Abstract
Insecticidal proteins from the soil bacterium Bacillus thuringiensis (Bt) are becoming a cornerstone of ecologically sound pest management. However, if pests quickly adapt, the benefits of environmentally benign Bt toxins in sprays and genetically engineered crops will be short-lived. The diamondback moth (Plutella xylostella) is the first insect to evolve resistance to Bt in open-field populations. Here we report that populations from Hawaii and Pennsylvania share a genetic locus at which a recessive mutation associated with reduced toxin binding confers extremely high resistance to four Bt toxins. In contrast, resistance in a population from the Philippines shows multilocus control, a narrower spectrum, and for some Bt toxins, inheritance that is not recessive and not associated with reduced binding. The observed variation in the genetic and biochemical basis of resistance to Bt, which is unlike patterns documented for some synthetic insecticides, profoundly affects the choice of strategies for combating resistance.
The common soil bacterium Bacillus thuringiensis (Bt) produces crystals containing insecticidal proteins (1). These toxins kill insects by binding to and creating pores in midgut membranes (2). Each of the many strains of Bt produces a characteristic set of crystal proteins (1). Each of these proteins is extremely toxic to certain insects yet is harmless to most other organisms, including people, wildlife, and even other insects (1). Because of their highly specific toxicity, Bt toxins offer tremendous benefits for insect pest management.
Populations of more than 500 species of insects and mites have evolved resistance to insecticides and acaricides (3); laboratory selection experiments show that many pests can also adapt to Bt toxins (4). So far, documented cases of resistance to Bt in open-field populations of pests are limited to one insect species, the diamondback moth (Plutella xylostella) (4). However, greatly increased use of Bt toxins, whether delivered by conventional sprays or by genetically engineered crops, raises the likelihood that pests will adapt (4–6). Transgenic corn and cotton that express the lepidopteran-active Bt toxins Cry1Ab and Cry1Ac, respectively, were grown commercially in the United States for the first time during 1996. In 1997, Bt-expressing corn, cotton, and potatoes were planted on more than 3 million hectares in the United States (7). This major increase in exposure to Bt toxins intensifies selection for resistance in pests.
The goal of resistance management is to delay resistance in pests; success requires knowledge about the genetic and biochemical basis of resistance (8). For example, the widely recommended refuge/high-dose strategy is based on the idea that refuges from exposure to Bt permit susceptible insects to survive and mate with resistant insects emerging from nearby Bt crops. This strategy is expected to work best if resistance is recessive, so that hybrid offspring from resistant and susceptible parents are killed by the toxin in the Bt-expressing crop (4, 5, 9, 10). Previously reported evidence shows that in some strains of diamondback moth, resistance to some Bt formulations and toxins is inherited as a partially or completely recessive trait (11–15). The resistant NO-QA strain of diamondback moth from Hawaii harbors a recessive mutation that confers resistance to at least four Bt toxins (14). Although various mechanisms of resistance are possible (13, 16, 17), reduced binding of toxin to midgut membranes is the only mechanism of resistance to Bt known in diamondback moth (13, 15).
A key question in resistance management and evolutionary theory is whether separate populations arrive at the same genetic solution when faced with similar selection (18, 19). If this occurs, either by gene flow that spreads adaptive mutations (20) or by independent evolution, optimal strategies for managing resistance can be devised by analyzing a few resistant populations.
We tested this hypothesis by comparing resistant strains of diamondback moth isolated from Hawaii (NO-QA), Pennsylvania (PEN), and the Philippines (PHI). Previous comparisons among independently published studies suggested that differences among strains might occur (15, 21, 22), yet it was not clear if the observed variation was genetically based or caused by variation in environmental conditions and techniques among laboratories. Direct experimental comparisons reported here revealed key differences among strains of diamondback moth in the genetic and biochemical basis of resistance to Bt toxins that profoundly affect the choice of resistance management strategies.
MATERIALS AND METHODS
Insects.
Three resistant strains of diamondback moth were started from field populations (21, 23, 24) that had been sprayed repeatedly with commercial formulations of Bt subsp. kurstaki such as Dipel and Thuricide, which contain several toxins including Cry1Aa, Cry1Ab, and Cry1Ac (25). Before we compared the resistant strains, each had been selected with Bt in the laboratory to reduce the frequency of susceptible individuals. The NO-QA strain from Hawaii, which was derived from approximately 80 individuals sampled in August 1989 from a watercress farm in Pearl City on the island of Oahu, had been selected with Dipel more than 20 times (23). The PEN strain from Pennsylvania, which was derived from 280 moths emerging from a collection of 608 larvae taken in July 1993 from two collard fields in Newtown, had been selected with a mixture of Cry1Ac and Cry1C three times followed by three selections with only Cry1Ac. The PHI strain from the Philippines (21), which was derived from 130 pupae sent to the University of València from a sample collected in March 1993 near Baguio City on the island of Luzon, had been selected eight times with Cry1Ab. In addition, to reduce heterogeneity observed within PHI, larvae from PHI were exposed to Cry1Ab during this study. Adult survivors of this exposure were used in single-pair crosses; the F1 progeny of survivors were tested in bioassays. As internal standards, we used the susceptible LAB-P strain from Hawaii (22) in bioassays and the susceptible LAB-V strain, which was obtained from The Netherlands and reared at the University of València (21), in binding experiments.
