Abstract
In transgenic and nontransgenic plants, viruses are both initiators and targets of a defense mechanism that is similar to posttranscriptional gene silencing (PTGS). Recently, it was found that potyviruses and cucumoviruses encode pathogenicity determinants that suppress this defense mechanism. Here, we test diverse virus types for the ability to suppress PTGS. Nicotiana benthamiana exhibiting PTGS of a green fluorescent protein transgene were infected with a range of unrelated viruses and various potato virus X vectors producing viral pathogenicity factors. Upon infection, suppression of PTGS was assessed in planta through reactivation of green fluorescence and confirmed by molecular analysis. These experiments led to the identification of three suppressors of PTGS and showed that suppression of PTGS is widely used as a counter-defense strategy by DNA and RNA viruses. However, the spatial pattern and degree of suppression varied extensively between viruses. At one extreme, there are viruses that suppress in all tissues of all infected leaves, whereas others are able to suppress only in the veins of new emerging leaves. This variation existed even between closely related members of the potexvirus group. Collectively, these results suggest that virus-encoded suppressors of gene silencing have distinct modes of action, are targeted against distinct components of the host gene-silencing machinery, and that there is dynamic evolution of the host and viral components associated with the gene-silencing mechanism.
In plants, posttranscriptional gene silencing (PTGS) is manifested as the reduction in steady-state levels of specific RNAs after introduction of homologous sequences in the plant genome. This reduction is caused by an increased turnover of target RNA species, with the transcription level of the corresponding genes remaining unaffected (reviewed in ref. 1). Recently, it was shown that PTGS involves systemic spread of a silencing signal directing sequence-specific RNA degradation (2, 3). Although the exact mechanism by which PTGS operates has yet to be elucidated, various findings that viruses can both initiate and be the targets of PTGS (4) led to the suggestion that PTGS is a natural mechanism by which plants recognize and combat foreign nucleic acids (5).
In support of the proposed relationship between PTGS and virus resistance, it was shown that some viruses induce an RNA-mediated defense (RMD) in nontransgenic plants. This induced defense is similar to PTGS in that it is characterized by nucleotide sequence-specific resistance against virus infection (6). In some but not all instances, the upper leaves of plants exhibiting this RMD were said to have recovered because they contained only low levels of viral RNA and were symptom-free (7, 8).
However, the ability of viruses to infect plants indicates that they have evolved to avoid or suppress the RMD. This idea was first prompted by analysis of potyviral synergistic interactions with other viruses (9). It was shown that this synergism was the result of suppression of a host defense mechanism by the Hc-protease (HcPro) encoded in the potyviral genome (10). Subsequent studies further established that HcPro was a suppressor of PTGS and provided a link between PTGS and antiviral defense (11–13). Presumably, the suppression acts against the RMD evoked above. A second protein, the 2b protein of cucumber mosaic virus (CMV), was also identified as a suppressor of PTGS in Nicotiana benthamiana (12). Interestingly, HcPro and the 2b proteins did not target the silencing mechanism in the same way; HcPro suppressed silencing in tissues where it was already established, whereas the 2b protein only affected silencing initiation (12, 14).
Although 2b and HcPro are dissimilar at the protein sequence level, they are both pathogenicity determinants of their respective viruses (15, 16). By extrapolation, we predicted that many viral pathogenicity determinants would be identified as suppressors of gene silencing and that, more generally, many viruses would have the ability to suppress PTGS. Here, we test these ideas by infecting N. benthamiana plants exhibiting PTGS of a green fluorescent protein (GFP) transgene with a range of viruses. Plants were also infected with potato virus X (PVX) vectors expressing previously identified viral pathogenicity determinants. If these wild-type and recombinant viruses produced suppressors of a PTGS-like resistance mechanism, we predicted that they would interfere with PTGS of GFP. The outcome of these experiments was consistent with our prediction and revealed that suppression of gene silencing is a widespread strategy among plant viruses. Our study led to the identification of three viral suppressors of PTGS and revealed an intriguing phenotype of silencing suppression that operates in the vicinity of the veins.
Materials and Methods
Plant Material.
Transgenic N. benthamiana plants carrying the GFP ORF were described previously (12).
PTGS Suppression Assay.
Leaves of seedlings of line 16c were infiltrated with a strain of Agrobacterium tumefaciens carrying a binary Ti plasmid vector, into which a functional 35S-GFP cassette had been inserted, as reported (17). After 15–20 days, when PTGS of GFP was achieved in the whole plant, a systemic leaf was inoculated with a wild-type or recombinant virus. This leaf is referred to as “inoculated leaf.” The challenged virus was then allowed to spread in the silenced plant, and two types of leaves were collected at 14 or 20 days postinoculation (DPI). “Old leaves” were infected leaves that had emerged before the virus had spread systemically, whereas “new leaves” were those emerging after the virus had moved systemically.
Wild-Type Viruses.
Isolates of alfalfa mosaic virus (AMV), foxtail mosaic virus (FoMV), narcissus mosaic virus (NMV), nandina virus X (NVX), viola mosaic virus (VMV), and tomato bushy stunt virus (TBSV) were obtained from Roger Hull from the John Innes Centre (JIC) collection (Norwich, U.K.). Cowpea mosaic virus (CPMV) was obtained from George Lomonosoff at JIC. African cassava mosaic virus (ACMV) was obtained from John Stanley at JIC. Tobacco rattle virus strain PPK20 was obtained from John Bol (Leiden University, Leiden, The Netherlands). Other viruses were obtained from a lab collection. These are tobacco mosaic virus (TMV)-U1 (18), PVX-UK3 (19), potato virus Y (PVY)N, CMV (12), tobacco black ring virus strain W22 (8), and rice yellow mottle virus (RYMV)-N (20).
