Abstract
Neuronal migration is a critical phase of brain development, where defects can lead to severe ataxia, mental retardation, and seizures. In the developing cerebellum, granule neurons turn on the gene for tissue plasminogen activator (tPA) as they begin their migration into the cerebellar molecular layer. Granule neurons both secrete tPA, an extracellular serine protease that converts the proenzyme plasminogen into the active protease plasmin, and bind tPA to their cell surface. In the nervous system, tPA activity is correlated with neurite outgrowth, neuronal migration, learning, and excitotoxic death. Here we show that compared with their normal counterparts, mice lacking the tPA gene (tPA−/−) have greater than 2-fold more migrating granule neurons in the cerebellar molecular layer during the most active phase of granule cell migration. A real-time analysis of granule cell migration in cerebellar slices of tPA−/− mice shows that granule neurons are migrating 51% as fast as granule neurons in slices from wild-type mice. These findings establish a direct role for tPA in facilitating neuronal migration, and they raise the possibility that late arriving neurons may have altered synaptic interactions.
Migration of neurons is a critical phase of brain development. Defects in this process can lead to severe ataxia (1), mental retardation, and seizures (2). Granule neurons in the developing cerebellum turn on tissue plasminogen activator (tPA) gene expression as they leave their germinative zone in the external granule cell layer (EGL) and begin to migrate inward into the cerebellar molecular layer on their way to the internal granule cell layer (3). Granule neurons both secrete tPA (4), an extracellular serine protease that converts the proenzyme plasminogen into the active protease plasmin, and bind tPA to their cell surface (5).
In the nervous system, tPA activity is correlated with neurite outgrowth (4), neuronal migration (6, 7), learning (8, 9), and excitotoxic cell death (10). Although tPA expression has been shown to correlate with neuronal migration (3) and serine protease inhibitors block neuronal migration in cell culture (6, 7, 11), a direct role for tPA in neuronal migration in the brain has not been established. Therefore, when adult mice missing the tPA gene demonstrated no readily distinguishable phenotype (12), we were eager to examine their developmental neuroanatomy.
The studies presented here show that during the most active phase of active granule cell migration, there are significantly more granule neurons in transit in the cerebellar molecular layer of the mice lacking the tPA gene. This increase in molecular-layer granule neurons is caused by a markedly retarded rate of neuronal migration through the molecular layer by granule neurons from the tPA-deficient mice.
Methods
Animals.
Breeding pairs of mice lacking either the tPA gene (tPA−/−) or the urokinase-type plasminogen activator (uPA) gene (uPA−/−) generated by homologous recombination in C57BL/6 mice with SV129 stem cells (12), and their outbred C57BL/SV129+/+ control counterparts, were the generous gift of Peter Carmeliet (Center for Molecular and Vascular Biology, University of Leuven, Leuven, Belgium). Mice having different grandmothers were bred in our colony and backcrossed to wild-type controls every 10 generations to prevent strain divergence. Similarly, tPA−/− mice placed on a >98% C57BL/6 background were purchased from The Jackson Laboratory along with C57BL/6 wild-type (+/+) control mice. Gene knockouts were confirmed by the absence of plasminogen activator activity. All animal procedures were according to protocols approved by the Institutional Animal Care and Use Committee.
Fixed Tissue Preparation and Cell Counting.
The whole brain was removed from mice at postnatal days P7, P10, P13,
and P16 and fixed in Cajal fixative, dehydrated, and then embedded in
polyester wax (13). Ten-micrometer sagittal sections were taken at the
midline and at  and
and  lateral to the midline, and
were stained with hematoxylin/eosin. All cerebellar folia were
counted in each of the 100 sections from 30 normal
(tPA+/+) or knockout
(tPA−/− and uPA−/−)
C57BL/129 mice, or the C57BL/6 tPA+/+ and
tPA−/− mice, of different ages, and the mean
(±SEM) number of granule neurons per mm2 of
molecular layer was determined. The hippocampus was used to determine
the degree of lateral displacement for the sagittal sections.
lateral to the midline, and
were stained with hematoxylin/eosin. All cerebellar folia were
counted in each of the 100 sections from 30 normal
(tPA+/+) or knockout
(tPA−/− and uPA−/−)
C57BL/129 mice, or the C57BL/6 tPA+/+ and
tPA−/− mice, of different ages, and the mean
(±SEM) number of granule neurons per mm2 of
molecular layer was determined. The hippocampus was used to determine
the degree of lateral displacement for the sagittal sections.