Bioassays.
We tested the protoxin form of six toxins in bioassays: Cry1Aa, Cry1Ab, Cry1Ac, Cry1C, Cry1F, and Cry1J. We obtained Cry1A proteins from recombinant Escherichia coli HB101, which expressed either Cry1Aa, Cry1Ab, or Cry1Ac. Protoxin preparations were purified as intracellular inclusion bodies isolated from lysed cells (14). We used liquid formulations containing Cry1C (MYX833-4C1) or Cry1F (MYX837-446) that had been expressed in and encapsulated by transgenic Pseudomonas fluorescens (Mycogen, San Diego). Cry1J was obtained from Ecogen strain EG7279 (22).
Larvae were reared on cabbage and tested with bioassays at the University of Hawaii (22). Groups of 10 third instars were allowed to eat cabbage leaf disks that had been dipped in distilled water dilutions of individual Bt toxins (22). In all tests, the concentration of Cry1A toxins and Cry1J was 10 mg protoxin per liter; Cry1C and Cry1F were tested at 10 ml formulated toxin per liter (22). Mortality was recorded after 5 days and adjusted for mortality of larvae that ate control disks dipped in distilled water only. In all treatments, including distilled water controls, we added a surfactant (0.2% Triton AG98, from Rohm & Haas).
Evaluation of Maternal Effects, Sex Linkage, and Dominance.
We performed reciprocal single-pair crosses (14) between adults from each of the three resistant strains and the susceptible LAB-P strain. In every single-pair cross, one virgin female and male were caged together for mating and egg production (14). Each resulting family was reared separately on cabbage (14). In bioassays of these single-pair F1 families, broods were split so that offspring from each family were exposed to one of four toxins (Cry1Aa, Cry1Ab, Cry1Ac, or Cry1F) or to distilled water only as a control.
We evaluated maternal effects and sex linkage by comparing the results of reciprocal crosses. We estimated h, the dominance of resistance, using the previously described single-concentration method (26, 27) as follows: h = (v12 − v22)/ (v11 − v22), where v11, v12, and v22 are the relative viabilities of larvae from parental resistant strains, F1 hybrid progeny (pooled across single-pair families for each resistant strain × LAB-P), and the susceptible LAB-P strain, respectively. Values of h range from 0 (completely recessive resistance) to 1 (completely dominant resistance). This method of estimating dominance assumes that no genetic variation in susceptibility occurs within the parental resistant or susceptible strains. Genetic variation within the resistant or susceptible parental strains can bias such estimates of dominance downward or upward, respectively (14, 26). Because previous work revealed the presence of alleles for resistance within the susceptible LAB-P strain at a frequency of about 0.11 (14), we viewed values of h as first approximations of dominance. To obtain a more detailed assessment of dominance and genetic variation within parental strains, we analyzed results from individual families separately.
Genetic Correlations and Number of Loci.
To estimate genetic correlations of resistance between pairs of toxins, we used family mean correlations (28) based on analysis of arcsine-transformed mortality from split broods of the single-pair F1 families (14). If resistance to each toxin in a pair is controlled by independently segregating genes, no significant genetic correlation is expected. Strong genetic correlations imply that resistance to the toxins in a pair is controlled by a single locus or tightly linked loci (14). We used two-way analysis of variance (ANOVA) of arcsine-transformed mortality data from split broods of the single-pair families to test for significant variation in mortality among families, among toxins, and family-by-toxin interactions. Significant variation in mortality among families indicates genetic heterogeneity within one or both of the parental strains in the cross. A significant family-by-toxin interaction shows that resistance is not controlled entirely by a single locus. Results from NO-QA × LAB-P were reported previously (14) and are summarized here for comparisons with PEN × LAB-P and PHI × LAB-P.
Interstrain Complementation Tests for Allelism.
