Abstract
The Bs2 resistance gene of pepper specifically recognizes and confers resistance to strains of Xanthomonas campestris pv. vesicatoria that contain the corresponding bacterial avirulence gene, avrBs2. The involvement of avrBs2 in pathogen fitness and its prevalence in many X. campestris pathovars suggests that the Bs2 gene may be durable in the field and provide resistance when introduced into other plant species. Employing a positional cloning strategy, the Bs2 locus was isolated and the gene was identified by coexpression with avrBs2 in an Agrobacterium-mediated transient assay. A single candidate gene, predicted to encode motifs characteristic of the nucleotide binding site–leucine-rich repeat class of resistance genes, was identified. This gene specifically controlled the hypersensitive response when transiently expressed in susceptible pepper and tomato lines and in a nonhost species, Nicotiana benthamiana, and was designated as Bs2. Functional expression of Bs2 in stable transgenic tomatoes supports its use as a source of resistance in other Solanaceous plant species.
Bacterial spot disease of tomato and pepper, caused by Xanthomonas campestris pv. vesicatoria (Xcv), can be devastating to commercial production of these crops in areas of the world with high humidity and heavy rainfall. Control of Xcv is based largely on the application of pesticide; however, genetic resistance has been described in pepper and tomato (1–4). In pepper, several single loci (Bs1, Bs2, and Bs3) that confer resistance in a “gene-for-gene” manner have been identified (5) and the corresponding avirulence genes (avrBs1, avrBs2, and avrBs3) have been cloned from Xcv and shown to be essential for controlling resistance (6, 7).
Of particular interest is the genetic interaction that is governed by the avirulence gene avrBs2 and the resistance (R) gene Bs2. The avrBs2 gene encodes a protein with homology to the Agrobacterium tumefaciens agrocinopine synthase and Escherichia coli UgpQ, suggesting a possible enzymatic function as a phosphodiesterase (8). Mutant Xcv strains in which the avrBs2 gene has been disrupted or replaced are less virulent on susceptible hosts, growing 10–100 times less than the wild-type strain (8, 9). A survey of various races of Xcv and other pathovars of X. campestris also has shown that avrBs2 is widespread (9). These studies suggest that avrBs2 plays a highly conserved role in the fitness of X. campestris, and the effectiveness of the Bs2 resistance gene in the field may be based on the fact that it controls both virulence and avirulence in Xcv (9).
To date, more than 15 R genes have been isolated (10). Although the genes that have been cloned are involved in specific interactions with a diverse array of pathogens, with the exception of the Pto kinase (11), they encode a number of common motifs at the protein level (12, 13). This is somewhat surprising given that the majority of the avirulence genes, which along with the R genes govern the specificity of these interactions, encode apparently unrelated proteins. The significance of the observed similarities among most R gene products and how these domains function to confer resistance is still unknown.
We report here the map-based cloning and characterization of the Bs2 gene of pepper. Confirmation of the isolation of the Bs2 gene was obtained by an Agrobacterium-mediated transient coexpression assay and stable transformation of tomato and Nicotiana benthamiana. Transgenic tomato plants expressing the pepper Bs2 gene suppress the growth of Xcv in an avrBs2-dependent manner, indicating that the gene is able to function in a heterologous system. The Bs2 gene is a member of the nucleotide binding site–leucine-rich repeat (NBS-LRR) class of R genes. Interestingly, the Bs2 gene exhibits restricted taxonomic functionality (RTF), conferring a resistance response only to related genera in the Solanaceae.
Materials and Methods
Plants, Bacterial Strains, and Disease Resistance Scoring.
Pepper cultivars (cv.) used were Early Calwonder (ECW; bs1/bs1, bs2/bs2, bs3/bs3) and the near-isogenic cv. ECW-20R (bs1/bs1, Bs2/Bs2, bs3/bs3) and ECW-123R (Bs1/Bs1, Bs2/Bs2, Bs3/Bs3). In addition, the susceptible tomato cv. VF36 and the nonhost N. benthamiana were used for functional assays. Disease resistance phenotypes were determined by hand infiltration of suspensions of Xcv (pepper race 4 strain P38 at 1 × 108 to 2 × 108) colony-forming units per ml in 10 mM MgCl2 as described (6). Resistance, indicated by a HR, was scored 24–48 hr postinfiltration.
Isolation of Genomic DNA and Recombinant DNA Methods.
Isolation of pepper genomic DNA was as described (14), except fresh tissue samples were used. Other molecular biological techniques were performed by using standard protocols (15, 16) unless otherwise noted.
Identification and Analysis of Yeast Artificial Chromosome (YAC) Clones.
After isolation of total genomic yeast DNA and plasmid rescue of the YAC ends, probes were isolated by digesting the rescued fragments with the cloning-site enzyme, EcoRI, and either SacI, BamHI, or SphI. Using previously identified recombinants (17), end probes were mapped and used to orient the YAC clones. One end probe, 22D8Sac7, was used to screen 47 EcoRI-digested YAC DNA pools (384 clones per pool) by DNA gel blot analysis (18), and the clone YCA92F5 was identified by colony hybridization.
cDNA Library Screening.
