Abstract
tRNA binding to the ribosomal P site is dependent not only on correct codon–anticodon interaction but also involves identification of structural elements of tRNA by the ribosome. By using a phosphorothioate substitution–interference approach, we identified specific nonbridging Rp-phosphate oxygens in the anticodon loop of tRNAPhe from Escherichia coli which are required for P-site binding. Stereo-specific involvement of phosphate oxygens at these positions was confirmed by using synthetic anticodon arm analogues at which single Rp- or Sp-phosphorothioates were incorporated. Identical interference results with yeast tRNAPhe and E. coli tRNAfMet indicate a common backbone conformation or common recognition elements in the anticodon loop of tRNAs. N-ethyl-N-nitrosourea modification–interference experiments with natural tRNAs point to the importance of the same phosphates in the loop. Guided by the crystal structure of tRNAPhe, we propose that specific Rp-phosphate oxygens are required for anticodon loop (“U-turn”) stabilization or are involved in interactions with the ribosome on correct tRNA–mRNA complex formation.
A central aspect of translation is the ribosome-mediated interaction of tRNA with mRNA. Proofreading and selection of the cognate tRNA at its mRNA codons occur at the ribosomal A site; tRNA binding to the A site is strictly mRNA-dependent (1). Codon–anticodon interaction also takes place at the ribosomal P site (2, 3), but tRNA P-site binding can also occur in the absence of mRNA (4). Thus, at least at this site, the ribosome is directly contacting the tRNA; indeed, specific bases in the 16S rRNA were recently shown to be involved (5). However, mRNA-independent binding is weak and tRNA and mRNA mutually enhance their binding to the ribosome.
There is little absolute sequence conservation between the various tRNAs, and many of the conserved bases were shown not to be important for protein synthesis (6, 7). The exception is the universally conserved 3′ CCA end, for which recent experimental evidence has indicated a base pairing contact to the 23S rRNA (8). Binding energy for tRNA P-site binding is provided solely by the 3′ CCA end and the anticodon arm region (9–11). Given the sequence variation of the anticodon region, it is likely that the ribosome recognizes its molecular shape, with the ribose–phosphate backbone best presenting possible binding determinants. Consistent with this view, an anticodon stem-loop RNA oligonucleotide was found to bind to 30S subunits, whereas its analogue containing 2′-deoxy groups did not (12). More recently, a specific 2′-hydroxyl group at position U33 in the anticodon loop of Escherichia coli tRNAPhe was found to be required for ribosomal P site binding (13).
Phosphorothioate-substituted tRNAs have been used previously to footprint the ribosome on the tRNA. In these experiments, the accessibility of phosphorothioates in ribosome-bound tRNAs toward iodine cleavage was monitored (14). Here, by using the substitution–interference analysis outlined below, we show that three Rp-phosphate oxygens in the anticodon loop of tRNA are required for functional mRNA-dependent binding to the ribosomal P site.
METHODS
Generation of Phosphorothioate-Containing tRNAs and Binding to Ribosomes.
tRNAs containing Rp-phosphorothioates were produced essentially as described (15), with conditions for T7 RNA polymerase transcription being 26 mM MgCl2, 50 mM Tris⋅HCl (pH 7.9), 2 mM spermidine, 5 mM DTT, 4 mM each rNTP, and 40 μM of one of the four α-S-triphosphate nucleotides (NTPαS, purchased from NEN). One percent-substituted tRNAs were chosen to avoid multiple interference in a single tRNA molecule. The resulting transcripts were eluted from 10% denaturing polyacrylamide gels, 5′-dephosphorylated, and [32P]-labeled by T4 polynucleotide kinase and [γ-32P]ATP, followed by a second gel purification. Templates for in vitro transcription were pSTtPhe (14) for tRNAPhe (E. coli), and p67YF0 for tRNAPhe (Saccharomyces cerevisiae, yeast) (16); both were linearized with BstNI. For tRNAfMet, a template was generated from a tRNAfMet precursor clone. This was accomplished by PCR amplification of the tRNA sequence with primers hybridizing to the 5′ end (5′-TAATACGACTCACTATAGGCGGGGTGGAGCAGCCTG3′, including the T7 RNA polymerase promoter, in italics) and 3′ end (5′-TGGTTGCGGGGGCCGGATTTG-3′) of the tRNA; note the guanosine substitution (instead of a cytosine) at the 5′ terminal position of the tRNA (underlined) for efficient in vitro transcription from the PCR product as the only base substitution compared with the wild-type sequence. The in vitro-synthesized tRNAfMet showed clear mRNA-dependent P site binding, comparing well with the in vivo-generated tRNA (data not shown). Templates for the MF-mRNA were generated by linearizing plasmid pTZ-MF [kindly provided by K. Nierhaus, (17)] with SspI.
