Abstract
The anti-idiotype approach is based on the assumption that an antibody specific for a receptor-binding domain of a ligand could be structurally related to the receptor. Therefore, a structural mimic of a receptor-binding domain, selected with an anti-ligand antibody, might be a functional substrate for the receptor. This hypothesis was addressed here by generating antibodies recognizing the Rev-nuclear export signal (NES). A functional NES is required for active export, presumably by interacting directly or indirectly with the nuclear pore complex. Anti-NES antibodies were used to isolate RNA mimics of the NES peptide from combinatorial RNA libraries. The RNA-mimics are exported actively, block Rev-dependent export of a reporter RNA, and inhibit cap-dependent U1 snRNA export in Xenopus oocytes, properties previously reported for NES-peptide conjugates.
Structurally and functionally similar surfaces or domains can be created by molecules with dissimilar primary sequences or even by molecules containing different types of building blocks. This is suggested by the large number of artificial ligands isolated from combinatorial libraries. The libraries can be antibodies, peptides, peptides displayed on phages, nucleic acids, or chemical compound libraries. The substrates used for ligand selection range from peptides, proteins, antibodies, and small molecules to transition state analogs (1–3). Nuclear export of RNPs and proteins requires the presence of nuclear export signals (NESs), which are thought to interact with transport factors, which provide the contact with the nuclear pore complex (4–6). Several proteins have been shown to contain a functional NES (7–9), one of which is the HIV-Rev protein containing a leucine-rich NES. The NES is essential for Rev-mediated export of RNA containing a Rev-binding site (10–13), probably by binding to an NES receptor and subsequent translocation. A common factor seems to be used for cap-dependent export of U snRNAs and NES-dependent export, because BSA-NES peptide conjugates inhibit not only Rev-mediated transport but also export of capped RNAs (7).
It should be possible, at least in principle, to isolate a mimic of a receptor-binding domain and analyze in a complex in vivo situation whether the mimic could be a functional substrate for the receptor. This was attempted in this study by generating antibodies recognizing the Rev-NES. The antibody was used to isolate RNA mimics of the NES peptide (export aptamers, XAPs) from combinatorial RNA libraries (14, 15), and the effect of XAPs on nuclear cytoplasmic transport in Xenopus oocytes was analyzed. It is suggested that anti-idiotype RNA and peptide conjugates could be functionally equivalent in Xenopus oocytes.
MATERIALS AND METHODS
Antibodies were raised in rabbits against a BSA-conjugate carrying peptides (C LPP LER LTL) corresponding to the Rev-NES (7) and were affinity-purified on peptide columns (Hitrap, Pharmacia). Five milligrams of peptide was used in the coupling step, 12 ml of serum was loaded, and 400 μg of antibody was obtained.
Aptamer Selection.
A detailed description for selection from constrained libraries has been reported recently (16). Briefly, for three selection cycles control IgGs (rabbit anti-human IgG; Sigma, I-2011) coupled to magnetic beads were used to deplete the library from aptamers binding to the constant region of rabbit IgGs, the depleted libraries were incubated with affinity-purified anti-NES antibody coupled to magnetic beads, and RNA was eluted by digestion of antibodies with proteinase K. Two additional cycles of selection were performed, differing from cycles 1–3 by the elution method; after being washed with NaK150 (50 mM Tris⋅HCl, pH 7.5/100 mM KCl/50 mM NaCl/1 mM MgCl2)/0.1%Nonidet P-40, beads were incubated for 15 min at 25°C with 2 mM NES peptide (C LPP LER LTL) in 10 mM Tris⋅HCl, pH 7.5/100 mM KCl/1 mM MgCl2. The composition of the selection buffer NaK150 is compatible with the requirements for G-quartets but also suitable for other RNA structures (16). Forty-eight clones eluted in cycle 5 from the library were analyzed individually by DNA-sequencing and immunoprecipitation of labeled RNA with anti-NES and control antibodies; 13/48 were anti-NES-specific, representing 9 individual sequences (see Table 1). For all experiments RNA was renatured by incubation for 10 min at 50°C in 10 mM Tris⋅HCl, pH 7.5/100 mM KCl/1 mM MgCl2. RNAs were uncapped unless stated otherwise (m7GpppG U1, ApppG Ad46).
Table 1.
RNA selected with the anti-NES antibodies (XAPs)
The XAP sequences corresponding to the initially variable part of the library are aligned. The common features are four groups of three G residues (in boldface type; note that one G residue of each group originated from the variable loops) and a maximum length of 3 nucleotides in loops 1–4. RNAs lacking a third G residue in one of the groups, or containing 4 nucleotides in one of the loops 2–4 are not binding detectably. Shown also are the sequences of variants (VGGG, VGUG, VGG) designed to investigate the importance of a G scaffold. There, the constant features were the loop sequences (in boldface type). In VTL the stem is closed by a stable tetraloop sequence; the constant sequence of the stems is not shown.
Export Competition.