A pepper leaf Lambda ZAP II cDNA library was constructed from ECW-20R infected with Xcv (avrBs2) following manufacturer’s instructions (Stratagene). About 1 × 106 plaques were screened with radiolabeled YCA22D8. Lifts were prehybridized with sheared total pepper genomic DNA, YAC vector DNA, and DNA from a YAC not at the Bs2 locus.
Cosmid Contig.
Sau3AI and TaqI partially digested YCA22D8 and YCA80H11 DNA was ligated to the vector pCLD04541 (19, 20). Transduced E. coli DH5α were plated on medium with tetracycline (10 μg/ml) and cycloheximide (50 μg/ml). Initially, colony lifts were probed with the A2 and F1 markers. Cosmid clones were analyzed by restriction digestion with EcoRI and BamHI. Single- and low-copy-number fragments were identified by probing cosmid blots with total pepper genomic DNA. EcoRI fragments not hybridizing to the probe were used to identify additional cosmids and for mapping. To confirm clone fidelity, fragments from each cosmid were used to probe DNA gel blots of the other YACs and pepper genomic DNA.
Physical Delimitation of the Locus.
Sequence information from regions internal to previously identified recombinants (17) was used to design primers for the L1 marker (5′-AAGGGCCTACATTGGTTACC-3′ and 5′-AGCCAAAGACCGGAATGACG-3′) and the R1 marker (5′-GAAAACTTCCACTGGTCTGC-3′ and 5′-CTCAGATAGACCTTGAAGG-3′). An F2 mapping population of 1,800 plants (from an F1 plant of an ECW × ECW-123R cross) was screened with L1 and R1 to identify recombinants between these markers and the Bs2 locus.
Sequencing of the Bs2 Locus.
pBluescript KS(+) libraries from the cosmids C17, C18, C19, C20, C21, and C22 were constructed by sonication of cosmids. One kb size selected DNA fragments were then subcloned into the EcoRV site of Bluescript KS(+). Colonies containing pepper genomic DNA clones were identified by hybridization and subjected to PCR by using T3 and T7 primers. Products purified with QIAquick PCR purification kit (Qiagen) were sequenced by using SK and KS primers and the ABI Prism FACS kit (dyedeoxy terminators; PE Biosystems, Foster City, CA). Sequencing reactions were analyzed by using the ABI 377 DNA sequencer (PE Biosystems). Sequence alignment and analysis was performed by using the sequencher 3.0 software (Gene Codes, Ann Arbor, MI). Identification of coding regions was performed by using the lasergene DNA sequence analysis software package (DNAstar, Madison, WI), and DNA sequence similarities were identified by using blast, identify, and psort (21–24).
5′ and 3′ Rapid Amplification of cDNA Ends (RACE) Analysis.
Total RNA was isolated from pepper leaf tissue by using Trizol reagent according to supplier’s instructions (GIBCO/BRL). RACE analysis was performed by using the GIBCO/BRL Life Technologies 5′ and 3′ race systems (Version 2.0) according to the manufacturer. For 5′ RACE, the first-strand primer was 5′-CCATCCCACACTTCACAACTCCA-3′. The nested gene-specific primers used for the PCR amplifications were 5′-CTGCTTCACCAATCATCTTAACCC-3′ and 5′-AACCTTCGACGCGCCTTTTTTTTC-3′. For 3′ RACE analysis, the first-strand cDNA was amplified by using the primer 5′-GTCTAGTCCTCGTCAGC-3′ and reamplification was performed by using a nested primer 5′-GTCCTTGAGCGCCTCATG-3′. The final PCR products of the 5′ and 3′ RACE reactions were cloned into the pCRII-TOPO vector (Invitrogen), and 5–10 independent clones were sequenced for each end.
Agrobacterium-Mediated Transient Coexpression Assay.