Recombinant Viruses.
The P1 protein sequence of an RYMV isolate from Nigeria (20) was amplified by using the following 5′ phosphorylated primers: ATG ACT CGG TTG GAA GTT C-3′ for the intact protein (P1) and ATC ACA CGG TTG TAA GGT TC-3′ for an untranslatable protein (mP1). The phosphorylated downstream primer used for amplification was CAT CCC GTG TCA GTC TG. The two PCR fragments were cloned into the EcoRV site of the PVX vector (p2C2S) (19). The orientation of RYMV PCR fragments was confirmed by colony-PCR using antisense primer in the vector sequence at the 3′ end of the p2C2S multiple cloning site (GTA GTT GAG GTA GTT GAC CC) and the two sense RYMV 5′ primers described above. PVX-AC2 and PVX-mAC2 (21) were provided by John Stanley. PVX-HS142 and PVX-HS160 (22) referred to as PVX-19k and PVX-m19k, respectively, were provided by Andrew Jackson, University of California, Berkeley.
In Vitro Transcription and Northern Blot Analysis.
In vitro transcription reactions to produce infectious recombinant PVX RNAs and inoculation were as described (19). Northern blot analysis was described previously (12).
GFP Imaging.
Visual detection of GFP was as described (12). Close-up images were obtained by using a Leica MZ FLIII dissecting stereomicroscope coupled to a fluorescence module. The filter set used for GFP imaging was the GFP-plus fluorescence set from Leica (excitation 480 nm, dichromatic beam splitters, 505 nm, Barrier filter 510 nm. Photographs were produced by using a Leica MPS60 device coupled to the stereomicroscope.
Results
Suppression of Gene Silencing by Diverse Plant Viruses.
A test for silencing suppression was based on a previously described experimental system (12). This system involves transgenic N. benthamiana plants carrying a highly expressed GFP transgene that makes them fluoresce bright green under UV illumination. Systemic silencing in these plants was induced by infiltration of lower leaves of transgenic seedlings with a strain of A. tumefaciens, as described (17). By 20 days postinfiltration, silencing of the GFP was extensive in all vegetative tissues of the plants, and, consequently, they appeared uniformly red under UV illumination. At this stage, there was no PTGS in the growing points of the plant, and silencing was maintained by being constantly initiated in nonsilenced cells located near or in the meristems (17). These silenced plants were then infected with a range of plant viruses, and, when systemic symptoms were observed, the extent of green fluorescence was assessed under UV illumination. In addition, Northern blot analysis was performed to assess the level of GFP mRNAs in infected tissues.
Our findings were that many but not all of the viruses tested suppressed gene silencing in N. benthamiana (Table 1). With several viruses, suppression occurred in old leaves (OL) that had emerged before the virus had spread, as well as in new emerging leaves (NL). This was reminiscent of the pattern of silencing suppression previously described for PVY (12). In contrast, TBSV suppressed gene silencing only in new emerging tissues, as was previously reported for CMV (12, 14). FoMV, alfalfa mosaic virus, or tobacco black ring virus were like PVX in that they were fully infectious but did not have any effect on GFP silencing. From the diversity of viruses tested in this analysis, we conclude that PTGS suppression is a property of many plant viruses. However, because the spatial pattern and degree of suppression varied extensively between viruses, it was likely that different mechanisms would be involved.
Table 1.
Suppression of PTGS of GFP mRNA caused by various plant viruses
| Virus group | Virus | Suppression of PTGS | Old leaves/New leaves | Whole leaf/Vein centric | Protein* | Other known functions† |
|---|---|---|---|---|---|---|
| Alfamovirus | ALMV | 0/9 | — | — | — | — |
| Comovirus | CpMV | 5/6 | OL and NL | Vein centric | ? | — |
| Cucumovirus | CMV | 20/20 | NL only | Whole Leaf | 2b | Host-specific long distance movement |
| Geminivirus | ACMV | 6/6 | OL and NL | Whole leaf | AC2 | Virion sense gene expression transactivator |
| Nepovirus | TBRV | 0/6 | — | — | — | — |
| Potexvirus | PVX | 0/9 | — | — | — | — |
| FoMV | 0/9 | — | — | — | — | |
| NMV | 8/9 | OL and NL | Whole leaf | ? | — | |
| NVX | 7/9 | OL and NL | Whole leaf | ? | — | |
| VMV | 7/9 | OL and NL | Whole leaf | ? | — | |
| Potyvirus | PVY/TEV | 10/10 | OL and NL | Whole leaf | HcPro | Genome amplification |
| TEV | Viral synergism | |||||
| Long distance movement | ||||||
| Polyprotein processing | ||||||
| Aphid transmission | ||||||
| Sobemovirus | RYMV | —‡ | —‡ | —‡ | P1 | Virus accumulation |
| Long distance movement | ||||||
| Tobamovirus | TMV | 4/6 | OL and NL | Vein centric | ? | — |
| Tobravirus | TRV | 7/9 | OL and NL | Whole leaf | ? | — |
| Tombusvirus | TBSV | 7/9 | NL only | Vein centric | 19K | Host-specific spread and symptom determinant |
PTGS of the GFP mRNA was induced in transgenic N. benthamiana by Agrobacterium infiltration, as described (17). After systemic infection, suppression of gene silencing was assessed under UV illumination over time and confirmed by RNA gel blot analysis. RNA samples were taken from either old leaves that had emerged before the virus had spread systemically (OL) or new leaves emerging after virus infection (NL). The total number of plants tested is indicated as well as the phenotype of suppression in leaves (affecting whole tissues or vein centric). Viruses were tested in duplicate independent experiments during the summer and the winter.
