Abstract
An organism identified as Pseudomonas diminuta was found to hydrolyze parathion. Cells grown for 48 h contained 3,400 U of parathion hydrolase activity per liter of broth. Expression of enzymatic activity was lost at a high frequency (9 to 12%) after treatment with mitomycin C. Hydrolase-negative derivatives were missing a plasmid present in the wild-type organism. The molecular mass of this plasmid (pCS1), as determined by electron microscopy, was about 44 × 106 daltons.
Full text
PDF
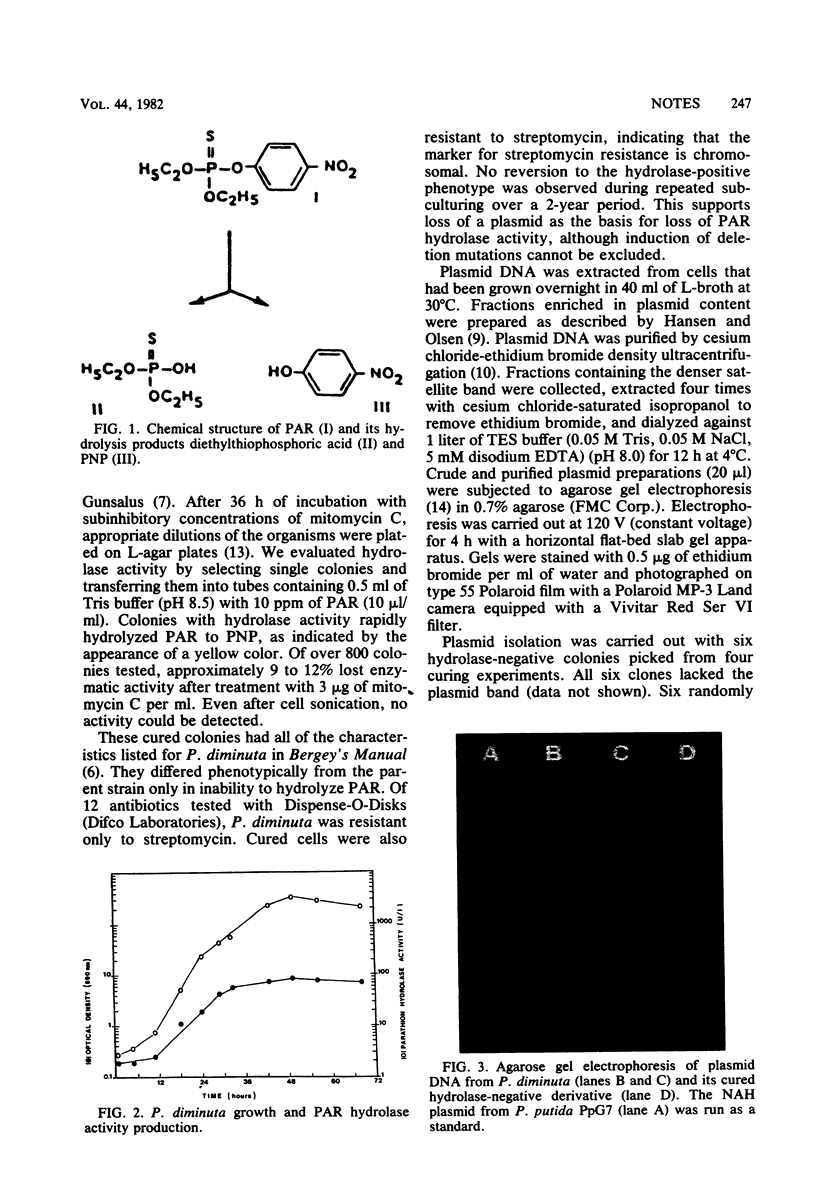
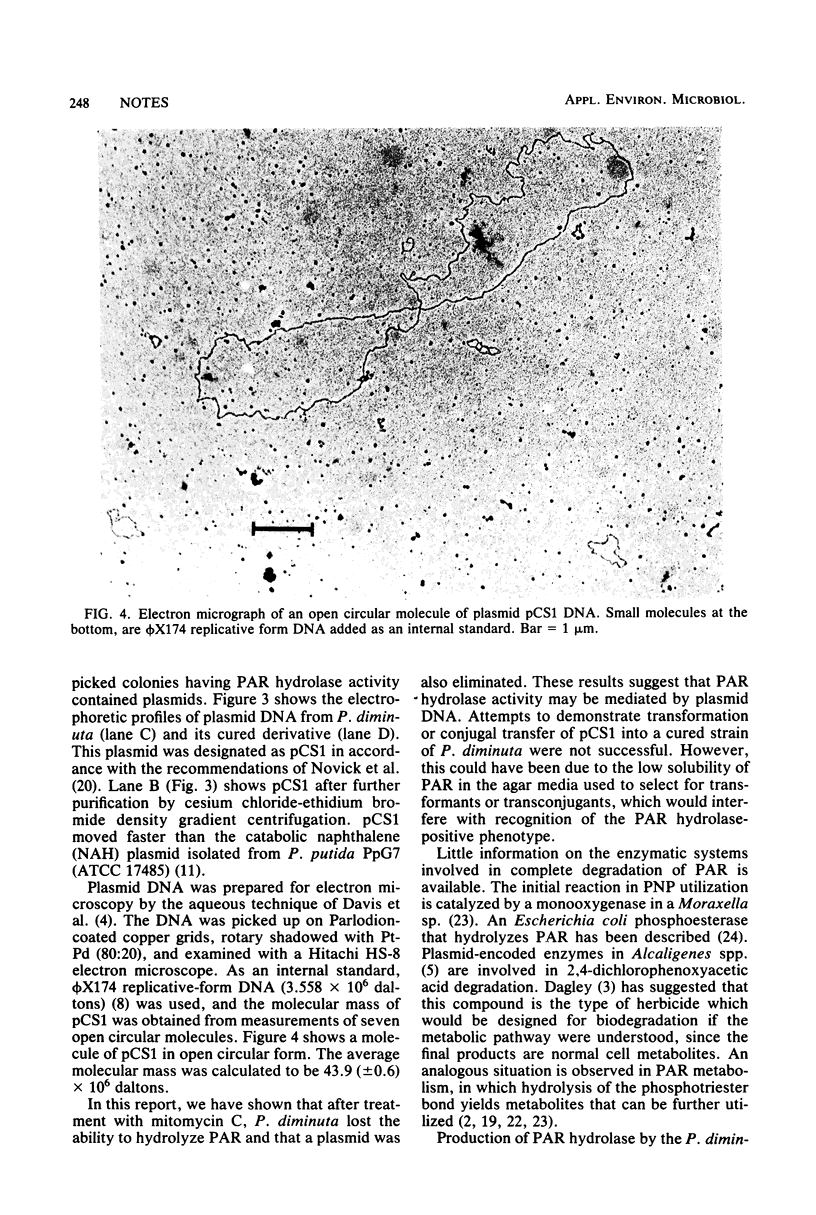

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brown P. R., Clarke P. H. Amino acid substitution in an amidase produced by an acetanilide-utilizing mutant of Pseudomonas aeruginosa. J Gen Microbiol. 1972 Apr;70(2):287–288. doi: 10.1099/00221287-70-2-287. [DOI] [PubMed] [Google Scholar]
- Cook A. M., Daughton C. G., Alexander M. Phosphorus-containing pesticide breakdown products: quantitative utilization as phosphorus sources by bacteria. Appl Environ Microbiol. 1978 Nov;36(5):668–672. doi: 10.1128/aem.36.5.668-672.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Pemberton J. M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981 Feb;145(2):681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn N. W., Gunsalus I. C. Transmissible plasmid coding early enzymes of naphthalene oxidation in Pseudomonas putida. J Bacteriol. 1973 Jun;114(3):974–979. doi: 10.1128/jb.114.3.974-979.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher H. W., Williams R. C. Electron microscopic visualization of nucleic acids and of their complexes with proteins. Annu Rev Biochem. 1979;48:649–679. doi: 10.1146/annurev.bi.48.070179.003245. [DOI] [PubMed] [Google Scholar]