Constructs for the transient assays were made in pMD1, a derivative of pBI121 (CLONTECH) in which the GUS reporter gene has been replaced with a synthetic polylinker (courtesy of M. Dixon, Sainsbury Laboratory, Norwich, England). The 35S-avrBs2 construct consists of the avrBs2 ORF (8) cloned between the CaMV 35S promoter and the nopaline synthase 3′ sequences of pMD1 (D.D. and B. Savidge, unpublished results). The Bs2 candidate gene construct was generated by ligating a 5′ and 3′ RACE product onto an internal, 2.3-kb SalI-EcoRI genomic fragment from a cosmid subclone (Fig. 1D). These Bs2 sequences then were cloned between the CaMV 35S promoter and the nopaline synthase 3′ sequences of pMD1. Clones were sequenced for verification. Constructs were mobilized into A. tumefaciens C58C1 (pCH32) by triparental matings using standard methods. The pCH32 plasmid (courtesy of A. Hamilton, Sainsbury Laboratory, Norwich, England) was constructed by cloning the VirE operon from pSW108 (25) into the PvuII site of pCH30, a derivative of the binary vector pCC113 (26). Cells were grown overnight on Luria agar (LA) containing rifampicin at 100 μg/ml, tetracycline at 5 μg/ml, and kanamycin at 50 μg/ml. Bacteria were collected and suspended in 10 mM MgCl2/10 mM Mes/150 μM acetosyringone to a final OD600 of 0.6. After a few hours of induction in acetosyringone, A. tumefaciens containing the 35S-Bs2 construct was mixed with an equal volume of cells containing the 35S-avrBs2 construct. This mixture of cells was hand-infiltrated into the intercellular leaf spaces of ECW, VF36, and N. benthamiana plants with a plastic transfer pipette. As controls, suspensions of cells containing only one construct were infiltrated in comparable areas of the same leaves.
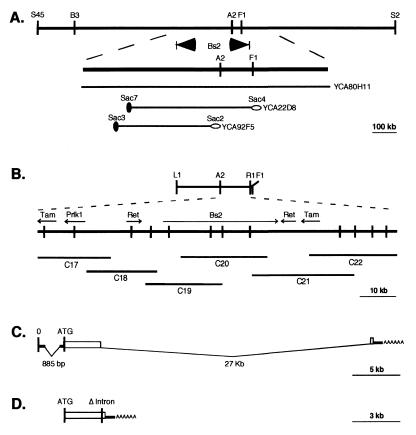
Figure 1.
Map-based cloning of the bacterial spot resistance gene Bs2 from pepper. (A) Contig of the three ECW-123R pepper library YAC clones (YCA80H11, YCA22D8, and YCA92F5) that span the Bs2 locus. (B) Contig of cosmid clones (C17-C22) that span the Bs2 region from the YAC contig. This region was sequenced. Arrows show ORFs and direction of transcription for Bs2, Prlk1, Tam (homologous to transposon TAM), and Ret (homologous to retrotransposons). (C) Bs2 transcript before splicing. (D) Bs2 ORF with deletion of untranslated upstream sequences and 27-kb intron that was used in 35S:Bs2 promoter constructs.
Plant Transformation.
The same 35S-Bs2 construct used in the transient assays was mobilized into the A. tumefaciens LBA4404 and used for the generation of stably transformed tomato (cv. VF36) and N. benthamiana plants by using standard procedures (27, 28). Putative transformants were analyzed by PCR using the following primers: Bs2 L1 (5′-CTGCTTCACCAATCATCTTAAC-3′) and Bs2 R1 (5′-TGAGACTAACAGGAAC-TGTACT-3′), which amplify a 0.5-kb fragment of the Bs2 gene. Tomato transformants were assayed for Bs2 function by infiltration of Xcv P38 (avrBs2−) and the isogenic Xcv P38:avrBs2 (avrBs2+). N. benthamiana transformants were assayed with X. campestris pv. campestris (Xcc) 8004 (avrBs2+) and the isogenic Xcc 8004:ΔavrBs2 (avrBs2−). The A. tumefaciens strain containing the 35S-avrBs2 construct also was used to detect functional expression of the Bs2 gene in transgenic lines by using the Agrobacterium transient assay. The concentrations of bacteria and assay conditions were as described above. Standard bacterial growth curve assays were performed by using transgenic tomato plants (7).
Results
Identification of an Expressed Sequence at the Bs2 Locus.
Two YAC clones, YCA22D8 and YCA80H11, were isolated previously by using the cosegregating AFLP marker A2 and shown to be approximately 550 kb and 1.2 Mb, respectively (29). Mapping of end probes derived from these clones indicated that they span the Bs2 locus (Fig. 1A). To identify expressed sequences in the region covered by the smaller clone, a cDNA library made from resistant pepper leaves infected with Xcv expressing avrBs2 was screened with YCA22D8 DNA. Seven cDNA clones were isolated and, based on cross-hybridization and sequence, were shown to represent members of a multigene family. Southern analysis of YCA22D8 indicated that only one member of this family is located within the Bs2 locus. This gene encodes a protein with a high degree of homology to Rlk1 of Arabidopsis (30). Preliminary northern analysis of this gene, which we have designated Prlk1, indicates that it is induced upon infection by an avirulent Xcv strain and to a lesser extent with a virulent strain (T. Tai and M. Whalen, unpublished). However, Prlk1 did not function in an avrBs2-dependent manner to induce a HR when assayed in the Agrobacterium-transient resistance assay discussed below. Given this result and the size of YCA22D8, it seemed likely that additional expressed sequences should be present in this region that would contain Bs2 activity.
Defining the Bs2 Locus.
