Abstract
Kinase suppressor of Ras (KSR) is an evolutionarily conserved component of Ras-dependent signaling pathways. Here, we find that murine KSR (mKSR1) translocates from the cytoplasm to the plasma membrane in the presence of activated Ras. At the membrane, mKSR1 modulates Ras signaling by enhancing Raf-1 activity in a kinase-independent manner. The activation of Raf-1 is mediated by the mKSR1 cysteine-rich CA3 domain and involves a detergent labile cofactor that is not ceramide. These findings reveal another point of regulation for Ras-mediated signal transduction and further define a noncatalytic role for mKSR1 in the multistep process of Raf-1 activation.
The Ras signaling cascade is a central pathway whereby many growth and developmental signals are transmitted from the cell surface to the nucleus. Recently, we have reported that a protein kinase named kinase suppressor of Ras (KSR) facilitates the transmission of signals from Ras to the Raf-1/mitogen and extracellular regulated kinase (MEK)/mitogen-activated protein kinase (MAPK) module (1). KSR was originally identified in genetic screens for downstream effectors of Ras, undertaken in Drosophila and Caenorhabditis elegans (2–4), and is an evolutionarily conserved component of Ras-dependent signaling pathways. Genetic studies in both Drosophila and C. elegans indicate that KSR plays a positive role in the transmission of Ras-mediated signals (2–4). These data are further supported by biochemical studies examining the function of mammalian KSR (1, 5). In Xenopus oocytes and cultured mammalian cells, murine KSR1 (mKSR1) cooperated with oncogenic RasV12 to promote meiotic maturation and cellular transformation, respectively (1). mKSR1 enhanced the biological activity of activated Ras by accelerating the kinetics of MEK1 and MAPK activation. Surprisingly, however, the cooperative function of mKSR1 was not contained within the catalytic domain but rather was localized to a 104-aa region that includes the conserved cysteine-rich CA3 domain (1).
In this report, we investigate the mechanism by which mKSR1 facilitates Ras-mediated signal transduction. We find that mKSR1 functions at the plasma membrane and modulates Ras signaling by augmenting Raf-1 activity in a kinase-independent manner. The cysteine-rich CA3 domain is required for both the stable interaction of mKSR1 at the plasma membrane and for the activation of Raf-1. In addition, we find that the activation of Raf-1 appears to involve a detergent-labile cofactor that is not ceramide. Furthermore, we are unable to corroborate recent findings suggesting that mKSR1 is a ceramide-activated protein kinase and that mKSR1 directly modulates Raf-1 activity by phosphorylation (6).
MATERIALS AND METHODS
Plasmids.
The mKSR1 CA3 domain construct was generated by PCR amplification of a DNA fragment corresponding to amino acids 319–390 of mKSR1, in which two copies of the polyoma virus-derived (Pyo) epitope tag (amino acids MEYMPME) were included in the 5′ primer immediately upstream of amino acid 319. The PCR product was then subcloned into a BamHI site in pcDNA3 (Invitrogen) and sequenced. The mKSR1 CA3 domain was released from pcDNA3 by BamHI digestion, and the fragment was subcloned into the BglII site of the SP64T polylinker. The B-Raf CRD domain (residues 235–280) was amplified by PCR using primers that included an AUG codon at the 5′ end of the domain and a FLAG epitope (IBI, Kodak) and a STOP codon at the 3′ end of the domain. The amplified DNA fragment was inserted into a pTZ18-derived plasmid. The Myr-KSR construct was generated by inserting a KpnI/BssHII double-stranded oligonucleotide encoding amino acids 1–16 of mouse c-Src (Myr; myristylation signal sequence) into the KpnI/BssHII cloning sites found immediately upstream of the Pyo tag in a pBacPak-8 (CLONTECH)/Pyo-mKSR1 construct (unpublished data). The resulting Myr-Pyo in-frame fusion product was sequenced, and a KpnI/NotI fragment (corresponding to Myr-Pyo-mKSR1) was then transferred into the KpnI/NotI cloning sites of pcDNA3.
RNA Transcription and Oocyte Microinjection.
Capped RNA was transcribed using the Message Machine kit (Ambion, Austin, TX). Buffer or RNA (≈30 ng) encoding the various mKSR1 constructs or a construct encoding the cysteine-rich domain of the B-Raf kinase (B-Raf CRD) was injected into defolliculated stage VI oocytes as described in ref. 1. After 8–12 hr, the oocytes were injected with RasV12 RNA and were subsequently scored for germinal vesicle breakdown (GVBD).
Cell Fractionation.