Ribosomes and ribosomal subunits were isolated from E. coli strains MRE600 and D10-can20-12E by using standard procedures (18). In all cases of tRNA binding to ribosomes, the tRNAs and the anticodon arms were activated in binding buffer (see below) for 1 min at 65°C and then for 5 min at 37°C before addition to the ribosome–mRNA complex. tRNA or anticodon arm analogue binding was performed in binding buffer containing 6 mM MgCl2, 150 mM NH4OAc, 50 mM Tris⋅HCl (pH 7.4), 2 mM spermidine, 0.05 mM spermine, and 2 mM DTT. 32P-labeled tRNA (0.2–1 pmol) or anticodon arm analogue was added to 10 pmol 70S or 30S subunits in the presence of 10 μg poly(U) for tRNAPhe or 0.75 μg (50 pmol) MF-mRNA for tRNAfMet, or in the absence of mRNA. The samples (50 μl total) were incubated for 5 min at 37°C and held for 20 min on ice. Ice-cold buffer (1 ml) was added, and the samples were filtered through a 0.45 μM cellulose nitrate filter (Sartorius). Under these conditions, 65–75% of the input tRNA was bound to ribosomes in the presence of mRNA. After washing five times with 1 ml binding buffer, the tRNA–ribosome samples were reeluted from the filter as described (13). Iodine cleavage of phosphorothioate-containing tRNAs (pto-tRNAs) was subsequently performed in the presence of 1 mM iodine (10 mM stock solution in ethanol) in a 80% formamide solution for 2 min at room temperature, followed by precipitation in the presence of 10 μg glycogen and subsequent separation on 10% denaturing polyacrylamide gels containing 7 M urea. Quantification was performed by scanning the gels on a PhosphorImager (Molecular Dynamics) and calculating the relative intensity of an individual band compared with the band in the unselected lane that was set to a value of 1.0. Positions 1–10 and 69–75 of the tRNA were not monitored because of the lack of resolution or possible nonquantitative recovery of small oligoribonucleotides after cleavage.
Purification of Phosphorothioate-Containing Anticodon Analogues and Kd Measurements.
HPLC purification of the two diastereoisomers of single-phosphorothioate-containing oligoribonucleotides was performed as described (19, 20). Briefly, 10 μg of gel-purified oligonucleotide was loaded at a flow rate of 1 ml/min onto a 250 mm C-18 reverse phase column that had been equilibrated with 2% acetonitrile and 0.1 M NH4OAc. The oligonucleotides were eluted by increasing the acetonitrile content to 20% in 20 min. For certain oligonucleotides, the column was heated to 45°C for separation of the two isomers. The two eluted peaks were 5′ 32P-labeled and the correct position of the phosphorothioate examined by iodine cleavage. The later-eluting oligoribonucleotide showed a higher resistance to partial digestion with snake venom phosphordiesterase (data not shown), consistent with the properties of the Sp diastereoisomer; therefore, the earlier eluting oligonucleotide was assumed to be Rp, in agreement with results described earlier (19, 20). Single-point (i.e., single-ribosome concentration) binding efficiencies shown in Fig. 3 were determined under the standard conditions described above. Kd determinations of these anticodon analogues to 30S ribosomal subunits were performed as described (13), except that the divalent metal ion concentration was 6 mM MgCl2.
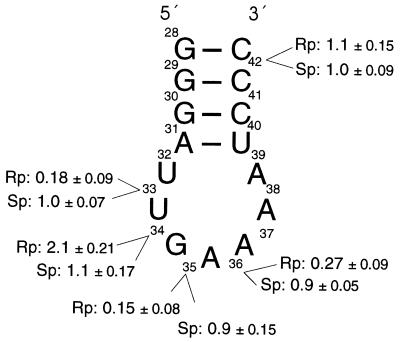
Figure 3.
Binding of anticodon arm analogues to 30S ribosomal subunits or 70S ribosomes. The sequence and secondary structure of the anticodon stem-loop region of tRNAPhe (E. coli) is shown. RNA oligonucleotides (15-mers) containing a single phosphorothioate substitution at the indicated positions were synthesized and the Rp- and Sp-containing diastereoisomers separated by reverse phase HPLC. The relative binding efficiencies of the various oligonucleotides to 30S subunits in the presence of poly(U) determined by filter binding assay are indicated as fraction of bound anticodon arm analogue compared with the unsubstituted anticodon arm analogue (1.0, single point measurements). For binding to 70S ribosomes, identical values (within the variations indicated) were determined.
N-Ethyl-N-Nitrosourea (ENU) Modification of tRNAs.
ENU modification of 5′ or 3′ 32P-labeled tRNA was performed as described by Vlassov et al. (21, 22) in the presence of 10 mM MgCl2, 50 mM Tris⋅HCl (pH 7.4). One-fifth volume of 32 mg/ml ENU (Sigma) in ethanol was added to the sample that was incubated for 30 min at 37°C. After precipitation, the modified tRNAs were bound to poly(U)-programmed 30S subunits or 70S ribosomes under conditions given above (6 mM MgCl2), filtered, and reeluted as described (13). Hydrolysis of ethylated phosphates occurred in gel-loading buffer (1× TBE/8 M urea).
RESULTS
Experimental Strategy.
