Abstract
Invasion of erythrocytes by malaria parasites is mediated by specific molecular interactions. Whereas Plasmodium vivax and Plasmodium knowlesi use the Duffy blood group antigen, Plasmodium falciparum uses sialic acid residues of glycophorin A as receptors to invade human erythrocytes. P. knowlesi uses the Duffy antigen as well as other receptors to invade rhesus erythrocytes by multiple pathways. Parasite ligands that bind these receptors belong to a family of erythrocyte-binding proteins (EBP). The EBP family includes the P. vivax and P. knowlesi Duffy-binding proteins, P. knowlesi β and γ proteins, which bind alternate receptors on rhesus erythrocytes, and P. falciparum erythrocyte-binding antigen (EBA-175), which binds sialic acid residues of human glycophorin A. Binding domains of each EBP lie in a conserved N-terminal cysteine-rich region, region II, which contains around 330 amino acids with 12 to 14 conserved cysteines. Regions containing binding residues have now been mapped within P. vivax and P. knowlesi β region II. Chimeric domains containing P. vivax region II sequences fused to P. knowlesi β region II sequences were expressed on the surface of COS cells and tested for binding to erythrocytes. Binding residues of P. vivax region II lie in a 170-aa stretch between cysteines 4 and 7, and binding residues of P. knowlesi β region II lie in a 53-aa stretch between cysteines 4 and 5. Mapping regions responsible for receptor recognition is an important step toward understanding the structural basis for the interaction of these parasite ligands with host receptors.
Invasion of erythrocytes by Plasmodium merozoites is mediated by specific molecular interactions between erythrocyte receptors and parasite ligands (1). Plasmodium vivax and Plasmodium knowlesi bind the Duffy blood group antigen to invade human erythrocytes (2, 3). Duffy-negative human erythrocytes are completely resistant to invasion by these parasites. In contrast, P. knowlesi can use the Duffy antigen as well as alternate receptors to invade rhesus erythrocytes by multiple pathways (4). Plasmodium falciparum, the most important parasite for human malaria, commonly uses sialic acid residues on glycophorin A as receptors to invade human erythrocytes (5, 6).
The parasite ligands that bind these receptors belong to the erythrocyte-binding protein (EBP) family (7). The EBP family includes the Duffy-binding proteins of P. vivax and P. knowlesi, P. knowlesi β and γ proteins, which bind alternate receptors on rhesus erythrocytes, and P. falciparum sialic acid-binding protein, also known as erythrocyte-binding antigen (EBA-175), which binds sialic acid residues of glycophorin A (7). Each EBP contains two cysteine-rich domains, region II and region VI, which contain conserved cysteines and hydrophobic amino acid residues. The functional binding domains of EBPs lie in region II, the conserved N-terminal cysteine-rich region (8–10). These functional domains are referred to as Duffy-binding-like (DBL) domains after region II of the P. vivax Duffy-binding protein, the first functional domain to be identified (8). Whereas region II of the P. vivax Duffy-binding protein specifically binds the human Duffy antigen, region II of the P. knowlesi Duffy-binding protein binds both human and rhesus Duffy antigens (8). The DBL domains (region II) of the P. knowlesi β and γ proteins bind alternate receptors on rhesus erythrocytes and may mediate invasion by Duffy antigen-independent pathways (8). Region II of P. falciparum EBA-175 contains two DBL domains, F1 and F2 (9). F2 binds sialic acid residues of glycophorin A (10).
DBL domains are also found in members of the P. falciparum erythrocyte membrane protein-1 (PfEMP-1) family, which are encoded by var genes and are expressed on the surface of P. falciparum-infected trophozoites and schizonts (11–13). Some members of the PfEMP-1 family mediate cytoadherence of P. falciparum trophozoites and schizonts to host endothelial cells or uninfected erythrocytes, phenomena that are implicated in cerebral malaria. DBL domains of PfEMP-1 have been shown to bind uninfected erythrocytes to mediate rosetting (14, 15) and may also mediate binding to endothelial receptors such as ICAM-1, CD31, thrombospondin, and chondroitin sulfate A.
DBL domains are thus found in parasite ligands that mediate erythrocyte invasion and cytoadherence, two processes that underlie malaria pathogenesis. To understand the structural basis of these receptor–ligand interactions, it is important to determine the three-dimensional structures of DBL domains and map regions within DBL domains that contain receptor-binding residues. In this report, we have mapped regions containing binding residues within DBL domains of two EBPs, namely, the P. vivax Duffy-binding protein and the P. knowlesi β protein. Chimeric DBL domains containing sequences from P. vivax region II fused to P. knowlesi β region II sequences were expressed on the surface of mammalian COS cells and tested for binding to normal and enzyme-treated human and rhesus erythrocytes. Binding residues of both DBL domains lie in their central regions. Identification of regions important for receptor recognition is a first step toward understanding the structural basis for the interaction of DBL domains with host receptors.