- Hansen J. B., Olsen R. H. Isolation of large bacterial plasmids and characterization of the P2 incompatibility group plasmids pMG1 and pMG5. J Bacteriol. 1978 Jul;135(1):227–238. doi: 10.1128/jb.135.1.227-238.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphreys G. O., Willshaw G. A., Anderson E. S. A simple method for the preparation of large quantities of pure plasmid DNA. Biochim Biophys Acta. 1975 Apr 2;383(4):457–463. doi: 10.1016/0005-2787(75)90318-4. [DOI] [PubMed] [Google Scholar]
- Johnston J. B., Gunsalus I. C. Isolation of metabolic plasmid DNA from Pseudomonas putida. Biochem Biophys Res Commun. 1977 Mar 7;75(1):13–19. doi: 10.1016/0006-291x(77)91282-7. [DOI] [PubMed] [Google Scholar]
- LENNOX E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology. 1955 Jul;1(2):190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- Laveglia J., Dahm P. A. Degradation of organophosphorus and carbamate insecticides in the soil and by soil microorganisms. Annu Rev Entomol. 1977;22:483–513. doi: 10.1146/annurev.en.22.010177.002411. [DOI] [PubMed] [Google Scholar]
- Meyers J. A., Sanchez D., Elwell L. P., Falkow S. Simple agarose gel electrophoretic method for the identification and characterization of plasmid deoxyribonucleic acid. J Bacteriol. 1976 Sep;127(3):1529–1537. doi: 10.1128/jb.127.3.1529-1537.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnecke D. M. Enzymatic hydrolysis of organophosphate insecticides, a possible pesticide disposal method. Appl Environ Microbiol. 