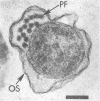
Abstract
Bovine rumen fluid contained relatively large numbers of spirochetes capable of fermenting polymers commonly present in plant materials. Polymers such as xylan, pectin, and arabinogalactan served as fermentable substrates for the spirochetes, whereas cellulose did not. Furthermore, spirochetes cultured from rumen fluid utilized as growth substrates hydrolysis products of plant polymers (e.g., D-xylose, L-arabinose, D-galacturonic acid, D-glucuronic acid, cellobiose), but did not ferment amino acids. Viable cell counts of spirochetes capable of fermenting individual plant polymers or their hydrolysis products yielded minimum values ranging from 0.2 X 10(6) to 4 X 10(6) cells per ml of rumen fluid. Thirteen strains of rumen spirochetes were characterized in terms of their fermentation products from glucose, the guanine plus cytosine content of their DNA, their ultrastructure, and their ability to ferment pectin, starch, or arabinogalactan. Of the 13 strains, 6 fermented glucose mainly to formate, acetate, and succinate, whereas the remaining 7 strains did not produce succinate, but instead formed ethanol, in addition to formate and acetate. The succinate-forming strains had two periplasmic (axial) fibrils per cell, measured 0.2 to 0.3 by 5 to 8 micrograms, had a guanine plus cytosine content of the DNA ranging from 36 to 38 mol%, and lacked the ability to ferment pectin, starch, or arabinogalactan. The ethanol-forming strains had from 8 to more than 32 periplasmic fibrils per cell, tended to be larger in cell size than the succinate-forming strains, and had a guanine plus cytosine content of the DNA ranging from 41 to 54 mol%. Some of the ethanol-forming strains fermented pectin, starch, or arabinogalactan. The results of this study indicate that the bovine rumen is inhabited by a physiologically and morphologically diverse population of spirochetes. It is likely that these spirochetes contribute significantly to the degradation of plant materials ingested by the ruminants.
Full text
PDF



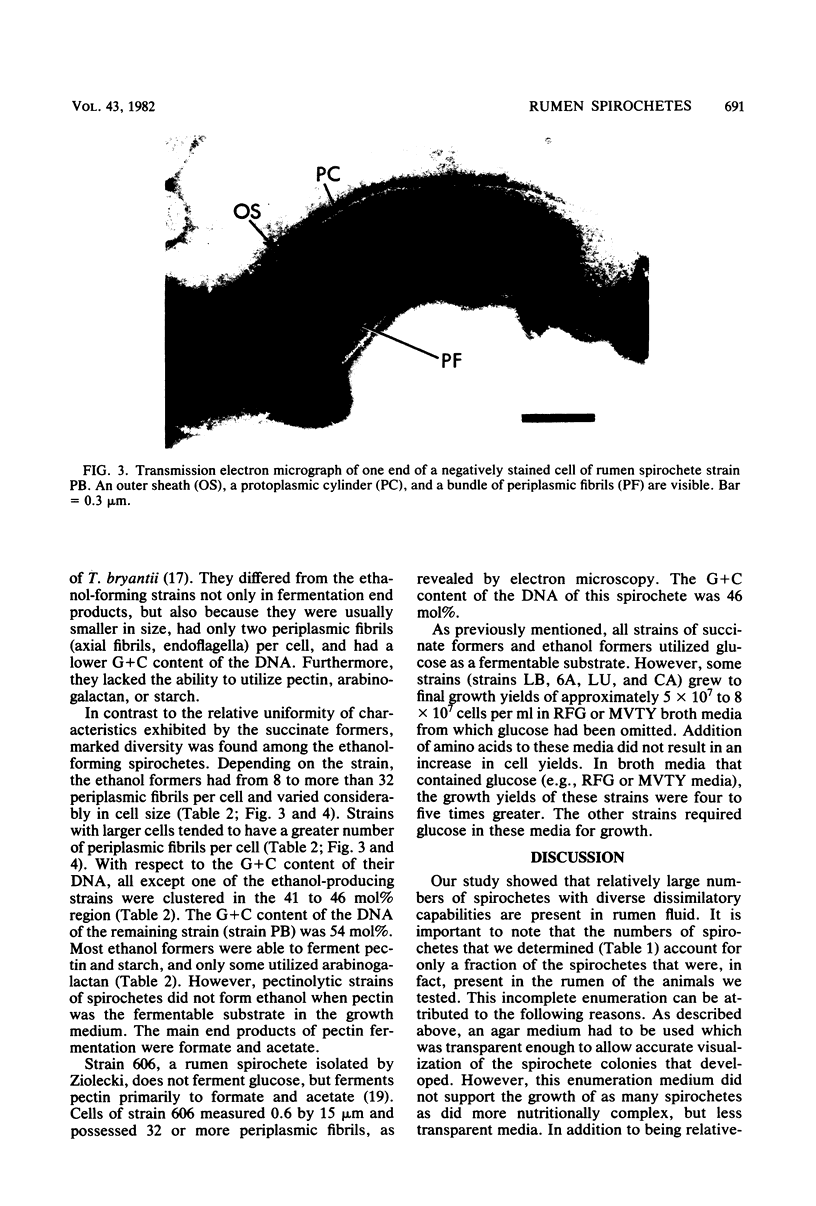


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bauchop T., Clarke R. T., Newhook J. C. Scanning electron microscope study of bacteria associated with the rumen epithelium of sheep. Appl Microbiol. 1975 Oct;30(4):668–675. doi: 10.1128/am.30.4.668-675.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breznak J. A., Canale-Parola E. Morphology and physiology of Spirochaeta aurantia strains isolated from aquatic habitats. Arch Microbiol. 1975 Sep 30;105(1):1–12. doi: 10.1007/BF00447104. [DOI] [PubMed] [Google Scholar]
- Canale-Parola E. Motility and chemotaxis of spirochetes. Annu Rev Microbiol. 1978;32:69–99. doi: 10.1146/annurev.mi.32.100178.000441. [DOI] [PubMed] [Google Scholar]