Unlabeled, uncapped RNA (4 nmol) was precipitated with ethanol and resuspended in 4 μl H2O, and 1 μl 5× renaturation buffer was added and incubated for 10 min at 50°C. Renatured RNA (2.5 μl) was mixed with either 0.5 μl of each of the labeled RNAs (m7U1, U6, tRNA) or with 1.5 μl Rev/Ad46 complexes (ApppG-capped Ad46 was incubated with recombinant Rev-protein in vitro as described in ref. 13 but using a 10-fold diluted Rev stock solution). In all experiments the final XAP concentration was 5 pmol/10 nl. The amount of Rev/Ad46 complex injected was sufficient to saturate the Rev-mediated export pathway. Injection of larger quantities did not increase the absolute amount of Ad46 transported into the cytoplasm (Fig. 3C; data not shown). For all injections nuclei were visualized by centrifugation and transport-analyzed as described (17).
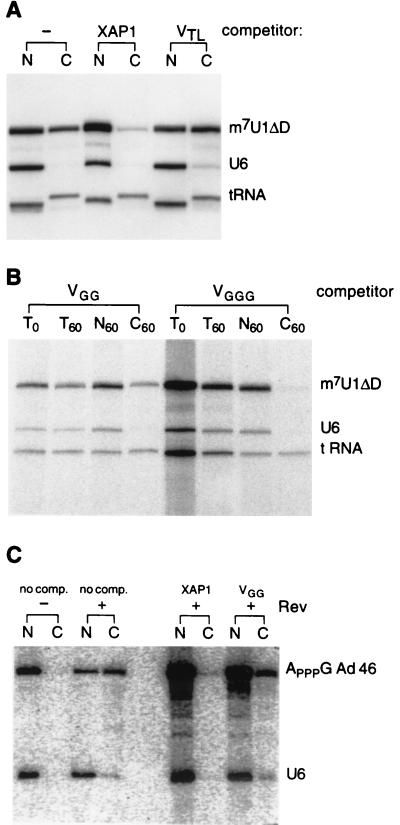
Figure 3.
Inhibition of RNA export in oocytes. (A) A mixture of radioactively labeled m7G-U1ΔD, U6, and tRNA was injected into the nucleus of oocytes in the absence (−) or presence of unlabeled, uncapped competitor RNA (XAP1, VTL). After 1 h oocytes were separated into nuclear (N) and cytoplasmic (C) fractions, and extracted RNA was analyzed on a denaturing acrylamide gel (the T7htRNA appears to be longer once transported to the cytoplasm; apparently nucleotides are added to the 3′ end before or during the translocation step). (B) As above. In addition, RNA was extracted from total oocytes immediately after injection (T0) and after 1 h (T60). (C) Labeled ApppG-capped Ad46 was preincubated in the presence (+) or absence (−) of Rev protein, mixed with labeled U6, and injected into the nucleus of oocytes either alone (no comp.) or together with unlabeled, uncapped competitor (XAP1, VGG). Oocytes were separated after 45 min into nuclear (N) and cytoplasmic (C) fractions, and extracted RNA was analyzed.
Genes.
The XAPs transcribed from the pol III genes differed from the RNAs obtained from the selection by the first 6 bp of the constant stem (5′-GAGCUU instead of 5′-GGGUUG and AAGCUU-3′ instead of CAACCC-3′). The pol III promoter was provided by a 490-bp BamHI/EcoRV fragment from the hU6EV gene (17); the 3′-flanking sequence was nucleotides 1–130 of the sequence 3′ to the hU6snRNA sequence isolated by PCR incorporating a HindIII site at the 5′ end and a ClaI site at the 3′ end. The genes for T7U1ΔD and T7U6 are those of ref. 18; T7htRNAmet used for the experiment of Fig. 3B was the one of ref. 7. The tRNA of Fig. 3A was expressed from a newly constructed T7htRNAmet gene. A T7 promoter sequence was inserted in front of a htRNAmet gene (19); the first 3 bases of the coding region were changed to 5′-GGG; the complementary sequence at the 3′-end (bases 69–71) was changed to CCC; and bases 72, 73, and +1 were changed to GGG-3′, generating a SmaI site.
Immunoprecipitation/Peptide Competition.
Bead-bound anti-NES (2 μg) was incubated for 20 min at 25°C with 100 μg peptide in NaK150 in a volume of 50 μl. Radioactively labeled RNA was added and incubation continued for 15 min before washing with NaK150/0.1%Nonidet P-40. The sequence of the control peptide was M VSK LSQ LQT ELK.
Native Gels.
In 20 μl NaK150 with 5–20 μg ytRNA/μg HeLa NXT (Computer Cell Culture Center, Belgium) or oocyte NXT (20), 10 units of RNasin and labeled/unlabeled XAPs (renatured) were incubated for 20 min at 20°C; 3 μl of 87% glycerol was added and the reaction was loaded on 6% acrylamide/0.1% Triton X-100/TBE gels (run at 7.5 V/cm at 4°C for 2.5 h). Labeled RNA was transcribed as described (18), but in the absence of cap analogs, and gel-purified, and 100 cps per reaction was used.