Plasmid DNAs (5 μg) were transiently transfected into 293 cells by the calcium phosphate method (1) or into COS cells with lipofectamine (6). Forty to forty-eight hours after transfection, cells were lysed in a ground glass homogenizer and fractionated into membranes and cytosol, or microinjected oocytes were fractionated into membrane and cytosolic fractions (7). Each fraction was assayed by immunoprecipitation for the presence of mKSR1 proteins or H-RasV12.
In Vitro Protein Kinase Assays and Ceramide-Level Determinations.
Raf-1 and MAP kinase (1), mKSR1 (6), and JNK (8) activities were assayed and ceramide levels were determined (9) as previously described.
RESULTS
The mKSR1 CA3 Domain Is Required for the Augmentation of Ras Signaling.
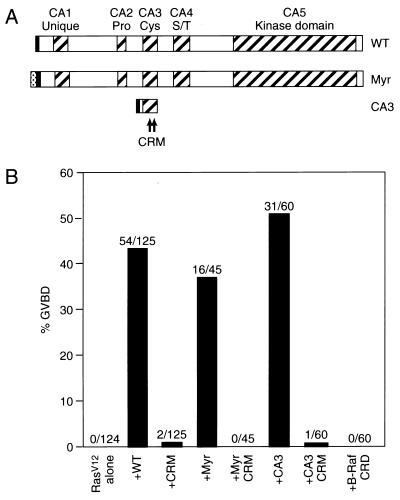
The CA3 domain of mKSR1 has sequence similarities with cysteine-rich domains (referred to as C1 domains) found in the protein kinase C (PKC) and the Raf family of protein kinases (10), and for these kinases the C1 domain plays an important role in their activation and signaling capabilities (11–17). To evaluate the significance of the CA3 domain to mKSR1 function, we disrupted the putative cysteine finger motif contained within this domain by mutating two of the conserved cysteine residues (C359S and C362S; referred to as CRM). The resulting mutant protein was then examined for its ability to augment Ras signaling in Xenopus oocyte meiotic maturation assays. As previously observed (1), expression of wild-type (WT) mKSR1 markedly accelerated oocyte maturation induced by RasV12 (Fig. 1B); however, CRM mKSR1 was unable to mediate this effect (Fig. 1B). Oocytes expressing CRM mKSR1 did undergo maturation with the same kinetics as did oocytes injected with RasV12 alone (unpublished observations), indicating that although disruption of the CA3 domain abolished mKSR1 cooperativity, it did not generate a dominant inhibitory protein. Interestingly, a construct encoding only the CA3 domain (amino acids 319–390) was fully competent to augment Ras signaling (Fig. 1B), and, as was observed for full-length mKSR1, incorporation of the CRM mutation into the CA3 domain protein abolished the cooperativity (Fig. 1B). The specificity of the mKSR1 CA3 domain to mediate this effect was demonstrated by the finding that expression of the C1 domain of the B-Raf kinase (B-Raf CRD) was unable to cooperate with RasV12 (Fig. 1B). Thus, the CA3 domain is necessary and sufficient for the functional interaction between mKSR1 and Ras that facilitates Ras signaling.
Figure 1.
The mKSR1 CA3 domain is required for the augmentation of Ras signaling. (A) Schematic representation of the WT, Myr, and CA3 mKSR1 proteins. The hatched boxes represent the five conserved areas (CA1–CA5) of the KSR family members. Pro, Cys, and S/T indicate that the corresponding CA regions are rich in proline, cysteine, or serine/threonine residues, respectively. The CRM mutation (C359S, C362S) is depicted by asterisks. For Myr mKSR1, the myristylation sequence from the src tyrosine kinase was inserted at the N terminus of WT mKSR1 (gray box). In addition, each construct contained a polyoma-derived (Pyo) epitope tag (solid box). (B) Induction of Xenopus oocyte meiotic maturation by the expression of RasV12 alone or by the coexpression of RasV12 with mKSR1 proteins. GVBD was scored when 0% of the oocytes expressing RasV12 alone and 40% of oocytes coexpressing RasV12 and WT mKSR1 had undergone GVBD. The percentage of oocytes undergoing GVBD is expressed as a solid bar, and the ratio of the number of oocytes undergoing GVBD to the total number injected is displayed above each bar. The numbers obtained represent a compilation of at least three independent experiments where equivalent amounts of the mKSR1 and RasV12 proteins were expressed.
The CA3 Domain Mediates the Ras-dependent Plasma Membrane Localization of mKSR1.