To detect functionally important phosphates in tRNA, the phosphorothioate-interference technique (23–26) was adapted for tRNA binding to the ribosomal P site. Substitution of the nonbridging phosphate oxygens with sulfur is thought to disrupt tertiary interactions and/or to interfere with the binding of magnesium ions (27, 28). Rp-pto-tRNAs were generated by in vitro transcription in the presence of one of the four α-S-triphosphate nucleotides (15) at 1% of the corresponding unsubstituted ribonucleotide. In vitro-transcribed tRNAs are substrates for aminoacylation and are active in translation (16, 29). The pto-tRNAs were labeled at the 5′ end with 32P and bound under P-site binding conditions to 70S ribosomes or 30S ribosomal subunits in the presence or absence of mRNA. Ribosome-bound tRNA was then selected by nitrocellulose filter binding. If introduction of a phosphorothioate at a certain position in the tRNA interferes with ribosomal binding, then tRNA molecules containing the phosphorothioate substitution will be eliminated from the binding-competent tRNA pool. The distribution of phosphorothioates in ribosome-bound tRNAs retained on the filter was then compared with that of the nonselected, total population of tRNA molecules. It is important to note that the iodine cleavage used to detect the positions of phosphorothioate interference was performed under denaturing conditions after the tRNA was removed from the ribosome.
In an independent approach, we used ENU, which ethylates randomly the Sp- or Rp-phosphate oxygens and offers the advantage that it can be used with natural tRNAs containing the posttranscriptional modifications (21). As in the method described above, the tRNAs were modified at a low level, selected by binding to the ribosomal P site and their modification pattern compared with the unselected population.
Interference of Phosphorothioates in tRNAPhe from E. coli.
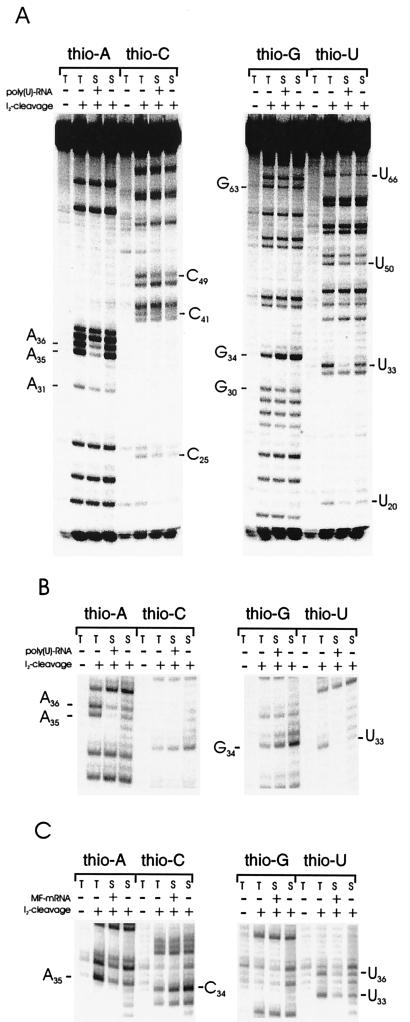
In vitro-transcribed tRNAPhe from E. coli containing Rp-phosphorothioates was previously shown to be capable of aminoacylation and binding to the ribosomal A and P sites (14); by using the 5′ [32P]-end-labeled tRNAPhe containing 1% phosphorothioates (pto-tRNAPhe), we obtained binding affinities similar to those of unsubstituted tRNA (data not shown, see Methods). Pto-substituted tRNAs were bound to E. coli 30S ribosomal subunits and selected by filter binding. Fig. 1A shows a representative gel-autoradiograph of such an experiment. Reduction in the amount (i.e., counterselection) of phosphorothioates can be seen at several positions (indicated in the figure) throughout the tRNA. Almost complete counterselection was found at positions U33, A35, and A36 in the anticodon loop, indicating a strong interference with binding. Moreover, interference at these positions was strongly mRNA-dependent. These results are summarized in Fig. 2 and quantified for positions in the anticodon loop in Table 1. Interestingly, at position G34 a strong enhancement in the lanes containing the selected tRNAs can be seen, indicating that a Rp-phosphorothioate at this position enhances 30S ribosomal binding, independent of mRNA.
Figure 1.
Phosphorothioate substitution–interference analyses. PhosphorImager printouts of selected and unselected tRNAs containing one of the four Rp-phosphorothioate nucleotides (thio-A, -C, -G, -U) separated on 10% polyacrylamide gels after iodine cleavage as indicated. The phosphorothioate-containing tRNAs were selected for P-site binding on 30S ribosomal subunits in the presence and absence of mRNA as indicated. Positions showing strong interference are indicated. T, unselected (total) phosphorothioate-containing tRNA population; S, selected (binding-competent) phosphorothioate-containing tRNA population. (A) tRNAPhe (E. coli). The sequence of the tRNA is shown in Fig. 2. (B) tRNAPhe from yeast. Only the bands corresponding to the anticodon region are presented, the sequence of the anticodon stem-loop region is 5′- CCAGACUGAAAAUCUGG-3′ (paired region are underlined, and anticodon in italics). (C) tRNAfMet (E. coli), anticodon region. The sequence of the anticodon stem-loop region is 5′-UCGGGCUCAUAACCCGA-3′. A specific mRNA containing a single AUG codon in the middle of a 46-nucleotide molecule (MF-mRNA) was used.