To determine if the locus or loci responsible for resistance to Bt toxins varied among populations, we performed complementation tests for allelism between resistant strains. Using bioassays, we tested the offspring of single-pair crosses from each pairwise combination of resistant strains. If two resistant strains are crossed, each with recessive alleles for resistance at separate loci, allelic complementation will restore susceptibility (the wild-type phenotype) in the progeny. However, if the recessive resistance alleles occur at the same locus in different populations, progeny will be resistant because they will inherit resistance alleles at the same locus from both parents. When resistance is not recessive, interstrain complementation tests are less useful because F1 progeny express some resistance even if the relevant loci differ among strains.
Binding Assays.
We performed binding assays with brush border membrane vesicles (BBMV) and four toxins: Cry1Aa, Cry1Ab, Cry1Ac, and Cry1C. BBMV were prepared from whole fourth instar insects (29) at the University of València by a slight modification of the differential magnesium precipitation method of Wolfersberger et al. (30). BBMV were frozen in liquid nitrogen and kept at −80°C until binding assays were conducted. The concentration of proteins in the BBMV preparations was determined by the method of Bradford (31) using BSA as standard.
Cry1A protoxins were obtained from recombinant Escherichia coli as described above. Cry1C provided by Ruud de Maagd was obtained from recombinant E. coli that expressed the cry1C gene from the 60.5 strain of B. thuringiensis subsp. entomocidus (40). Protoxins were solubilized and trypsin-activated using standard procedures (41). Labeling of trypsin-activated Cry1A toxins (25 μg each) with [125I]NaI (1 mCi for Cry1Ab and Cry1Ac; 0.5 mCi for Cry1Aa; 1 Ci = 37 GBq) was carried out with the chloramine-T method (32). Trypsin-activated Cry1C (25 μg) was labeled with [125I]NaI (0.5 mCi) using Iodo-Gen (Pierce) (33). Labeled toxins were separated from free iodine using BioGel P30 (Bio-Rad) columns. Specific activities were 0.63 mCi/mg for Cry1Aa, 1.9 mCi/mg for Cry1Ab, 2.1 mCi/mg for Cry1Ac, and 0.15 mCi/mg for Cry1C.
Binding experiments were performed in a final volume of 0.1 ml in binding buffer (8 mM Na2HPO4/2 mM KH2PO4/150 mM NaCl, pH 7.4/0.1% BSA) containing 1.1 nM 125I-labeled Cry1Aa, 1.5 nM 125I-labeled Cry1Ab, 1.0 nM 125I-labeled Cry1Ac, or 0.3 nM 125I-labeled Cry1C (34). After incubation for 30 min for Cry1A toxins or 90 min for Cry1C, bound toxins were separated from free toxins by filtration in glass-fiber filters (GF/F, Whatman). Filters were washed with 5 ml cold binding buffer, and the radioactivity was measured in a 1282 Compugamma CS Gamma Counter (LKB). Nonspecific binding values obtained with a 150- to 1,000-fold excess of nonlabeled toxin were subtracted from each data point.
RESULTS
Resistance and Cross-Resistance Spectrum.
The NO-QA and PEN strains were extremely resistant to Cry1Aa, Cry1Ab, Cry1Ac, Cry1F, and Cry1J (Table 1). Because neither strain had been exposed to Cry1F or Cry1J, decreased susceptibility to these toxins represents cross-resistance. In contrast, PHI was only partially resistant to the three Cry1A toxins and susceptible to Cry1F and Cry1J (Table 1). All strains were susceptible to Cry1C (Table 1).
Table 1.
Responses to Bt toxins by resistant strains (NO-QA, PEN, and PHI) and a susceptible strain (LAB-P) of diamondback moth
| Strain | Mean mortality, %*
|
|||||
|---|---|---|---|---|---|---|
| Cry1Aa | Cry1Ab | Cry1Ac | Cry1C | Cry1F | Cry1J | |
| LAB-P | 96.1 | 99.0 | 98.0 | 96.8 | 100.0 | 96.1 |
| NO-QA | 0.7 | 0.2 | 0.7 | 73.5 | 0.0 | 0.0 |
| PEN | 0.0 | 1.2 | 0.0 | >90† | 6.8 | <10† |
| PHI | 11.5 | 39.1 | 40.2 | 100.0 | 100.0 | 97.5 |
Mortality after 5 days was adjusted for control mortality. The concentration of each toxin was 10 mg toxin per liter except for Cry1C and Cry1F, which were tested at 10 ml formulated toxin per liter. Each test was replicated at least four times. Average total sample size was 142 larvae per toxin per strain.
Mortality of PEN in response to Cry1C and Cry1J was estimated from a related study (unpublished) in which artificial diet bioassays with PEN showed no resistance to Cry1C and >1,000-fold resistance to Cry1J relative to a susceptible strain.
Evaluation of Maternal Effects, Sex Linkage, and Dominance.
For each resistant strain, susceptibility did not differ between progeny derived from reciprocal crosses (e.g., PHI female × LAB-P male vs. LAB-P female × PHI male). These results indicate that maternal effects and sex linkage were not evident; resistance was autosomally inherited.