Materials and Methods
Plasmids for Expression of Chimeric DBL Domains on COS Cell Surface.
Plasmid pRE4, which contains the gene encoding Herpes simplex virus glycoprotein D (HSV gD) under control of the Rous sarcoma virus long terminal repeat promoter in a mammalian expression vector, has been described earlier (16). Plasmids pHVDR22, pHKADR22, pHKBDR22, and pHKGDR22, which are designed to express region II of P. vivax and P. knowlesi EBPs fused to the secretory signal sequence and transmembrane domain of HSV gD, were constructed by using pRE4 as described previously (8). Similar methods have been used to express chimeric DBL domains containing P. vivax region II sequences fused to sequences from P. knowlesi β region II on the surface of COS cells. DNA fragments encoding stretches of P. vivax region II and P. knowlesi β region II were amplified by PCR by using Pyrococcus furiosus DNA polymerase (Stratagene), ligated to yield DNA fragments encoding chimeric DBL domains and cloned in frame with the signal sequence and transmembrane segment of HSV gD in plasmid pRE4 (Fig. 1a). Plasmids designed to express chimeric DBL domains are described below (Fig. 2). Supplemental material describing methods for plasmid construction is posted on the PNAS web site (see www.pnas.org).
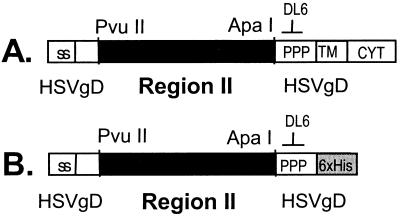
Figure 1.
Expression of parasite DBL domains in COS cells. (A) Construct used to express region II of EBPs on COS cell surface. The signal sequence (ss) and transmembrane segment (TM) of HSV gD are used to target region II to the cell surface. DNA-encoding region II is cloned in the PvuII and ApaI sites within the gene for HSV gD. Monoclonal antibody DL6, directed against the proline-rich segment (PPP) of HSV gD, is used to detect the fusion protein. CYT, HSV gD cytoplasmic domain. (B) Construct used to express region II of EBPs as secreted proteins in COS cells. TM and CYT of HSV gD are replaced with a six-histidine (6 x His) tag.
Figure 2.
Binding specificity of region II of P. vivax and P. knowlesi EBPs and chimeric binding domains. Region II of P. vivax (V) and P. knowlesi (Kα) Duffy-binding proteins, P. knowlesi β (Kβ) and γ (Kγ) proteins and chimeric domains (CH1 to CH7) containing P. vivax region II sequences (black) fused to P. knowlesi β region II sequences (white) were expressed on the surface of COS cells and tested for binding to erythrocytes (RBCs). P. vivax- and P. knowlesi-binding domains contain 12 conserved cysteines (C1 to C12). Normal Duffy-positive (Duffy Pos) and Duffy-negative (Duffy Neg) human RBCs, rhesus RBCs, and RBCs treated with neuraminidase (Neu) or chymotrypsin (Chym) were tested for binding to transfected COS cells. Number of rosettes scored in 50 fields at ×100 is reported for each construct. +, 20–50 rosettes; −, no rosettes were seen; +, 1–10 rosettes.
CH1.
Plasmid CH1 is designed to express a chimeric DBL domain containing amino acids 198 to 401 of P. vivax Duffy-binding protein fused to amino acids 399 to 517 of P. knowlesi β protein.
CH2.
Plasmid CH2 is designed to express a chimeric DBL domain containing amino acids 198 to 379 of P. vivax Duffy-binding protein fused to amino acids 377 to 517 of P. knowlesi β protein.
CH3.
Plasmid CH3 is designed to express a chimeric DBL domain containing amino acids 199 to 364 of P. knowlesi β protein fused to amino acids 368 to 522 of P. vivax Duffy-binding protein.
CH4.
Plasmid CH4 is designed to express a chimeric DBL domain containing amino acids 258 to 401 of P. vivax Duffy-binding protein flanked by amino acids 199 to 254 of P. knowlesi β protein at the N terminus and amino acids 399 to 517 of P. knowlesi β protein at the C terminus.
CH5.
Plasmid CH5 is designed to express a chimeric DBL domain containing amino acids 304 to 401 of P. vivax Duffy-binding protein flanked by amino acids 199 to 300 of P. knowlesi β protein at the N terminus and amino acids 399 to 517 of P. knowlesi β protein at the C terminus.
CH6.
Plasmid CH6 is designed to express a chimeric DBL domain containing amino acids 199 to 300 of P. knowlesi β protein fused to amino acids 304 to 522 of P. vivax Duffy-binding protein.
CH7.
Plasmid CH7 is designed to express a chimeric DBL domain containing amino acids 255 to 300 of P. knowlesi β protein flanked by amino acids 198 to 257 of P. vivax Duffy-binding protein at the N terminus and amino acids 304 to 522 of P. vivax Duffy-binding protein at the C terminus.