A chromosome walk was initiated using cosmid libraries made from YCA22D8 and YCA80H11. Clones from the region were identified using AFLP markers, YAC ends, and Prlk1 as probes. Additional probes were obtained from the cosmids and used to construct a contig of about 300 kb spanning the Bs2 locus (Fig. 1A). Two PCR-based markers, L1 and R1, were used to screen 1,800 progeny from an F2 population. Nine new recombinants were detected, and fine mapping with these recombinants delimited the Bs2 locus to a region of about 100 kb spanned by cosmids C17 through C22 (Fig. 1B).
Sequence Analysis Identifies a Bs2 Gene Candidate.
To identify all the ORFs in the region, plasmid libraries of the cosmid contig clones were made and used for sequencing the locus. Six coding regions were identified including two retroelements, two Tam-like transposons, a sequence with homology to Prlk1 gene, and a gene encoding a tripartite NBS and a LRR motif (Fig. 1B). The NBS-LRR gene was tentatively designated as the Bs2 gene. Southern analysis indicated that it is a member of a multigene family (Fig. 2A) and is the only one at the locus as defined by the recombinants. Comparison of several clones obtained from 5′ RACE with the genomic sequence of the candidate Bs2 gene revealed the presence of an 885-bp intron in the 5′ untranslated region (Fig. 1C). The 5′ splice site of the intron is located 52 bases downstream from the transcriptional start. A TATA box is located 34 bases upstream of the transcriptional start. 3′ RACE analysis revealed the presence of a 27-kb intron followed by sequence encoding 10 aa and a 280-bp untranslated 3′ region containing a polyadenylation site (Fig. 1C).
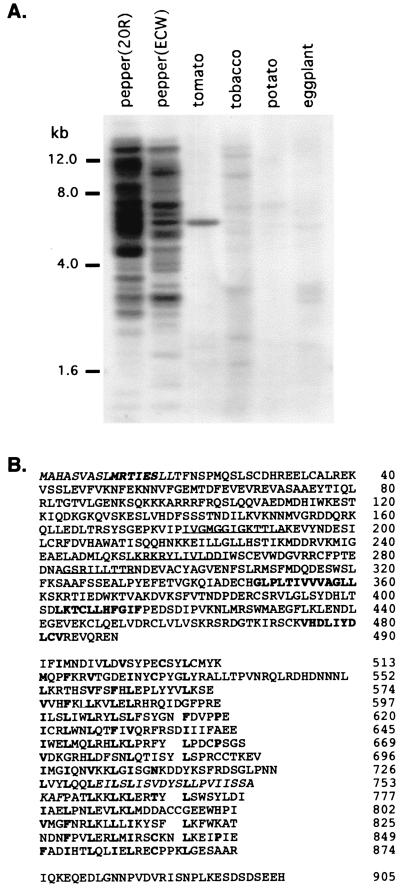
Figure 2.
(A) Homologs of the Bs2 gene in DNA gel blots of various Solanaceous plants; EcoRI cut genomic plant DNA probed with the Bs2 0.5-kb PCR product of Bs2 L1 and Bs2 R1. (B) Predicted amino acid sequence of the Bs2 gene product. Residues of significance are indicated by italics (hydrophobic domains), bold italics (mt targeting sequence), underlining (NBS), and bold (disease resistance signatures including GLPL, CFLY, MHD, and LRR).
The sequence of the Bs2 gene product is most similar to Rx virus resistance gene from potato (38% identity; ref. 31). The predicted protein has a putative NBS domain consisting of P-loop, kinase 2, and kinase 3a sequences followed by a domain with unknown function including sequences similar to the conserved sequence motifs GLPL, CFLY (CLLH in BS2), and MHD (VHD in Bs2; Fig. 2B). The putative LRR domain of Bs2 has 14–15 imperfect copies of the repeat and matches the cytoplasmic LRR consensus sequence (32). Bs2 has a hydrophobic N terminus containing a consensus sequence for mitochondrial sorting (24) and is similar to the human apoptotic protease activating factor-1 (APAF-1) through its central NBS domain (33). Unlike some other R proteins that contain a cytoplasmic LRR domain, Bs2 does not appear to contain either leucine zipper or TIR motifs (10).
An Agrobacterium-Mediated Transient Coexpression Assay Confirms the Bs2 Gene Isolation.