For various isoforms of PKC, the C1 domain is required for the localization of these kinases to the plasma membrane in response to activating agents (18–20). By analogy, therefore, we investigated the role of the CA3 domain in mediating the intracellular localization of mKSR1. Expressed alone in oocytes (unpublished observations) and in transiently transfected 293 cells (Fig. 2A), WT mKSR1 was found to be exclusively cytosolic. In the presence of activated RasV12, however, a portion of the WT protein was observed in the membrane fraction (Fig. 2A). In contrast, CRM mKSR1 was not detected in the membrane fraction even when coexpressed with RasV12 (Fig. 2B). These findings indicate that an intact CA3 domain is required for the stable interaction of mKSR1 at the plasma membrane and further support the idea that the functional interaction observed between mKSR1 and Ras occurs at the plasma membrane.
Figure 2.
The mKSR1 CA3 domain is required for the Ras-dependent membrane localization of mKSR1. (A) 293 cells transiently expressing WT and Myr mKSR1 or coexpressing WT and RasV12 were fractionated into membrane and cytosolic fractions. The mKSR1 proteins were immunoprecipitated and examined by immunoblot analysis using αPyo antibody. (B) 293 cells coexpressing RasV12 and either WT or CRM mKSR1 proteins were analyzed as in A.
Because constitutive membrane localization activates Raf-1 (21, 22) and, like Raf-1, a fraction of mKSR1 translocates to the plasma membrane in the presence of activated Ras, we next examined whether membrane localization was sufficient to activate mKSR1. A myristylated version of mKSR1 (Myr-KSR) was generated that constitutively localized mKSR1 to the plasma membrane in the absence of activated Ras (Fig. 2A). Although Myr-KSR did accelerate RasV12-induced maturation (Fig. 1B), expression of Myr-KSR alone in Xenopus oocytes was unable to promote oocyte maturation (unpublished observations). In addition, the CRM mutation abolished the cooperative effect of Myr-KSR (Fig. 1B). These results suggest that constitutive membrane localization does not generate an activated form of mKSR1 and cannot restore cooperativity to proteins containing the CRM mutation. Therefore, in addition to its role in membrane localization, the mKSR1 CA3 domain appears to provide yet another function required for the augmentation of Ras signaling.
The CA3 Domain Stimulates Membrane-Bound Raf-1 Activity.
Drosophila genetic epistasis experiments have positioned KSR to function either upstream of or in parallel to Raf (2), suggesting that KSR might be a kinase involved in Raf-1 activation. In addition, in mammalian cells, the isolated kinase domain of mKSR1 has been found to physically interact with Raf-1 at the plasma membrane (1). However, we have been unable to demonstrate that Raf-1 is a direct substrate of mKSR1 in vitro, and in previous studies examining mKSR1 function in Xenopus oocytes, no modulation in Raf-1 activity was detected, although enhanced activation of MEK1 and MAPK was observed (1). Nevertheless, it is possible that mKSR1 may influence Raf-1 activity as it is being activated at the membrane, which may not be readily detectable when Raf-1 activity is monitored from detergent-lysed cells in immune complex kinase assays. Therefore, a more careful examination of the activity of membrane-associated Raf-1 was performed. Oocytes expressing Raf-1 or Raf-1 and the CA3 domain were injected with RNA encoding activated RasV12. Detergent-free or detergent-solubilized membrane fractions were then prepared (7) immediately after (0 min) or 150 min after the injection of RasV12 RNA, a time before MEK1 activation and when the cooperative effect of mKSR1 would be expected to influence Raf-1 activity. At the 0-min time point, before RasV12 expression, no Raf-1 protein or activity was observed in either membrane preparation (Fig. 3A). At the 150-min point, when processed Ras and the CA3 domain were present in the membrane fractions (Fig. 3B), equivalent levels of Raf-1 activity (as measured by the phosphorylation of kinase-inactive MEK1 on Ser-218 and Ser-222) was detected in detergent-solubilized membranes, regardless of the presence of CA3 (Fig. 3A). Interestingly, under detergent-free conditions, a 5-fold elevation in MEK1 phosphorylation was observed in membranes containing CA3. The enhancement in activity was not merely a consequence of the extraction procedure, because Raf-1 activity was equivalent in both membrane preparations lacking CA3 (Fig. 3A). Thus, the mKSR1 CA3 domain appears to augment the activity of membrane-bound Raf-1 in a detergent-sensitive manner, suggesting the existence of a detergent-labile cofactor.
Figure 3.