The identification of the 2b protein and HcPro as PTGS suppressors is described in refs. 11–13. The identification of AC2, P1, and the 19K protein as PTGS suppressors is described in this study.
†Appropriate references can be found in refs. 16 (2b), 21 (AC2), 13 and 15 (HcPro), 24 (P1), and 22 [19K (19-kDa)].
‡RYMV is not infectious in N. benthamiana. The P1 protein has been identified as a PTGS suppressor by expression from the PVX vector.
The Geminivirus-Encoded AC2 Protein Is a Suppressor of Gene Silencing.
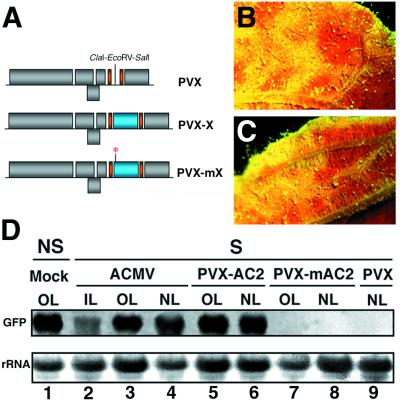
As shown in Table 1, infection of ACMV led to suppression of GFP silencing at about 3 wk postinoculation in both fully expanded and new emerging infected tissues (Fig. 1B). Correspondingly, Northern blot analysis revealed that GFP mRNA levels were high in both types of tissues and that suppression also occurred in inoculated leaves, although to a lower extent (Fig. 1D, lanes 1–4). Therefore, these results were consistent with a suppressor of PTGS encoded in the ACMV genome.
Figure 1.
Suppression of PTGS by ACMV and PVX-AC2. (A) Schematic representation of the PVX vector used to express various pathogenicity determinants tested in this study (referred to as “X”). Individual sequences were inserted into the P2C2S PVX vector using the ClaI–EcoRV–SalI multiple cloning site (19), leading to “PVX-X”. Expression of the inserts (X, depicted as a blue box) and the PVX coat protein is controlled by duplicated coat protein promoters (indicated by a solid orange bar). Mutant versions of all pathogenicity determinants, referred to as “mX,” were also used in this study (mutation indicated by a red asterisk). (B) Close-up image of an ACMV-infected leaf from a GFP-silenced N. benthamiana. (C) Close-up image of a PVX-AC2-infected leaf from a GFP-silenced N. benthamiana. Photos from B and C were taken under UV illumination from a dissecting microscope at 15 DPI. The red tissue corresponds to chlorophyll fluorescence under UV and, thus, is indicative of gene silencing of GFP. The green fluorescent tissue that sometimes appears yellow is from expression of GFP and, thus, indicates suppression of gene silencing. (D) Northern blot analysis of RNA extracted at 20 DPI from either mock-infected, nonsilenced (NS), or silenced (S) N. benthamiana infected with ACMV, PVX-AC2, PVX-mAC2, or PVX. RNA samples were taken either from inoculated leaves (IL), old leaves that had emerged before the virus had spread systemically (OL), or from new leaves emerging after virus infection (NL). Equal amounts of each RNA sample (10 μg) were assayed by RNA gel blotting by using a 32P-labeled GFP cDNA as probe. Ethidium bromide staining of ribosomal RNA (rRNA) shows equal loading of the samples.
To identify this putative suppressor, we exploited previous findings that a PVX vector expressing the AC2 protein (PVX-AC2) produced necrotic symptoms that were much more severe than those of wild-type PVX, suggesting that AC2 suppressed a host defense mechanism (21). From the above results, it was likely that AC2 was a suppressor of RMD.
The test of this hypothesis was to infect GFP-silenced plants with PVX-AC2 (Fig. 1A). As a control, plants were also inoculated with PVX-mAC2 (Fig. 1A) in which a single point mutation introduces a premature stop codon in the AC2 ORF (21). At about 2 wk postinoculation, PVX-AC2-infected plants exhibited severe symptoms, as reported (21). Under UV illumination, most of the infected tissues, including leaves that had emerged prior to virus inoculation, were green fluorescent (Fig. 1C), and GFP mRNA levels were similar to those in nonsilenced GFP plants (Fig. 1D, lanes 5 and 6). In contrast, PVX-mAC2 did not produce severe symptoms and did not suppress GFP silencing (Fig. 1D, lanes 7–8). From these results, we conclude that the AC2 protein encoded in the ACMV genome is a suppressor of maintenance of PTGS in N. benthamiana.
Vein-Specific Suppression of Silencing by the 19-kDa (“19K”) Protein of TBSV.
