Abstract
Natural selection is expected to eliminate genetic incompatibilities from interbreeding populations. We have identified a globally distributed incompatibility in the primarily selfing species Caenorhabditis elegans that has been maintained despite its negative consequences for fitness. Embryos homozygous for a naturally occurring deletion of the zygotically acting gene, zeel-1, arrest if their sperm parent carries an incompatible allele of a second, paternal effect locus, peel-1. The two interacting loci are tightly linked, with incompatible alleles occurring in linkage disequilibrium in two common haplotypes. These haplotypes exhibit elevated sequence divergence, and population genetic analyses of this region indicate that natural selection is preserving both haplotypes in the population. Our data suggest that long-term maintenance of a balanced polymorphism has permitted the incompatibility to persist despite gene flow across the rest of the genome.
Caenorhabditis elegans is a globally distributed species of free-living, bacteria-eating nematode. Although rare males contribute at a low rate to outcrossing, C. elegans occurs primarily as inbred, self-fertilizing hermaphrodites (1-4). A wild isolate from Hawaii, CB4856, has been identified among well-studied isolates as the most divergent at the sequence level from the standard laboratory strain, N2, derived from an isolate from Bristol, England (5-7). As a result of this sequence divergence, the Hawaiian strain is widely used to map mutations induced in the Bristol background.
Genetic incompatibility between Bristol and Hawaii
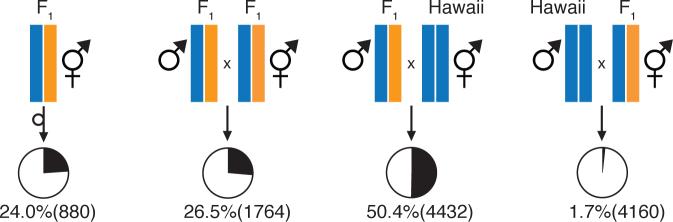
We generated recombinant inbred lines (RILs) from the tenth generation of an advanced intercross between Bristol and Hawaii to study natural genetic variation in C. elegans, and we genotyped the RILs at 1450 single nucleotide polymorphism markers (8). We noted that a region on the left arm of chromsome I exhibited a dramatic deficit of Hawaii alleles among the RILs. Of 239 RILs, only five carried the Hawaii allele at the most skewed marker, and simulations of the intercross pedigree indicated that this allele frequency skew could not have arisen by drift, suggesting that selection had acted during construction of the RILs (Fig. S1). We then crossed Hawaii to a Bristol strain carrying a visible marker located 10cM from the most skewed RIL marker, and we examined F2 progeny produced by self-fertilizing F1 hermaphrodites. Surprisingly, approximately 25% of F2 progeny arrested as embryos, and embryonic lethality segregated opposite the visible marker (Table 1). F2 lethality was not an effect of the marker: self-fertilizing F1 hermaphrodites derived from reciprocal crosses between Hawaii and wildtype Bristol produced 25% dead embryos, as did F1 hermaphrodites mated to F1 males (Fig. 1).
Table 1.
F2 lethality segregates opposite visible marker bli-3, located 10 cM from the most skewed RIL marker. Genotypes of F2 progeny from selfing Bristol/bli-3 and Hawaii/bli-3 hermaphrodites were scored. Embryonic lethality from selfing Hawaii/bli-3 hermaphrodites was slightly greater than 25% because the visible marker introduces a small percentage of lethality.
| F2 genotype | Bristol/bli-3 hermaphrodite | Hawaii/bli-3 hermaphrodite |
|---|---|---|
| bli-3/bli-3 | 21.7%(128) | 21.8%(128) |
| bli-3/+ | 49.9%(295) | 42.0%(246) |
| +/+ | 24.5%(145) | 5.1%(30) |
| arrested embryos | 3.9%(23) | 31.1%(182) |
Fig. 1.
Paternal effect-by-zygotic lethality. Percent embryonic lethality (total) was scored from crosses shown. Orange and blue indicate Bristol and Hawaii haplotypes, respectively. Pie charts show the proportions of embryos that hatched (white) and failed to hatch (black). F1 individuals were derived from reciprocal Bristol x Hawaii crosses. Embryonic lethality from selfing Bristol and Hawaii hermaphrodites, reciprocal Bristol x Hawaii, and reciprocal Bristol x F1 crosses was less than 0.8% (n > 240).