Inserts encoding chimeric DBL domains in plasmids CH4 and CH7 were sequenced in both directions to confirm fusion boundaries and to rule out introduction of errors by PCR. Boundaries between P. vivax and P. knowlesi sequences were confirmed by sequencing junction regions within inserts of other plasmids.
COS Cell Culture, Transfection, and Erythrocyte-Binding Assays.
COS7 cells (American Type Culture Collection) were cultured as described previously (8). COS7 cells growing in 35-mm-diameter wells (40–60% confluent) were transfected with 2.5 μg of plasmid DNA by using Lipofectin (GIBCO/BRL). Mouse monoclonal antibody DL6 (mAb DL6, gift from Roselyn Eisenberg and Gary Cohen, University of Pennsylvania, Philadelphia, PA) directed against HSV gD sequences (Fig. 1) was used to detect expression of the fusion proteins as described previously (8). Transfected COS cells expressing DBL domains on the surface were used in erythrocyte-binding assays as described earlier (8). The number of COS cells covered with erythrocyte rosettes was scored in 50 fields at ×100 by using an inverted microscope. Binding of normal and enzyme-treated erythrocytes to untransfected COS cells was tested to rule out nonspecific binding.
Erythrocytes and Their Treatment with Enzymes.
Human and rhesus blood was collected in 10% citrate phosphate dextrose, stored at 4°C for up to 2 wk, and washed three times in RPMI 1640 (GIBCO/BRL) before use. Duffy phenotypes were determined by standard methods by using two antisera, anti-Fya and anti-Fyb (Ortho Diagnostics). Duffy-positive human erythrocytes with Fy(a+b+) phenotype were used. Washed human and rhesus red cells were treated with chymotrypsin and neuraminidase, as described previously (17). An agglutination assay by using lectin MAA (Roche Molecular Biochemicals), which detects α(2–3)-linked sialic acids, was used to confirm complete removal of sialic acid residues from neuraminidase-treated erythrocytes.
Expression of DBL Domains as Soluble Proteins Secreted from COS Cells, Metabolic Labeling, Immunoprecipitation, and Erythrocyte-Binding Assays.
Plasmids pVs, pKβs, and pCH4s were designed to express P. vivax region II, P. knowlesi β region II, and chimeric DBL domain CH4 as secreted proteins in COS7 cells. They contain DNA fragments encoding DBL domains fused to DNA sequences encoding HSV gD signal peptide at the 5′ end and amino acids 268 to 340 of HSV gD followed by a 6-histidine tag at the 3′ end (Fig. 1b). The fusion constructs are cloned downstream of the cytomegalovirus immediate–early promoter in vector pCINEO (Promega). Supplemental material describing methods used for plasmid construction is posted on the PNAS web site (see www.pnas.org).
Transfected COS cells were washed with sterile phosphate-buffered saline 48 hr posttransfection, starved for 1 hr in methionine and cysteine-deficient DMEM (GIBCO/BRL), and cultured in deficient medium supplemented with [35S]Met/[35S]Cys (Dupont NEN) for 8–10 hr. Radio-labeled culture supernatants were centrifuged to remove cell debris and stored at −80°C.
Expression of recombinant DBL domains was detected in culture supernatants by immunoprecipitation. Radio-labeled culture supernatants were incubated with mAb DL6 on ice for 1 hr and with Protein A Sepharose CL-4B beads (Pharmacia) at room temperature for 1 hr. The beads were washed once with 0.5% BSA in 0.5% Triton X-100/0.15M NaCl/1 mM EDTA/50 mM Tris, pH 7.4 (NETT) and twice with NETT buffer. Bound proteins were eluted by boiling, separated by SDS/PAGE, and detected by autoradiography.
Radio-labeled supernatants containing recombinant DBL domains were used in erythrocyte-binding assays as described earlier (18). Briefly, radio-labeled supernatants were incubated with either human (Duffy positive and Duffy negative) or rhesus erythrocytes to allow binding. Erythrocytes with bound proteins were collected by centrifugation through dibutyl phthalate (Sigma), bound proteins were eluted with 300 mM NaCl, separated by SDS/PAGE, and detected by autoradiography. Erythrocyte-binding assays were performed in the presence of the chemokine, melanoma growth stimulating activity (MGSA, gift from Richard Horuk, Berlex, Inc., Richmond, CA), or peptide HPEP35 (see below) to test their effect on binding.
Peptide Synthesis.
The sequence of peptide HPEP35 corresponds to amino acids 10 to 44 of the human Duffy blood group antigen, previously identified as the binding site for P. vivax on the human Duffy blood group antigen (19). HPEP35 was synthesized by using an Applied Biosystems Model 430A peptide synthesizer, cleaved by anhydrous hydrogen fluoride treatment, and purified by RP-HPLC. Mass spectrometric and RP-HPLC analyses confirmed the purity of HPEP35.