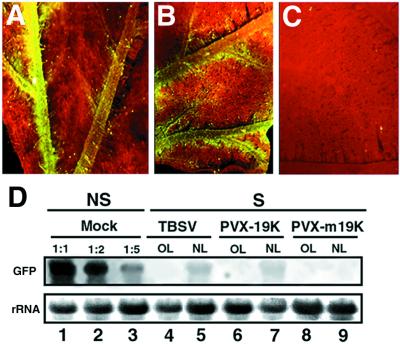
N. benthamiana infected with TBSV showed reversion of PTGS at about 3 wk postinoculation, when symptoms were fully systemic (Table 1). As in CMV-infected plants, the restoration of green fluorescence occurred only in new emerging infected leaves. However, this suppression of silencing was weaker than with CMV, so that the green fluorescence was barely detectable under UV illumination from a hand-held lamp. Also unlike CMV, TBSV suppressed PTGS only in and around the veins (Fig. 2A). Vein-specific reversion of GFP was more evident when detached, new emerging leaves were observed under a dissecting microscope (Fig. 2A). Northern blot analysis showed that GFP RNAs were more abundant in the new leaves of the infected plants than in old leaves or in mock-inoculated, nonsilenced plants. However, the GFP RNA in the new leaves was <20% of the level in mock-inoculated plants (Fig. 3D, lanes 4 and 5).
Figure 2.
Vein-specific suppression of PTGS caused by TBSV and PVX-19K. (A) Close-up image of a TBSV-infected leaf from a GFP-silenced N. benthamiana. (B) Close-up image of a PVX-19K-infected leaf from a GFP-silenced N. benthamiana. (C) Close-up image of a PVX-m19K-infected leaf from a GFP-silenced N. benthamiana. Photographs A, B, and C were taken under UV illumination from a dissecting microscope at 20 DPI. (D) Northern blot analysis of RNA extracted at 20 DPI from silenced (S) N. benthamiana infected with PVX-19K or PVX-m19K. RNA samples were taken either from old leaves (OL) or from new emerging leaves (NL). Equal amounts of each RNA sample (15 μg) were assayed by RNA gel blotting by using a 32P-labeled GFP cDNA as probe. Lanes 1–3 show a dilution series of GFP RNAs from a nonsilenced (NS) plant into total RNA from a nontransformed (NT) plant. GFP RNA was diluted to a one-half (1:2) or to one-fifth (1:5) of the reference sample (1:1). Ethidium bromide staining of ribosomal RNA (rRNA) shows equal loading of the samples.
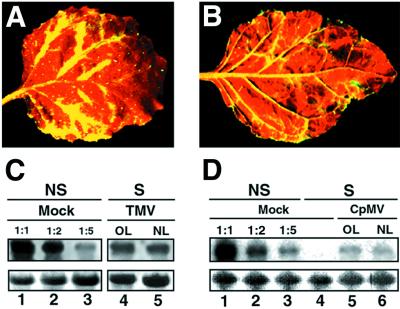
Figure 3.
Suppression of PTGS caused by TMV and CPMV occurs preferentially in the vicinity of the veins. (A) Close-up image of a TMV-infected leaf from a GFP-silenced N. benthamiana. (B) Close-up image of a CPMV-infected leaf from a GFP-silenced N. benthamiana. Photographs A and B were taken under UV illumination from a hand-held lamp at 20 DPI. (C) Northern blot analysis of RNA extracted at 20 DPI from silenced (S) N. benthamiana infected with TMV. RNA samples were taken either from old leaves (OL) or from new emerging leaves (NL). Equal amounts of each RNA sample (15 μg) were assayed by RNA gel blotting by using a 32P-labeled GFP cDNA as probe. Samples were separated on the same agarose gel and blotted on the same filter that was used in Fig. 2, thus allowing the use of the same GFP RNA dilution series as a reference. (D) Northern blot analysis of RNA extracted at 20 DPI from silenced (S) N. benthamiana infected with CPMV. Equal amounts of each RNA sample (15 μg) were assayed by RNA gel blotting by using a 32P-labeled GFP cDNA as probe. Mock control lanes 1–3 were prepared as in Fig. 2. Ethidium bromide staining of ribosomal RNA at the bottom shows equal loading of the samples.
It has been reported that the 19K protein of TBSV is a pathogenicity determinant. For example, a PVX vector expressing the 19K protein (pHS142), referred to here as PVX-19K (Fig. 1A), induced severe symptoms on N. benthamiana (22). In addition, inactivation of the 19K protein in TBSV had an attenuating effect on the lethal apical necrotic symptom phenotype that is usually elicited in plants by TBSV (22). Collectively, these data indicate that the TBSV 19K protein possesses attributes of a suppressor of gene silencing. To test this hypothesis, silenced GFP plants were inoculated with PVX-19K (Fig. 1A). As a control, plants were also inoculated with pHS160 (referred to here as PVX-m19K) carrying a nontranslatable form of the 19K protein (Fig. 1A, and ref. 22). By 2 wk postinoculation, plants infected with PVX-19K exhibited very severe symptoms, whereas PVX-m19K-infected plants had mild mosaic symptoms, as reported (22). Suppression of silencing occurred in PVX-19K-infected plants, but was manifested only in new emerging tissues and was most pronounced in the veins (Fig. 2B). However, symptoms of PVX-19K were visible on all areas of the leaves (data not shown). Similar tissues infected with PVX-m19K remained uniformly red fluorescent (Fig. 2C). Northern blot analysis of RNA extracted from new emerging, infected leaves showed that only low levels of GFP RNAs could be detected in PVX-19K-infected tissues (Fig. 2D, lanes 6 and 7) and that GFP RNAs were below the level of detection in PVX-m19K-infected tissues (Fig. 2D, lanes 8 and 9). Taken together, these results suggest that the 19K protein of TBSV is a weak suppressor of PTGS in N. benthamiana that operates in the vicinity of the vein tissues of new emerging leaves.