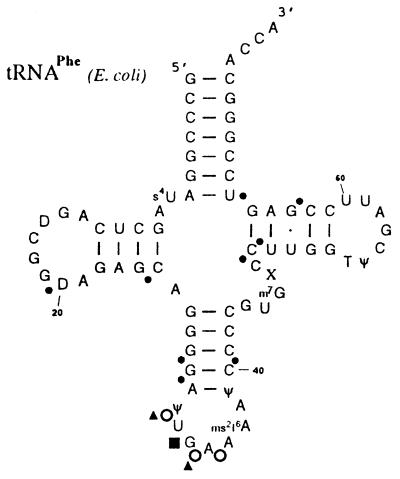
Figure 2.
Secondary structure of natural tRNAPhe (E. coli) showing nucleotides at which substitution of the 5′ Rp-phosphate oxygens with sulfur interferes with P-site binding at 30S subunits. Note that for phosphorothioate–interference experiments, in vitro transcribed tRNAs lacking the indicated natural modifications were used, whereas for ENU modification–interference experiments natural tRNAs were taken. Large and small circles indicate strong and weak interference, respectively; open circles indicate interference strongly dependent on mRNA presence. The filled rectangle indicates enrichment of tRNAs containing a Rp-phosphorothioates at position 34. Triangles indicate positions of interference when ethylated by ENU.
Table 1.
Positions in the anticodon loop region of tRNAPhe (E. coli) determined by random substitution–interference analyses at which introduction of a 5′-Rp phosphorothioate interferes with P-site binding
| 30S | 30S − mRNA | 70S | 70S − mRNA | |
|---|---|---|---|---|
| U33 | 0.30 ± 0.06 | 0.53 ± 0.06 | 0.55 ± 0.09 | 0.49 ± 0.15 |
| G34 | 1.95 ± 0.17 | 2.05 ± 0.08 | 0.96 ± 0.08 | 1.02 ± 0.15 |
| A35 | 0.29 ± 0.10 | 0.82 ± 0.12 | 0.84 ± 0.05 | 0.88 ± 0.11 |
| A36 | 0.39 ± 0.12 | 0.65 ± 0.10 | 0.84 ± 0.09 | 0.78 ± 0.11 |
The mean values (± variation) of relative band intensity of individual nucleotides compared to those in the unselected tRNA lane (1.0) are shown. Selection was performed on 30S subunits or 70S ribosomes in the presence and absence (−mRNA) of mRNA.
It was shown previously that only the anticodon stem-loop region interacts with the 30S subunit (10, 30). Because strong interference was found to be restricted to the anticodon loop, we next asked whether there are differences between subunit and whole ribosome, and repeated the same experiments by using 70S ribosomes. An interference pattern similar to that seen for the 30S subunits was indeed observed (data not shown). However, a strong quantitative difference was found at the positions in the anticodon loop (U33, G34, A35, and A36; Table 1); interference was much less pronounced when the tRNA was bound to 70S ribosomes and the mRNA-dependent interference is not seen as with 30S subunits.
We also tested the interference of phosphorothioates in the presence of manganese or cadmium ions that, as “soft” metals, have been shown to suppress the substitution if coordination of a magnesium ion to the substituted phosphate oxygen is affected (31); this effect is commonly referred to “manganese rescue” (24–26). However, no change in the interference pattern in the presence of manganese or cadmium ions was detected (data not shown).
Similarly, the same degree of interference was observed for positions in the anticodon loop when selection was performed at high magnesium ion concentration with 30S subunits (25 mM MgCl2, data not shown), indicating that increasing the magnesium ion concentration cannot compensate the effect of the substitution.
Interference of Phosphorothioates in tRNAPhe from Yeast and tRNAfMet from E. coli.
If there is a sequence-independent requirement of specific phosphate oxygens in tRNAs for P-site binding we expected to see the same or a similar interference pattern in different tRNAs. We therefore generated pto-tRNAPhe from yeast by in vitro transcription as described above and tested this tRNA for interference with P-site binding to 30S subunits in the presence and absence of poly(U) as mRNA. As can be seen in Fig. 1B, this tRNA shows an identical interference pattern with the equivalent tRNA from E. coli with strong interference visible at positions U33, A35, and A36. The same interference pattern was detected when 70S ribosomes were used, although the interference was less pronounced at the positions in the anticodon loop (data not shown), comparable to the situation with the E. coli tRNA.
To examine a tRNA for a different amino acid, we next used a template for initiator tRNA (tRNAfMet) from E. coli and synthesized in vitro a phosphorothioate-containing tRNA. In this case, a specific mRNA was used, namely one in which a single AUG codon, followed by a single UUC codon (phenylalanine), was located in the middle of a 46-nucleotide molecule transcribed in vitro. Selection was performed on 30S subunits only. This mimics very closely an initiating ribosome with initiator tRNA bound to the ribosomal P site.