The mKSR1 CA3 domain augments Raf-1 activity in a detergent-sensitive manner. Xenopus oocytes expressing Raf-1 alone (−) or coexpressing Raf-1 and the mKSR1 CA3 domain (+) were injected with RasV12 RNA. Immediately after (0 min) or 150 min after RasV12 injection, oocytes were lysed in hypotonic buffer and membranes were isolated. (A) Raf-1 proteins were immunoprecipitated from membrane fractions resuspended in RIPA buffer (+ detergent) or phosphate-buffered saline (− detergent), and in vitro kinase assays were performed using kinase-inactive MEK as a substrate (1). Phosphorylation of MEK1 on Ser-218 and Ser-222 was determined by tryptic peptide mapping analysis. (B) RasV12 and CA3 proteins were immunoprecipitated from membrane (P100) and cytosolic fractions (S100) isolated at 150 min after injection with RasV12 RNA and were examined by immunoblot analysis using Ras and Pyo antibody, respectively. The migration of processed (Pro) and unprocessed (UnPro) Ras proteins is indicated.
Ceramide Does Not Modulate mKSR1 Activity.
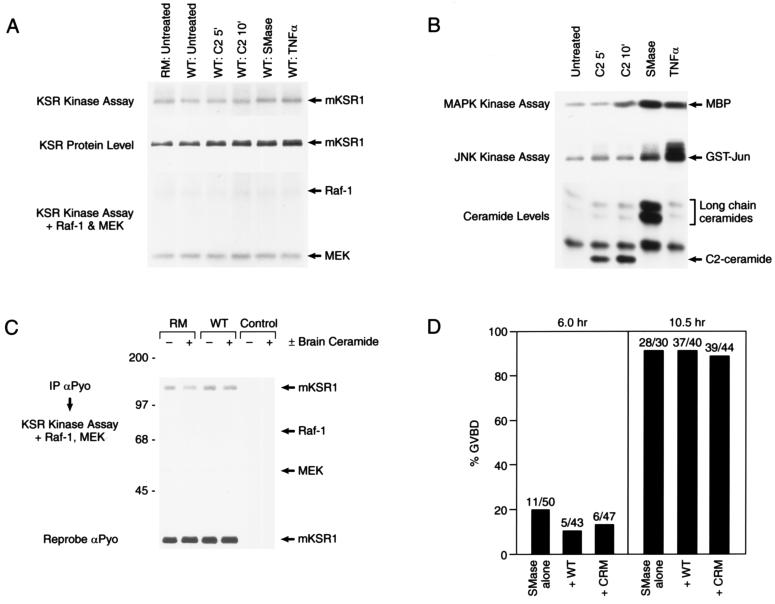
The cysteine-rich CA3 domain of mKSR1 resembles the atypical class of C1 domains (10). Proteins containing atypical C1 domains do not bind diacylglycerol or phorbol esters; however, they have been reported to bind other lipid second messengers, such as phosphatidic acid, phosphatidylinositol 3,4,5-P3, and ceramide (23–27). Because a recent report has suggested a role for ceramide in mKSR1 activation (6), we next investigated whether the cooperative effect mediated by the CA3 domain might involve ceramide. Cos cells transiently expressing mKSR1 were treated with C2 ceramide or were treated with sphingomyelinase or tumor necrosis factor α (TNFα), both of which generate ceramide in vivo (28). In response to the various treatments, endogenous levels of ceramide were elevated and changes in JNK and MAPK activity were detected (Fig. 4B), demonstrating that a biological response had been elicited by these factors (9, 29). When mKSR1 was examined in immune complex kinase assays, phosphorylation of mKSR1 was observed, although no significant change was detected in response to increased levels of ceramide (Fig. 4A). In addition, both kinase-inactive (RM) and WT mKSR1 proteins were phosphorylated, indicating the presence of a contaminating kinase activity associated with the mKSR1 immunoprecipitates (Fig. 4A). When exogenous Raf-1 was added to the mKSR1 immunoprecipitates, no phosphorylation of Raf-1 or modulation of Raf-1 activity was detected (Fig. 4A), suggesting that Raf-1 is not a direct substrate of mKSR1. Nevertheless, because the cooperative effect of mKSR1 observed in oocytes was detergent-sensitive and might not be observed under immunoprecipitation conditions, the effect of adding purified brain ceramide directly to mKSR1 and Raf-1 in vitro was examined. Again, mKSR1 phosphorylation was not increased, and no effect of mKSR1 on Raf-1 activity was observed in the presence of ceramide (Fig. 4C). Finally, the effect of mKSR1 on sphingomyelinase/ceramide-mediated oocyte maturation was examined (30). Like expression of RasV12, sphingomyelinase treatment did promote oocyte maturation; however, unlike RasV12-mediated GVBD, the presence of WT mKSR1 did not accelerate sphingomyelinase-induced maturation, nor was an effect of CRM mKSR1 observed (Fig. 4D). Taken together, our data strongly indicate that the activational effect mediated by the mKSR1 CA3 domain does not involve ceramide. Further, our data do not support the recent findings that mKSR1 is a ceramide-activated protein kinase and that Raf-1 is a direct substrate of mKSR1 (6).