Other Examples in Which Suppression of PTGS Occurs Preferentially in or Near the Veins.
As part of our survey, we investigated the effect of TMV and CPMV, type members of the tobamovirus and comovirus groups, respectively. Inoculation of the corresponding viruses onto GFP-silenced plants led to suppression of gene silencing that affected both new emerging and already expanded silenced tissues, thus indicating that maintenance of PTGS was alleviated (Table 1, and Fig. 3 A and B). However, as shown previously for TBSV and PVX-19K, suppression was mostly manifested near or in the veins with most tissues of the lamina remaining silenced (i.e., red fluorescent), although symptoms of the respective viruses were observed on the whole leaf lamina (data not shown). This phenotype did not change over time, even when infected leaves were fully expanded and completely infected. With both viruses, green fluorescence in the vicinity of the veins was very strong, and this effect was clearly apparent under UV illumination from a hand-held lamp (Fig. 3 A and B). Northern blot analysis of RNAs extracted from infected leaves showed that GFP RNA accumulation was restored in those tissues but at a low level when compared with the abundance of GFP RNA extracted from similar tissues of nonsilenced, noninfected plants (Fig. 3 C and D). This was probably because of dilution of the vein tissue into the most abundant silenced tissues of the lamina. Therefore, this molecular analysis was consistent with the particular phenotype of silencing suppression observed under UV illumination.
A Pathogenicity Determinant from RYMV Suppresses PTGS in the Nonhost N. benthamiana Species.
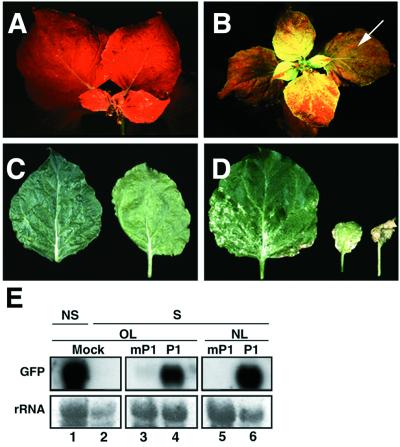
RYMV is a sobemovirus exhibiting a very narrow host range. It only systemically infects monocotyledonous species belonging to the Oryzae, Phalaridae, and Eragrostidae tribes (23). Recent studies have characterized the P1 protein of RYMV as an important pathogenicity determinant in rice (24). To test whether it would suppress gene silencing in a RYMV nonhost species, the P1 ORF was introduced into the PVX vector and GFP-silenced N. benthamiana were infected with the resulting recombinant virus (PVX-P1; Fig. 1A). As a control, a PVX vector carrying a nontranslatable form of P1 (PVX-mP1; Fig. 1A) was also inoculated. At about 2 wk postinoculation, tissues infected with PVX-P1 exhibited severe chlorosis and white necrosis (Fig. 4D). Under UV illumination, these tissues, including leaves that had emerged prior to virus inoculation, appeared green fluorescent (Fig. 4B). Accordingly, in young infected tissues, GFP mRNA levels were similar to those in nonsilenced GFP plants (Fig. 4E, lane 6). GFP mRNAs could also be detected in infected leaves that had emerged prior to virus inoculation, although to a lower extent (Fig. 4E, lane 4). In contrast, neither severe symptoms nor reversion of GFP silencing was caused by PVX-mP1 infection (Fig. 4 A, C, and E, lanes 3 and 5). From these data, we conclude that the P1 protein of RYMV is a suppressor of maintenance of PTGS in N. benthamiana, although it is encoded in the genome of a virus that is not infectious on Nicotiana species.
Figure 4.
Severe symptoms and suppression of PTGS caused by PVX expressing the RYMV P1 protein. (A) UV illumination of a GFP-silenced N. benthamiana infected with PVX-mP1 at 14 DPI. (B) UV illumination of a GFP-silenced N. benthamiana infected with PVX-P1 at 14 DPI. Reversion of silencing occurs in both new emerging tissues and old leaves (indicated by an arrow). (C) Mild mosaic symptoms caused by PVX-mP1 at 14 DPI. (D) Severe necrotic symptoms caused by PVX-P1 at 14 DPI. (E) Northern blot analysis of RNA extracted at 14 DPI from either mock-infected, nonsilenced (NS), or silenced (S) N. benthamiana infected with PVX-P1 or PVX-mP1. RNA samples were taken from either old leaves (OL) or new emerging leaves (NL). Equal amounts of each RNA sample (10 μg) were assayed by RNA gel blotting by using a 32P-labeled GFP cDNA as probe. Ethidium bromide staining of ribosomal RNA (rRNA) shows equal loading of the samples.
Strong Variations in the Ability to Suppress PTGS in N. benthamiana Are Observed Between Highly Related Members of the Potexvirus Group.