[3H]Thymidine Birthdating.
tPA+/+ and tPA−/− mice received two i.p. injections of [3H]thymidine (each 25 μCi; 1 μCi = 37 kBq) 8 h apart on P7. The cerebella were removed on P10, fixed, sectioned, dipped in NTB-2 (Kodak) emulsion, and exposed for 3 weeks at 4°C. The emulsion was developed in Kodak Microdol and counterstained with Giemsa. Radiolabeled cells were counted as above in 100 sections.
Migration Microscopy.
Cerebella were removed from P8 mice, placed in cold saline, and sliced into 400-μm sagittal sections, which were rinsed and placed on rehydrated collagen-coated coverslips, covered with a drop of basal Eagle’s medium, and then incubated at 37°C for 10 min in 5% CO2/95% air. The pia was gently removed from the edges of the tissue and small crystals of 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate (DiI) were placed adjacent to the tissue edge and incubated for 45 min at 37°C. Using a dissection microscope, the DiI crystals were removed and the coverslip was placed in a Dvorak–Stotler chamber and viewed with a Bio-Rad 600 confocal microscope with the lowest laser setting and attenuated 90% with neutral density filters to reduce cell damage. Digital images were collected at 20- to 30-min intervals. Digital images of DiI fluorescently labeled migrating neurons were also collected at 20- to 30-min intervals on an inverted Zeiss 35IM microscope with a ×63 Plan-neofluar objective, a Dage CCD 72 digital camera, and National Institutes of Health image software on a Power Mac.
Results
The most active phase of granule neuron migration in the mouse cerebellum occurs during the second postnatal week (14, 15). Fixed tissue sections of mouse cerebella from P7, P10, P13, and P16 were examined. The striking feature in these cerebellar sections from the tPA−/− mice was the markedly increased number of elongated migrating granule cells in the molecular layer of P7, P10, and P13 mice, compared with their genetically normal counterparts (Fig. 1). Electron microscopy shows that migrating granule neurons in both tPA−/− and normal (tPA+/+) cerebella have similar morphologies and leading processes as they traverse the molecular layer (data not shown). However, by the end of the granule cell migration period at P16, the molecular layers of both tPA−/− and tPA+/+ mice have similar cell numbers.
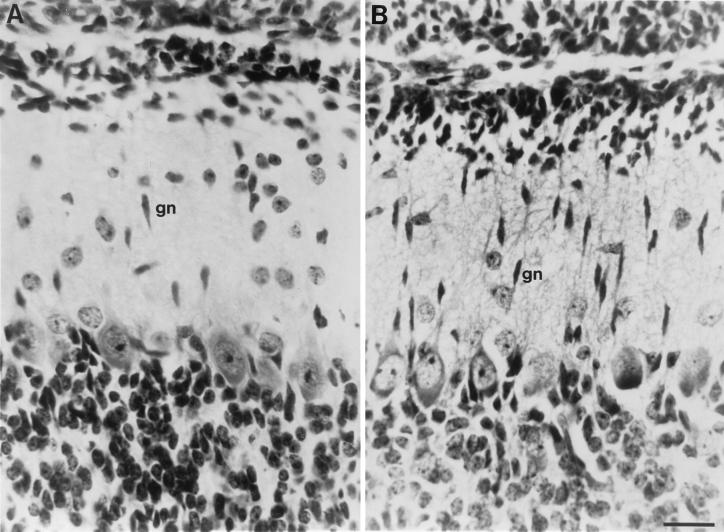
Figure 1.
Granule cells in the molecular layer of P10 cerebella from tPA+/+ and tPA−/− mice. (A) Sagittal section from tPA+/+ mouse showing elongated, presumptive migrating, granule neurons (gn) in the cell sparse molecular layer. (B) Sagittal section of an identical folia and similar lateral displacement as in A, from a tPA−/− mouse showing large numbers of elongated migratory granule neurons in the cerebellar molecular layer. About 2- to 3-fold more granule neurons are seen in the molecular layer of tPA−/− mice, where granule cell migration appears delayed. (Bar = 50 μm.)