The selected pto-tRNAsfMet showed the same pattern of interference in the anticodon loop as the other tRNAs tested (U33, A35, and U36; Fig. 1C). Again, interference at these nucleotides was strongly dependent on mRNA presence, indicating that a phosphorothioate at these positions prevents formation of a mRNA-dependent complex at the anticodon loop. Interestingly, for both tRNAs tested, a much more intense band at position 34 can be detected in the lanes with the selected tRNAs (G34 for tRNAPhe in Fig. 1B, C34 for tRNAfMet in Fig. 1C).
Anticodon Oligonucleotides Bearing Single Phosphorothioate Groups at Specific Positions.
Short oligoribonucleotides (15-mers) comprising the anticodon stem-loop region have been shown previously to mimic mRNA-dependent tRNA binding to the 30S subunit (10, 12, 13). To confirm the results obtained by using randomly substituted phosphorothioates, we tested chemically synthesized oligoribonucleotides comprising positions 28–42 of tRNAPhe (from E. coli) containing a single phosphorothioate at positions U33, G34, A35, A36, or C42. Oligonucleotides containing Rp and Sp phosphorothioate diastereoisomers were separated by reverse-phase HPLC (19, 20).
The various Rp- and Sp-phosphorothioate-containing oligoribonucleotides were 5′ [32P]-end-labeled and tested for binding to 30S ribosomal subunits by filter binding. An unsubstituted RNA anticodon arm analogue was used for comparison. The oligonucleotides containing Rp-phosphorothioates at U33, A35, and A36 all showed strongly decreased relative binding affinities (Fig. 3). In contrast, their Sp counterparts exhibited the same binding affinities as the unsubstituted RNA, as did oligoribonucleotides containing an Sp phosphorothioate at position G34 or the Rp or Sp phosphorothioate at position C42. Introduction of a Rp-phosphorothioate at position G34 led to a 2-fold increase in binding affinity. The results of the single nucleotide substitution experiments confirmed in detail those of the modification–interference experiments described above. Indeed, the Kd values of Rp-substituted RNA oligonucleotides at U33, A35, and A36 were increased by more than 20-fold, whereas introduction of a Rp phosphorothioate at position G34 led to a 7-fold decrease in Kd, compared with the unsubstituted RNA oligonucleotide. The exact values are given in Table 2.
Table 2.
Kd values of the all-RNA anticodon stem-loop analogue (derived from E. coli tRNAPhe) and single phosphorothioate-substituted analogues (15-mer oligonucleotides) to 30S ribosomal subunits in the presence of poly(U) and 6 mM MgCl2
| Analogue | Kd |
|---|---|
| All-RNA | 1.2 (±0.2) × 10−6 M |
| Rp-thioU33 | 3.1 (±0.4) × 10−5 M |
| Rp-thioG34 | 3.0 (±0.2) × 10−7 M |
| Rp-thioA35 | 3.3 (±0.1) × 10−5 M |
| Rp-thioA36 | 2.0 (±0.4) × 10−5 M |
As expected, the binding affinities of these oligonucleotides to 70S ribosomes were identical to those observed for binding to the 30S subunits (Fig. 3).
Interference of ENU-Modified tRNAs.
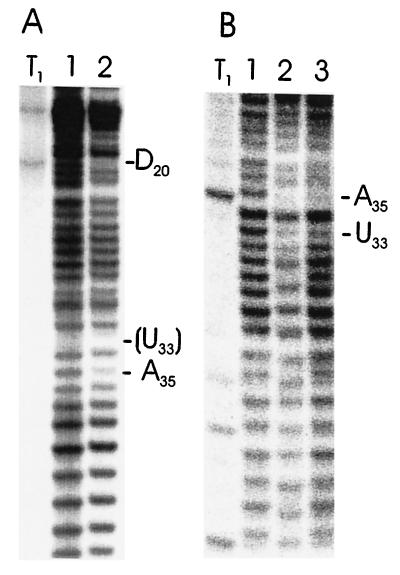
As an independent approach, we used ENU, which ethylates randomly the Sp- or Rp-phosphate oxygens of nucleic acids (21, 22). We performed the modification under conditions where the tRNA adopts its native conformation and used these ENU-modified tRNAs for modification–interference experiments. tRNAPhe and tRNAfMet, both from E. coli, were tested by this approach to detect interference with P-site binding. The overall accessibility of the tRNA phosphates monitored by ENU (Fig. 4) is identical to that previously reported (21, 22). However, in the selected lanes, a clear reduction in band intensity can be seen at positions U33 and A35 in tRNAPhe (E. coli), as well as a reduction at position A35 and D20 in tRNAfMet. This finding indicates that ethylation of these phosphates interferes with binding. The position U33 of tRNAfMet cannot be monitored, as position C32 bearing a natural modified 2′-O-methyl group prevents hydrolysis of the ethylated phosphate 5′ to U33.
Figure 4.