1976 Jul;32(1):7–13. doi: 10.1128/aem.32.1.7-13.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnecke D. M., Hsieh D. P. Microbial decontamination of parathion and p-nitrophenol in aqueous media. Appl Microbiol. 1974 Aug;28(2):212–217. doi: 10.1128/am.28.2.212-217.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnecke D. M., Hsieh D. P. Pathways of microbial metabolism of parathion. Appl Environ Microbiol. 1976 Jan;31(1):63–69. doi: 10.1128/aem.31.1.63-69.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick R. P., Clowes R. C., Cohen S. N., Curtiss R., 3rd, Datta N., Falkow S. Uniform nomenclature for bacterial plasmids: a proposal. Bacteriol Rev. 1976 Mar;40(1):168–189. doi: 10.1128/br.40.1.168-189.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pemberton J. M., Fisher P. R. 2,4-D plasmids and persistence. Nature. 1977 Aug 25;268(5622):732–733. doi: 10.1038/268732a0. [DOI] [PubMed] [Google Scholar]
- SIMPSON J. R., EVANS W. C. The metabolism of nitrophenols by certain bacteria. Biochem J. 1953 Jul 17;55(320TH):xxiv–xxiv. [PubMed] [Google Scholar]
- Spain J. C., Wyss O., Gibson D. T. Enzymatic oxidation of p-nitrophenol. Biochem Biophys Res Commun. 1979 May 28;88(2):634–641. doi: 10.1016/0006-291x(79)92095-3. [DOI] [PubMed] [Google Scholar]
- Zech R., Wigand K. D. Organophosphate-detoxicating enzymes in E. coli. Gelfiltration and isoelectric focusing of DFPase, paraoxonase and unspecific phosphohydrolases. Experientia. 1975 Feb 15;31(2):157–158. doi: 10.1007/BF01990678. [DOI] [PubMed] [Google Scholar]