Results
Binding Specificity of P. vivax and P. knowlesi EBPs.
The binding domain of each EBP lies in the N-terminal conserved cysteine-rich region, region II (8, 10). Region II of P. vivax and P. knowlesi EBPs was expressed on the surface of mammalian COS cells and tested for binding to normal and enzyme-treated human and rhesus erythrocytes. The secretory signal sequence and transmembrane segment of HSV gD were used to target region II to the surface of transfected mammalian cells (Fig. 1a). Immunofluorescence assays by using mAb DL6, which binds HSV gD sequences in the fusion proteins, confirmed the expression of region II on the COS cell surface.
P. vivax region II binds Duffy-positive human erythrocytes but not Duffy-negative human erythrocytes or rhesus erythrocytes (Fig. 2). Neuraminidase treatment of Duffy-positive human erythrocytes results in enhanced binding to P. vivax region II. Although untreated rhesus erythrocytes do not bind P. vivax region II, neuraminidase-treated rhesus erythrocytes bind the P. vivax domain at low levels.
Region II of the P. knowlesi Duffy antigen-binding protein (encoded by the P. knowlesi α gene) binds Duffy-positive human erythrocytes and rhesus erythrocytes but not Duffy-negative human erythrocytes or chymotrypsin-treated rhesus erythrocytes that have lost the Duffy antigen (Fig. 2). Neuraminidase treatment of human as well as rhesus erythrocytes results in enhanced binding to P. knowlesi α region II.
Region II of P. knowlesi β and γ proteins binds rhesus erythrocytes but not human erythrocytes (Fig. 2). Moreover, P. knowlesi β and γ region II bind chymotrypsin-treated rhesus erythrocytes, indicating that they do not bind the rhesus Duffy antigen. Neuraminidase treatment of rhesus erythrocytes reduces binding to P. knowlesi β region II by greater than 90%, suggesting that sialic acid residues on rhesus erythrocytes serve as receptors for P. knowlesi β region II. Neuraminidase treatment of rhesus erythrocytes does not affect binding to P. knowlesi γ region II.
Mapping Regions Containing Binding Residues Within Region II of P. vivax Duffy-Binding Protein and P. knowlesi β Protein.
Chimeric DBL domains containing P. vivax region II sequences fused to P. knowlesi β region II sequences were expressed on the surface of COS cells and tested for binding to erythrocytes (Fig. 2). Both P. vivax region II and P. knowlesi β region II contain 12 conserved cysteines. Chimeric DBL domains were designed to contain each of these cysteines to allow correct folding. Immunofluorescence assays by using mAb DL6 confirmed expression of each chimeric domain on the surface of COS cells.
Chimera CH1, which contains P. vivax region II sequences from Cys-1 to Cys-7 fused to P. knowlesi β region II sequences from Cys-7 to Cys-12, binds erythrocytes with the same specificity as P. vivax region II. Both domains bind Duffy-positive human erythrocytes but not Duffy-negative human erythrocytes or rhesus erythrocytes, indicating that binding residues for the Duffy antigen lie between Cys-1 and Cys-7 of P. vivax region II. Neuraminidase treatment of Duffy-positive human erythrocytes results in enhanced binding, and neuraminidase treatment of rhesus erythrocytes allows weak binding to both P. vivax region II and CH1.
Chimera CH2 contains P. vivax region II sequences from Cys-1 to Cys-6 fused to P. knowlesi β region II sequences from Cys-6 to Cys-12. CH2 does not bind Duffy-positive human erythrocytes. Presence of the stretch from Cys-6 to Cys-7 of P. vivax region II appears to be necessary for binding to human erythrocytes. Neuraminidase treatment allows Duffy-positive human erythrocytes to bind weakly to CH2, suggesting that some binding residues for the Duffy antigen may lie between Cys-1 and Cys-6 of P. vivax region II.
Chimera CH3 contains P. knowlesi β region II sequences from Cys-1 to Cys-6 fused to P. vivax region II sequences from Cys-6 to Cys-12. CH3 binds erythrocytes with the same specificity as P. knowlesi β region II. Both bind normal as well as chymotrypsin-treated rhesus erythrocytes. In addition, neuraminidase treatment of rhesus erythrocytes reduces binding to CH3 and P. knowlesi β region II by greater than 90%. The binding site for sialic acid residues on rhesus erythrocytes thus lies between Cys-1 and Cys-6 of P. knowlesi β region II.