ENU modification–interference analyses. PhosphorImager printout of polyacrylamide gels showing separation of selected or nonselected bases of E. coli tRNAfMet (A) or tRNAPhe (B) with ENU modification after cleavage (lanes: 1, total, unselected tRNA; 2, selection with 30S subunits; 3, selection with 70S ribosomes). Positions of interference are indicated. The ethylated phosphate at nucleotide U33 in tRNAfMet is lacking (shown in parentheses) as 2′-O-methylation of base C32 prevents hydrolysis between these bases. T1, partial T1-digest of [32P]-end-labeled tRNAs. tRNAPhe was 5′ and tRNAfMet was 3′ [32P]-end-labeled, respectively.
Remarkably however, in both tRNAs tested, the phosphates at position 36 do not show interference with binding when modified by ENU. A lack of interference at position 36 was also obtained with tRNAPhe from yeast, in which only position 36 of the three of interest can be monitored (C32 and G34 bear 2′-O-methyl groups; data not shown). For tRNAPhe, the selection on whole (70S) ribosomes also shows clear reduction of selected ENU-modified phosphates at positions U33 and A35; this result is therefore different from that seen in the phosphorothioate–interference experiments. In summary, the results point to the importance for ribosomal P-site binding of phosphates 5′ to positions 33 and 35 in the anticodon loop of tRNAs containing the natural base modifications.
As the ethylated phosphate renders the backbone of the RNA more labile to hydrolysis, it might be argued that cleavage could have occurred before selection. However, as can be seen in the figure, fragments as short as 20 nucleotides (and shorter, data not shown) were resolved on the gel; these short fragments comprising the first 20 nucleotides of the tRNA would not have bound to 30S subunits. Therefore, the cleavage must have occurred after the selection.
DISCUSSION
The phosphorothioate substitution–interference approach revealed several Rp-phosphate oxygens in tRNA as being important for P-site binding, with the most pronounced ones in the anticodon loop region (positions 33, 35, and 36). Fig. 5 shows this region in the three-dimensional structure from yeast tRNAPhe. Interference with binding to 30S subunit is strongly dependent on the presence of mRNA. The interference caused by Rp-phosphorothioates in the anticodon loop of tRNAPhe with ribosomal P-site binding was proven by RNA oligonucleotides containing a single Rp-phosphorothioate and revealed an exact correlation between the two approaches. Substitution at the Sp locations of these positions as well as substitution at G34 (the Sp isomer only) did not affect binding and demonstrates the high stereo-specific interference. The similar interference patterns found in different tRNAs imply either sequence-independent recognition by the ribosome and/or the adoption of a similar conformation of the anticodon regions of the tRNAs tested during P-site binding.
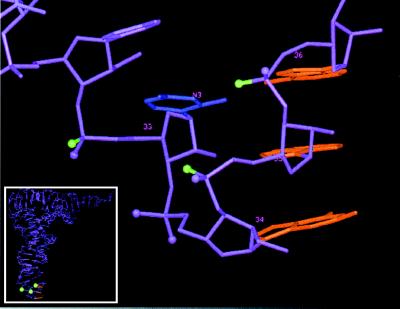
Figure 5.
Three-dimensional structure of the anticodon loop of tRNAPhe from yeast. The three bases of the anticodon are in red. The green balls depict Rp-phosphate oxygens identified in this study which when substituted with sulfur interfere with tRNA P-site binding; those in magenta do not interfere with binding [except at G34 (Rp), at which binding was enhanced]. The base presented in blue is U33; the labeled atom N3 is proposed to interact with the Rp-phosphate oxygen of A36 (32). The complete tRNA is seen in the Inset.
Dabrowski et al. (14) have also used phosphorothioate-containing tRNAs; however, they did not detect the interference that we identified. As we could directly support our findings by using specifically modified oligonucleotides, we conclude that our selection method used to separate the unbound tRNA from ribosomes and the subsequent quantification method is more sensitive and accurate.
The phosphorothioates in the anticodon stem and loop region showed increased interference when selected on 30S subunits compared with 70S ribosomes (Table 1). A possible explanation is the additional binding energy provided by the 3′ CCA end of tRNA during interaction with the 50S subunit (8), which reduces the requirement on the interaction between the anticodon region and 30S subunits. Support for this view is provided by single-phosphorothioate-substituted anticodon arm analogues lacking the 3′ CCA end that showed identical binding affinities to 30S ribosomal subunits and 70S ribosomes (Fig. 3). Alternatively, as the tRNA–mRNA complex, once bound to 70S ribosomes, is required to move during translocation, binding to 30S subunits might be tighter to prevent this movement until the 50S subunit associates. mRNA-dependent interference was seen only with 30S subunits and not when full-length tRNA was bound to 70S, possibly indicating differences in the composition of the P site between 30S subunits and whole ribosomes.
It is interesting to note that substitution of the Rp-phosphate oxygen of position 34 with sulfur results in increased binding of tRNA and anticodon analogues to ribosomes. It vividly demonstrates that the fit of the anticodon region into the 30S ribosomal P site has evolved for specific and sufficient binding; however, increasing the tRNA binding strength would probably impede its dissociation required for efficient translocation.