RESULTS AND DISCUSSION
The purpose of this work was to interfere with protein–protein interactions in vivo; therefore, a stable RNA structure was desirable for the XAPs. One way of achieving this is to incorporate stable secondary structure elements into the fixed sequences of the RNA library. Although this might reduce the structural complexity of the library compared with libraries without constraints, it has been shown that specifically and tightly binding aptamers can be isolated from constrained libraries (16, 21, 22). Based on the knowledge gained from previous selections (16), a new library was constructed (G2QCL, Fig. 1). This library was chosen because stem-based G-quartets seem to be very stable in oocytes (J.H., unpublished data) and because this library should be compatible with many different stem-based structural motifs. G2QCL was used to select aptamers with the anti-NES antibody. The antibody reacts on Western blots with recombinant Rev-protein but only weakly with the transport-deficient mutant RevM10 (Fig. 2D). Nine individual XAPs were isolated (Table 1); the common features are four groups of three G residues separated by one to three nucleotides located at the top of the 18-bp stem presumably formed by the fixed library sequences. Whether the nine different sequences shown in Table 1 represent different sequence solutions for a similar structural motif, or whether they result from the utilization of a polyclonal serum for the selection has not been investigated.
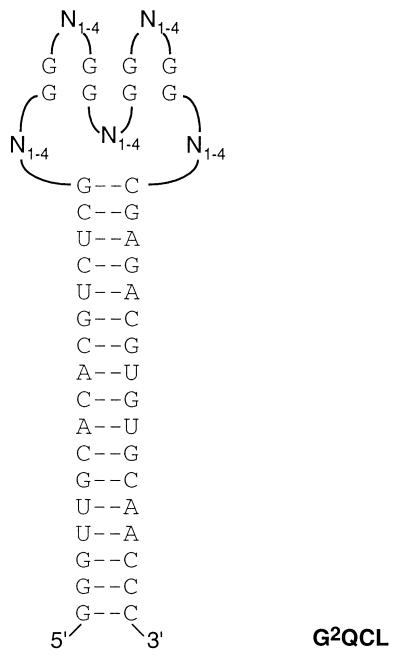
Figure 1.
Structure of the RNA library G2QCL. The invariable sequence required for binding of the PCR primers was designed to form an 18-bp stem to favor formation of a stable secondary structure core element. The random part consisted of 5 loops (N1–4), variable in length and sequence (1–4 bases per loop), separated by 4 groups of 2 G residues. In the text the loops are named 1–5, in 5′-3′ direction of the RNA.
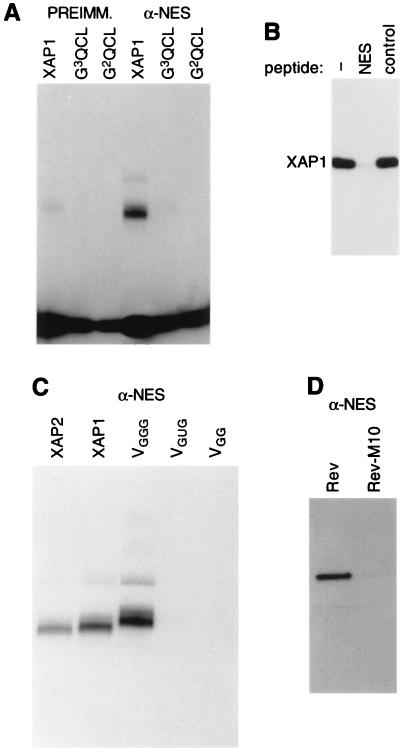
Figure 2.
Binding specificity. (A) Immunoprecipitation of radioactively labeled XAP1 or RNA transcribed from the unselected libraries G2QCL or G3QCL with preimmune serum or anti-NES antibodies, analyzed on a denaturing acrylamide gel (label at bottom is free GTP). (B) Immunoprecipitation of radioactively labeled XAP1 with anti-NES antibodies that had been preincubated with either buffer (−), NES peptide (NES), or control peptide (control) and analyzed on a denaturing acrylamide gel. (C) Immunoprecipitation of radioactively labeled XAPs or variants with anti-NES antibodies analyzed on a denaturing acrylamide gel. (D) Binding specificity of anti-NES antibodies analyzed by Western blotting of recombinant Rev wild type and Rev M10.
Because the groups of G residues might have the potential to form a three-layered G-quartet structure, RNAs (VGGG, VGG, VGUG) differing only in this scaffold motif were analyzed. These variants had been constructed as part of an attempt to explore systematically quartet-based scaffold motifs (J.H., unpublished data). The loop sequences were derived from a mixed G/A-quartet isolated with an anti-ferritin antibody (16). Because the loop sequences appeared to be very similar to the selected consensus for anti-NES G-quartet loops, the variants turned out to be useful tools. However, the anti-ferritin and the anti-NES antibody recognize distinct features of the aptamers. None of the variants or XAPs is recognized by the anti-ferritin antibody. The two G residues in loop 3 and the residues AG in loop 5 can be deleted in the variants without affecting anti-NES binding. VGG might lack one of the three G-quartet layers, whereas VGUG might contain a U-quartet in between two G-quartets. Mixed G- and U-quartets were reported for an unusually stable RNA tetraplex structure analyzed by NMR (23). The variant VTL contains a stable tetraloop sequence to close the constant stem structure.