Figure 4.
Augmentation of Ras signaling by mKSR1 does not involve ceramide. (A) Cos cells were transiently transfected with constructs encoding wild-type (WT) or kinase-inactive (RM) mKSR1. At 60 hr posttransfection, serum starved cells were left untreated or were stimulated with 20 μM C2 ceramide for 5 or 10 min, 100 milliunits/ml sphingomyelinase (SMase) for 20 min or 10 nM tumor necrosis factor α (TNFα) for 20 min. KSR proteins were immunoprecipitated using αPyo antibody, and mKSR1 immune complex kinase assays were performed in vitro as described by Zhang et al. (ref. 19; Top). Immunoprecipitated mKSR1 was detected by immunoblot analysis (Middle). To observe phosphorylation of Raf-1 or modulation of Raf-1 activity, purified activated Raf-1, coexpressed in Sf9 cells in the presence of RasV12 and v-src, and kinase-inactive MEK1 were added to the mKSR1 immune complex kinase assays previously described (ref. 19; Bottom). (B) Cos cells were treated as in A and endogenous ceramide levels, JNK activity, and MAPK activity were determined. Ceramide levels were normalized to the untreated control. C2-ceramide levels were elevated 2.9- and 3.7-fold at 5 and 10 min, respectively, and long-chain ceramide levels were elevated 12-fold by SMase and 1.9-fold by TNFα. (C) Purified brain ceramide (100 nM) (+) or diluent (−) was added in vitro to mKSR proteins immunoprecipitated from transfected Cos cells and immune complex kinase assays performed in the presence of activated Raf-1 and kinase-inactive MEK1 as previously described (ref. 19; Top). Immunoprecipitated mKSR1 was detected by immunoblot analysis (Middle). (D) Oocytes preinjected with buffer or RNA encoding WT and CRM mKSR1 constructs were treated with 250 milliunits sphingomyelinase. GVBD was then scored 6 and 10.5 hr after treatment.
DISCUSSION
In this report, we have investigated the mechanism whereby mKSR1 facilitates Ras signaling and have examined the importance of the CA3 domain to mKSR1 function. We find that mKSR1 is a cytoplasmic protein that translocates to and functions at the plasma membrane in the presence of activated Ras. The translocation of mKSR1 to the plasma membrane in a Ras-dependent manner is consistent with the recent report that mKSR1 localizes to the membrane in response to serum stimulation of quiescent NIH 3T3 cells (5). The signal that induces the translocation of mKSR1 has not been elucidated; however, unlike Raf-1, which localizes to the plasma membrane by directly binding activated Ras (31), we have been unable to detect a specific direct interaction between mKSR1 and Ras (data not shown). Once at the membrane, our data indicates that mKSR1 modulates Ras signaling, at least in part, by augmenting Raf-1 activity in a detergent-sensitive, kinase-independent manner (Fig. 3). The cysteine-rich CA3 domain is essential for the ability of mKSR1 to facilitate Ras signaling (Fig. 1) and functions to both enhance Raf-1 activity (Fig. 3) and to stably localize mKSR1 to the plasma membrane (Fig. 2).
One model to explain the biological activity of the CA3 domain is that both the translocation of mKSR1 and the activation of Raf-1 involves an interaction with a lipid second messenger. Support for this model comes from the observations that lipid second messenger production is increased in response to Ras activation (32–34) and that a lipid cofactor has been implicated in the Raf-1 activation process (15, 35). Raf-1 activity has been shown to be augmented in vitro by the addition of membranes from v-Ras- and v-Src-transformed fibroblasts (35) and by the addition of lipids extracted from membranes of SF9 expressing c-H-Ras and activated Src (15). Furthermore, proteins containing C1 domain have been shown to interact with lipids (10). The CA3 domain of mKSR1 resembles atypical C1 domains, found in the atypical forms of PKC (ζ and λ/ι), Raf-1, n-chimaerin, as well as the nonkinase oncoproteins Vav and Lfc (10). Like typical C1 domains, the structure of atypical C1 domains is coordinated by two molecules of zinc. Yet, unlike typical C1 domains, atypical C1 domains do not bind diacylglycerol or phorbol esters. However, other lipid second messengers, such as phosphatidic acid, phosphatidylinositol 3,4,5-P3, and phosphatidylserine, have been implicated to interact with PKCζ, PKCλ/ι and Raf-1 (23–25, 36–38). Therefore, the mKSR1 CA3 domain may modulate Raf-1 activity by increasing the local concentration of one of the above or a yet to be identified stimulatory lipid cofactor.