The ENU modification–interference analyses provide additional evidence for the importance of phosphates 5′ to positions 33 and 35 for P-site binding. However, as ENU is able to modify either of the two nonbridging phosphate oxygens, it is not however possible to distinguish between an interference effect at the Rp- or the Sp-phosphate oxygens. In contrast to the phosphorothioate experiments, position 36 in three different tRNAs did not show ENU interference with P site binding. An explanation for this discrepancy might lie in the hydrogen bonding between the Rp-phosphate oxygen of A36 and the N3 position of base U33 in the crystal structure of tRNAPhe from yeast, as part of the anticodon loop (“U-turn”) stabilization (ref. 32; compare Fig. 5). If the Rp-phosphate oxygen of A36 interacts with U33 in the structure in solution, then only the Sp-phosphate oxygen will be modified by ENU and most likely binding will not be affected. This interpretation is consistent with the noninterference result of a phosphorothioate at the Sp location at position 36 identified by our oligonucleotide approach. In contrast, substitution of the Rp-phosphate oxygen of A36 with sulfur interferes with binding (Fig. 3) and would prevent hydrogen bonding to the U33–N3 position.
The two other Rp-phosphate oxygens in the anticodon loop at positions U33 and A35—identified by phosphorothioate and ENU interference—could also be involved in intramolecular interactions. The interference of the sulfur-substituted Rp-phosphate-oxygen at position U33 found under all conditions (i.e., with and without a possible stabilization by 50S subunits or mRNA) would support this concept. However, as seen in Fig. 5, the Rp phosphorothioate of U33 and, to a lesser extent, A35 point out of the loop in the crystal structure. Therefore, these phosphates could also be more directly involved in interacting with the ribosome and could act as acceptors of hydrogen bonds donated by the ribosome. Possible hydrogen bond donors could be specific bases in the rRNAs, identified as being involved in tRNA P-site binding by footprinting and interference analyses (5, 33). As we found much less interference at the anticodon loop phosphates for mRNA-independent P-site binding, we conclude that these phosphate oxygens must be specifically involved in formation or recognition of the codon–anticodon complex by the ribosome. mRNA binding to the ribosome per se is only weak, and, to our knowledge, apart from the well-known Shine–Dalgarno interaction during initiation, no evidence for a direct contact between mRNA and ribosome has been presented. Furthermore, it has been suggested that the tRNAs stabilize the mRNA at the A and P site (34, 35). Sterically, it is rather unlikely that the identified phosphate oxygens in the anticodon loop interact with the mRNA, as they point in the opposite direction away from the anticodon bases. Therefore, they might mediate interaction of the correctly formed codon–anticodon complex with the 30S subunit and provide the necessary stabilizing binding energy. A similar conclusion was reached recently by Potapov and coworkers on the involvement of specific 2′-hydroxyl groups in mRNAs (36).
Regardless of the actual role of these phosphate oxygens, a specific molecular conformation in the anticodon loop required for mRNA-dependent P-site binding involves the specific backbone residues identified in this report.
Acknowledgments
We are grateful to Renée Schroeder for continuous support and encouragement. We thank Christian Berens, Knud H. Nierhaus, Harry F. Noller, Renée Schroeder, Tim Skern, Barbara Streicher, and Eric Westhof for stimulating discussions. Beatrice Clouet d’Orval provided helpful information for HPLC purification of phosphorothioate-containing oligonucleotides. This work was supported by grants from the Austrian Science Foundation (FWF, Biotech-2, 930345) and the Austrian National Bank (5461) to U.v.A.
ABBREVIATIONS
- pto-tRNA
phosphorothioate-containing tRNA
- ENU
N-ethyl-N-nitrosourea.