Chimera CH4 contains amino acids from the central region of P. vivax region II (Cys-4 to Cys-7) flanked by P. knowlesi β sequences at the amino (Cys-1 to Cys-4) and carboxyl ends (Cys-7 to Cys-12). CH4 binds Duffy-positive but not Duffy-negative human erythrocytes, indicating that binding residues for the human Duffy antigen lie between Cys-4 and Cys-7 of P. vivax region II. Surprisingly, CH4 also binds rhesus erythrocytes. Moreover, CH4 binds neuraminidase-treated rhesus erythrocytes and does not bind chymotrypsin-treated rhesus erythrocytes, suggesting that CH4 may bind the rhesus Duffy antigen. This possibility is explored further later.
Chimera CH5 contains the central region of P. vivax region II (Cys-5 to Cys-7) flanked by P. knowlesi β region II sequences at the amino (Cys-1 to Cys-5) and carboxyl (Cys-7 to Cys-12) ends. CH5 does not bind Duffy-positive human erythrocytes, suggesting that the amino acid stretch from Cys-4 to Cys-5 of P. vivax region II is necessary for binding the Duffy antigen. CH5 binds rhesus erythrocytes with the same specificity as P. knowlesi β region II, suggesting that the binding site for sialic acid residues lies between Cys-1 and Cys-5 of P. knowlesi β region II.
Chimera CH6 contains P. knowlesi β region II sequences from Cys-1 to Cys-5 fused to P. vivax region II sequences from Cys-5 to Cys-12. Chimera CH7 contains P. knowlesi β region II sequences from Cys-4 to Cys-5 flanked by P. vivax region II sequences at the amino (Cys-1 to Cys-4) and carboxyl (Cys-5 to Cys-12) ends. CH6 and CH7 have the same binding specificity as P. knowlesi β region II, suggesting that the binding site for sialic acid residues on rhesus erythrocytes lies between Cys-4 and Cys-5 of P. knowlesi β region II.
Functional Erythrocyte-Binding Assays by Using Soluble DBL Domains Secreted from COS Cells.
P. vivax region II, P. knowlesi β region II, and chimeric DBL domain CH4 were expressed as secreted proteins in COS cells and tested for binding to erythrocytes. The signal sequence of HSV gD was used to target DBL domains to the COS cell secretory pathway (Fig. 1b). Transfected COS cells were metabolically labeled with [35S]Met/[35S]Cys. Radio-labeled DBL domains of approximately 67 kDa were detected in the culture supernatants by immunoprecipitation with mAb DL6 (Fig. 3). The functional activity of secreted DBL domains was tested as follows.
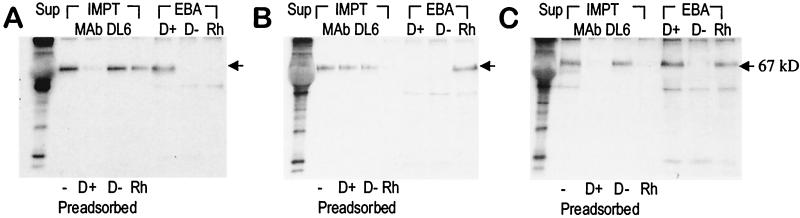
Figure 3.
Expression of P. vivax region II, P. knowlesi β region II, and chimeric domain CH4 as secreted proteins in COS cells. COS cells transfected with constructs designed to express P. vivax region II (A), P. knowlesi β region II (B), and chimera CH4 (C) as secreted proteins were metabolically labeled with [35S]Met/[35S]Cys. Recombinant domains were detected in culture supernatants (Sup) by immunoprecipitation (IMPT) with monoclonal antibody DL6 (Mab DL6) directed against HSV gD sequences in the fusion proteins. Supernatants were used either directly for immunoprecipitation (−) or after preabsorption with Duffy-positive human erythrocytes (D+), Duffy-negative human erythrocytes (D−), or rhesus erythrocytes (Rh). Radio-labeled culture supernatants were also used directly in erythrocyte-binding assays (EBA) with D+, D−, and Rh erythrocytes.
Radio-labeled supernatants were preabsorbed with either human or rhesus erythrocytes before immunoprecipitation with mAb DL6 (Fig. 3). Whereas preabsorption with Duffy-positive human erythrocytes results in removal of the recombinant protein from supernatants containing radio-labeled P. vivax region II, preabsorption with Duffy-negative human erythrocytes or rhesus erythrocytes has no effect. In contrast, in the case of supernatants containing radio-labeled P. knowlesi β region II, preabsorption with rhesus erythrocytes results in removal of the recombinant protein, but preabsorption with human erythrocytes (Duffy positive or Duffy negative) has no effect. In the case of supernatants containing radio-labeled CH4, preabsorption with both Duffy-positive human erythrocytes and rhesus erythrocytes leads to removal of recombinant CH4 from the supernatant, but preabsorption with Duffy-negative human erythrocytes has no effect.
The ability of recombinant DBL domains to bind erythrocytes was also directly tested in erythrocyte-binding assays (Fig. 3). Recombinant P. vivax region II binds Duffy-positive but not Duffy-negative human erythrocytes or rhesus erythrocytes. P. knowlesi β region II binds only rhesus erythrocytes, and chimera CH4 binds Duffy-positive human erythrocytes and rhesus erythrocytes but not Duffy-negative human erythrocytes.