Lethality caused by paternal effect-by-zygotic interaction
Segregation of embryonic lethality opposite the visible marker implied that the arrested embryos represented those homozygous for the Hawaii allele of a locus linked to the marker, on the left arm of chromosome I. Because embryonic lethality within the Hawaii strain itself is very low (less than 1%), we reasoned that F2 lethality reflected an incompatibility between the Hawaii allele of this locus and an element in the Bristol genome. We also reasoned that it did not reflect two synthetically lethal alleles segregating in the F2 population because such an interaction would affect less than one quarter of F2 embryos (up to 3/16, depending on linkage and dominance). One-quarter lethality is expected, however, if the incompatibility involves an interaction between the genotype of the zygote and a maternal or paternal effect.
To test this possibility, Hawaii × Bristol F1 males and hermaphrodites were separately backcrossed to Hawaii individuals, and lethality was scored among the resulting embryos. We observed 50% lethality when F1 males were mated to Hawaii hermaphrodites, but less than 2% lethality in the reciprocal cross (Fig. 1). Thus, lethality depends on both paternal and zygotic genotype, but is independent of maternal cytoplasm. (Note that both Hawaii and F1 hermaphrodites produced dead embryos, 50% and 25%, respectively, when mated to F1 males.) In sum, lethality appears to result from a paternal effect-by-zygotic interaction whereby embryos homozygous for the Hawaii allele of a zygotically acting locus fail to hatch when the sperm parent – male or hermaphrodite – is a Hawaii × Bristol heterozygote. An interaction between a paternal effect and a zygotically acting gene is surprising because sperm-supplied factors are expected to act during fertilization and first cleavage (9), while early embryogenesis is primarily controlled by maternally contributed factors, and zygotic transcription is not known to occur before the 4-cell stage (10).
Tight linkage of the zygotically acting and paternal effect loci
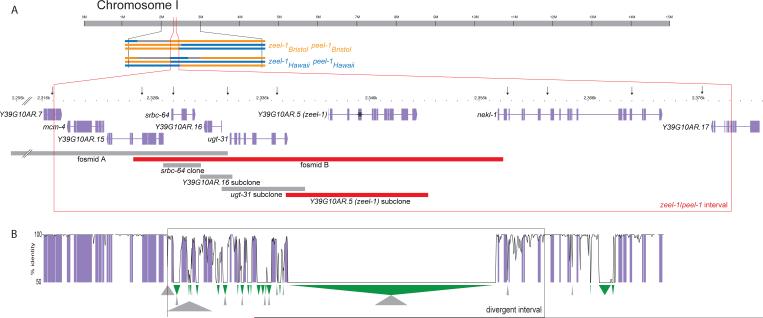
To understand the genetic basis of the incompatibility, we used the RILs to map both the zygotically acting locus, zeel-1 (zygotic epistatic embryonic lethal-1), and the paternal effect locus, peel-1 (paternal effect epistatic embryonic lethal-1). We crossed RILs to each of Bristol and Hawaii and scored lethality among embryos laid by self-fertilizing F1 hermaphrodites and Hawaii hermaphrodites mated to F1 males. The pattern of lethality among F2 and backcross embryos was consistent with each RIL carrying either the Bristol alleles of both zeel-1 and peel-1 or the Hawaii alleles of both. We identified only one genomic interval in which all lines of the former class carried the Bristol haplotype and all lines of the latter class carried the Hawaii haplotype. Thus, both zeel-1 and peel-1 map to this interval, a 62 kb region on chromosome I (position 2,317,234 to 2,379,249) (Fig. 2A). We confirmed tight linkage between the two loci; they do not segregate independently among backcross progeny (Table S1).
Fig. 2.
zeel-1 and peel-1 map to the same 62 kb interval. A. Colored bars represent Bristol (orange) and Hawaii (blue) haplotypes carried by six RILs used to map zeel-1 and peel-1. Grey bars show regions between undetermined RIL breakpoints. RIL genotypes inferred from mapping crosses are shown to the right of the haplotypes. Red box indicates interval to which zeel-1 and peel-1 map. Gene annotations derive from WormBase (ws170) gene models for Bristol sequence. Bars indicate coverage of fosmid A (WRM0614dC06), fosmid B (WRM0633bE09), and subclones used in transgenic complementation, with rescuing transgenes in red. Asterisk indicates the frameshift introduced into the Y39G10AR.5 subclone. Arrows indicate markers used to genotype wild isolates. All markers are in complete linkage disequilibrium. B. mVista alignment of Hawaii and Bristol sequence across a portion of the zeel-1/peel-1 interval (35). We used default parameters and Bristol as the reference sequence. Black box surrounds the interval of elevated divergence between Bristol and Hawaii. Large (>50 bp) deletions (green) and insertions (grey) in Hawaii relative to the Bristol sequence are shown below the alignment.