The quantitative difference in molecular-layer granule neurons between
tPA−/− and wild-type control mice is readily
apparent in Table 1. Because different regions of the
cerebellum develop at different rates, positionally matched sagittal
sections at the midline and at  and
and  of the lateral
distance were compared by counting the molecular-layer granule neurons
in all folia at each time point. The tPA−/−
mice show about twice as many granule cells in the molecular layer
throughout the developing cerebellum in P7, P10, and P13 mice as did
control mice. Although our initial studies were performed on
tPA−/− or tPA+/+
C57BL/129 hybrid mice from the homologous recombination stem cells,
recently, this tPA−/− knockout has been put
onto a relatively (>98%) pure C57BL/6 background. Analysis of these
C57BL/6 tPA−/− mice and wild-type C57BL/6
mice shows a similar increase in granule cell number (Table 1). These
findings show that the increase in granule cell number is not due to a
region-specific sampling or mouse strain effect.
of the lateral
distance were compared by counting the molecular-layer granule neurons
in all folia at each time point. The tPA−/−
mice show about twice as many granule cells in the molecular layer
throughout the developing cerebellum in P7, P10, and P13 mice as did
control mice. Although our initial studies were performed on
tPA−/− or tPA+/+
C57BL/129 hybrid mice from the homologous recombination stem cells,
recently, this tPA−/− knockout has been put
onto a relatively (>98%) pure C57BL/6 background. Analysis of these
C57BL/6 tPA−/− mice and wild-type C57BL/6
mice shows a similar increase in granule cell number (Table 1). These
findings show that the increase in granule cell number is not due to a
region-specific sampling or mouse strain effect.
Table 1.
Granule cells in the cerebellar molecular layer of normal and tPA-knockout mice
| Age | Genotype | Cells/mm2
molecular layer
|
||
|---|---|---|---|---|
| Midline |
 lateral
lateral |
 lateral lateral |
||
| P7 | tPA+/+ | 1,008 ± 38 | 924 ± 39 | 683 ± 33 |
| tPA−/− | 1,747 ± 55 | 1,842 ± 73 | 1,600 ± 67 | |
| P10 | tPA+/+ | 746 ± 27 | 1,036 ± 44 | 724 ± 27 |
| tPA−/− | 1,523 ± 36 | 1,381 ± 34 | 1,342 ± 37 | |
| C57BL tPA+/+ | 885 ± 40 | 652 ± 30 | 1,031 ± 60 | |
| C57BL tPA−/− | 1,272 ± 75 | 1,738 ± 92 | 1,389 ± 91 | |
| uPA−/− | 836 ± 32 | ND | 852 ± 32 | |
| P13 | tPA+/+ | 328 ± 13 | 349 ± 13 | 435 ± 14 |
| tPA−/− | 657 ± 26 | 561 ± 19 | 207 ± 17 | |
| P16 | tPA+/+ | 124 ± 10 | 167 ± 12 | 273 ± 24 |
| tPA−/− | 149 ± 17 | 94 ± 9 | 94 ± 6 | |
Elongated migrating granule neurons in the cerebellar molecular layer were counted in sagittal 10-μm sections of whole mouse brain stained with hematoxylin/eosin. Significantly more (P < 0.001) granule neurons were seen in the molecular layer of tPA−/− mice at all times between P7 and P13 than in their tPA+/+ counterparts. Only at the end of the granule cell migratory period (P16) did the two genotypes show similar numbers of granule cells in the molecular layer. tPA-knockout mice maintained on a more pure (>98%) C57BL/6 genetic background and their normal counterparts were also compared at P10, and showed results similar to those of the transgenic hybrids. An additional control was the P10 uPA−/− knockout mouse, which mimicked the normal control mice, thus confirming the tPA gene specificity of this effect.