References
- 1.Noller H F. Annu Rev Biochem. 1991;60:191–227. doi: 10.1146/annurev.bi.60.070191.001203. [DOI] [PubMed] [Google Scholar]
- 2.Lührmann R, Eckhardt H, Stöffler G. Nature (London) 1979;280:423–425. doi: 10.1038/280423a0. [DOI] [PubMed] [Google Scholar]
- 3.Wurmbach P, Nierhaus K H. Proc Natl Acad Sci USA. 1979;76:2143–2147. doi: 10.1073/pnas.76.5.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grajevskaja R A, Odinzov V B, Saminsky E M, Bresler S E. FEBS Lett. 1973;33:11–14. doi: 10.1016/0014-5793(73)80147-4. [DOI] [PubMed] [Google Scholar]
- 5.von Ahsen U, Noller H F. Science. 1995;267:234–237. doi: 10.1126/science.7528943. [DOI] [PubMed] [Google Scholar]
- 6.Uhlenbeck O C, Lowary P T, Wittenberg W L. Nucleic Acids Res. 1982;10:3341–3352. doi: 10.1093/nar/10.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nazarenko I A, Harrington K M, Uhlenbeck O C. EMBO J. 1994;13:2464–2471. doi: 10.1002/j.1460-2075.1994.tb06531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Samaha R S, Green R, Noller H F. Nature (London) 1995;377:309–314. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- 9.Sprinzl M, Wagner T, Lorenz S, Erdmann V A. Biochemistry. 1976;15:3031–3039. doi: 10.1021/bi00659a015. [DOI] [PubMed] [Google Scholar]
- 10.Rose S J, Lowary P T, Uhlenbeck O C. J Mol Biol. 1983;167:103–117. doi: 10.1016/s0022-2836(83)80036-9. [DOI] [PubMed] [Google Scholar]
- 11.Nekhai S A, Parfenov D V, Saminsky E M. Biochim Biophys Acta. 1994;1218:481–484. doi: 10.1016/0167-4781(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 12.Koval’chuke O V, Potapov A P, El’skaya A V, Potapov V K, Krinetskaya N F, Dolinnaya N G, Shabarova Z A. Nucleic Acids Res. 1991;19:4199–4201. doi: 10.1093/nar/19.15.4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.von Ahsen U, Green R, Schroeder R, Noller H F. RNA. 1997;3:49–56. [PMC free article] [PubMed] [Google Scholar]
- 14.Dabrowski M, Spahn C M T, Nierhaus K H. EMBO J. 1995;14:4872–4882. doi: 10.1002/j.1460-2075.1995.tb00168.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Griffiths A D, Potter B V L, Eperon I C. Nucleic Acids Res. 1987;10:4145–4162. doi: 10.1093/nar/15.10.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sampson J R, Uhlenbeck O C. Proc Natl Acad Sci USA. 1988;85:1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Triana-Alonso F J, Chakraburtty K, Nierhaus K H. J Biol Chem. 1995;270:20473–20478. doi: 10.1074/jbc.270.35.20473. [DOI] [PubMed] [Google Scholar]
- 18.Rheinberger H J, Geigenmuller U, Wedde M, Nierhaus K H. Methods Enzymol. 1988;164:658–670. doi: 10.1016/s0076-6879(88)64076-6. [DOI] [PubMed] [Google Scholar]
- 19.Slim G, Gait M J. Nucleic Acids Res. 1991;19:1183–1187. doi: 10.1093/nar/19.6.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maschhoff K L, Padgett R A. Nucleic Acids Res. 1993;21:5456–5462. doi: 10.1093/nar/21.23.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vlassov V V, Giege R, Ebel J P. FEBS Lett. 1980;120:12–16. doi: 10.1016/0014-5793(80)81034-9. [DOI] [PubMed] [Google Scholar]
- 22.Vlassov V V, Giege R, Ebel J-P. Eur J Biochem. 1981;119:51–59. doi: 10.1111/j.1432-1033.1981.tb05575.x. [DOI] [PubMed] [Google Scholar]
- 23.Waring R B. Nucleic Acids Res. 1989;17:10281–10293. doi: 10.1093/nar/17.24.10281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harris M E, Pace N R. RNA. 1995;1:210–218. [PMC free article] [PubMed] [Google Scholar]
- 25.Chanfreau G, Jaquier A. Science. 1994;266:1383–1387. doi: 10.1126/science.7973729. [DOI] [PubMed] [Google Scholar]
- 26.Christian E L, Yarus M. Biochemistry. 1993;32:4475–4480. doi: 10.1021/bi00068a001. [DOI] [PubMed] [Google Scholar]
- 27.Eckstein F. Annu Rev Biochem. 1985;54:367–402. doi: 10.1146/annurev.bi.54.070185.002055. [DOI] [PubMed] [Google Scholar]
- 28.Jaffe E K, Cohn M. Biochemistry. 1987;17:652–657. doi: 10.1021/bi00597a014. [DOI] [PubMed] [Google Scholar]
- 29.Harrington K M, Nazarenko I A, Dix D B, Thompson R C, Uhlenbeck O C. Biochemistry. 1993;32:7617–7622. doi: 10.1021/bi00081a003. [DOI] [PubMed] [Google Scholar]
- 30.Hüttenhofer A, Noller H F. Proc Natl Acad Sci USA. 1992;89:7851–7855. doi: 10.1073/pnas.89.17.7851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pecoraro V L, Hermes J D, Cleland W W. Biochemistry. 1984;23:5262–5271. doi: 10.1021/bi00317a026. [DOI] [PubMed] [Google Scholar]
- 32.Quigley G J, Rich A. Science. 1976;194:796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- 33.Moazed D, Noller H F. J Mol Biol. 1990;211:135–145. doi: 10.1016/0022-2836(90)90016-F. [DOI] [PubMed] [Google Scholar]
- 34.Beyer D, Skripin E, Wadzack J, Nierhaus K H. J Biol Chem. 1994;269:30713–30717. [PubMed] [Google Scholar]
- 35.Hartz D, Binkley J, Hollingsworth T, Gold L. Genes Dev. 1990;4:1790–1800. doi: 10.1101/gad.4.10.1790. [DOI] [PubMed] [Google Scholar]
- 36.Potapov A P, Triana-Alonso F J, Nierhaus K H. J Biol Chem. 1995;270:17680–17684. doi: 10.1074/jbc.270.30.17680. [DOI] [PubMed] [Google Scholar]