Incomplete penetrance of the incompatibility
The penetrance of the incompatibility, i.e. the extent of lethality among zeel-1Hawaii homozygotes sired by peel-1 heterozygotes, was complete when oocytes were fertilized by male sperm but incomplete when they were fertilized by hermaphrodite sperm. We collected embryos from self-fertilizing F1 hermaphrodites and from F1 hermaphrodites mated to F1 males and genotyped surviving progeny at the zeel-1 locus. Among self-progeny, approximately 10% of zeel-1Hawaii homozygotes survived to hatching, although most had retarded development and abnormal morphologies (8). In contrast, none survived when fertilized by F1 males. Penetrance of the incompatibility also appeared complete among embryos from F1 males backcrossed to Hawaii hermaphrodites, as these broods lacked the deformed larvae characteristic of surviving zeel-1Hawaii homozygotes.
The morphological defects of surviving zeel-1Hawaii homozygotes were highly variable and often similar to the terminal phenotype of observed in arrested embryos, which usually showed tissue differentiation but no elongation past the two-fold stage. Nevertheless, some zeel-1Hawaii homozygotes matured to adulthood and produced progeny. These progeny were entirely wildtype, implying that the paternal effect is not caused by heritable defects such as DNA damage or aneuploidy in zeel-1Hawaii sperm.
Globally distributed incompatibility
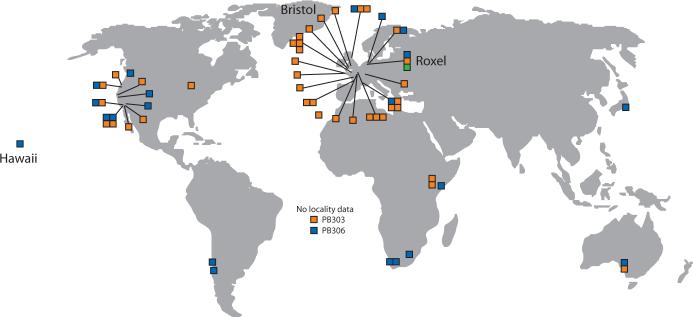
To determine the distribution of alleles causing the Bristol-Hawaii incompatibility in the global C. elegans population, we phenotyped 62 wild isolates from 40 localities. From each locality, we phenotyped only strains known to be genetically distinct. We crossed each strain to Bristol and Hawaii and scored lethality among embryos from self-fertilizing F1 hermaphrodites, F1 males backcrossed to Hawaii hermaphrodites, and F1 males backcrossed to hermaphrodites of the wild isolate itself. All but one wild isolate produced a pattern of lethality consistent with carrying either the Bristol alleles of both peel-1 and zeel-1 (Bristol-compatible strains) or the Hawaii alleles of both (Hawaii-compatible strains). The exception, a strain collected from Roxel, Germany, was compatible with both Bristol and Hawaii, showing no lethality in crosses to either strain. The global distribution of Bristol-compatible and Hawaii-compatible strains demonstrates that the two classes are not geographically isolated (Fig. 3), consistent with an absence of large-scale population structure in C. elegans (1, 5, 11-13). Both classes were found in many localities, and individual samples of compost from two localities in northern Germany contained both Bristol- and Hawaii-compatible strains, indicating that the two classes co-occur at the most local level (11).
Fig. 3.
Strains compatible with Bristol and incompatible with Hawaii (orange) and strains compatible with Hawaii and incompatible with Bristol (blue) are globally distributed. Strains compatible with both Bristol and Hawaii (green) derive from a locality in Roxel, Germany. For each locality, only strains known to be genetically distinct are shown. Strain names and localities are presented in Table S7.
Molecular signatures of balancing selection
The interval to which zeel-1 and peel-1 map, contains a region of dramatically elevated sequence divergence between the Bristol and Hawaii haplotypes. This region spans 33 kb of Bristol sequence and includes four full genes and part of a fifth (Fig. 2B). The Hawaii haplotype contains a 19 kb deletion spanning the gene Y39G10AR.5. Divergence within coding segments of the remaining genes averages 5%, fifty-fold higher than previous genome-wide estimates of pairwise divergence from both coding and non-coding sequence (6, 7). Non-coding segments in this region are largely unalignable and contained many insertions and deletions, mainly comprised of repetitive elements. The left boundary of the divergent interval is abrupt and is marked by a 1 kb insertion in Hawaii. Genomic divergence within the 13 kb immediately outside the insertion is 0.1%. The right boundary is less abrupt, with divergence falling gradually to 0.7% across 4 kb.