As reported previously for NO-QA × LAB-P (14), mortality of F1 larvae from PEN × LAB-P showed that resistance to Cry1Aa, Cry1Ab, Cry1Ac, and Cry1F was recessive in PEN (Table 2 and Fig. 1). Estimates of dominance (h) based on mean mortality pooled across families ranged from 0.06 to 0.13 for PEN (Table 2), which indicates almost complete recessiveness of resistance to each of the four toxins.
Table 2.
Mean mortality of F1 larvae from single-pair hybrid crosses and dominance (h)*
| Cross | Cry1Aa
|
Cry1Ab
|
Cry1Ac
|
Cry1F
|
||||
|---|---|---|---|---|---|---|---|---|
| Mortality, % | h | Mortality, % | h | Mortality, % | h | Mortality, % | h | |
| NO-QA × LAB-P | 75.6 | 0.26 | 88.5 | 0.12 | 82.0 | 0.18 | 79.0 | 0.20 |
| PEN × LAB-P | 85.2 | 0.13 | 89.6 | 0.11 | 88.0 | 0.11 | 92.3 | 0.06 |
| PHI × LAB-P | 38.5 | 0.70 | 87.1 | 0.22 | 64.1 | 0.48 | ND† | ND† |
We estimated h, the dominance of resistance, as follows: h = (v12 − v22)/(v11 − v22), where v11, v12, and v22 are the relative viabilities of larvae from parental resistant strains (Table 1), F1 progeny (n = 243–472 F1 larvae per toxin for each resistant strain × LAB-P), and LAB-P (Table 1), respectively. Values of h range from 0 (completely recessive resistance) to 1 (completely dominant resistance). Concentrations were 10 mg Cry1A toxin per liter and 10 ml formulated Cry1F per liter.
Not determined because PHI was susceptible to Cry1F.
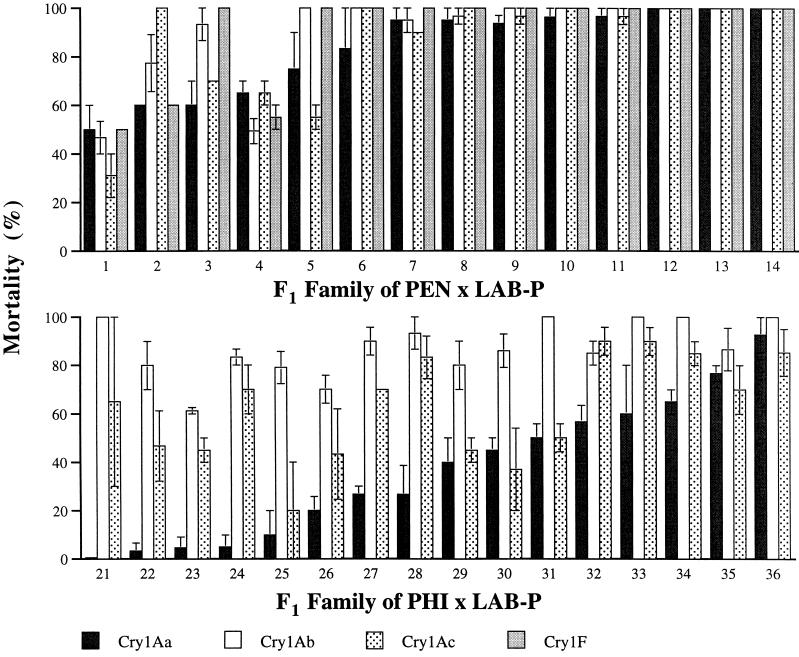
Figure 1.
Responses to Bt toxins from split broods of hybrid F1 progeny from single-pair crosses between strains of diamondback moth. (Upper) Resistant strain PEN × susceptible strain LAB-P (n = 1,605 larvae). (Lower) Resistant strain PHI × LAB-P (n = 1,451 larvae). Families are numbered from lowest (left) to highest mortality caused by Cry1Aa. Two or three groups of 9–11 larvae from each family were tested against each toxin (n = 59–121 larvae per family). Concentrations were 10 mg Cry1A toxin per liter and 10 ml formulated Cry1F per liter. Bars show mean mortality ± 1 SE. Genetic correlations were estimated from arcsine-transformed mortality data as described previously (14). Two-way ANOVA of the arcsine-transformed mortality data revealed significant effects of family (P < 0.0001 for both PEN and PHI), toxin (P = 0.0011 for PEN and P < 0.0001 for PHI), and family-by-toxin interaction (P < 0.0001 for PEN and P = 0.0002 for PHI).