Binding Specificity of Chimeric DBL Domain CH4.
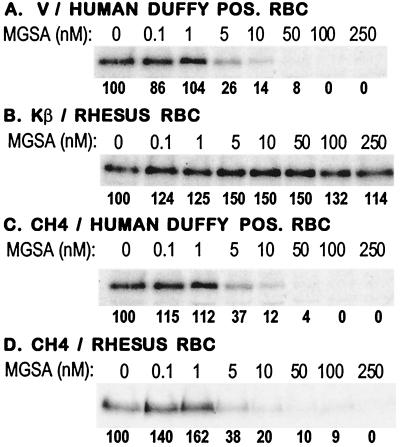
The chemokine MGSA binds both human and rhesus Duffy antigens (18, 19). To determine whether CH4 binds the Duffy antigen on both human and rhesus red cells, erythrocyte-binding assays were performed with culture supernatants containing radio-labeled P. vivax region II, P. knowlesi β region II, and CH4 in the presence of MGSA. MGSA inhibits the binding of P. vivax region II to human erythrocytes but has no effect on the binding of P. knowlesi β region II to rhesus erythrocytes (Fig. 4 a and b). MGSA also blocks the binding of CH4 to human as well as rhesus erythrocytes (Fig. 4 c and d), indicating that CH4 binds both the human and rhesus Duffy antigens.
Figure 4.
Inhibition of erythrocyte binding by DBL domains with MGSA. (A–D) Erythrocyte-binding assays were performed by using radio-labeled culture supernatants containing P. vivax region II (V), P. knowlesi β region II (Kβ), and CH4 with Duffy-positive (POS) human erythrocytes (RBCs) or rhesus RBCs in the presence of the chemokine MGSA. Binding at each MGSA concentration is expressed as a percentage of binding in the absence of MGSA.
The binding site for P. vivax maps to a 35-aa stretch of the N-terminal extracellular region of the Duffy antigen (19). Erythrocyte-binding assays were performed in the presence of HPEP35, a 35-aa peptide derived from the binding site used by P. vivax on the Duffy antigen. HPEP35 inhibits the binding of P. vivax region II to human erythrocytes but has no effect on the binding of P. knowlesi β region II to rhesus erythrocytes (Fig. 5 a and b). HPEP35 also inhibits the binding of CH4 to human as well as rhesus erythrocytes (Fig. 5 c and d), indicating that both CH4 and P. vivax region II bind the same epitope on the Duffy antigen.
Figure 5.
Inhibition of erythrocyte binding by DBL domains with peptide HPEP35. (A–D) Erythrocyte-binding assays were performed by using radio-labeled culture supernatants containing P. vivax region II (V), P. knowlesi β region II (Kβ), and chimeric domain CH4 with Duffy-positive (POS) human erythrocytes (RBCs) or rhesus RBCs in the presence of HPEP35, a 35-aa peptide derived from the sequence of the P. vivax-binding site on the human Duffy antigen. Binding at each HPEP35 concentration is expressed as a percentage of binding in the absence of HPEP35.
Discussion
The conserved N-terminal cysteine-rich regions of EBPs, region II, serve as receptor-binding domains. Region II of P. vivax Duffy-binding protein binds Duffy-positive human erythrocytes but not Duffy-negative human erythrocytes or rhesus erythrocytes (Fig. 2). N-glycanase-treated rhesus erythrocytes bind P. vivax region II, suggesting that glycosylation of the rhesus Duffy antigen inhibits binding to the P. vivax domain (19). The binding site for P. vivax region II has been mapped to a 35-aa stretch of the N-terminal extracellular regions (amino acids 10 to 44) of the human and rhesus Duffy antigens (19). Removal of sialic acid residues may allow access to binding sites on the peptide backbone of the Duffy antigen. This might explain the enhanced binding of neuraminidase-treated human erythrocytes and the low level binding of neuraminidase-treated rhesus erythrocytes to P. vivax region II.
P. knowlesi depends completely on the Duffy antigen for invasion of human erythrocytes but can use multiple pathways to invade rhesus erythrocytes (4). Three members of the EBP family have been identified from P. knowlesi (7). Region II of the P. knowlesi Duffy-binding protein (encoded by the P. knowlesi α gene) binds both the human and rhesus Duffy antigens. Region II of P. knowlesi β and γ proteins binds alternate receptors on rhesus erythrocytes and may be responsible for the Duffy antigen-independent invasion pathways. Treatment of rhesus erythrocytes with neuraminidase leads to greater than 90% reduction in binding to P. knowlesi β region II, indicating that sialic acid residues on rhesus erythrocytes serve as receptors for P. knowlesi β. The receptor for P. knowlesi γ remains to be identified.