Plasminogen activator knockout mice (tPA−/− or uPA−/−) do not show compensatory up-regulation of the other plasminogen activator. Because there is virtually no urokinase in mouse cerebellum (3), it is not surprising that urokinase knockouts (uPA−/−) show normal granule cell migration. Although other proteases in brain such as the kallikreins (16) can activate plasminogen, they are less effective activators. Taken together, these findings suggest a role for tPA specifically in this migratory event.
To more directly address whether the increased cell number in the molecular layer was because of migrating granule neurons, [3H]thymidine birthdating (17) was used to label granule neurons on P4 and P7 during their last mitoses in the EGL before their inward migration. Cerebella fixed and sectioned 3 days after thymidine labeling also showed (Fig. 2) the same 2-fold increase in the number of radiolabeled cells in the molecular layer of the tPA−/− cerebellum (Fig. 2B). Most birthdated granule neurons had already completed their transit across the molecular layer of the tPA+/+ cerebellum (Fig. 2A), which takes about 2 days from their last mitosis (15). This result indicates that the increased cell numbers in the molecular layer are caused by a migratory granule cell population originating in the EGL several days earlier.
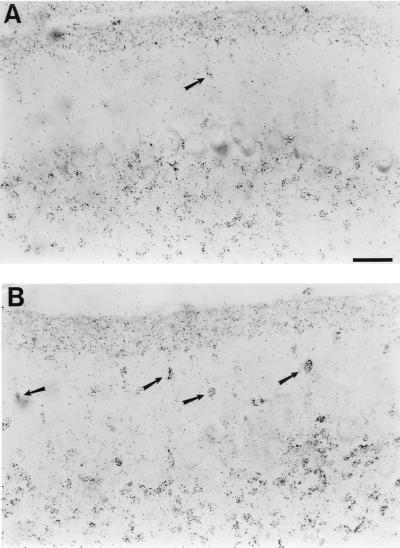
Figure 2.
[3H]Thymidine-birthdated granule neurons in the molecular layer of P10 cerebella from tPA+/+ and tPA−/− mice. (A) Autoradiography of a sagittal section of a tPA+/+ mouse cerebellum showing relatively few birthdated migratory granule neurons still remaining in the molecular layer. Most granule cells have already moved into the internal granule layer beneath the Purkinje neurons. (B) Autoradiography of a similar section from a tPA−/− mouse cerebellum showing many more birthdated migratory granule neurons still in the molecular layer. (Bar = 50 μm.)
The possibility that the increased number of granule neurons in the molecular layer reflected a greater number of granule cells being generated in the EGL of tPA−/− mice, or alternatively, a reduced level of cell death in the tPA−/− mice, was explored. Quantitative DNA analysis of cerebella from P11 mice showed nearly identical amounts in the tPA−/− (159 ± 6 μg of DNA per cerebellum) and tPA+/+ (162 ± 2 μg of DNA per cerebellum) mice, whereas P15 tPA+/+ mice contained about 10% more DNA (198 μg of DNA per cerebellum) than the tPA−/− mice (178 μg of DNA per cerebellum). Furthermore, the internal granule cell layer of adult mice from both genotypes had similar thickness and granule cell numbers, thereby also supporting the finding that similar numbers of granule neurons were generated in these mice. DNA TUNEL labeling to assess programmed (apoptotic) cell death (18) in tPA−/− and tPA+/+ mice also showed similar numbers of dying cells (data not shown). These findings rule out increased cell production or decreased cell death as accounting for the elevated number of molecular-layer granule neurons in tPA−/− mice.
