sufficient to generate a highly pathogenic SHIV. Other studies have reported that neutralization-resistant virus variants frequently arise during chronic SIVmac, SIVsm, and SHIV infections of Asian macaques (21–24). In the case of the HIV-1DH12 isolate, the principal neutralization epitope has been mapped to the five noncontiguous variable regions (V1-V5) of gp120, using a series of HIV-1 constructs carrying chimeric gp120 molecules (Michael Cho, personal communication). Thus, the multiple changes that accumulated within the V1 and V2 regions of SHIVDH12R gp120 (Fig. 2) are very likely responsible for the loss of neutralization sensitivity.
Our inability to detect neutralizing antibody against SHIVDH12R at any time during the SHIV infection of macaque 565Z was not the result of the envelope of SHIVDH12R being less immunogenic. As noted above, several rhesus monkeys, inoculated with less than 100 TCID50, of virus have suffered rapid and irreversible CD4+ cell loss, yet have developed neutralizing antibody against SHIVDH12R (Y.E., T.I., and M.M., unpublished work). Thus the absence of neutralizing activity directed against SHIVDH12R in animal 565Z very likely reflects the inability of an immunocompromised macaque to mount a neutralizing antibody response against emerging SHIVs bearing variant envelope glycoproteins. This finding is very reminiscent of the situation with HIV-1-infected individuals in whom neutralization of autologous isolates has been difficult to demonstrate. For example, a recent study reported that sera from long-term nonprogressors were able to neutralize contemporary autologous virus whereas only one of nine HIV-1 isolates from progressors or short-term nonprogressors was neutralized by autologous serum (25). Historically this was also the case for the original source of the HIV-1DH12 isolate: autologous antibody-positive serum obtained from this AIDS patient at the time of virus isolation exhibited no neutralizing activity directed against HIV-1DH12 (Ron Willey, personal communication), yet naïve chimpanzees or chimpanzees persistently infected with nonpathogenic HIV-1SF2, rapidly developed high titers of autologous neutralizing antibody after a challenge or superchallenge with HIV-1DH12 (10, 26).
SHIVDH12R represents a potentially useful reagent that can be used as a challenge virus in vaccine experiments or for the study of cellular and immunologic events attending the rapid and irreversible depletion of CD4+ T lymphocytes in vivo. Unlike some highly pathogenic SHIVs, SHIVDH12R stock can be readily prepared in either rhesus or human PBMCs and progeny virions can be titered in PBMCs or MT4 cells. Thus far, SHIVDH12R has induced marked CD4+ cell loss in all i.v.-inoculated rhesus macaques (Fig. 1 B and C).
Acknowledgments
We thank Drs. Ali Azadegan, Carole Clarke, and Randy Elkins and Messrs. George Coleman, Charles Thornton, and Russ Byrum for invaluable assistance in the animal experiments; Drs. Michael Eckhaus, Georgina Miller, and David Green for pathological analyses; Dr. Michael Cho for providing vaccinia-expressed gp120; Dr. Vanessa Hirsch for providing SIV infected animal plasma; and Mr. Ron Willey for helpful suggestions.
Abbreviations
- SIV
simian immunodeficiency virus
- SHIV
simian/human immunodeficiency virus
- PBMC
peripheral blood mononuclear cell
References
- 1.Shibata R, Maldarelli F, Siemon C, Matano T, Parta M, Miller G, Fredrickson T, Martin M A. J Infect Dis. 1997;176:362–373. doi: 10.1086/514053. [DOI] [PubMed] [Google Scholar]