XAPs Compete with the NES Peptide for Antibody Binding.
Binding specificity of XAPs was analyzed by immunoprecipitation. XAP1 bound specifically to the anti-NES antibody, whereas RNA transcribed from the unselected library G2QCL did not (Fig. 2A, XAP1 and G2QCL). Furthermore, RNA transcribed from G3QCL, a different library that has not been used for the anti-NES selection, was not immunoprecipitated (Fig. 2A, G3QCL). G3QCL differs from G2QCL by the presence of three fixed G residues per group, rather than only two per group. Apparently the presence of four groups of three G residues alone is not sufficient for stable interaction with the anti-NES antibody. However, this does not prove that the selected loop sequences contain essential contact points for the antibody. Proper folding of the G-quartet might depend on loop structures formed only by specific loop sequences. To demonstrate that XAPs were binding to the antigen-binding site, the antibody was preincubated with NES peptide or an unrelated control peptide, and radioactively labeled XAP1 was added subsequently. Although a control peptide did not interfere with XAP1 immunoprecipitation (Fig. 2B, control), the NES peptide abolished immunoprecipitation of XAP1 (Fig. 2B, NES). Next, the scaffold variants VGGG, VGG, and VGUG were tested for binding to the anti-NES antibody. VGGG was immunoprecipitated like XAP1 and XAP2; in contrast, VGG and VGUG were not immunoprecipitated (Fig. 2C). This indicated that altering the G-scaffold either weakens or interferes with binding to the antibody or alters the folding of the variants. Because no differences in affinity or binding specificity of XAP1–9 for the anti-NES antibody were observed, subsequent experiments were performed only with XAP1, XAP2, and the scaffold variants.
XAPs Inhibit cap- and Rev-Dependent RNA Export.
It has been suggested that cap-dependent export of U1snRNA and Rev-dependent export of a reporter RNA (pAd46) might share a common export factor, because BSA-coupled NES peptides inhibited export of both RNAs (7). The export of tRNA was not inhibited, indicating that an independent pathway is used by tRNA. If the XAPs would be structural mimics of the NES peptide, XAPs as well might be able to inhibit RNA export specifically.
To test this idea, radioactively labeled m7GpppG-capped U1ΔD, an U1snRNA mutant that is exported like U1wt but, unlike U1wt, is unable to return to the nucleus, was injected into the nucleus of oocytes together with uncapped competitor RNAs. U6snRNA was coinjected as a marker for nuclear injection (U6snRNA is specifically retained in the nucleus), and tRNA was coinjected as a control for unspecific inhibition of transport. When no competitor was injected, a significant amount of U1 and tRNA had been transported into the cytoplasm, whereas U6 was exclusively nuclear, indicating that no RNA had been injected into the cytoplasm (Fig. 3A, −). In contrast, coinjection of XAP1 inhibited specifically U1 export (Fig. 3A, XAP1). Coinjection of VTL did not affect RNA export (Fig. 3B, VTL), suggesting that a G-quartet structure is essential for inhibition. As a further demonstration of specificity of export inhibition the scaffold variants VGG and VGGG were coinjected with the transport substrates. Again, as observed with XAP1, VGGG inhibited specifically export of U1 but not export of tRNA (Fig. 3B, VGGG), whereas VGG, a variant not binding to the anti-NES antibody, did not affect RNA export (Fig. 3B, VGG). Next, Rev-dependent transport of Ad46 was analyzed (13). Ad46 preincubated with Rev was transported into the cytoplasm, whereas in the absence of Rev, Ad46 was exclusively nuclear (Fig. 3C, no comp., +/− Rev). Coinjection of XAP1 inhibited export of Ad46 (Fig. 3C, XAP1), whereas coinjection of VGG did not (Fig. 3C, VGG), indicating that XAP1 and Rev compete for a common transport factor (twice as much U6/Ad46/Rev were injected in the presence of competitor RNA compared with Fig. 3C Left; however, the absolute amount of Ad46 in the cytoplasm does not increase because transport of rev/Ad46 complexes was already saturated by the amount injected in the absence of competitor). The inhibition experiments show a clear correlation between the ability to bind to the anti-NES antibody and inhibition of export via the NES-/cap-dependent pathway. RNA binding to the antibody blocks U1 and Ad46 export; RNA not binding to the antibody does not inhibit transport.
Export of XAPs Transcribed in Vivo.