Ceramide is another lipid second messenger that has been reported to interact with proteins containing atypical C1 domains (26, 37), yet our studies strongly indicate that the stimulatory effect of mKSR1 on Raf-1 activity does not involve ceramide (Fig. 4). These findings are in contrast to those of a recent study (6) that identifies mKSR1 to be a ceramide-activated protein (CAP) kinase and that finds Raf-1 to be a direct substrate of mKSR1 activity. In the experiments presented here, the addition of ceramide either in vivo or in vitro had no effect on mKSR1 phosphorylation as measured by immune complex kinase assays (Fig. 4A). In addition, when Raf-1 was added to mKSR1 as an exogenous substrate, no phosphorylation of Raf-1 or modulation in Raf-1 activity was observed (Fig. 4A), suggesting that Raf-1 is not a substrate of mKSR1. Furthermore, if mKSR1 is a CAP kinase that phosphorylates and activates the Raf-1/MEK1/MAPK signaling module, then one would predict that overexpression of mKSR1 in Xenopus oocytes would accelerate the kinetics of sphingomyelinase/ceramide-mediated meiotic maturation. However, no cooperativity was observed between mKSR1 and sphinomyelinase treatement in oocytes (Fig. 4D), even though mKSR1 readily cooperated with RasV12 in this assay (ref. 1; Fig. 1). Moreover, the functional interaction between mKSR1 and activated Ras supports the genetic data indicating that KSR1 is a positive effector of Ras-dependent signaling in both Drosophila (2) and C. elegans (3, 4).
In contrast to a model evoking a stimulatory lipid cofactor, the CA3 domain of mKSR1 may activate Raf-1 by facilitating the binding of an auxiliary protein that augments Raf-1 activity. The best-characterized example of a membrane-localized protein that modulates Raf-1 activity is Ras (39). The Ras-dependent activation of Raf-1 involves the interaction of Raf-1 with posttranslationally modified, GTP-bound Ras (16, 40–42), and although mKSR1 does not appear to directly interact with Ras GTP, the CA3 domain might still play a role in promoting or stabilizing the Ras/Raf-1 interaction. In addition to Ras, other proteins have been shown to interact with C1 domains (43–45) and may be involved in mediating the CA3 domain effect.
Finally, we cannot exclude the possibility that the CA3 domain functions to stabilize Raf-1 in an active conformation within the context of the membrane environment in a manner that is independent of a lipid or protein cofactor. However, the inability of myristylated CRM mKSR1 to cooperate with RasV12 (Fig. 1) suggests that residence in the membrane alone is not sufficient to explain the biological effects of mKSR1. Thus, further investigation is required to determine the precise mechanism by which the mKSR1 CA3 domain enhances Raf-1 activity. Regardless of the precise mechanism involved in mediating the CA3 domain effect, however, this study does reveal another level of complexity involved in the transmission of signals from Ras to the MAPK module and further defines a noncatalytic role for mKSR1 in the multistep process of Raf-1 activation (39).
Acknowledgments
We thank Robert M. Stephens for providing the B-Raf CRD construct, Olivier Cuvillier for expert advice concerning lipid preparation, Karen Mathes for technical assistance, and members of the Morrison and Rubin laboratories for advice during the course of this work. This work was supported in part by the National Cancer Institute, Department of Health and Human Services, under contract with Advanced BioScience Laboratories (N.R.M., A.C., and D.K.M.), the Medical Research Council of Canada (M.T.), and the Howard Hughes Medical Institute (G.M.R.).