We genotyped the wild isolates with markers located throughout the interval and found that all Hawaii-compatible strains carry Hawaii-like haplotypes, while all Bristol-compatible strains carry Bristol-like haplotypes (Table S2). The doubly compatible strain carries a Bristol-like haplotype. Linkage disequilibrium among markers within the divergent interval is complete, but breaks down for markers 165 kb to the left and 78 kb to the right of the interval.
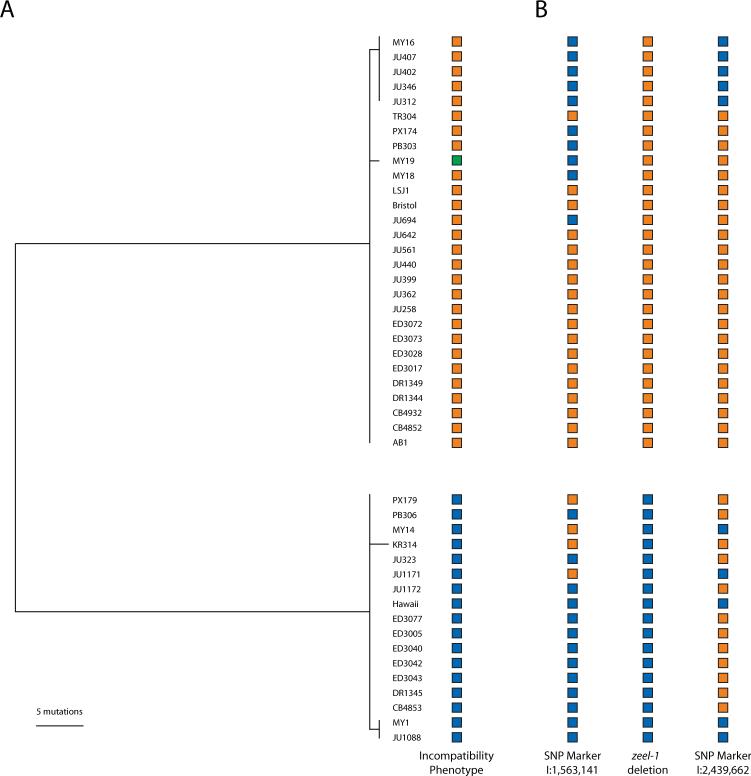
To understand the cause of elevated polymorphism in the zeel-1/peel-1 interval, we sequenced the exons and adjacent regions of srbc-64, a gene located within the divergent interval, from 45 genetically distinct wild isolates. Elevated polymorphism may result from an elevated mutation rate, an ancient coalescence of neutral alleles, or long-term balancing selection maintaining divergent haplotypes against loss by genetic drift, thereby permitting them to accumulate more mutations than expected under neutrality (14). Under balancing selection, most mutations will differentiate the two major haplotype classes, creating an excess of intermediate frequency alleles. Among the srbc-64 sequences, we observed 80 polymorphic sites but only 6 distinct haplotypes, far fewer than expected for a neutral sample (P < 0.0001). Furthermore, the allele frequency spectrum was strongly skewed toward intermediate frequency alleles (Tajima's D = 3.56, P < 0.0001). These data are consistent with long-term maintenance by balancing selection.
The srbc-64 sequences form two distinct, deeply branching clades, reflecting the Bristol-and Hawaii-compatible classes (Fig. 4A). This topology is not representative of the genome as a whole. Markers on both sides of the divergent interval exhibit evidence of genetic exchange between the two classes (Fig. 4B), and phylogenies constructed from mitochondrial sequence and nuclear sequences located elsewhere in the genome do not resolve Bristol- and Hawaii-compatible strains into two distinct clades (5, 15), indicating that gene flow occurs across the rest of the genome.
Fig. 4.
Signature of balancing selection on the zeel-1/peel-1 interval. A. Haplotypes of 1193 base pairs of the srbc-64 gene, excluding gapped sites, are split into two deeply divergent clades, one compatible with Bristol (orange) and one with Hawaii (blue). Doubly-compatible MY19 (green) has a haplotype similar to Bristol. B. Recombination has separated the zeel-1 deletion polymorphism from marker SNPs on both sides of the divergent interval.