Nuclear export of RNA is believed to be an active process dependent on the presence of export signals. One of these signals is the m7G-cap structure. This was demonstrated by expressing U1snRNAs with the U6 promoter, a RNA pol III promoter. The pol III U1snRNAs, lacking the pol II-specific cap structure and therefore lacking the export signal, were not exported from the nucleus (17). If the XAPs were binding to the NES receptor, they should be exported even when transcribed by RNA pol III, because a cap structure would not be required anymore for active transport. To test this, XAPs and variants were expressed in oocytes with the hU6 promoter, in analogy to the earlier work with U1snRNA (Fig. 4A). Whereas U6snRNA was present only in the nucleus, most of XAP1 and XAP2 was present in the cytoplasm (Fig. 4B, XAP1 and XAP2). Identical results were obtained when pol III VGGG genes were injected (data not shown). Curiously, the variants VGG and VGUG were also found in the cytoplasm (Fig. 4B, VGG and VGUG). Because these variants were not binding detectably to the anti-NES antibody in vitro, either immunoprecipitation required stronger interactions than NES-receptor-mediated export in oocytes, or very small RNAs might be able to leave the nucleus passively.
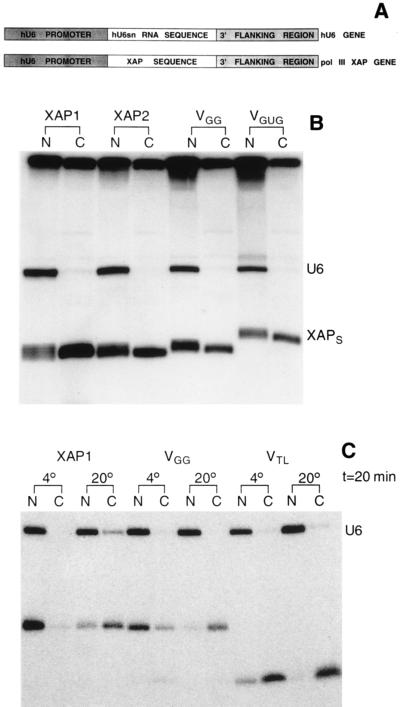
Figure 4.
Expression of XAPs in vivo. (A) Construction of the pol III XAP genes. Restriction sites were introduced at the 3′ end of the hU6 promoter and at the 5′ end of the 3′ flanking region of the hU6snRNA gene. The first 6 bp of the XAP-stem sequence were changed, with respect to the sequence shown in Fig. 2, to facilitate the incorporation of a restriction site sequence into the XAP stem. In addition, the start site was changed to GAG because a higher level of expression was obtained, compared with clones starting with GGG. (B) Intracellular localization of XAPs transcribed by RNA pol III. Equal amounts of hU6 genes and pol III XAP-genes (XAP1, XAP2, VGG, VGUG) were injected into the nucleus of oocytes together with [α-32P]GTP as described (17). Twelve hours later oocytes were separated into nuclear (N) and cytoplasmic (C) fractions, and extracted RNA was analyzed on a denaturing acrylamide gel. (C) Temperature sensitivity of XAP export. XAP1, VGG, and VTL were transcribed in vitro (all uncapped) and injected into the nucleus of oocytes together with U6snRNA. Oocytes were kept for 20 min at 4° or at 20° and separated into nuclear (N) and cytoplasmic (C) fractions, and extracted RNA was analyzed on a denaturing acrylamide gel.
Temperature Sensitivity of XAP Export.
Rev-mediated export, like active transport across the nuclear membrane in general, is temperature sensitive. By analogy to what is known about nuclear import, it is assumed that at low temperature export factors still bind the substrate but the translocation step is blocked. To distinguish between active transport and diffusion, XAPs were injected into the nucleus of oocytes, and export at low and normal temperature was analyzed.
In oocytes kept at low temperature XAP1 was present only in the nucleus (Fig. 4C, 4°), whereas at normal temperature about 75% of XAP1 was present in the cytoplasm (Fig. 4C, XAP1, 20°; some leakage had occurred at the time of injection, indicated by the presence of U6 in the cytoplasm, estimated to be about 5%). Identical observations were made when export of variants VGUG and VGGG was analyzed; these RNAs were exclusively found in the nucleus at low temperature (data not shown). A similar, but not identical, result was obtained when injecting VGG. At low temperature about 5% of VGG was found in the cytoplasm (Fig. 4C, VGG, 4°), and at normal temperature more than 90% of VGG was cytoplasmic (Fig. 4C, VGG, 20°). This suggested that, although some VGG left the nucleus by a diffusion-mediated process, transport was predominantly active. That small RNAs not having either nuclear retention signals or positive transport signals could be able to leave the nucleus by diffusion is strongly suggested by the intracellular localization found for VTL. At low temperature more than 90% was present in the cytoplasm (Fig. 4C, VTL). In summary, RNAs interacting strongly with the anti-NES antibody (VGGG, XAP1, XAP2) and capable of inhibiting U1 export were retained in the nucleus when active transport was blocked. In contrast, RNA not binding to the anti-NES antibody and unable to inhibit U1 export (VTL) apparently left the nucleus by a diffusion-mediated process. Unexpectedly, VGG seems to be transported actively although not binding to the antibody nor inhibiting U1 export. Antibody and NES receptor might have overlapping but not identical requirements for binding. In addition, the affinity of VGG for the receptor might be too low to compete with genuine substrates for binding but sufficient for interaction in the absence of competition (see below). These observations could indicate that nuclear retention at low temperature might have been due to binding to the NES receptor.