ABBREVIATIONS
- KSR
kinase suppressor of Ras
- PKC
protein kinase C
- WT
wild type
- GVBD
germinal vesicle breakdown
- MEK
mitogen and extracellular regulated kinase
References
- 1.Therrien M, Michaud N R, Rubin G M, Morrison D K. Genes Dev. 1996;10:2684–2695. doi: 10.1101/gad.10.21.2684. [DOI] [PubMed] [Google Scholar]
- 2.Therrien M, Chang H C, Solomon N M, Karim F D, Wassarman D A, Rubin G M. Cell. 1995;83:879–888. doi: 10.1016/0092-8674(95)90204-x. [DOI] [PubMed] [Google Scholar]
- 3.Sundaram M, Han M. Cell. 1995;83:889–901. doi: 10.1016/0092-8674(95)90205-8. [DOI] [PubMed] [Google Scholar]
- 4.Kornfeld K, Hom D B, Horvitz H R. Cell. 1995;83:903–913. doi: 10.1016/0092-8674(95)90206-6. [DOI] [PubMed] [Google Scholar]
- 5.Xing H, Kornfeld K, Muslin A J. Curr Biol. 1997;7:294–300. doi: 10.1016/s0960-9822(06)00152-7. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Yao B, Delikat S, Bayoumy S, Lin X-H, Basu S, McGinley M, Chan-Hui P-Y, Lichenstein H, Kolesnick R. Cell. 1997;89:63–72. doi: 10.1016/s0092-8674(00)80183-x. [DOI] [PubMed] [Google Scholar]
- 7.Evans J P, Kay B K. In: Methods in Cell Biology. Kay B K, Peng H B, editors. Vol. 36. San Diego, CA: Academic; 1991. pp. 142–143. [Google Scholar]
- 8.Verheij M, Bose R, Lin H X, Yao B, Jarvis W D, Grant S, Birrer M J, Szabo E, Zon L I, Kyriakis J M, Haimovitz-Friedman A, Fuks Z, Kolesnick R N. Nature (London) 1996;380:75–79. doi: 10.1038/380075a0. [DOI] [PubMed] [Google Scholar]
- 9.Cullivier O, Pirianov G, Kleuser B, Vanek P G, Coso O A, Gutkind J S, Spiegel S. Nature (London) 1996;381:800–803. doi: 10.1038/381800a0. [DOI] [PubMed] [Google Scholar]
- 10.Hurley J H, Newton A C, Parker P J, Blumberg P M, Nishizuka Y. Protein Sci. 1997;6:477–480. doi: 10.1002/pro.5560060228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruder J T, Heidecker G, Rapp U R. Genes Dev. 1992;6:545–556. doi: 10.1101/gad.6.4.545. [DOI] [PubMed] [Google Scholar]
- 12.Zhang X-F, Settleman J, Kyriakis J M, Takeuchi-Suzuki E, Elledge S J, Marshall M S, Bruder J T, Rapp U R, Avruch J. Nature (London) 1993;364:308–313. doi: 10.1038/364308a0. [DOI] [PubMed] [Google Scholar]
- 13.Ono Y, Fujii T, Igarashi K, Kuno T, Tanaka C, Kikkawa U, Nishizuka Y. Proc Natl Acad Sci USA. 1989;86:4868–4871. doi: 10.1073/pnas.86.13.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shieh H-L, Hansen H, Zhu J, Riedel H. Mol Carcinogen. 1995;12:166–176. doi: 10.1002/mc.2940120308. [DOI] [PubMed] [Google Scholar]
- 15.Dent P, Reardon D B, Morrison D K, Sturgill T W. Mol Cell Biol. 1995;15:4125–4135. doi: 10.1128/mcb.15.8.4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo Z, Diaz B, Marshall M S, Avruch J. Mol Cell Biol. 1997;17:46–53. doi: 10.1128/mcb.17.1.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton, A. C. Curr. Opin. Cell Biol. 9, 161–167.