To identify regions within DBL domains that contain binding residues, chimeric DBL domains containing sequences from P. vivax region II fused to sequences from P. knowlesi β region II were expressed on the surface of COS cells and tested for binding to erythrocytes. The binding profile of chimera CH1 was identical to that of P. vivax region II, suggesting that binding residues for the Duffy antigen lie between Cys-1 and Cys-7 of P. vivax region II. Although chimera CH2 does not bind Duffy-positive human erythrocytes, neuraminidase treatment allows binding at low levels. The region from Cys-1 to Cys-6 of P. vivax region II must therefore contain some binding residues for the Duffy antigen. The region from Cys-6 to Cys-7 may also contain some binding residues for the Duffy antigen. Absence of the binding residues that lie between Cys-6 and Cys-7 may reduce binding affinity, resulting in the inability of CH2 to bind Duffy-positive human erythrocytes. Alternatively, all the binding residues for the Duffy antigen may lie between Cys-1 and Cys-6 of P. vivax region II. However, replacement of P. vivax sequences from Cys-6 to Cys-7 with P. knowlesi β sequences in CH2 may prevent correct folding of the binding site, reducing binding affinity. The present study cannot distinguish between these possibilities.
Chimera CH4 binds Duffy-positive human erythrocytes, indicating that binding residues for the Duffy antigen lie between Cys-4 and Cys-7 of P. vivax region II. Chimera CH5, which contains P. vivax region II sequences from Cys-5 to Cys-7 flanked by P. knowlesi β region II sequences, does not bind Duffy-positive human erythrocytes, suggesting that the presence of the entire stretch from Cys-4 to Cys-7 of P. vivax region II is necessary for binding.
Surprisingly, CH4 also binds rhesus erythrocytes. Moreover, CH4 binds neuraminidase-treated rhesus erythrocytes and does not bind chymotrypsin-treated rhesus erythrocytes, suggesting that it binds the rhesus Duffy antigen. The chemokine MGSA, which binds the Duffy antigen, inhibits the binding of soluble CH4 to rhesus erythrocytes, demonstrating that CH4 binds the rhesus Duffy antigen. Moreover, peptide HPEP35, which was derived from the binding site for P. vivax on the human Duffy antigen, inhibits the binding of P. vivax region II and CH4 to human and rhesus erythrocytes. CH4 and P. vivax region II thus bind the same epitope on the Duffy antigen.
Unlike P. vivax region II and CH1, CH4 does not require removal of sialic acid residues to bind the rhesus Duffy antigen. Whereas the N terminus of CH1 contains amino acid sequences from Cys-1 to Cys-7 of P. vivax region II, the N terminus of CH4 contains P. knowlesi β region II sequences from Cys-1 to Cys-4 fused to P. vivax region II sequences from Cys-4 to Cys-7. Replacement of P. vivax sequences with P. knowlesi β sequences at the N terminus (Cys-1 to Cys-4) seems to allow CH4 to bind normal rhesus erythrocytes, suggesting that amino acid sequences outside the binding site may influence the fine specificity of binding of DBL domains.
Chimera CH7, which contains amino acid residues from Cys-4 to Cys-5 of P. knowlesi β region II flanked by P. vivax region II sequences, binds rhesus erythrocytes with the same specificity as P. knowlesi β region II. The binding site for sialic acid residues on rhesus erythrocytes thus lies between Cys-4 and Cys-5 of P. knowlesi β region II.
Binding residues of P. vivax region II and P. knowlesi β region II thus lie in the central regions of these binding domains. Contact residues for the Duffy antigen lie in a 170-aa stretch between Cys-4 and Cys-7 of P. vivax region II. In the case of P. knowlesi β region II, a 53-aa stretch from Cys-4 to Cys-5 contains binding residues for sialic acids on rhesus erythrocytes. Site-directed mutagenesis will be used to identify contact residues within these central regions.
Sequence polymorphism studies with P. vivax field isolates from Papua New Guinea found that the central stretch of region II (between Cys-4 and Cys-7) is a hypervariable region (20). This stretch has a higher amino acid substitution rate than other parts of the P. vivax Duffy-binding protein, suggesting that this region may be under immune pressure. However, most of the amino acid substitutions in this region were found to be conservative. This study provides evidence that the central stretch of P. vivax region II is important for receptor recognition. Functional constraints may be responsible for the limited variation observed in the central stretch of region II in P. vivax field isolates.
It remains to be seen whether the binding residues of DBL domains of other EBPs and PfEMP-1 also lie in their central regions. Structural studies with DBL domains should reveal the three-dimensional structures of DBL domains and their binding pockets. An understanding of the structural basis for the interaction of these parasite ligands with host receptors may enable development of novel methods to block these interactions and inhibit erythrocyte invasion or reverse cytoadherence.