XAP Binding Activities in Nuclear Extracts.
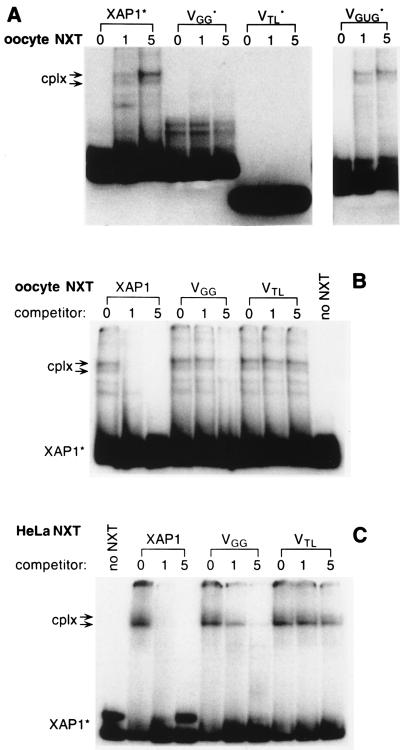
The results of the experiments presented so far would be compatible with the idea that the XAPs were binding to an NES receptor in oocytes. It was of interest to see whether XAP-specific binding activities were detectable in nuclear extracts. To this end, radioactively labeled XAPs and XAP variants were incubated in oocyte nuclear extracts, and complexes formed were analyzed on nondenaturing acrylamide gels. When examining XAP1, XAP2, VGGG, and VGUG, routinely two complexes of reduced electrophoretic mobility were observed, the slower migrating complex being more abundant (Fig. 5A, cplx). Faster-migrating complexes seen below the two prominent bands in oocyte nuclear extracts are likely to be a result of proteolytic degradation, because their presence or absence was highly variable with different extract preparations. No complexes were visible with the tetraloop stem VTL (Fig. 5A, VTL) or with VGG (Fig. 5A, VGG).
Figure 5.
Detection of XAP-binding activities in extracts. (A) Radioactively labeled, uncapped (indicated by ∗) XAP1, VGG, VTL, and VGUG were incubated with increasing amounts (lanes 1, 7 μg; lanes 5, 35 μg) of oocyte nuclear extracts (NXT), and specific complexes formed (cplx) were analyzed on nondenaturing acrylamide gels. (B) RNA competition. Oocyte NXT (15 μg) was preincubated with increasing amounts (lanes 1, 170 pmol; lanes 5, 850 pmol) of unlabeled XAP1, VGG, and VTL for 20 min at 20°. Radioactively labeled XAP1* was added and incubated for an additional 15 min at 20°, and complexes formed (cplx) were analyzed on nondenaturing acrylamide gels. (C) Same as in B, but with HeLa NXT (15 μg).
A competition assay was employed to establish that complex formation of XAPs in nuclear extracts correlated with RNA export inhibition seen in oocytes. Oocyte nuclear extract was preincubated with unlabeled XAPs or variants for 20 min before adding radioactively labeled XAP1. At the lowest concentration used, unlabeled XAP1 practically abolished complex formation of the labeled XAP1 added subsequently (Fig. 5B, XAP1, lane1), whereas unlabeled VGG had to be added in much higher concentrations (Fig. 5B, VGG, lane 5). In contrast, VTL was completely ineffective in sequestering XAP-binding activities (Fig. 5C, VTL). The results of this competition experiment would indicate that XAP1 and VGG were binding to the same factor(s) but with different affinities. This is also suggested by the fact that no complex formation was observed when labeled VGG was incubated in nuclear extracts. Complexes formed by VGG might not have been sufficiently stable to survive electrophoretic analysis. However, an excess of unlabeled VGG could interfere with complex formation of XAP1, because the two-step incubation renders the competition experiment more sensitive than direct complex analysis. The unlabeled competitor has to sequester the factor(s) only during the short second incubation step, when labeled XAP1 is present. Subsequent disintegration of VGG complexes during electrophoresis would not allow XAP1 complex formation. Therefore, the ability of the variants to compete for complex formation of XAP1 in nuclear extracts correlates with their capability of inhibiting cap- and Rev-dependent RNA export and also with the observed nuclear retention at low temperature. XAP1 was the strongest competitor for complex formation in vitro and exclusively nuclear in oocytes at low temperature. VGG was a weaker competitor in vitro, and some VGG left the nucleus at low temperature, whereas VTL was not competing in vitro nor retained in the nucleus.