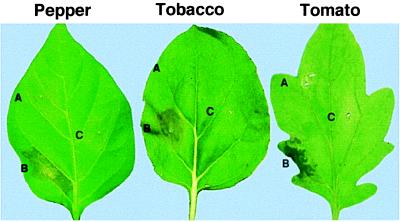
Infiltration of A. tumefaciens containing a CaMV 35S-avrBs2 construct into pepper leaves results in the induction of a HR only in plants containing the Bs2 gene (D.D. and B. Savidge, unpublished results). Based on this result, a transient coexpression assay was developed to identify the candidate gene. In this assay, an A. tumefaciens strain containing the 35S-avrBs2 construct and an Agrobacterium strain containing a 35S-candidate Bs2 construct were mixed together and infiltrated into the intracellular leaf space of susceptible pepper or nonhost plants. Plants infiltrated with strains containing either the 35S-avrBs2 or the 35S-candidate Bs2 construct alone did not exhibit any response on susceptible pepper plants. However, plants that were infiltrated with a mixture of the strains developed a HR 24–48 hr postinfiltration (Fig. 3). The HR reactions were characteristic of avrBs2-induced HR in peppers with the Bs2 gene. Similar reactions were observed when avrBs2 and Bs2 were transiently expressed in other Solanaceous plants including potato and eggplant; however, coinfiltration of non-Solanaceous species including Arabidopsis, turnip, cucumber, and broccoli did not result in a HR (data not shown). Furthermore, homology to Bs2 was observed only in these Solanaceous plants (Fig. 2A). Finally, no response was observed when A. tumefaciens strains containing 35S-avrBs2 and 35S-Prlk1 constructs were coinfiltrated in pepper, tomato, or N. benthamiana (data not shown). The requirement of the candidate Bs2 gene and the avrBs2 gene for the elicitation of the HR in susceptible pepper and tomato plants and the nonhost N. benthamiana clearly confirms the identity of the Bs2 gene.
Figure 3.
Pepper (ECW), tobacco (N. benthamiana), and tomato (VF36) are shown 48 hr postinoculation with Agrobacterium containing binary plasmid 35S:avrBs2 (A), mixture of 35S:avrBs2 (B), and 35S:Bs2 (C).
Expression of the Bs2 Gene in Tomato Confers Resistance to Bacterial Spot Disease.
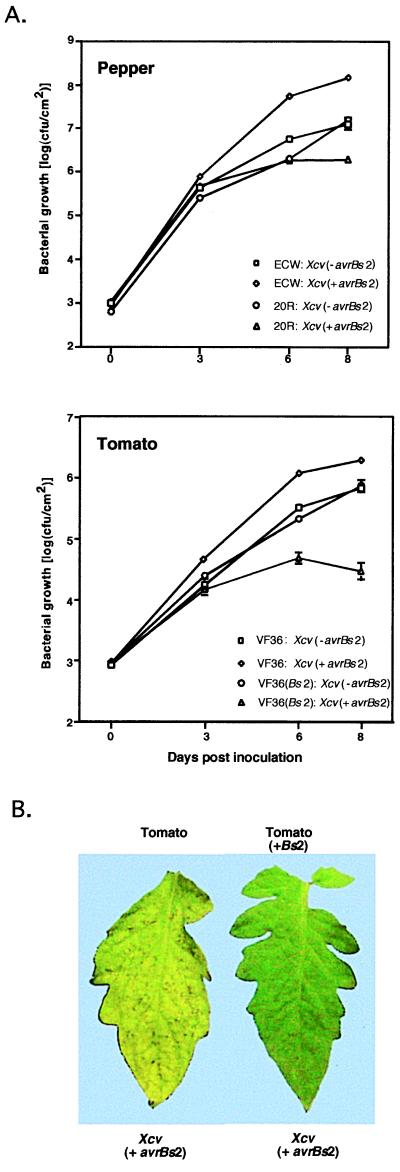
Tomato and N. benthamiana plants expressing the Bs2 gene were generated by Agrobacterium-mediated plant transformation. A total of 16 tomato and 14 N. benthamiana primary transformants (T1) were obtained and shown to contain the Bs2 transgene by PCR. All the T1 plants exhibited Bs2 activity as indicated by the specific induction of a HR upon infiltration with either Agrobacterium- or Xcv-expressing avrBs2. Secondary transformants (T2) derived from selfing T1 plants of each species were assessed for the presence of the transgene by PCR and functional activity by bacterial infiltration. Functional activity cosegregated with the presence of the Bs2 transgene for three different VF36 T2 populations and three different N. benthamiana T2 populations. To determine the effect of the Bs2 gene on bacterial growth, bacterial growth curve assays were performed on selected T2 tomato plants by using avirulent (avrBs2+) and virulent (avrBs2−) strains of Xcv (Fig. 4A). These assays indicate that the Bs2 gene suppresses the growth of Xcv-expressing avrBs2 in tomato to levels characteristic of resistant plants. In addition, the effect of the Bs2 gene can be observed by the lack of chlorotic disease symptoms on leaves inoculated with Xcv (Fig. 4B). In the case of the nonhost N. benthamiana, infiltration of Xcc containing avrBs2, which normally exhibits no response 24–48 hr postinfiltration, resulted in the elicitation of a HR in the transformed N. benthamiana plants whereas an isogenic Xcc strain lacking the avrBs2 gene did not elicit a HR (data not shown).
Figure 4.