- 18.Schwartz J H. Proc Natl Acad Sci USA. 1993;90:8310–8313. doi: 10.1073/pnas.90.18.8310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang G, Kazanietz M G, Blumberg P M, Hurley J H. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
- 20.Szallasi Z, Bogi K, Gohari S, Biro T, Acs P, Blumberg P M. J Biol Chem. 1996;271:18299–18301. doi: 10.1074/jbc.271.31.18299. [DOI] [PubMed] [Google Scholar]
- 21.Stokoe D, MacDonald S G, Cadwallader K, Symons M, Hancock J F. Science. 1994;264:1463–1467. doi: 10.1126/science.7811320. [DOI] [PubMed] [Google Scholar]
- 22.Leevers S J, Paterson H F, Marshall C F. Nature (London) 1994;369:411–414. doi: 10.1038/369411a0. [DOI] [PubMed] [Google Scholar]
- 23.Nakanishi H, Exton J H. J Biol Chem. 1992;267:16347–16354. [PubMed] [Google Scholar]
- 24.Nakanishi H, Brewer K A, Exton J H. J Biol Chem. 1993;268:13–16. [PubMed] [Google Scholar]
- 25.Lozano J E, Berra E, Municio M M, Diaz-Meco M T, Dominguez I, Sanz L, Moscat J. J Biol Chem. 1994;269:19200–19202. [PubMed] [Google Scholar]
- 26.Huwiler A, Brunner J, Hummel R, Ver Voordeldonk M, Stabel S, Van Den Bosch H, Pfeilschifter J. Proc Natl Acad Sci USA. 1996;93:6959–6963. doi: 10.1073/pnas.93.14.6959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muller G, Ayoub M, Storz P, Rennecke J, Fabbro D, Pfizenmaier K. EMBO J. 1995;14:1961–1969. doi: 10.1002/j.1460-2075.1995.tb07188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hannun Y A. J Biol Chem. 1994;269:3125–3128. [PubMed] [Google Scholar]
- 29.Kyriakis J M, Banerjee P, Nikolakaki E, Dai T, Rubie E A, Ahmad M F, Avruch J, Woodgett J R. Nature (London) 1994;369:156–160. doi: 10.1038/369156a0. [DOI] [PubMed] [Google Scholar]
- 30.Strum J C, Swenson K I, Turner J E, Bell R M. J Biol Chem. 1995;270:13541–13547. doi: 10.1074/jbc.270.22.13541. [DOI] [PubMed] [Google Scholar]
- 31.Moodie S A, Wolfman A. Trends Genet. 1994;10:44–48. doi: 10.1016/0168-9525(94)90147-3. [DOI] [PubMed] [Google Scholar]
- 32.Wolfman A, Macara I G. Nature (London) 1987;325:359–361. doi: 10.1038/325359a0. [DOI] [PubMed] [Google Scholar]
- 33.Rodriguez-Viciana P, Warne P H, Dhand R, Vanhaesebroeck B, Gout I, Fry M J, Waterfield M D, Downward J. Nature (London) 1994;370:527–532. doi: 10.1038/370527a0. [DOI] [PubMed] [Google Scholar]
- 34.Martin A, Duffy P A, Liossis C, Gomez-Munoz A, O’Brien L, Stone J C, Brindley D N. Oncogene. 1997;14:1571–1580. doi: 10.1038/sj.onc.1200987. [DOI] [PubMed] [Google Scholar]
- 35.Dent P, Sturgill T W. Proc Natl Acad Sci USA. 1994;91:9544–9548. doi: 10.1073/pnas.91.20.9544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ghosh S, Zie W Q, Quest A F G, Mabrouk G M, Strum J C, Bell R M. J Biol Chem. 1994;269:10000–10007. [PubMed] [Google Scholar]
- 37.Mott H R, Carpenter J W, Zhong S, Ghosh S, Bell R M, Campbell S L. Proc Natl Acad Sci USA. 1996;93:8312–8317. doi: 10.1073/pnas.93.16.8312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh S, Strum J C, Sciorra V A, Daniel L, Bell R M. J Biol Chem. 1996;271:8472–8480. doi: 10.1074/jbc.271.14.8472. [DOI] [PubMed] [Google Scholar]
- 39.Morrison D K, Cutler R E., Jr Curr Opin Cell Biol. 1997;9:174–179. doi: 10.1016/s0955-0674(97)80060-9. [DOI] [PubMed] [Google Scholar]
- 40.Hu C, Kariya K, Tamada M, Akasaka K, Shirouzu M, Kataoka T. J Biol Chem. 1995;270:30274–30277. doi: 10.1074/jbc.270.51.30274. [DOI] [PubMed] [Google Scholar]
- 41.Lerner E C, Qian Y, Blaskovich M A, Fossum R D, Vogt A, Sun J M, Cox A D, Der C J, Hamilton A D, Sebti S M. J Biol Chem. 1995;270:26802–26806. doi: 10.1074/jbc.270.45.26802. [DOI] [PubMed] [Google Scholar]
- 42.Stokoe D, McCormick F. EMBO J. 1997;16:2384–2396. doi: 10.1093/emboj/16.9.2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Michaud N R, Fabian J R, Mathes K D, Morrison D K. Mol Cell Biol. 1995;15:3390–3397. doi: 10.1128/mcb.15.6.3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diaz-Meco M T, Municio M M, Sanchez P, Lozano J, Moscat J. Mol Cell Biol. 1996;16:105–114. doi: 10.1128/mcb.16.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diaz-Meco M T, Municio M M, Frutos S, Sanchez P, Lozano J, Sanz L, Moscat J. Cell. 1996;86:777–786. doi: 10.1016/s0092-8674(00)80152-x. [DOI] [PubMed] [Google Scholar]