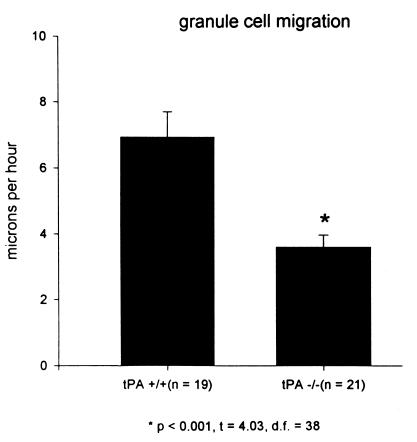
The static analyses of increased granule cell number in the cerebellar molecular layer described above suggest that the rate of granule cell migration through the molecular layer of tPA−/− mice cerebella may be slower than that of their normal counterparts. Therefore, a dynamic or real-time analysis of granule neuron migration through the molecular layer of cerebellar slice cultures from tPA−/− and their normal [tPA+/+] counterpart mice was assessed by confocal microscopy and digital imaging of DiI-labeled granule neurons (Fig. 3). A cerebellar slice from a P8 tPA+/+ mouse shows a granule neuron (indicated by the arrow) migrating inward from the EGL and leaving behind a trailing axon. The granule cell migrates at a rate of 15 μm/h over 100 min. Fig.3 Right shows a granule neuron in a P8 tPA−/− cerebellum moving inward through the molecular layer at a rate of 5.6 μm/h over a 140-min period. Although a cleaner image was obtained by confocal microscopy, we, like other workers (H. Komuro, personal communication), experienced increased cell damage and reduced efficiency even at a low laser setting and >90% attenuation with neutral density filters. Therefore, more images were obtained by fluorescence microscopy; the findings were similar. In a P8 cerebellar slice from a normal [tPA+/+] mouse (Fig. 4 Left), granule neurons migrate away from the pial surface toward a stationary DiI crystal at rates of 10.9 and 14.7 μm/h. Granule neurons in the molecular layer of a P8 cerebellar slice from a tPA−/− mouse (Fig. 4 Right) migrate inward at rates of 2.3 and 3.6 μm/h. An analysis of some 20 granule cells in P8 cerebella of each genotype and visualization by confocal or fluorescence microscopy is shown in Fig. 5; granule neurons in tPA+/+ mice migrated at an average of 7.3 ± 0.79 μm/h compared with rates of 3.7 ± 0.36 μm/h in tPA−/− mice. Thus, granule neurons in tPA−/− mice migrate through the molecular layer at about one-half the rate measured for migration of granule neurons in normal tPA+/+ mice.
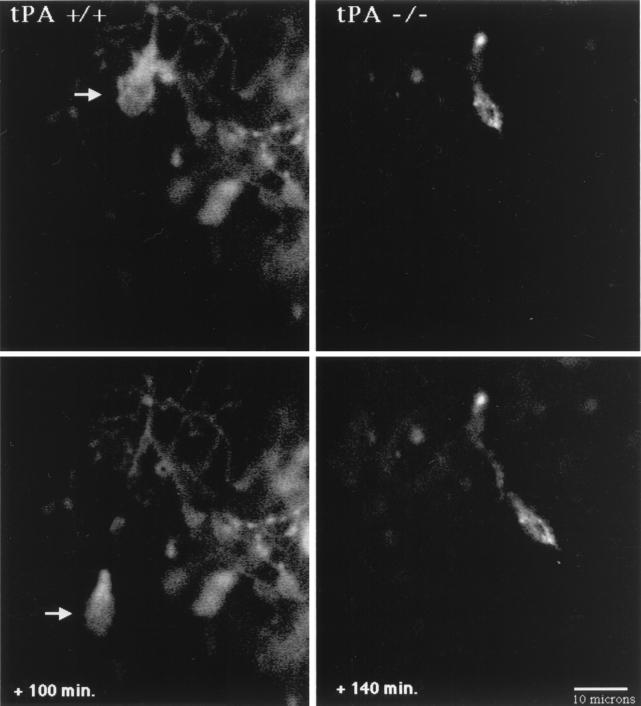
Figure 3.
Confocal microscopy images of migrating granule neurons in cerebellar slices from tPA+/+ and tPA−/− P8 mice. In the tPA+/+ mouse (Left), a DiI-labeled granule neuron (arrow) migrates inward at 15 μm/h from the EGL over the next 100 min, leaving behind its trailing axon. The P8 cerebellar slice from a tPA−/− mouse (Right) shows an inwardly migrating granule neuron that moves through the molecular layer at 5.6 μm/h over the next 140 min. Digital images were collected at 20- to 30-min intervals. (Bar = 10 μm.)