The exceptional divergence between the Bristol and Hawaii haplotypes does not appear to be due to diversifying selection favoring amino acid-changing substitutions, nor relaxed selection allowing degeneration of their protein-coding sequences. Despite synonymous divergence orders of magnitude above that observed for genes outside the interval, genes within the interval exhibit ratios of non-synonymous to synonymous substitution of less than 0.2, consistent with purifying selection (Table 2). Given a direct estimate of the mutation rate (µ = 2.1 × 10−8 per site per generation) (16), the estimated divergence at synonymous sites implies that the incompatible haplotypes diverged roughly 3.5 million generations ago.
Table 2.
Genes in interval of elevated divergence between the Bristol and Hawaii haplotypes exhibit signatures of purifying selection.
| Gene | Number of Sites | dN | dS | ω |
|---|---|---|---|---|
| To the left of the divergent interval | ||||
| Y39G10AR.7 | 1422 | 0.001 | 0.000 | — |
| mcm-4 | 2469 | 0.000 | 0.004 | — |
| Y39G10AR.15 | 2112 | 0.001 | 0.000 | — |
| Inside the divergent interval | ||||
| srbc-64 | 870 | 0.018 | 0.147 | 0.124 |
| Y39G10AR.16* | 681 | 0.019 | 0.106 | 0.181 |
| ugt-31 | 1563 | 0.022 | 0.158 | 0.140 |
| On the right boundary of the divergent interval | ||||
| nekl-1† | 2949 | 0.001 | 0.019 | 0.050 |
dN, nonsynonymous substitutions per nonsynonymous site. dS, synonymous substitutions per synonymous site. ω, the dN/dS ratio. These quantities were estimated by maximum likelihood with PAML (34).
The Hawaii and Bristol sequences have different predicted splice sites on both sides of intron 3, resulting in several predicted amino acid residues that are not shared between the two alleles. We considered only the shared exonic sequences.
nekl-1 cDNAs from Bristol and Hawaii differ slightly from the predicted exon structure. We used the exon structure from our cDNA clones (8).
Identification of zeel-1
We cloned zeel-1 by introducing two fosmids carrying Bristol genomic DNA that together covered 45 kb of the 62 kb zeel-1 interval into the Hawaii background (Fig. 2A). To test for rescue, we crossed transgenic individuals to Bristol and scored lethality among embryos from three crosses: self-fertilizing F1 hermaphrodites, F1 males backcrossed to Hawaii hermaphrodites, and Bristol x Hawaii F1 males backcrossed to transgenic hermaphrodites. In the second cross, the transgene is inherited through the sperm, whereas in the third it is inherited through the oocyte. All crosses showed a reduction in embryonic lethality for transformants carrying fosmid B but not in those carrying fosmid A (Fig. 5). Ability of the transgene to mediate rescue when inherited through either sperm or oocyte implied that rescue occurs through the zygotic transcription of zeel-1Bristol. We genotyped surviving self-cross and male backcross progeny and found that survival of zeel-1Hawaii homozygotes required inheritance of the transgene, further supporting that zeel-1 acts zygotically (Table S3).
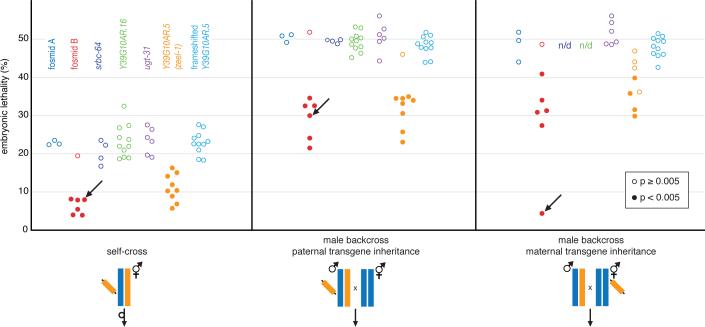
Fig. 5.
Transgenic complementation identifies Y39G10AR.5 as zeel-1. Embryonic lethality was scored in three crosses: selfing F1 hermaphrodites carrying the transgene, Hawaii hermaphrodites x F1 males carrying the transgene, and Hawaii hermaphrodites carrying the transgene x F1 males. Orange and blue bars designate Bristol and Hawaii haplotypes, respectively. Diagonal segments represent transgenes. Within crosses, each circle represents the result for an independent transgenic line (n/d, not determined). For each result, we scored at least 50 embryos, typically 200−300. Filled circles mark results showing a significant reduction in lethality (χ2 one-sided P < 0.005). Most transgenes were not integrated and were therefore not transmitted to all progeny. Arrows designate the single integrated line. In the male backcross with maternal transgene inheritance, where the integrated transgene was transmitted to all progeny, embryonic lethality was 4% (n = 298).