In contrast to the recessive inheritance of resistance to all three Cry1A toxins observed in NO-QA and PEN, results with PHI show recessive inheritance of resistance to Cry1Ab (h = 0.22), but not to Cry1Aa (h = 0.70) or Cry1Ac (h = 0.48) (Table 2 and Fig. 1). Mortality caused by Cry1Aa ranged from 0 to 10% (mean = 4.6%) in 5 of the 16 single-pair F1 families derived from PHI × LAB-P (Fig. 1). These results show that PHI harbored at least one dominant mutation conferring resistance to Cry1Aa. Mortality caused by Cry1Ac to the 16 F1 families from PHI × LAB-P ranged from 21.1 to 90% (Fig. 1) with a mean of 64.1%, which suggests control by one or more semidominant mutations.
Genetic Correlations and Number of Loci.
As observed previously for NO-QA (14), PEN showed strong genetic correlations in resistance between all six pairwise combinations of Cry1Aa, Cry1Ab, Cry1Ac, and Cry1F (mean r = 0.75, df = 12, P < 0.05 for each pair). The simplest interpretation of this evidence is that, like NO-QA (14), PEN has a mutation at a single locus that confers resistance to these four toxins.
In contrast, resistance to Cry1F in PHI was not correlated with resistance to the three Cry1A toxins (Table 1). Further, in F1 families from PHI × LAB-P, genetic correlations in resistance between the three pairwise combinations of Cry1A toxins were weak (mean r = 0.51) and the correlation between resistance to Cry1Aa and Cry1Ab was not significant (r = 0.43, df = 14, P = 0.096). Family 21 derived from PHI × LAB-P, in which mortality was 0% for Cry1Aa and 100% for Cry1Ab (Fig. 1), provides striking evidence for independent genetic control of resistance to these two toxins in PHI (Fig. 1).
Mortality varied significantly among the 14 single-pair families derived from PEN × LAB-P: for families 1–5, mean mortality was 68.2%; for families 6–14, mean mortality was 98.3% (Fig. 1). This type of variation among families is similar to that observed previously from NO-QA × LAB-P and is consistent with the previous conclusion that some LAB-P parents were heterozygous for a multitoxin resistance gene (14). However, significant family-by-toxin interactions in mortality of split broods from PEN × LAB-P (Fig. 1) and NO-QA × LAB-P (14) suggest that, in addition to a multitoxin resistance mutation, PEN and NO-QA contain one or more other loci influencing resistance. For PEN × LAB-P, this interaction reflects, in part, unexpectedly low mortality caused by Cry1Ac in families 1, 3, and 5 and unexpectedly high mortality caused by Cry1Ac in family 2 (Fig. 1).
Interstrain Complementation Tests for Allelism.
F1 progeny from crosses between NO-QA and PEN were resistant to the three Cry1A toxins and Cry1F (range in mortality = 0–1.7%, n = 118–301 larvae per toxin). These results suggest that NO-QA and PEN share a genetic locus that controls multitoxin resistance. Further, these data and the responses of the parental strains (Table 1) suggest that NO-QA and PEN were almost completely homozygous for resistance at this locus.
Responses to Cry1Ab, the only toxin to which PHI showed recessive inheritance, varied among F1 progeny from PHI × NO-QA and PHI × PEN. Of 30 single-pair families (total n = 1070 larvae), 20 had mortality ranging from 0 to 15% (mean = 3.3%) and 10 had mortality ranging from 20 to 67% (mean = 43.7%). The 20 families with little or no mortality show that resistance to Cry1Ab in PHI was controlled at least in part by allelic variation at the same locus that controls multitoxin resistance in PEN and NO-QA. It appears that the 20 families with mortality close to 0% represent crosses in which the PHI parent was homozygous for resistance to Cry1Ab, whereas the others represent crosses in which the PHI parent was heterozygous. This explanation is supported by the correspondence between estimates of the frequency of Cry1Ab-resistant homozygotes in PHI based on the survival of PHI larvae (0.609, n = 652 larvae, Table 1) and the frequency of F1 families from PHI × NO-QA and PHI × PEN with low mortality (20/30 = 0.667) (X2 = 0.42, df = 1, P > 0.5).
Binding Assays.