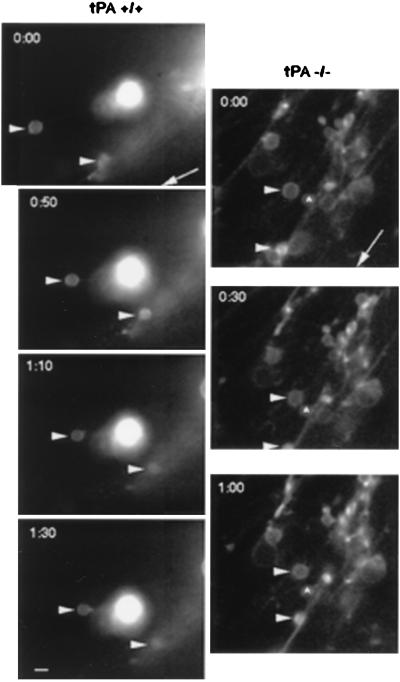
Figure 4.
DiI-labeled granule cell migration in the molecular layer of P8 cerebellar slices. Digital images of DiI fluorescently labeled migrating granule neurons in P8 cerebellar slices from tPA+/+ or tPA−/− mice were collected every 20–30 min. (Left) The two labeled cells (arrowheads) in the tPA+/+ molecular layer are migrating inward along radial processes toward a stationary DiI fragment and away from the pial surface (whose direction is indicated by the arrow) at rates of 11 μm/h for the upper cell and 15 μm/h for the lower cell. (Right) Granule neurons (arrowheads) in the tPA−/− molecular layer show slower rates of inward migration along radial processes (2.3 μm/h for the upper cell and 3.6 μm/h for the lower cell) relative to a stationary cell (∧). (Bar = 7 μm.)
Figure 5.
Granule neurons migrate slower in cerebellar sections from tPA−/− mice. DiI-labeled granule neurons were followed at 20-min intervals by fluorescence microscopy and digital video. About 20 cells were followed in eight cerebellar slices from either tPA+/+ or tPA−/− mice. Granule neurons in the tPA−/− cerebella migrated significantly (P < 0.001) slower, at an average rate of 3.7 μm/h, than the 7.3 μm/h average for granule neurons in the tPA+/+ cerebellar slices.
These findings indicated that tPA gene expression is required for maintaining the maximal rate of granule neuron migration during cerebellar development. Our previous studies (3, 19) with the mouse cerebellum have shown that tPA mRNA is not expressed by mitotic cells in the EGL; however, tPA expression is induced in postmitotic granule neurons that are about to enter the molecular layer and those granule cells actively traversing the molecular layer. tPA mRNA levels are down-regulated after the granule neurons complete their migration and are in the internal granule cell layer. Furthermore, tPA protein is readily identifiable on migrating granule neurons in the molecular layer of P8 cerebella (S.P.H. and N.W.S., unpublished results), and tPA activity is highest in the P7–P14 cerebellum (3). Thus, high levels of tPA mRNA and protein expression occur at the time of most active granule cell migration. This correlation supports a role for tPA function in neuronal migration.
Earlier studies (6, 7) showed that serine protease inhibitors that block tPA activity could inhibit mouse granule neuron migration both in cell culture and in vivo, suggesting that the catalytic site of tPA is required for its effect on granule cell migration. tPA has only two well-characterized substrates, plasminogen and pro-hepatocyte growth factor/scatter factor (HGF/SF). tPA can cleave pro-HGF/SF into active HGF/SF (20) in some brain regions (21); however, this growth factor and its receptor c-met are not readily detected in the cerebellar molecular layer (21). Although the primary substrate of tPA, plasminogen, was thought to be strictly a liver protein, more recent data show that plasminogen mRNA is present in adult hippocampal neurons (22, 23), and both neonatal and adult mouse cerebellum express plasminogen mRNA and protein (M.E.B. and N.W.S., unpublished results). Plasminogen mRNA levels in 8-and 14-day cerebella from tPA+/+ (0.5 ng/μg of RNA) and tPA−/− (0.45 ng/μg of RNA) mice suggest that similar amounts of plasminogen are available. Furthermore, thrombospondin, which facilitates tPA activation of plasminogen to plasmin, is found on the surface of the Bergmann radial glial fibers (24), on which many granule neurons migrate through the molecular layer (25). Plasmin generation on the surface of granule neurons may facilitate their migration by transient cleavage of cell–cell and cell–matrix adhesions during cell movement.