To identify zeel-1, we individually subcloned the four predicted genes carried by fosmid B (Fig. 2A), introduced each into Hawaii, and tested for rescue. Only one subclone, containing predicted open reading frame Y39G10AR.5, rescued lethality, indicating that this gene is zeel-1 (Fig. 5). zeel-1 belongs to a previously uncharacterized, Caenorhabditis-specific family of genes with homology to zyg-11, the substrate-recognition subunit of the CUL-2-based E3 ubiquitin ligase complex (17). zeel-1 is located within the divergent interval and is deleted in Hawaii and all Hawaii-compatible wild isolates (Table S2). To test whether transgenic rescue requires ZEEL-1 protein, we generated a second zeel-1 subclone identical to the first except that it contained a frame shift (via a four base pair insertion) truncating the protein at 25% of its length. When bombarded into Hawaii, the frame shifted transgene failed to rescue, indicating that rescue requires ZEEL-1 protein (Fig. 5).
Analysis of peel-1
Transgenic worms carrying Bristol-library fosmids, which together cover seven of the nine predicted genes in the peel-1 interval, failed to induce the paternal effect lethality of zeel-1Hawaii homozygotes, as did subclones of the four genes within the divergent interval (8). Putatively null alleles of srbc-64, nekl-1, and Y39G10AR.17, as well as RNAi targeting the remaining genes, did not abolish the paternal effect (Tables S4, S5). These negative results are equivocal, due to potential germline silencing of transgenes, a possible requirement for chromosomal heterozygosity of peel-1 in the spermatogenic germline, and the ineffectiveness of RNAi against sperm-expressed genes (18).
To find potential peel-1 mutations, we examined the doubly-compatible wild strain MY19 from Roxel, Germany. MY19 shows no lethality in crosses with Bristol and carries an intact zeel-1 sequence, but it also fails to induce paternal effect lethality in crosses with Hawaii, suggesting that it carries a Hawaii-like allele of peel-1 or a suppressor of the paternal effect. To test for the existence of an unlinked suppressor, we mapped the inability of MY19 to induce lethality of zeel-1Hawaii homozygotes relative to a marker 10 cM from the peel-1 interval (8). Absence of the paternal effect in MY19 mapped 10 cM from the marker (Table S6), suggesting that MY19 might carry a mutation in peel-1. We sequenced all coding regions contained within the peel-1 interval in MY19, as well as most non-coding regions located within the interval of elevated divergence between Bristol and Hawaii.
The MY19 haplotype is nearly identical to Bristol and contains only 12 non-synonymous polymorphisms, four of which fall in genes for which putative null alleles failed to abolish the paternal effect. Of the remaining polymorphisms, seven fall in Y39G10AR.15, a gene with spermatogenic germline expression (19) but located outside the divergent interval. The sequence of Y39G10AR.16, another gene with spermatogenic expression (19), but located within the divergent interval, contains no non-synonymous polymorphisms. Notably, the coding sequence of zeel-1MY19 is identical to zeel-1Bristol, suggesting that the paternal effect does not arise from the activity of zeel-1 itself or that if it does, MY19 and Bristol differ in their regulatory regions.
Discussion
We discovered a genetic incompatibility in C. elegans that causes lethality of embryos homozygous for a naturally occurring deletion of zeel-1 when sired by individuals heterozygous for the Bristol and Hawaii alleles of a tightly linked paternal effect gene, peel-1. Paternal effect mutations are rare, and only one has been described in C. elegans (20). The interaction between a paternal effect and a zygotically acting gene is surprising given that sperm-supplied factors are expected to act before zygotic transcription begins (9, 10). Zygotically expressed ZEEL-1 may be a molecular antidote to a sperm-carried PEEL-1 protein that is otherwise toxic during development. Analogous maternal effect-by-zygotic interactions have been described in Tribolium (21).