(A) Kinetics of bacterial growth in tomato plants transformed with 35S:Bs2. Plants were vacuum-infiltrated with Xcv (with or without avrBs2) at a concentration of 1 × 105 colony-forming units (cfu)/ml. Bacterial concentrations in plant leaves were assayed after 0, 3, 6, and 8 days. Data points represent the mean of three replicate experiments plus or minus SE. (B) Disease symptoms on tomato (VF36) transformed with or without 35S:Bs2 2 days postinoculation of Xcv(avrBs2) at 1 × 106 cfu/ml.
Discussion
In agriculture, the most efficient form of protection against pathogens is genetic resistance. Like most traits manipulated by breeders, the genetic resistance typically employed is based on single, dominant or semidominant genes. These R genes usually confer race-specific resistance, and, as such, their effectiveness and durability are based on their interaction with complementary pathogen avirulence genes. The breakdown in effectiveness of a R gene often occurs as a result of the emergence of strains no longer expressing the specific avirulence gene product. In the case of the pepper Bs2 gene, the avirulence gene avrBs2 has been shown to be involved in the fitness of the Xcv pathogen and to be highly conserved among other X. campestris pathovars (9). Because the fitness contribution of avrBs2 may offset the virulence of strains lacking avrBs2, the Bs2 gene may provide effective, durable field resistance. Although there have been some reports of Bs2 resistance breaking down in the field (34), these peppers are typically hybrids that are heterozygous at the Bs2 locus and have an intermediate level of resistance (R. E. Stall, personal communication). Because the avrBs2 gene is in other X. campestris pathovars that infect different plant species, transfer of the Bs2 gene into these species may serve as a novel source of resistance. Evidence from our laboratory (D.D. and B.J.S., unpublished observations) indicates that avrBs2 also contributes to the fitness of these other X. campestris pathovars, which suggests that if the Bs2 gene is functional in other plant species, it may provide a durable resistance there as well.
In this study, we have used a positional cloning approach to isolate the Bs2 gene. Although expression library screening using spanning YACs has been employed successfully to identify genes of interest (11), only one candidate, a putative receptor-like kinase gene Prlk1, was isolated when the spanning YAC clone YCA22D8 was used as a probe. The presence of such a gene in the Bs2 region was of interest given that at least one R gene, Xa21 from rice, has been shown to be a receptor-like kinase, although its putative receptor domain consists of a LRR motif (35). Other receptor-like kinases also have been implicated in disease resistance (36, 37), and, recently, a gene from Brassica oleracea, SRF2, was shown to be induced upon wounding and infection with X. campestris pv. campestris (38). Interestingly, Northern blot analysis has suggested that the Prlk1 gene is induced upon infection with Xcv (T.H.T. and M.C.W., unpublished results).
To ensure the detection of all the putative ORFs at the Bs2 locus, a sequencing approach was taken that resulted in the identification of an ORF encoding a putative protein with very high homology to the NBS-LRR class of R genes. RACE analyses revealed the presence of a short intron in the 5′ untranslated region and an extremely long (27-kb) intron positioned near the end of the coding region.
To verify that this was the Bs2 gene, a transient coexpression assay was used with susceptible pepper and tomato cultivars and the nonhost N. benthamiana. In this assay, functional activity was defined as the induction of a HR in an avrBs2-dependent manner. Development of the assay provided a rapid method for testing Bs2 gene function. Application of the assay to various plant species indicates that the Bs2 gene that originates from pepper, a member of the Solanaceae, functions only in Solanaceous plants. This phenomenon, which we refer to as RTF, has also been observed when the RPS2 gene of Arabidopsis thaliana was shown to be nonfunctional in transgenic tomatoes (D.D. and B.J.S., unpublished observations). It remains to be determined whether RTF can be overcome to allow cloned R genes, such as Bs2, to provide resistance in unrelated species.
Elicitation of a race-specific HR is a hallmark of many R genes; however, the HR itself has been separated from resistance (39). To show that the candidate Bs2 gene actually confers resistance by suppressing the growth of Xcv, transgenic tomatoes were generated and bacterial growth assays were performed. The results show that the candidate Bs2 gene functions in tomato to suppress the growth of an Xcv strain expressing avrBs2 to levels indicative of resistance, and infected leaves showed no symptoms of disease (Fig. 4). These results confirm the identity of the Bs2 gene and demonstrate its use as a novel source of genetic resistance to bacterial spot disease in tomato.