In another report (26), protease inhibitors demonstrated no apparent effect on rat granule cell migration in cerebellar slices, suggesting that tPA catalytic activity may not be required. However, mice and rats use different promoters and show different patterns of tPA expression (27). Purkinje cells express the most tPA in rats, with little tPA in granule cells (9, 26), whereas in mice, Purkinje neurons rarely express tPA and most of the tPA is associated with granule neurons (3). These differences among species may explain why tPA activity is involved in mouse, but not rat, granule cell migration.
Granule cell migration involves numerous molecules playing specific roles in cell motility, cell–cell recognition, cell adhesion, and cell signaling, including L1 (28), astrotactin (29), ion channels (30), and neuregulin/erbB4 (31), etc. Current data suggest that tPA probably plays a more general role facilitating periodic cell detachment during migration by its activation of cell surface-associated plasminogen to plasmin. However, we must also note that tPA may play a signaling role in migration by acting on a cell-surface receptor (5, 32) independent of the catalytic ability of tPA.
Thus, these findings show that the absence of tPA leads to retarded cerebellar granule cell migration. Only future physiological and behavioral studies can tell whether this delay in granule neuron arrival in the mouse internal granule cell layer has an effect on the degree or functional specificity of synaptic connections made with mossy fiber inputs to the cerebellum.
Acknowledgments
We thank Dr. P. Carmeliet for his generous gift of C57BL/129 tPA−/− mice, Dr. W. Proctor for the cerebellar slice technology, and Ms. Jayne Ferguson for expert technical assistance. This research was supported by grants from the National Institute of Neurological Disorders and Stroke and the National Science Foundation.
Abbreviations
- tPA
tissue plasminogen activator
- uPA
urokinase-type plasminogen activator
- EGL
external granule cell layer
- Pn
postnatal day n
- DiI
1,1′-dioctadecyl-3,3,3′,3′-tetramethylindocarbocyanine perchlorate
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Patil N, Cox D, Bhat B, Faham M, Meyers R, Peterson A. Nat Genet. 1995;11:126–129. doi: 10.1038/ng1095-126. [DOI] [PubMed] [Google Scholar]
- 2.Mizuguchi M, Takashima S, Kakita A, Yamada M, Ikeda K. Am J Pathol. 1995;147:1142–1151. [PMC free article] [PubMed] [Google Scholar]
- 3.Friedman G C, Seeds N W. J Comp Neurol. 1995;360:658–670. doi: 10.1002/cne.903600410. [DOI] [PubMed] [Google Scholar]
- 4.Krystosek A, Seeds N W. Proc Natl Acad Sci USA. 1981;78:7810–7814. doi: 10.1073/pnas.78.12.7810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verrall S, Seeds N W. J Cell Biol. 1989;109:265–271. doi: 10.1083/jcb.109.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moonen G, Graw-Wagemans M P, Selak I. Nature (London) 1982;298:753–755. doi: 10.1038/298753a0. [DOI] [PubMed] [Google Scholar]
- 7.Seeds N W, Haffke S, Christensen K, Schoonmaker J. Adv Exp Med Biol. 1990;265:169–178. doi: 10.1007/978-1-4757-5876-4_16. [DOI] [PubMed] [Google Scholar]
- 8.Qian Z, Gilbert M, Colicos M, Kandel E, Kuhl D. Nature (London) 1993;361:453–456. doi: 10.1038/361453a0. [DOI] [PubMed] [Google Scholar]
- 9.Seeds N W, Williams B L, Bickford P C. Science. 1995;270:1992–1994. doi: 10.1126/science.270.5244.1992. [DOI] [PubMed] [Google Scholar]