- 2.Matano T, Shibata R, Siemon C, Connors M, Lane H C, Martin M A. J Virol. 1998;72:164–169. doi: 10.1128/jvi.72.1.164-169.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carmichael A, Jin X, Sissons P, Borysiewicz L. J Exp Med. 1993;177:249–256. doi: 10.1084/jem.177.2.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein M R, van Baalen C A, Holwerda A M, Kerkhof Garde S R, Bende R J, Keet I P, Eeftinck-Schattenkerk J K, Osterhaus A D, Schuitemaker H, Miedema F. J Exp Med. 1995;181:1365–1372. doi: 10.1084/jem.181.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rinaldo C, Huang X L, Fan Z F, Ding M, Beltz L, Logar A, Panicali D, Mazzara G, Liebmann J, Cottrill M, et al. J Virol. 1995;69:5838–5842. doi: 10.1128/jvi.69.9.5838-5842.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schmitz J E, Kuroda M J, Santra S, Sasseville V G, Simon M A, Lifton M A, Racz P, Tenner-Racz K, Dalesandro M, Scallon B J, et al. Science. 1999;283:857–860. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 7.Committee on Care and Use of Laboratory Animals. Guide for the Care and Use of Laboratory Animals. Bethesda: Natl. Inst. Health; 1985. , DHHS Publ. No. (NIH) 85–23, revised ed. [Google Scholar]
- 8.Dewar R L, Highbarger H C, Sarmiento M D, Todd J A, Vasudevachari M B, Davey R T, Jr, Kovacs J A, Salzman N P, Lane H C, Urdea M S. J Infect Dis. 1994;170:1172–1179. doi: 10.1093/infdis/170.5.1172. [DOI] [PubMed] [Google Scholar]
- 9.Hirt B. J Mol Biol. 1967;14:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 10.Willey R L, Shibata R, Freed E O, Cho M W, Martin M A. J Virol. 1996;70:6431–6436. doi: 10.1128/jvi.70.9.6431-6436.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reed L J, Muench H. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 12.Cho M W, Lee M K, Carney M C, Berson J F, Doms R W, Martin M A. J Virol. 1998;72:2509–2515. doi: 10.1128/jvi.72.3.2509-2515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. p. 53. [Google Scholar]
- 14.Joag S V, Li Z, Foresman L, Stephens E B, Zhao L J, Adany I, Pinson D M, McClure H M, Narayan O. J Virol. 1996;70:3189–3197. doi: 10.1128/jvi.70.5.3189-3197.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reimann K A, Li J T, Veazey R, Halloran M, Park I W, Karlsson G B, Sodroski J, Letvin N L. J Virol. 1996;70:6922–6928. doi: 10.1128/jvi.70.10.6922-6928.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karlsson G B, Halloran M, Li J, Park I W, Gomila R, Reimann K A, Axthelm M K, Iliff S A, Letvin N L, Sodroski J. J Virol. 1997;71:4218–4225. doi: 10.1128/jvi.71.6.4218-4225.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stephens E B, Mukherjee S, Liu Z Q, Sheffer D, Lamb-Wharton R, Leung K, Zhuge W, Joag S V, Li Z, Foresman L, et al. J Virol. 1998;72:5207–5214. doi: 10.1128/jvi.72.6.5207-5214.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang Y J, Dragic T, Cao Y, Kostrikis L, Kwon D S, Littman D R, KewalRamani V N, Moore J P. J Virol. 1998;72:9337–9344. doi: 10.1128/jvi.72.11.9337-9344.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shibata R, Igarashi T, Haigwood N, Buckler-White A, Ogert R, Ross W, Willey R, Cho M W, Martin M A. Nat Med. 1999;5:204–210. doi: 10.1038/5568. [DOI] [PubMed] [Google Scholar]
- 20.Igarashi T, Brown C, Azadegan A, Haigwood N, Dimitrov D, Martin M A, Shibata R. Nat Med. 1999;5:211–216. doi: 10.1038/5576. [DOI] [PubMed] [Google Scholar]
- 21.Burns D P, Collignon C, Desrosiers R C. J Virol. 1993;67:4104–4113. doi: 10.1128/jvi.67.7.4104-4113.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rudensey L M, Kimata J T, Long E M, Chackerian B, Overbaugh J. J Virol. 1998;72:209–217. doi: 10.1128/jvi.72.1.209-217.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnson P R, Hamm T E, Goldstein S, Kitov S, Hirsch V M. Virology. 1991;185:217–228. doi: 10.1016/0042-6822(91)90769-8. [DOI] [PubMed] [Google Scholar]
- 24.Igarashi T, Kuwata T, Takehisa J, Ibuki K, Shibata R, Mukai R, Komatsu T, Adachi A, Ido E, Hayami M. J Gen Virol. 1996;77:1649–1658. doi: 10.1099/0022-1317-77-8-1649. [DOI] [PubMed] [Google Scholar]
- 25.Pilgrim A K, Pantaleo G, Cohen O J, Fink L M, Zhou J Y, Zhou J T, Bolognesi D P, Fauci A S, Montefiori D C. J Infect Dis. 1997;176:924–932. doi: 10.1086/516508. [DOI] [PubMed] [Google Scholar]
- 26.Shibata R, Siemon C, Cho M W, Arthur L O, Nigida S M, Jr, Matthews T, Sawyer L A, Schultz A, Murthy K K, Israel Z, et al. J Virol. 1996;70:4361–4369. doi: 10.1128/jvi.70.7.4361-4369.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]