The putative protein encoded by the Bs2 gene is most similar to the Rx virus resistance gene of potato (31) and then to various R gene products including Prf, Mi-1, and I2C from tomato (40–42) and RPP8 from Arabidopsis (43). Bs2 has several structural features that are common to many R gene products including a putative NBS domain (P-loop, kinase 2, and kinase 3a sequences), a domain with unknown function including sequences similar to GLPL, CFLY, and MHD sequence motifs, and a cytoplasmic LRR domain (Fig. 2B). The N-terminal region of Bs2 is hydrophobic and contains a mitochondrial sorting signal sequence (MRTIES) but does not contain an apparent leucine zipper (LX6L) motif. Bs2 is similar to APAF-1 throughout its central NBS domain, suggesting a link to mammalian regulators of apoptosis (33). Given the modular nature of proteins containing caspase recruitment domains (CARD; ref. 44), it is not surprising that the similarity of Bs2 and APAF-1 does not extend to the N-terminal CARD or to the C-terminal WD-40 regulatory domain. The homology of NBS-LRR type R genes with APAF-1 has been noted previously (45). The presence of a mitochondrial sorting sequence in Bs2 may be consistent with a role similar to that of APAF-1 in interacting with mitochondrial factors to regulate cell death (46). Identifying proteins that interact with BS2 will aid in understanding the relevance of this similarity.
The cloning of Bs2 is major step in understanding its interaction with avrBs2 and should contribute to the elucidation of the mechanism(s) involved in race-specific disease resistance. The ability of the Bs2 gene to function in tomato provides a novel source of resistance against devastating outbreaks of bacterial spot disease in this economically important crop. Understanding the molecular basis for the RTF exhibited by Bs2 and devising strategies to overcome it will be key to using Bs2 in non-Solanaceous species.
Acknowledgments
We thank Mary Beth Mudgett for helpful suggestions and critical reading of the manuscript. This work was supported by a grant to B.J.S. from Diatech Ltd. T.H.T. was supported by a National Science Foundation Predoctoral Fellowship and by a Chancellor’s Opportunity Fellowship from the University of California.
Abbreviations
- NBS-LRR
nucleotide binding site–leucine-rich repeat
- RTF
restricted taxonomic functionality
- ECW
Early Calwonder
- YAC
yeast artificial chromosome
- HR
hypersensitive reaction
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF202179).
References
- 1.Cook A A, Stall R E. Phytopathology. 1963;53:1060–1062. [Google Scholar]
- 2.Cook A A, Guevara Y G. Plant Dis. 1984;68:329–330. [Google Scholar]
- 3.Kim B S, Hartmann R W. Plant Dis. 1985;69:233–235. [Google Scholar]
- 4.Jones J B, Scott J W. Plant Dis. 1986;70:337–339. [Google Scholar]
- 5.Hibberd A M, Bassett M J, Stall R E. Phytopathology. 1987;77:1304–1307. [Google Scholar]
- 6.Swanson J, Kearney B, Dahlbeck D, Staskawicz B J. Mol Plant–Microbe Interact. 1988;1:5–9. [PubMed] [Google Scholar]
- 7.Minsavage G V, Dahlbeck D, Whalen M C, Kearney B, Bonas U, Staskawicz B J, Stall R E. Mol Plant–Microbe Interact. 1990;3:41–47. [Google Scholar]
- 8.Swords K M M, Dahlbeck D, Kearney B, Roy M, Staskawicz B J. J Bacteriol. 1996;178:4661–4669. doi: 10.1128/jb.178.15.4661-4669.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kearney B, Staskawicz B J. Nature (London) 1990;346:385–386. doi: 10.1038/346385a0. [DOI] [PubMed] [Google Scholar]
- 10.Hammond-Kosack K, Jones J D G. Annu Rev Plant Physiol Plant Mol Biol. 1997;48:575–607. doi: 10.1146/annurev.arplant.48.1.575. [DOI] [PubMed] [Google Scholar]
- 11.Martin G B, Brommenschenkel S H, Chunwongse J, Frary A, Ganal M W, Spivey R, Wu T, Earle E D, Tanksley S D. Science. 1993;262:1432–1436. doi: 10.1126/science.7902614. [DOI] [PubMed] [Google Scholar]
- 12.Baker B, Zambryski P, Staskawicz B J, Dinesh-Kumar S P. Science. 1997;276:726–733. doi: 10.1126/science.276.5313.726. [DOI] [PubMed] [Google Scholar]
- 13.Staskawicz B J, Ausubel F M, Baker B, Ellis J G, Jones J D G. Science. 1995;268:661–667. doi: 10.1126/science.7732374. [DOI] [PubMed] [Google Scholar]
- 14.Tai T H, Tanksley SD. Plant Mol Biol Rep. 1990;8:297–303. [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J G, Struhl K. Current Protocols in Molecular Biology. New York: Greene & Wiley; 1987. [Google Scholar]
- 17.Tai, T., Dahlbeck, D., Peleman, J., Stall, R. E. & Staskawicz, B. J. (1999) Theor. Appl. Genet., in press.