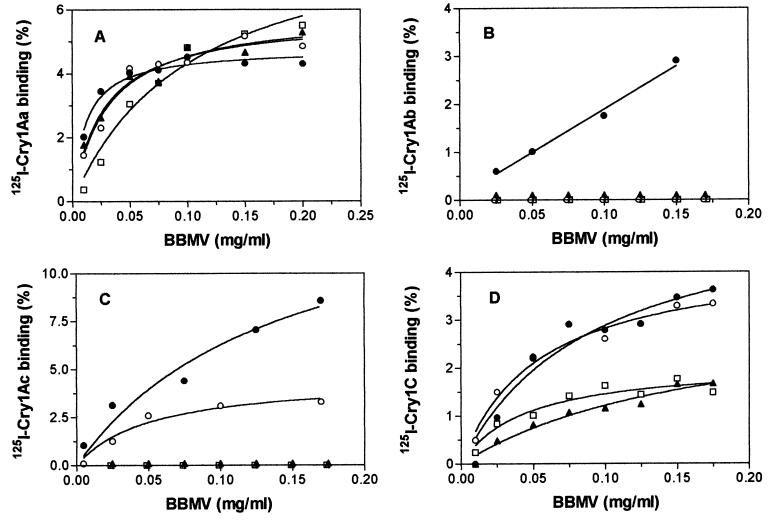
Binding tests with BBMV and 125I-labeled Bt toxins (Fig. 2) showed greatly reduced specific binding of Cry1Ab in the three resistant strains compared with the susceptible strain. In contrast, specific binding of Cry1Ac occurred in PHI but not in NO-QA or PEN. No obvious differences among strains were observed in the binding curves for Cry1Aa. Specific binding of Cry1C occurred in all four strains of diamondback moth.
Figure 2.
Specific binding of 125I-labeled Cry1Aa (A), Cry1Ab (B), Cry1Ac (C), and Cry1C (D) as a function of brush border membrane vesicle protein concentration for four strains of diamondback moth: LAB-V (•), NO-QA (□), PEN (▴), and PHI (○).
DISCUSSION
Our results show that the genetic and biochemical basis of resistance to Bt toxins varied among strains derived from widely separated populations of diamondback moth. Resistance to Bt in the PHI strain from the Philippines differed from that observed in the NO-QA strain from Hawaii and the PEN strain from Pennsylvania.
Similar to results from previous studies, we found correspondence between binding and bioassay data in some but not all cases. As expected, the susceptible strain bound all four toxins and all four strains bound Cry1C. Each of the three resistant strains showed greatly reduced binding of Cry1Ab, which implies that reduced binding of toxin to midgut membranes conferred resistance to Cry1Ab in these strains. The apparent lack of binding of Cry1Ab in PHI despite partial susceptibility to Cry1Ab in this strain might have been caused, in part, by a lower frequency of heterozygotes in the subpopulation tested in binding studies compared with the subpopulation tested in bioassays. Lack of specific binding was also associated with resistance to Cry1Ac in NO-QA and PEN but not in PHI.
Although the binding results reported here reveal some qualitative differences between strains, we view them cautiously because the technique used here provides only limited resolution. For example, the data reported here show no obvious differences among strains in binding of Cry1Aa. Nonetheless, competitive binding analyses in progress indicate that diamondback moth has two binding sites for Cry1Aa and that the one shared with Cry1Ab and Cry1Ac is altered in NO-QA and PEN but not in PHI (Ferré, unpublished data). Likewise, competitive binding tests will be needed to more fully understand results with other toxins.
In light of findings that Cry1Aa, Cry1Ab, Cry1Ac, and Cry1F compete for a common binding site in diamondback moth (21, 35), the data on inheritance of resistance and toxin binding reported here suggest that, in NO-QA and PEN, a recessive mutation or mutations at one locus reduce binding to this shared binding site. Although the results indicate the presence of a multitoxin resistance gene in NO-QA and PEN, significant family-by-toxin interactions in mortality observed in hybrid F1 offspring suggest that one or more additional loci influence resistance in both of these strains.
Results from complementation tests show that resistance to Cry1Ab in the PHI strain is controlled at least in part by allelic variation at the same locus that controls multitoxin resistance in NO-QA and PEN. However, the mutation in PHI at this locus does not confer resistance to Cry1F or Cry1Aa. Thus in PHI, mutations at two or more independent loci were needed to attain resistance to the three Cry1A toxins, and at least one additional mutation would be required for resistance to Cry1F. The binding data suggest that at least one mechanism other than reduced binding confers resistance to Cry1Aa and Cry1Ac in PHI. The pattern observed here in PHI and in resistant strains of diamondback moth from Malaysia (36) shows that binding of Cry1Ab can be altered without affecting binding of Cry1Aa or Cry1Ac.
The evidence shows that the multitoxin resistance mutations in NO-QA and PEN confer the same spectrum of resistance, exhibit the same patterns of dominance and genetic correlations, occur at the same locus, and share the same biochemical mechanism of resistance. The multitoxin resistance mutations in NO-QA and PEN are similar but not necessarily identical. In any case, the recent appearance of resistance to Bt and geographical isolation between source locations in Hawaii and Pennsylvania suggest that resistance arose independently in these populations. If so, this differs from worldwide spread of a single mutation by migration, which is the scenario reported for resistance to organophosphate insecticides in mosquitoes (20).