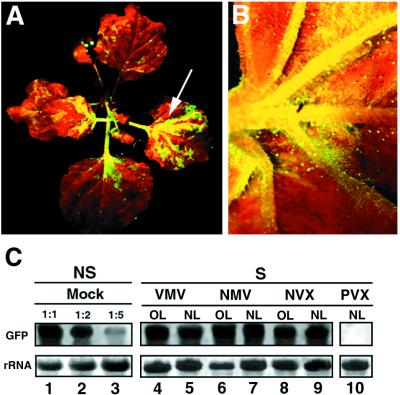
PVX and FoMV, both members of the potexvirus group, had no effect on PTGS of GFP in N. benthamiana (Table 1; and Fig. 5C, lane 10). In contrast, infection with other potexviruses, NMV, NVX, and VMV, led to suppression of gene silencing in N. benthamiana. This suppression was manifested in leaves that were expanded prior to inoculation as well as in young developing tissues (Fig. 5 A–C). The suppression was as strong as with HcPro, 2b, and AC2, and the levels of GFP mRNA in infected tissues were similar to those in mock-inoculated, nonsilenced plants (Fig. 5C).
Figure 5.
Effect of various potexviruses on PTGS of GFP. (A) UV illumination of a GFP-silenced N. benthamiana infected with NMV at 20 DPI. Reversion of silencing occurs in new emerging tissues as well as in old leaves (indicated by an arrow). (B) Close-up image of a NVX-infected leaf from a GFP-silenced N. benthamiana. This photograph was taken under UV illumination from a dissecting microscope at 20 DPI. (C) Northern blot analysis of RNA extracted at 20 DPI from silenced (S) N. benthamiana infected with VMV, NMV, NVX, or PVX. RNA samples were taken either from old leaves (OL) or from new emerging leaves (NL). Equal amounts of each RNA sample (15 μg) were assayed by RNA gel blotting by using a 32P-labeled GFP cDNA as probe. Samples were separated on the same agarose gel and blotted on the same filter that was used in Fig. 2, thus allowing the use of the same GFP RNA dilution series as a reference. Ethidium bromide staining of ribosomal RNA (rRNA) shows equal loading of the samples.
The inocula of these related viruses had been quantified using the local lesion host Chenopodium amaranticolor (25) and diluted, so that they would be comparable to a PVX inoculum used as an internal reference (40 lesions per leaf). Following infection, we confirmed that these viruses gave similar types of symptoms. Thus, the variation in the suppressor of silencing activity reflected intrinsic properties of the viruses rather than the degree of infection. Surprisingly, the variable suppressor activity did not correlate with the nucleotide sequence similarity of these viruses. PVX and FoMV, which did not suppress silencing, are only distant relatives. In contrast, NVX and VMV, which produced strong suppressors, are respectively 93% and 97% identical to PVX at the nucleotide level, based on sequence analysis of a region spanning the coat protein and the three movement proteins (A. Bendhamane and D.C.B., unpublished data). NMV, which also produced a suppressor, is only a distant relative of PVX. Therefore, there is extreme variation in the ability to suppress PTGS in closely related members of a single virus group.
Discussion
Suppression of PTGS as a General Strategy.
We predicted that many viruses would encode proteins that are suppressors of an RMD mechanism and that these proteins would also suppress PTGS (12). The likely candidate suppressors were viral proteins that, like the 2b protein or HcPro, were originally characterized as pathogenicity determinants. Consistent with this hypothesis, the ACMV AC2, the RYMV P1, and the TBSV 19K pathogenicity factors all suppress PTGS of a GFP transgene. It is therefore likely that the activity of these proteins in pathogenicity of the encoding virus is associated with suppression of RMD. The ability of these proteins to enhance symptoms of PVX vectors is most likely explained in the same way. The finding that a DNA geminivirus, ACMV, encodes a suppressor was not surprising because other geminiviruses are known to induce PTGS, and presumably RMD, in transgenic and nontransgenic plants (26, 27).
Each virus produced a characteristic pattern of silencing suppression. Some, like potyviruses, suppressed in young and old leaves. Others were like CMV and affected only young leaves. There was also variation in the tissue specificity with ACMV, VMV, NMV, NVX, and PVX-P1 affecting all tissues, whereas TBSV, TMV, and CPMV specifically suppressed silencing in tissues that were in or close to the veins. We do not think that these differences reflect the tissue tropism of these viruses. Similar patterns were reproduced when various suppressors were expressed from a PVX vector that has been shown to express foreign proteins uniformly throughout infected leaves (19). A more likely explanation depends jointly on the mode of action of the suppressor and the component of the gene-silencing mechanism that is targeted. For example, if a suppressor can degrade a component required for maintenance of gene silencing, it will have an effect in both new and old leaves. However, if the suppressor blocks synthesis or activation of a component required for silencing, the suppression would be restricted to new emerging leaves, where silencing would be established in the presence of the viral suppressor. In old leaves, the component would have been formed in the absence of the suppressor and, consequently, would be unaffected when the virus would infect the plant.
The suppression of silencing in veins, for example with the 19K protein of TBSV, could indicate that this protein is stable or expressed only in the veins or that it is targeted against a component of the PTGS mechanism that is qualitatively or quantitatively different between vascular and nonvascular tissue. Alternatively, the suppressor could be targeted against the systemic signal of PTGS. We have shown that this signal is phloem-transmitted and that, in recipient leaves, it is primarily located in and near the veins (17). Of these alternative explanations for suppression of silencing in veins, we consider that those involving vein-specific components or stability of the suppressors are unlikely because, in all cases, PTGS suppression extended into cells outside the vascular bundle and appeared to reflect movement of the signal rather than a precisely vein-specific silencing process. For this reason, we propose that the suppressors of TMV, CPMV, and TBSV are likely targeted against the systemic signal of silencing and may therefore represent a viral adaptation to systemic RMD.