The Bristol haplotype of the zeel-1/peel-1 interval gains a transmission advantage by inducing lethality of embryos not inheriting it. This segregation distortion is characteristic of genic drive, in which selection at the level of alleles within an individual (genic selection) acts independently of selection at the level of individuals within a population (genotypic selection). Aside from this similarity, however, the C. elegans incompatibility does not conform to the expectations of genic drive. In drive systems, driver haplotypes are expected to fix within populations and are observed only where genotypic selection against them prevents fixation (22). Where this countervailing force is absent, drivers become fixed in a population and are detectable only in interpopulation crosses (23-27). In C. elegans, homozygotes of both haplotypes are fit, and incompatible haplotypes co-occur globally. One explanation for the long- term maintenance of both haplotypes is that genotypic selection favoring the Hawaii haplotype counterbalances genic drive favoring the other. Because C. elegans is primarily selfing, and drive can only influence transmission in heterozygotes, this model would require a precise and stable level of outcrossing. Although a lack of knowledge of C. elegans population biology prevents us from rejecting this model, we favor an alternative explanation not subject to such constraints.
While the drive model implies that balancing selection is a consequence of the incompatibility, an alternative is that the incompatibility is an incidental side-effect of balancing selection. For example, balancing selection on pathogen resistance genes with costs in the absence of infection may maintain two haplotypes within a population for much longer than would be possible under neutral drift (28, 29). If the haplotypes occur largely as homozygotes within a primarily selfing species, mutations arising on one haplotype are rarely tested against mutations on the other. Alleles fixed by drift or positive selection within the genetic context of their own haplotype but incompatible with alleles of another are not purged from the population but instead maintained alongside the balanced polymorphism, which acts as an incompatibility trap. Since incompatible alleles are only deleterious after outcrossing, selection favoring the balanced polymorphism may be of lesser magnitude than selection against the incompatibility. Because long-term maintenance of incompatible alleles depends on tight linkage to the balanced polymorphism, the interacting loci are necessarily tightly linked to one another as well.
Our model, like genic drive, explains the occurrence of long-lived, incompatible alleles at tightly linked loci, but it better incorporates the low level of outcrossing and the global cooccurrence of both haplotypes. While the cause of balancing selection remains unknown, all genes within the divergent interval carry multiple non-synonymous differences between the haplotypes, and several are known or predicted to interact with signals from the environment (30-32). The deletion of zeel-1 may be analogous to presence/absence polymorphisms of pathogen resistance genes in Arabidopsis, known targets of balancing selection (28, 29).
The C. elegans incompatibility suggests that long-term balancing selection in selfing species may facilitate the sympatric accumulation and maintenance of Dobzhansky-Muller type incompatibilities involving tightly linked loci. In the Dobzhansky-Muller model of speciation, incompatibilities emerge from the deleterious interactions of alleles that are neutral or advantageous in their own genetic backgrounds. While classic models predict incompatible alleles to occur in allopatric populations (33), the C. elegans incompatibility occurs within interbreeding populations and does not appear to precipitate speciation, as gene flow between the incompatible classes occurs throughout the rest of the genome. The C. elegans incompatibility may represent an example of incidental linkage between developmentally and ecologically important genes driving the evolution of development.
Supplementary Material
References and notes
- 1.Barriere A, Felix MA. Curr Biol. 2005 Jul 12;15:1176. doi: 10.1016/j.cub.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 2.Hodgkin J, Doniach T. Genetics. 1997 May;146:149. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barriere A, Felix MA. Genetics. 2007 Jun;176:999. doi: 10.1534/genetics.106.067223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivasundar A, Hey J. Curr Biol. 2005 Sep 6;15:1598. doi: 10.1016/j.cub.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 5.Denver DR, Morris K, Thomas WK. Mol Biol Evol. 2003 Mar;20:393. doi: 10.1093/molbev/msg044. [DOI] [PubMed] [Google Scholar]
- 6.Swan KA, et al. Genome Res. 2002 Jul;12:1100. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wicks SR, Yeh RT, Gish WR, Waterston RH, Plasterk RH. Nat Genet. 2001 Jun;28:160. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- 8.Supplementary Online Material.