- De Ley J. Reexamination of the association between melting point, buoyant density, and chemical base composition of deoxyribonucleic acid. J Bacteriol. 1970 Mar;101(3):738–754. doi: 10.1128/jb.101.3.738-754.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinsdale D., Morris E. J., Bacon J. S. Electron microscopy of the microbial populations present and their modes of attack on various cellulosic substrates undergoing digestion in the sheep rumen. Appl Environ Microbiol. 1978 Jul;36(1):160–168. doi: 10.1128/aem.36.1.160-168.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt S. C., Leadbetter E. R. Fine structure of Sporocytophaga myxococcoides. Arch Mikrobiol. 1967 Jun 21;57(3):199–213. doi: 10.1007/BF00405947. [DOI] [PubMed] [Google Scholar]
- Holt S. C., Tanner A. C., Socransky S. S. Morphology and ultrastructure of oral strains of Actinobacillus actinomycetemcomitans and Haemophilus aphrophilus. Infect Immun. 1980 Nov;30(2):588–600. doi: 10.1128/iai.30.2.588-600.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCowan R. P., Cheng K. J., Bailey C. B., Costerton J. W. Adhesion of bacteria to epithelial cell surfaces within the reticulo-rumen of cattle. Appl Environ Microbiol. 1978 Jan;35(1):149–155. doi: 10.1128/aem.35.1.149-155.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SABATINI D. D., BENSCH K., BARRNETT R. J. Cytochemistry and electron microscopy. The preservation of cellular ultrastructure and enzymatic activity by aldehyde fixation. J Cell Biol. 1963 Apr;17:19–58. doi: 10.1083/jcb.17.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers A. A., Arthur R., Kuritza A. Digestion of larch arabinogalactan by a strain of human colonic Bacteroides growing in continuous culture. J Agric Food Chem. 1981 May-Jun;29(3):475–480. doi: 10.1021/jf00105a009. [DOI] [PubMed] [Google Scholar]
- Stanton T. B., Canale-Parola E. Enumeration and selective isolation of rumen spirochetes. Appl Environ Microbiol. 1979 Nov;38(5):965–973. doi: 10.1128/aem.38.5.965-973.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton T. B., Canale-Parola E. Treponema bryantii sp. nov., a rumen spirochete that interacts with cellulolytic bacteria. Arch Microbiol. 1980 Sep;127(2):145–156. doi: 10.1007/BF00428018. [DOI] [PubMed] [Google Scholar]
- Tomerska H. Decomposition of pectin in vitro by pure strains of rumen bacteria. Acta Microbiol Pol B. 1971;3(2):107–115. [PubMed] [Google Scholar]
- Ziołecki A. Isolation and characterization of large treponemes from the bovine rumen. Appl Environ Microbiol. 1979 Jan;37(1):131–135. doi: 10.1128/aem.37.1.131-135.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziołecki A., Wojciechowicz M. Small pectinolytic spirochetes from the rumen. Appl Environ Microbiol. 1980 Apr;39(4):919–922. doi: 10.1128/aem.39.4.919-922.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]