Diamondback moth resistance to Bt in Florida (15, 37) is similar to that from Hawaii and Pennsylvania described here. Because of their proximity, the probability of a common origin of resistance is greater for Florida and Pennsylvania than for the three widely separated sites compared in our analysis.
The PHI strain differed from NO-QA and PEN in cross-resistance to Cry1F and Cry1J, dominance of resistance to Cry1Aa and Cry1Ac, and binding of Bt toxins. The differences in dominance and cross-resistance profoundly affect the appropriate choice of tactics for managing resistance. For example, the widely recommended refuge/high-dose strategy (4, 5, 9, 10) is expected to work best when resistance is recessive, as seen in NO-QA and PEN, but may fail when resistance is not recessive, as seen with Cry1Aa and Cry1Ac in PHI. The dominant resistance to Cry1Aa observed in PHI is especially noteworthy because other cases of resistance to Bt toxins in Lepidoptera are not dominant (4, 13). Further, the success of rotations or combinations of toxins depends critically on patterns of cross-resistance, which differed among strains. Previous results with the tobacco budworm (Heliothis virescens) indicate that variation in resistance to Bt can occur among laboratory-selected strains (38, 39). Results with diamondback moth reported here show that variation in key resistance traits occurs among conspecific field populations and must be considered in the design of resistance-management strategies.
Acknowledgments
We thank T. Dennehy, P. Follett, B. Oppert, and Y. Park for comments and suggestions, and D. Coyle, K. Johnson, and N. Finson for technical assistance. Mycogen and Ruud de Maagd generously provided toxins. Funding was provided by the U.S. Department of Agriculture (TSTAR 95-34135-1771, NRI-CGP 96-35302-3470, and WRPIAP 97RA0304/0305-WR96-16) and the European Community (ECLAIR project AGRE-0003).
ABBREVIATIONS
- Bt
Bacillus thuringiensis
- BBMV
brush border membrane vesicles
References
- 1.Entwistle P, Bailey M J, Cory J, Higgs S, editors. Bacillus thuringiensis: An Environmental Biopesticide. New York: Wiley; 1993. [Google Scholar]
- 2.Gill S S, Cowles E A, Pietrantonio P V. Annu Rev Entomol. 1992;37:615–636. doi: 10.1146/annurev.en.37.010192.003151. [DOI] [PubMed] [Google Scholar]
- 3.Georghiou G P, Lagunes-Tejeda A. The Occurrence of Resistance to Pesticides in Arthropods. Rome: Food Agric. Org. U.N.; 1991. [Google Scholar]
- 4.Tabashnik B E. Annu Rev Entomol. 1994;39:47–79. [Google Scholar]
- 5.McGaughey W H, Whalon M E. Science. 1992;258:1451–1455. doi: 10.1126/science.258.5087.1451. [DOI] [PubMed] [Google Scholar]
- 6.Gould F, Anderson A, Jones A, Sumerford D, Heckel D, Lopez J, Micinski S, Leonard R, Laster M. Proc Natl Acad Sci USA. 1997;94:3519–3523. doi: 10.1073/pnas.94.8.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wadman M. Nature (London) 1997;388:817. [Google Scholar]
- 8.National Research Council. Pesticide Resistance: Strategies and Tactics for Management. Washington, DC: Natl. Acad. Press; 1986. [Google Scholar]
- 9.Georghiou G P, Taylor C E. J Econ Entomol. 1977;70:319–323. doi: 10.1093/jee/70.3.319. [DOI] [PubMed] [Google Scholar]
- 10.Liu Y B, Tabashnik B E. Proc R Soc Lond B. 1997;264:605–610. [Google Scholar]
- 11.Hama H, Suzuki K, Tanaka H. Appl Entomol Zool. 1992;27:355–362. [Google Scholar]
- 12.Tabashnik B E, Schwartz J M, Finson N, Johnson M W. J Econ Entomol. 1992;85:1046–1055. [Google Scholar]
- 13.Ferré J, Escriche B, Bel Y, Van Rie J. FEMS Microbiol Lett. 1995;132:1–7. [Google Scholar]
- 14.Tabashnik B E, Liu Y-B, Finson N, Masson L, Heckel D G. Proc Natl Acad Sci USA. 1997;94:1640–1644. doi: 10.1073/pnas.94.5.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tang J D, Gilboa S, Roush R T, Shelton A M. J Econ Entomol. 1997;90:732–741. [Google Scholar]