- 9.L'Hernault SW. WormBook. The C. elegans Research Community; Feb 20, 2006. Spermatogenesis. doi/10.1895/wormbook.1.30.1, http://www.wormbook.org. [Google Scholar]
- 10.Gonczy P, Rose LS. WormBook. The C. elegans Research Community; Oct 15, 2005. Asymmetric cell division and axis formation in the embryo. doi/10.1895/wormbook.1.85.1, http://www.wormbook.org. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haber M, et al. Mol Biol Evol. 2005 Jan;22:160. doi: 10.1093/molbev/msh264. [DOI] [PubMed] [Google Scholar]
- 12.Koch R, van Luenen HG, van der Horst M, Thijssen KL, Plasterk RH. Genome Res. 2000 Nov;10:1690. doi: 10.1101/gr.gr-1471r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sivasundar A, Hey J. Genetics. 2003 Jan;163:147. doi: 10.1093/genetics/163.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Charlesworth D. PLoS Genet. 2006 Apr;2:e64. doi: 10.1371/journal.pgen.0020064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutter AD. Genetics. 2006 Jan;172:171. doi: 10.1534/genetics.105.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denver DR, Morris K, Lynch M, Thomas WK. Nature. 2004 Aug 5;430:679. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
- 17.Vasudevan S, Starostina NG, Kipreos ET. EMBO Rep. 2007 Mar;8:279. doi: 10.1038/sj.embor.7400895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ahringer J. WormBook. The C. elegans Research Community; Apr 6, 2006. Reverse genetics. doi/10.1895/wormbook.1.30.1, http://www.wormbook.org. [Google Scholar]
- 19.Reinke V, Gil IS, Ward S, Kazmer K. Development. 2004 Jan;131:311. doi: 10.1242/dev.00914. [DOI] [PubMed] [Google Scholar]
- 20.Browning H, Strome S. Development. 1996 Jan;122:391. doi: 10.1242/dev.122.1.391. [DOI] [PubMed] [Google Scholar]
- 21.Beeman RW, Friesen KS, Denell RE. Science. 1992 Apr 3;256:89. doi: 10.1126/science.1566060. [DOI] [PubMed] [Google Scholar]
- 22.Hurst GD, Werren JH. Nat Rev Genet. 2001 Aug;2:597. doi: 10.1038/35084545. [DOI] [PubMed] [Google Scholar]
- 23.Atlan A, Mercot H, Landre C, Montchamp-Moreau C. Evolution Int J Org Evolution. 1997;51:1886. doi: 10.1111/j.1558-5646.1997.tb05111.x. [DOI] [PubMed] [Google Scholar]
- 24.Beeman RW, Friesen KS. Heredity. 1999 May;82(Pt 5):529. doi: 10.1038/sj.hdy.6885150. [DOI] [PubMed] [Google Scholar]
- 25.Fishman L, Willis JH. Genetics. 2005 Jan;169:347. doi: 10.1534/genetics.104.032789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orr HA, Irving S. Genetics. 2005 Feb;169:671. doi: 10.1534/genetics.104.033274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reed FA, Reeves RG, Aquadro CF. Evolution Int J Org Evolution. 2005 Jun;59:1280. [PubMed] [Google Scholar]
- 28.Stahl EA, Dwyer G, Mauricio R, Kreitman M, Bergelson J. Nature. 1999 Aug 12;400:667. doi: 10.1038/23260. [DOI] [PubMed] [Google Scholar]
- 29.Tian D, Araki H, Stahl E, Bergelson J, Kreitman M. Proc Natl Acad Sci U S A. 2002 Aug 20;99:11525. doi: 10.1073/pnas.172203599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shapira M, et al. Proc Natl Acad Sci U S A. 2006 Sep 19;103:14086. doi: 10.1073/pnas.0603424103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas JH. Genome Res. 2006 Aug;16:1017. doi: 10.1101/gr.5089806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas JH. Genetics. 2006 Jan;172:127. doi: 10.1534/genetics.104.040030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orr HA, Presgraves DC. Bioessays. 2000 Dec;22:1085. doi: 10.1002/1521-1878(200012)22:12<1085::AID-BIES6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z. Comput Appl Biosci. 1997 Oct;13:555. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 35.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. Nucleic Acids Res. 2004 Jul 1;32:W273. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.We thank the Caenorhabditis Genetics Center, M.-A. Felix, A. Barriere, E. Dolgin, and H. Van Epps for strains, R. Maruyama and A. Singson for advice, S. Skrovanek for lab assistance, and H. Coller, D. Gresham, R. Gosh, L. Moyle, J. Shapiro, and E. Smith for comments on the manuscript. Supported by a NDSEG fellowship to HSS, a Jane Coffin Childs Fellowship to MVR, NIH grants R37 MH059520 and R01 HG004321 and a James S. McDonnell Foundation Centennial Fellowship to LK, and NIH grant GM071508 to the Lewis-Sigler Institute. GenBank sequence accession numbers EU163897- EU163940
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.