Although TMV, TBSV, and CPMV are able to suppress PTGS only in or near the veins, they are nevertheless able to accumulate at a high level throughout the infected leaf. It is likely, therefore, that these viruses have secondary strategies for counteracting the effects of RMD. These strategies may involve evasion, so that the process is not activated, or escape from the antiviral mechanism. Luteoviruses, which are typically restricted to the phloem (25), may provide an interesting example of viruses that are unable to either suppress, evade, or escape from the effect of RMD outside the veins. Consistent with this idea, it has been reported that the level of potato leafroll luteovirus (PLRV) increased up to 12-fold in Nicotiana species that were coinfected with NMV, tobacco rattle virus (TRV), or PVY (28). It now seems likely that this increase was due, at least in part, to the ability of PLRV to spread beyond the veins as a result of suppression of RMD in the double-infected plants. Here, we show that NMV, TRV, and PVY are all able to suppress maintenance of PTGS in N. benthamiana (Table 1). In contrast, coinfection with alfalfa mosaic or tobacco black ring viruses that are unable to suppress PTGS (Table 1) did not alter PLRV concentration in leaves (28).
Gene Silencing Activation/Suppression as a Coevolutive Mechanism?
It is striking that the viral suppressors of silencing are so diverse. So far, we have been unable to identify any common structural features in these proteins, and we conclude that the suppressor function has evolved independently several times as a strategy to counteract the effects of RMD. In some instances, it is conceivable that some suppressors have converged toward the same function and, thus, are targeted against similar components of the silencing machinery. For example, the RYMV P1 protein shares striking functional similarities with the potyviral-encoded HcPro protein. First, when produced from the PVX vector, both proteins are suppressors of maintenance of PTGS in N. benthamiana. In addition, both proteins are required for efficient accumulation of viral RNAs in protoplasts and long distance movement in their respective host (15, 24).
Because RMD is likely to have a central role in plant–virus interactions, one can also anticipate that there will be a dynamic evolution of plant components required for the mechanism and, accordingly, of the virus-encoded components necessary to overcome it. The poty- and potexvirus groups probably represent different stages in this dynamic evolution. In the potyvirus group, the HcPro of tobacco etch virus (TEV) (11, 13), PVY (12), and pea seedborne mosaic virus (O.V., unpublished data) are suppressors of GFP silencing in Nicotiana species. In these viruses, the suppressor seems to be a conserved function, and its corresponding target is also likely to be conserved in different plants. Similarly, the target of the RYMV P1 protein may be conserved from rice to tobacco. In contrast, the potexvirus strategy for counteracting RMD and PTGS is apparently in a state of evolutionary flux. Presumably, PVX and FoMV, as opposed to VMV and NVX, do not have a functional suppressor of silencing in N. benthamiana and, on that host, must use alternative strategies to escape or evade the mechanism, as proposed above. However, it might be expected that on other hosts, PVX and FoMV would produce functional suppressors and, conversely, VMV and NVX would not. The test of this coevolution hypothesis would require a suitable set of host plant species exhibiting PTGS, rather than the single GFP N. benthamiana line used here.
In due course, it may transpire that the balance between RMD activation and suppression will strongly influence virus–host interactions (29). For example, if a virus cannot suppress, evade, or escape the effects of the mechanism, the inoculated plant will be considered as a nonhost because there will only be subliminal infection. Similarly, if the virus is able to suppress the mechanism but cannot block the signal of silencing, it is likely that local or systemic spread of the virus will be impaired. Probably the best prospect for understanding this proposed adaptative process involves characterization of mutants impaired in PTGS (30) and identification of host components interacting with viral suppressors. In addition, the increasing body of evidence that PTGS also operates in animals raises the fascinating possibility that silencing suppression has also been adopted by animal viruses (31).
Acknowledgments
We thank John Stanley, Andrew Jackson, and Herman Scholthof for providing PVX vectors; Andy Maule and our colleagues in the Sainsbury Laboratory for stimulating discussions; and A. Bendhamane for providing sequence data. We are grateful to the Gatsby Charitable Foundation for continuing support of the Sainsbury Laboratory. O.V. is supported by a European Union Training and Mobility of Researchers cat. 20 grant (ERB4001GT974039) from the European Commission.