- 10.Tsirka S E, Gualandris A, Amari D, Strickland S. Nature (London) 1995;377:340–344. doi: 10.1038/377340a0. [DOI] [PubMed] [Google Scholar]
- 11.Seeds N W, Verral S, Friedman G, Hayden S, Gadotti D, Haffke S, Christensen K, Gardner B, McGuire P, Krystosek A. Ann N Y Acad Sci. 1992;667:32–40. doi: 10.1111/j.1749-6632.1992.tb51592.x. [DOI] [PubMed] [Google Scholar]
- 12.Carmeliet P, Schoonjans L, Kieckens L, Ream B, Degen J, Bronson R, DeVos R, van den Oord J, Collen D, Mulligan R. Nature (London) 1994;368:419–424. doi: 10.1038/368419a0. [DOI] [PubMed] [Google Scholar]
- 13.Sidman R L, Mottla T, Feder N. Stain Technol. 1961;36:279–284. doi: 10.3109/10520296109113291. [DOI] [PubMed] [Google Scholar]
- 14.Miale I L, Sidman R L. Exp Neurol. 1961;4:277–296. doi: 10.1016/0014-4886(61)90055-3. [DOI] [PubMed] [Google Scholar]
- 15.Komuro H, Rakic P. J Neurosci. 1995;15:1110–1120. doi: 10.1523/JNEUROSCI.15-02-01110.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raidoo D M, Ramsaroop R, Naidoo S, Bhoola K. Immunopharmacology. 1996;32:39–47. doi: 10.1016/0162-3109(96)00007-0. [DOI] [PubMed] [Google Scholar]
- 17.Fujita S. J Cell Biol. 1967;32:277–287. doi: 10.1083/jcb.32.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hines M D, Allen-Hoffman B. Promega Notes. 1996;59:30–36. [Google Scholar]
- 19.Seeds N W, Friedman G, Hayden S, Thewke D, Haffke S, McGuire P, Krystosek A. Semin Neurosci. 1996;8:405–412. [Google Scholar]
- 20.Mars W M, Zarnegar R, Michalopoulos G K. Am J Pathol. 1993;143:949–958. [PMC free article] [PubMed] [Google Scholar]
- 21.Thewke D P, Seeds N W. J Neurosci. 1996;16:6933–6944. doi: 10.1523/JNEUROSCI.16-21-06933.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sappino A-P, Mandani R, Huarte J, Belin D, Kiss J, Wohlwend A, Vassalli J. J Clin Invest. 1993;92:679–685. doi: 10.1172/JCI116637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsirka S E, Rogove A, Bugge T, Degen J, Strickland S. J Neurosci. 1997;17:543–552. doi: 10.1523/JNEUROSCI.17-02-00543.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O’Shea K S, Rheinheimer J, Dixit V M. J Cell Biol. 1990;110:1275–1283. doi: 10.1083/jcb.110.4.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sotelo C, Alvarado-Mallart R-M, Frain M, Vernet M. J Neurosci. 1994;14:124–133. doi: 10.1523/JNEUROSCI.14-01-00124.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ware J H, DiBenedetto A, Pittman R. J Neurobiol. 1995;28:9–22. doi: 10.1002/neu.480280103. [DOI] [PubMed] [Google Scholar]
- 27.Holmberg M, Leonardsson G, Ny T. Eur J Biochem. 1995;231:466–474. doi: 10.1111/j.1432-1033.1995.tb20720.x. [DOI] [PubMed] [Google Scholar]
- 28.Lindner J, Zinsser G, Werz W, Goridis C, Bizzini B, Schachner M. Brain Res. 1986;377:298–304. doi: 10.1016/0006-8993(86)90872-3. [DOI] [PubMed] [Google Scholar]
- 29.Zheng T, Heintz N, Hatten M. Science. 1996;272:417–419. doi: 10.1126/science.272.5260.417. [DOI] [PubMed] [Google Scholar]
- 30.Kumoro H, Rakic P. Science. 1993;260:95–97. doi: 10.1126/science.8096653. [DOI] [PubMed] [Google Scholar]
- 31.Rio G, Reiff H, Qi P, Corfas G. Neuron. 1997;19:39–50. doi: 10.1016/s0896-6273(00)80346-3. [DOI] [PubMed] [Google Scholar]
- 32.Bu G, Maksymovitch E, Nerbonne J, Schwartz A. J Biol Chem. 1994;269:18521–18528. [PubMed] [Google Scholar]