Finally, given the conservation of transport factors between species, complexes formed by XAP1 were studied in HeLa nuclear extracts. As before, two prominent complexes were observed, the only visible difference being that in HeLa extracts the lower band was prominent rather than the upper band as in oocyte nuclear extracts. Specificity was confirmed as above by preincubation of the extract with various competitor RNAs (Fig. 5C), demonstrating that HeLa nuclear extracts also contain a binding activity that might represent an NES receptor.
Anti-idiotype RNA.
Antibodies have been used previously to select aptamers from combinatorial RNA libraries with antipeptide sera (22) or with monoclonal antibodies raised against proteins (16, 24). In all cases the RNA competed with the peptide or protein antigen for binding, implying that the aptamers were directed against the antigen-binding site. However, because the structures of the antibody–RNA and antibody–peptide complexes have not been solved, it is only certain that peptides and RNAs had overlapping binding sites.
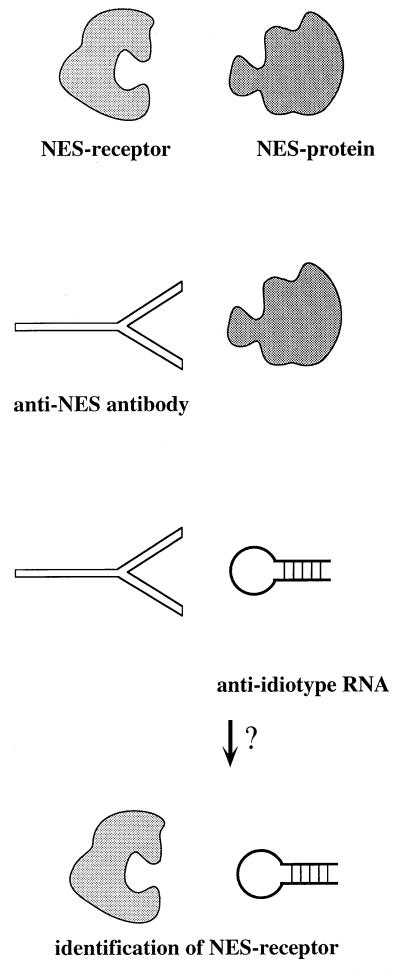
The approach reported here goes one step further. The anti-NES antibody is directed against a domain involved in protein–protein interactions, the binding of Rev to an NES receptor. That the XAPs would have competed with NES peptides for binding to the antibody was anticipated; the interesting question was whether the peptide mimic XAP would have been able to recognize an NES receptor. This should be possible only if the variable loop of the antibody formed a structure similar to that of the receptor domain normally recognizing the NES (Fig. 6). This reasoning is also the underlying principle for experiments using anti-idiotype antibodies, which have been used successfully as specific probes for receptors (25, 26). However, the validity of the experiments for the identification of unknown receptors to known ligands remained controversial because of undesired cross-reactivities of generated antibodies and because of experiments addressing the probability to produce an anti-idiotype antibody mimicking the receptor-binding domain of the ligand not only functionally but also structurally (27, 28). To which extent anti-idiotype RNA will be prone to artifacts remains to be tested.
Figure 6.
Model incorporating the experimental observations. The XAPs could act like anti-idiotype RNAs, because the antigen-binding site of the anti-NES peptide antibody might be structurally related to the NES-binding domain of an NES receptor.
The specific export inhibition by XAPs in oocytes strongly suggests that the anti-NES antibody shares a structural similarity with an NES receptor. The NES (or the nuclear localization signal, NLS) might be a particularly favorable target for anti-idiotype approaches, because rather than a discrete sequence, the requirement for receptor binding could be a more general character, e.g., a hydrophobic (or basic for the NLS) surface of a certain shape or dimension. For the nuclear pore complex this could be an efficient way of regulating transport, because it would allow a limited number of nuclear pore receptors to interact with a large number of soluble adapters binding to specific subsets of substrates. It is intriguing that a three-layered G-quartet structure was selected as a mimic of the NES, because both have a strong hydrophobic character. The planes of the G-quartet might stack on the constant stem and enable intercalation of or interaction with aromatic and hydrophobic amino acid side chains of the NES receptor. Apart from several aptamers isolated from combinatorial libraries (2, 16), G-quartets are formed, for example, by telomeric DNA sequences (29). There, the unusual stability of G-quartets might help to protect telomeres from degradation. Interstrand RNA G-quartets have been proposed as a way to align two HIV genomic RNA molecules in a parallel fashion for packaging (30). Although several Rev-interacting clones exhibiting structural homology to nucleoporins were isolated in two-hybrid screens with the Rev-NES, none of the proteins could be shown to interact directly with Rev when purified proteins were examined (31). In this respect the detection of XAP-binding activities in oocyte- and HeLa-nuclear extract might be a welcome addition to the currently used experimental systems to identify NES receptors.
Acknowledgments
We thank Riccardo Cortese for support at Istituto di Ricerche di Biologia Moleculare, Irene Bozzoni and her lab for hospitality and helpful discussions, Elisa Caffarelli for oocyte extracts, Manuela Emili for artwork, Elsebet Lund for exchanging data, and Angus Lamond and Iain Mattaj for valuable comments on the manuscript.