- 18.Mendez M J, Klapholz S, Brownstein B H, Gemmill R M. Genomics. 1991;10:661–665. doi: 10.1016/0888-7543(91)90449-o. [DOI] [PubMed] [Google Scholar]
- 19.Bent A F, Kunkel B N, Dahlbeck D, Brown K L, Schmidt R, Giraudat J, Leung J, Staskawicz B J. Science. 1994;265:1856–1860. doi: 10.1126/science.8091210. [DOI] [PubMed] [Google Scholar]
- 20.Jones J D G, Shlumukov L, Carland F, English J J, Scofield S R, Bishop G, Harrison K. Transgenic Res. 1992;1:285–297. doi: 10.1007/BF02525170. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 22.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nevill-Manning C G, Wu T D, Brutlag D L. Proc Natl Acad Sci USA. 1998;95:5865–5871. doi: 10.1073/pnas.95.11.5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winans S C, Allenza P, Stachel S E, McBride K E, Nester E W. Nucleic Acids Res. 1987;15:825–837. doi: 10.1093/nar/15.2.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen C Y, Wang L, Winans S C. Mol Gen Genet. 1991;230:302–309. doi: 10.1007/BF00290681. [DOI] [PubMed] [Google Scholar]
- 27.McCormick S. In: Plant Tissue Culture Manual. Lindsey K, editor. B6. Dordrecht, The Netherlands: Kluwer; 1991. pp. 1–9. [Google Scholar]
- 28.Horsch R B, Fry J, Hoffmann N, Neidermeyer J, Rogers S G, Fraley R T. In: Plant Molecular Biology Manual. Gelvin S B, Schilperoort R A, editors. A5. Dordrecht, The Netherlands: Kluwer; 1988. pp. 1–9. [Google Scholar]
- 29.Tai, T. & Staskawicz, B. J. (1999) Theor. Appl. Genet., in press.
- 30.Walker J C. Plant J. 1993;3:451–456. doi: 10.1111/j.1365-313x.1993.tb00164.x. [DOI] [PubMed] [Google Scholar]
- 31.Bendahmane A, Kanuya K, Baulcombe D. Plant Cell. 1999;11:781–791. doi: 10.1105/tpc.11.5.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones D A, Jones J D G. Adv Bot Res. 1997;24:89–167. [Google Scholar]
- 33.Zou H, Henzel W J, Liu X, Lutschg A, Wang X. Cell. 1997;90:405–413. doi: 10.1016/s0092-8674(00)80501-2. [DOI] [PubMed] [Google Scholar]
- 34.Kousik C S, Ritchie D F. Plant Dis. 1998;82:181–186. doi: 10.1094/PDIS.1998.82.2.181. [DOI] [PubMed] [Google Scholar]
- 35.Song W Y, Wang G-L, Chen L-L, Kim H-S, Pi L-Y, Holsten T, Gardner J, Wamg B, Zhai W-X, Zhu L-H, et al. Science. 1995;270:1804–1806. doi: 10.1126/science.270.5243.1804. [DOI] [PubMed] [Google Scholar]
- 36.Feuillet C, Schachermayr G, Keller B. Plant J. 1997;11:45–52. doi: 10.1046/j.1365-313x.1997.11010045.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Zafian P, Choudhary M, Lawton M. Proc Natl Acad Sci USA. 1996;93:2598–2602. doi: 10.1073/pnas.93.6.2598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pastuglia M, Roby D, Dumas C, Cock J M. Plant Cell. 1997;9:49–60. doi: 10.1105/tpc.9.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu I C, Parker J, Bent A F. Proc Natl Acad Sci USA. 1998;95:7819–7824. doi: 10.1073/pnas.95.13.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Salmeron J M, Oldroyd G E D, Rommens C M T, Scofield S R, Kim H-S, Lavelle D T, Dahlbeck D, Staskawicz B J. Cell. 1996;86:123–133. doi: 10.1016/s0092-8674(00)80083-5. [DOI] [PubMed] [Google Scholar]
- 41.Milligan S B, Bodeau J, Yaghoobi J, Kaloshian I, Zabel P, Williamson V M. Plant Cell. 1998;10:1307–1319. doi: 10.1105/tpc.10.8.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ori N, Eshed Y, Paran I, Presting G, Aviv D, Tanksley S, Zamir D, Fluhr R. Plant Cell. 1997;9:521–532. doi: 10.1105/tpc.9.4.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDowell J M, Dhandaydham M, Long T A, Aarts M G M, Goff S, Holub E B, Dangl J L. Plant Cell. 1998;10:1861–1874. doi: 10.1105/tpc.10.11.1861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bertin J, Nir W-J, Fischer C M, Tayber O V, Errada P R, Grant J R, Keilty J J, Gosselin M L, Robinson K E, Wong G H W, et al. J Biol Chem. 1999;274:12955–12958. doi: 10.1074/jbc.274.19.12955. [DOI] [PubMed] [Google Scholar]
- 45.van der Biezen E A, Jones J D G. Curr Biol. 1998;8:226–227. doi: 10.1016/s0960-9822(98)70145-9. [DOI] [PubMed] [Google Scholar]
- 46.Mignotte B, Vayassiere J-L. Eur J Biochem. 1998;252:1–15. doi: 10.1046/j.1432-1327.1998.2520001.x. [DOI] [PubMed] [Google Scholar]