- 16.Oppert B, Kramer K, Beeman R W, Johnson D, McGaughey W H. J Biol Chem. 1997;272:23473–23476. doi: 10.1074/jbc.272.38.23473. [DOI] [PubMed] [Google Scholar]
- 17.Forcada C, Alcacer E, Garcera M D, Martinez R. Arch Insect Biochem Physiol. 1996;31:257–272. doi: 10.1002/(SICI)1520-6327(199909)42:1<51::AID-ARCH6>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 18.McKenzie J A, Batterham P. Trends Ecol Evol. 1994;9:166–169. doi: 10.1016/0169-5347(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 19.ffrench-Constant R H, Steichen J, Rocheleau T A, Aronstein K, Roush R T. Proc Natl Acad Sci USA. 1993;90:1957–1961. doi: 10.1073/pnas.90.5.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Raymond M, Callaghan A, Fort P, Pasteur N. Nature (London) 1991;350:151–153. doi: 10.1038/350151a0. [DOI] [PubMed] [Google Scholar]
- 21.Ballester V, Escriche B, Ménsua J L, Riethmacher G W, Ferré J. Biocontrol Sci Tech. 1994;4:437–443. [Google Scholar]
- 22.Tabashnik B E, Malvar T, Liu Y-B, Finson N, Borthakur D, Shin B-Y, Park S-H, Masson L, de Maagd R A, Bosch D. Appl Environ Microbiol. 1996;62:2839–2844. doi: 10.1128/aem.62.8.2839-2844.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabashnik B E, Finson N, Groeters F R, Moar W J, Johnson M W, Luo K, Adang M J. Proc Natl Acad Sci USA. 1994;91:4120–4124. doi: 10.1073/pnas.91.10.4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chang W X Z, Tabashnik B E, Artelt B, Malvar T, Ballester V, Ferré J, Roderick G. Ann Entomol Soc Amer. 1997;90:590–595. [Google Scholar]
- 25.Abbott Laboratories. Bt Products Manual. N. Chicago, IL: Abbott; 1992. [Google Scholar]
- 26.Liu Y B, Tabashnik B E. Appl Environ Microbiol. 1997;63:2218–2223. doi: 10.1128/aem.63.6.2218-2223.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hartl D L. A Primer of Population Genetics. 2nd Ed. Sunderland, MA: Sinauer Associates; 1992. [Google Scholar]
- 28.Via S. Evolution. 1984;38:896–905. doi: 10.1111/j.1558-5646.1984.tb00360.x. [DOI] [PubMed] [Google Scholar]
- 29.Escriche B, Silva F J, Ferré J. J Invert Pathol. 1995;65:318–320. [Google Scholar]
- 30.Wolfersberger M, Luthy P, Maurer A, Parenti P, Sacchi F V, Giordana B, Hanozet G M. Comp Biochem Physiol. 1987;86A:301–308. [Google Scholar]
- 31.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 32.Van Rie J, Jansens J S, Hofte H, Degheele D, van Mellaert H. Appl Environ Microbiol. 1990;56:1378–1385. doi: 10.1128/aem.56.5.1378-1385.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hofmann C, Lüthy R, Hütter R, Pliska V. Eur J Biochem. 1988;173:85–91. doi: 10.1111/j.1432-1033.1988.tb13970.x. [DOI] [PubMed] [Google Scholar]
- 34.Ferré J, Real M D, Van Rie J, Jansens S, Peferoen M. Proc Natl Acad Sci USA. 1991;88:5119–5123. doi: 10.1073/pnas.88.12.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Granero F, Ballester V, Ferré J. Biochem Biophys Res Commun. 1996;224:779–783. doi: 10.1006/bbrc.1996.1099. [DOI] [PubMed] [Google Scholar]
- 36.Wright D J, Iqbal M, Granero F, Ferré J. Appl Environ Microbiol. 1997;63:1814–1819. doi: 10.1128/aem.63.5.1814-1819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tang J D, Shelton A M, Van Rie J, de Roeck S, Moar W J, Roush R T, Peferoen M. Appl Environ Microbiol. 1996;62:564–569. doi: 10.1128/aem.62.2.564-569.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gould F, Martinez-Ramirez A, Anderson A, Ferré J, Silva F J, Moar W J. Proc Natl Acad Sci USA. 1992;89:7986–7990. doi: 10.1073/pnas.89.17.7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gould F, Anderson A, Reynolds A, Bumgarner L, Moar W. J Econ Entomol. 1995;88:1545–1559. [Google Scholar]
- 40.Visser B, Van der Salm T, Van den Brink W, Folkers G. Mol Gen Genet. 1988;212:219–224. [Google Scholar]
- 41.Masson L, Préfontaine G, Péloquin L, Lau P C K, Brousseau R. Biochem J. 1990;269:507–512. doi: 10.1042/bj2690507. [DOI] [PMC free article] [PubMed] [Google Scholar]