Supplementary Material
Acknowledgments
We thank Drs. Roselyn Eisenberg and Gary Cohen for providing plasmid pRE4 and mAb DL6, Dr. Richard Horuk for providing MGSA, Sachchidanand and Prof. Virendra S. Chauhan for peptide synthesis, and Dr. Sandip Basu for providing access to the National Institute of Immunology primate facility. We are grateful to Drs. Jude N. Okoyeh and Jamieu Ogunbanwo for generously providing valuable reagents for use in this study. This work was supported by a grant from the Human Frontiers Science Program.
Abbreviations
- EBP
erythrocyte-binding protein
- DBL
Duffy-binding-like
- HSV gD
Herpes simplex virus glycoprotein D
- MGSA
melanoma growth stimulating activity
- PfEMP-1
P. falciparum erythrocyte membrane protein-1
References
- 1.Chitnis C E, Sinnis P, Miller L H. In: Malaria: Molecular and Clinical Aspects. Wahlgren M, Perlmann P, editors. London: Harwood; 1999. pp. 249–285. [Google Scholar]
- 2.Miller L H, Mason S J, Dvorak J A, McGinniss M H, Rothman I K. Science. 1975;189:561–563. doi: 10.1126/science.1145213. [DOI] [PubMed] [Google Scholar]
- 3.Miller L H, Mason S J, Clyde D F, McGinniss M H. N Engl J Med. 1976;295:302–304. doi: 10.1056/NEJM197608052950602. [DOI] [PubMed] [Google Scholar]
- 4.Haynes J D, Dalton J P, Klotz F W, McGinniss M H, Hadley T J, Hudson D E, Miller L H. J Exp Med. 1988;167:1873–1881. doi: 10.1084/jem.167.6.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller L H, Haynes J D, McAuliffe F M, Shiroishi T, Durocher J R, McGinniss M H. J Exp Med. 1977;146:277–281. doi: 10.1084/jem.146.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pasvol G, Wainscoat J S, Weatherall D J. Nature (London) 1982;297:64–66. doi: 10.1038/297064a0. [DOI] [PubMed] [Google Scholar]
- 7.Adams J H, Sim B K, Dolan S A, Fang X, Kaslow D C, Miller L H. Proc Natl Acad Sci USA. 1992;89:7085–7089. doi: 10.1073/pnas.89.15.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chitnis C E, Miller L H. J Exp Med. 1994;180:497–506. doi: 10.1084/jem.180.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sim B K, Orlandi P A, Haynes J D, Klotz F W, Carter J M, Camus D, Zegans M E, Chulay J D. J Cell Biol. 1990;111:1877–1884. doi: 10.1083/jcb.111.5.1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sim B K, Chitnis C E, Wasniowska K, Hadley T J, Miller L H. Science. 1994;264:1941–1944. doi: 10.1126/science.8009226. [DOI] [PubMed] [Google Scholar]
- 11.Baruch D I, Pasloske B L, Singh H B, Bi X, Ma X C, Feldman M, Taraschi T F, Howard R J. Cell. 1995;82:77–87. doi: 10.1016/0092-8674(95)90054-3. [DOI] [PubMed] [Google Scholar]
- 12.Su X Z, Heatwole V M, Wertheimer S P, Guinet F, Herrfeldt J A, Peterson D S, Ravetch J A, Wellems T E. Cell. 1995;82:89–100. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- 13.Smith J D, Chitnis C E, Craig A G, Roberts D J, Hudson-Taylor D E, Peterson D S, Pinches R, Newbold C I, Miller L H. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rowe J A, Moulds J M, Newbold C I, Miller L H. Nature (London) 1997;388:292–295. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- 15.Chen Q, Barragan A, Fernandez V, Sundstrom A, Schlichtherle M, Sahlen A, Carlson J, Datta S, Wahlgren M. J Exp Med. 1998;187:15–23. doi: 10.1084/jem.187.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cohen G H, Wilcox W C, Sodora D L, Long D, Levin J Z, Eisenberg R J. J Virol. 1988;62:1932–1940. doi: 10.1128/jvi.62.6.1932-1940.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camus D, Hadley T J. Science. 1985;230:553–556. doi: 10.1126/science.3901257. [DOI] [PubMed] [Google Scholar]
- 18.Horuk R, Chitnis C E, Darbonne W C, Colby T J, Rybicki A, Hadley T J, Miller L H. Science. 1993;261:1182–1184. doi: 10.1126/science.7689250. [DOI] [PubMed] [Google Scholar]
- 19.Chitnis C E, Chaudhuri A, Horuk R, Pogo A O, Miller L H. J Exp Med. 1996;184:1531–1536. doi: 10.1084/jem.184.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kappe S H, Curley G P, Noe A R, Dalton J P, Adams J H. Mol Biochem Parasitol. 1997;89:137–114. doi: 10.1016/s0166-6851(97)00113-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.