ABBREVIATIONS
- NES
nuclear export signal
- NXT
nuclear extract
- XAP
export aptamer
References
- 1.Fedor M J. Struct Biol. 1994;5:267–269. doi: 10.1038/nsb0594-267. [DOI] [PubMed] [Google Scholar]
- 2.Gold L, Polisky B, Uhlenbeck O, Yarus M. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- 3.Kenan D J. Trends Biol Sci. 1994;19:57–64. doi: 10.1016/0968-0004(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 4.Gerace L. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 5.Koepp D M, Silver P A. Cell. 1996;87:1–4. doi: 10.1016/s0092-8674(00)81315-x. [DOI] [PubMed] [Google Scholar]
- 6.Görlich D, Mattaj I W. Science. 1996;271:1513–1518. doi: 10.1126/science.271.5255.1513. [DOI] [PubMed] [Google Scholar]
- 7.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 8.Michael W M, Choi M, Dreyfuss G. Cell. 1995;83:415–422. doi: 10.1016/0092-8674(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 9.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 10.Emerman M, Vazeux R, Peden K. Cell. 1989;57:1155–1165. doi: 10.1016/0092-8674(89)90053-6. [DOI] [PubMed] [Google Scholar]
- 11.Felber B K, Hadzopoulou-Cladaras M, Cladaras C, Copeland T, Pavlakis G N. Proc Natl Acad Sci USA. 1989;86:1495–1499. doi: 10.1073/pnas.86.5.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Malim M H, Hauber J, Le S Y, Maizel J, Cullen B R. Nature (London) 1989;338:254–257. doi: 10.1038/338254a0. [DOI] [PubMed] [Google Scholar]
- 13.Fischer U, Meyer S, Teufel M, Heckel C, Lührmann R, Rautmann G. EMBO J. 1994;13:4105–4112. doi: 10.1002/j.1460-2075.1994.tb06728.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellington A D, Szostak J W. Nature (London) 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 15.Tuerk C, Gold L. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 16.Hamm J. Nucleic Acids Res. 1996;24:2220–2227. doi: 10.1093/nar/24.12.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hamm J, Mattaj I W. Cell. 1990;63:109–118. doi: 10.1016/0092-8674(90)90292-m. [DOI] [PubMed] [Google Scholar]
- 18.Hamm J, Darzynkiewicz E, Tahara S M, Mattaj I W. Cell. 1990;62:569–577. doi: 10.1016/0092-8674(90)90021-6. [DOI] [PubMed] [Google Scholar]
- 19.Zasloff M, Rosenberg M, Santos T. Nature (London) 1982;300:81–84. doi: 10.1038/300081a0. [DOI] [PubMed] [Google Scholar]
- 20.Gandini-Attardi D, Baldi I M, Mattoccia E, Tocchini-Valentini G P. Methods Enzymol. 1990;181:510–517. doi: 10.1016/0076-6879(90)81148-n. [DOI] [PubMed] [Google Scholar]
- 21.Giver L, Bartel D, Zapp M, Pawul A, Green M R, Ellington A D. Nucleic Acids Res. 1993;21:5509–5516. doi: 10.1093/nar/21.23.5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tsai D E, Kenan D J, Keene J D. Proc Natl Acad Sci USA. 1992;89:8864–8868. doi: 10.1073/pnas.89.19.8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cheong C, Moore P B. Biochemistry. 1992;31:8406–8414. doi: 10.1021/bi00151a003. [DOI] [PubMed] [Google Scholar]
- 24.Doudna J A, Cech T R, Sullenger B A. Proc Natl Acad Sci USA. 1995;92:2355–2359. doi: 10.1073/pnas.92.6.2355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sege K, Peterson P A. Proc Natl Acad Sci USA. 1978;75:2443–2447. doi: 10.1073/pnas.75.5.2443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gaulton G N, Greene M I. Annu Rev Immunol. 1986;4:253–280. doi: 10.1146/annurev.iy.04.040186.001345. [DOI] [PubMed] [Google Scholar]
- 27.Flügge U I, Weber A, Fischer K, Lottspecht F, Eckerskorn C, Waegemann K, Soll J. Nature (London) 1991;353:364–367. [Google Scholar]
- 28.Davis S J, Schockmel G A, Somoza C, Buck D W, Healey D G, Rieber E P, Reiter C, Williams A F. Nature (London) 1992;358:76–79. doi: 10.1038/358076a0. [DOI] [PubMed] [Google Scholar]
- 29.Rhodes D, Giraldo R. Curr Opin Struct Biol. 1995;5:311–322. doi: 10.1016/0959-440x(95)80092-1. [DOI] [PubMed] [Google Scholar]
- 30.Sundquist W I, Heaphy S. Proc Natl Acad Sci USA. 1993;90:3393–3397. doi: 10.1073/pnas.90.8.3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]