Abbreviations
- GFP
green fluorescent protein
- PTGS
posttranscriptional gene silencing
- RMD
RNA-mediated defense
- HcPro
Hc-protease
- DPI
days postinoculation
- PVX
potato virus X
- CMV
cucumber mosaic virus
- PVY
potato virus Y
- ACMV
African cassava mosaic virus
- TBSV
tomato bushy stunt virus
- TMV
tobacco mosaic virus
- CPMV
cowpea mosaic virus
- RYMV
rice yellow mottle virus
- NMV
narcissus mosaic virus
- NVX
nandina virus X
- VMV
viola mosaic virus
- FoMV
foxtail mosaic virus
- OL
old leaves
- NL
new emerging leaves
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Depicker A, Van Montagu M. Curr Opin Cell Biol. 1997;9:372–382. doi: 10.1016/s0955-0674(97)80010-5. [DOI] [PubMed] [Google Scholar]
- 2.Palauqui J-C, Elmayan T, Pollien J-M, Vaucheret H. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voinnet O, Baulcombe D C. Nature (London) 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- 4.Baulcombe D C. Curr Opin Plant Biol. 1999;2:109–113. doi: 10.1016/S1369-5266(99)80022-3. [DOI] [PubMed] [Google Scholar]
- 5.Baulcombe D C. Plant Mol Biol. 1996;32:79–88. doi: 10.1007/BF00039378. [DOI] [PubMed] [Google Scholar]
- 6.Ratcliff F, MacFarlane S, Baulcombe D C. Plant Cell. 1999;11:1207–1216. doi: 10.1105/tpc.11.7.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Covey S N, Al-Kaff N S, Langara A, Turner D S. Nature (London) 1997;385:781–782. [Google Scholar]
- 8.Ratcliff F, Harrison B D, Baulcombe D C. Science. 1997;276:1558–1560. doi: 10.1126/science.276.5318.1558. [DOI] [PubMed] [Google Scholar]
- 9.Vance V. Virology. 1991;182:486–494. doi: 10.1016/0042-6822(91)90589-4. [DOI] [PubMed] [Google Scholar]
- 10.Pruss G, Ge X, Shi X M, Carrington J C, Vance V B. Plant Cell. 1997;9:859–868. doi: 10.1105/tpc.9.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anandalakshmi R, Pruss G J, Ge X, Marathe R, Smith T H, Vance V B. Proc Natl Acad Sci USA. 1998;95:13079–13084. doi: 10.1073/pnas.95.22.13079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brigneti G, Voinnet O, Li W X, Ji L H, Ding S W, Baulcombe D C. EMBO J. 1998;17:6739–6746. doi: 10.1093/emboj/17.22.6739. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Kasschau K D, Carrington J C. Cell. 1998;95:461–470. doi: 10.1016/s0092-8674(00)81614-1. [DOI] [PubMed] [Google Scholar]
- 14.Beclin C, Berthome R, Palauqui J-C, Tepfer M, Vaucheret H. Virology. 1998;252:313–317. doi: 10.1006/viro.1998.9457. [DOI] [PubMed] [Google Scholar]
- 15.Kasschau K D, Cronin S, Carrington J C. Virology. 1997;228:251–262. doi: 10.1006/viro.1996.8368. [DOI] [PubMed] [Google Scholar]
- 16.Ding S W, Li W-X, Symons R H. EMBO J. 1995;14:5762–5772. doi: 10.1002/j.1460-2075.1995.tb00265.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Voinnet O, Vain P, Angell S, Baulcombe D C. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- 18.Marano M R, Baulcombe D. Plant J. 1998;13:537–546. [Google Scholar]
- 19.Chapman S N, Kavanagh T A, Baulcombe D C. Plant J. 1992;2:549–557. doi: 10.1046/j.1365-313x.1992.t01-24-00999.x. [DOI] [PubMed] [Google Scholar]
- 20.Pinto Y M, Kok R A, Baulcombe D C. Nat Biotechnol. 1999;17:702–707. doi: 10.1038/10917. [DOI] [PubMed] [Google Scholar]
- 21.Hong Y, Saunders K, Stanley J. Virology. 1997;228:383–387. doi: 10.1006/viro.1996.8403. [DOI] [PubMed] [Google Scholar]
- 22.Scholthof H B, Scholthof K B G, Jackson A O. Plant Cell. 1995;7:1157–1172. doi: 10.1105/tpc.7.8.1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakker W. Agricultural Research Report No. 829. Wageningen, The Netherlands: Centre for Agricultural Publishing and Documentation; 1974. [Google Scholar]
- 24.Bonneau C, Brugidou C, Chen L, Beachy R N, Fauquet C. Virology. 1998;244:79–86. doi: 10.1006/viro.1998.9100. [DOI] [PubMed] [Google Scholar]
- 25.Matthews R E F. Plant Virology. San Diego: Academic; 1991. [Google Scholar]
- 26.Kjemtrup S, Sampson K S, Peele C G, Nguyen L V, Conkling M A, Thompson W F, Robertson D. Plant J. 1998;14:91–100. doi: 10.1046/j.1365-313X.1998.00101.x. [DOI] [PubMed] [Google Scholar]
- 27.Atkinson R G, Bieleski L R F, Gleave A P, Jannsen B J, Morris B A M. Plant J. 1998;15:593–604. doi: 10.1046/j.1365-313x.1998.00211.x. [DOI] [PubMed] [Google Scholar]
- 28.Barker H. Ann Appl Biol. 1989;115:71–78. [Google Scholar]
- 29.Carrington J C, Whitham S A. Curr Opin Plant Biol. 1998;1:336–341. doi: 10.1016/1369-5266(88)80056-6. [DOI] [PubMed] [Google Scholar]
- 30.Elmayan T, Balzergue S, Beon F, Bourdon V, Daubremet J, Guenet Y, Mourrain P, Palauqui J C, Vernhettes S, Vialle T, et al. Plant Cell. 1998;10:1747–1757. doi: 10.1105/tpc.10.10.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Birchler J A, Pal-Bhadra M, Bhadra U. Nat Genet. 1999;21:148–149. doi: 10.1038/5926. [DOI] [PubMed] [Google Scholar]