Abstract
LEF-1 (lymphoid enhancer-binding factor 1) is a cell type-specific member of the family of high mobility group (HMG) domain proteins that recognizes a specific nucleotide sequence in the T cell receptor (TCR) α enhancer. In this study, we extend the analysis of the DNA-binding properties of LEF-1 and examine their contributions to the regulation of gene expression. We find that LEF-1, like nonspecific HMG-domain proteins, can interact with irregular DNA structures such as four-way junctions, albeit with lower efficiency than with specific duplex DNA. We also show by a phasing analysis that the LEF-induced DNA bend is directed toward the major groove. In addition, we find that the interaction of LEF-1 with a specific binding site in circular DNA changes the linking number of DNA and unwinds the double helix. Finally, we identified two nucleotides in the LEF-1-binding site that are important for protein-induced DNA bending. Mutations of these nucleotides decrease both the extent of DNA bending and the transactivation of the TCRα enhancer by LEF-1, suggesting a contribution of protein-induced DNA bending to the function of TCRα enhancer.
Keywords: lymphoid enhancer-binding factor 1, T cell receptor α enhancer
Regulation of DNA transactions such as transcription, recombination, and replication often involve proteins that induce various structural distortions in DNA. These protein-induced changes in DNA structure that include bending and unwinding have a possible role in the assembly of higher-order nucleoprotein structures (1–4). High mobility group (HMG) proteins in eukaryotes have been shown to bend the DNA helix and have been implicated as “architectural elements” in the assembly of stereospecific protein:DNA complexes (reviewed in refs. 3 and 4). HMG domain proteins share an 85-aa structural motif, termed the HMG domain, which recognizes DNA through the minor groove and induces a sharp bend in the DNA double helix (5–12). Several HMG domain proteins were also found to interact with irregular DNA structures in a sequence-nonspecific manner. In particular, HMG-1 interacts with kinks in the DNA generated by cis-platin adducts and with four-way junction DNA (7, 13).
HMG domain proteins can be grouped into two classes according to the number of HMG domains present in the proteins and the specificity of nucleotide sequence recognition (3, 14). Members of the first class of proteins, represented by HMG1 and UBF, contain multiple HMG domains (15–17). These proteins recognize DNA with a very modest sequence specificity and bind irregular DNA structures with high affinity (16–18). LEF-1 and the testing-determining factor SRY are representatives of the other class of HMG domain proteins that recognize variants of the consensus nucleotide sequence 5′-CTTTGAA through a single HMG domain (5, 6, 10, 19–22). In prokaryotes, HU protein and the integration host factor (IHF) represent functional analogs of HMG domain proteins and share similar DNA-binding properties despite differences in the structural fold (23, 24). A functional role for protein-induced DNA bending has been demonstrated in the assembly of a higher-order nucleoprotein complex at the attP site of bacteriophage lambda (2). In this context, IHF, which bends the DNA helix, facilitates the recognition of nonadjacent binding sites by Int protein. A role of IHF-induced DNA bending was inferred from experiments in which the IHF binding site was substituted by heterologous DNA-bending proteins or an intrinsic DNA bend (25–27). The HMG domain of LEF-1, which bends the DNA helix by 120°–130°, was also found to substitute for IHF in the assembly of a functional intasome (6). However, evidence for a role of LEF-induced DNA bending in the assembly and function of a multiprotein enhancer complex is limited.
LEF-1 regulates the minimal TCRα enhancer together with at least three other proteins: PEPB2α (CBFα, AML-1), Ets-1, and members of the ATF/CREB family (28). LEF-1 binds in the center of the enhancer and stabilizes a ternary complex composed of PEBP2α and Ets-1 at one end of the enhancer. Stabilization of the ternary complex is also dependent on an ATF/CREB protein bound at the other side of the enhancer, raising the possibility that bending by LEF-1 may juxtapose ATF/CREB- and Ets-1-binding sites (28). Support for a potential role of DNA bending was inferred from transfection experiments in which the HMG domain allowed a partial activation of enhancer function (28, 29). In addition, LEF-1 has a context-dependent activation domain that contributes to maximal function of the TCRα enhancer (30–32).
In this study, we extend our analysis of the role of DNA bending by LEF-1 for the regulation of TCRα enhancer function. We identified mutations in the LEF-1-binding site that decrease the angle of protein-induced DNA bending and reduce TCRα enhancer function in transfection experiments. Moreover, we show that LEF-1 binds to specific linear duplex DNA with higher affinity than to four-way junction DNA. Finally, we show that the LEF-1-induced DNA bend has a specific directionality and that binding of LEF-1 to circular DNA results in a significant unwinding of the double helix.
MATERIALS AND METHODS
Plasmid Construction to Determine the Direction of the Bend Angle.
Double-stranded oligonucleotides (phase 0, 5′-cGTAGGGCACCCTTTGAAGCTCTC CCCtcgac; phase 1, 5′-cGTAGGGCACCCTTTGAAGCTCTCCCtctcgac; phase 3, 5′-cGTAGGGCACCCTTTGAAGCTCTCCCCtcgctcgac; phase 5, 5′-cGTAGGGCACCC TTTGAAGCTCTCCCCtcgacctcgac; phase 7, 5′-cGTAGGGCACCCTTTGAAGCTC TCCCCtcgacacctcgac; phase 9, 5′-cGTAGGGCACCCTTTGAAGCTCTCCCctcgacac tgctcgac; phase 11, 5′-cGTAGGGCACCCTTTGAAGCTCTCCCCtcgacactgacctcgac; only the upper strand is shown) consisting of a LEF-1-binding site and a spacer of variable length (lowercase) were ligated into plasmid pTZ19U (United States Biochemical) cleaved with SacI and SalI. A DNA fragment containing six helically phased A tracts was isolated from plasmid pBEND1 (gift of S. Goodman and H. Nash, National Institutes of Health, Bethesda, MD) by restriction with XbaI and ligated into XbaI-cleaved plasmid pTZ18U (United States Biochemical). The final phasing constructs were generated by ligation of PstI and AlwNI DNA fragments containing the LEF-1-binding site and the gene for β-lactamase with a PstI and AlwNI DNA fragment containing the A tract DNA and the origin of replication. The final constructs were verified by DNA sequencing. For the phasing analysis, a 475-bp PvuII DNA fragment was isolated from each construct and radiolabeled with T4 polynucleotide kinase and [γ-32P]ATP. The different labeled DNA fragments were incubated with purified LEF-HMG domain peptide and analyzed in a gel electrophoretic mobility-shift assay.
Construction of Synthetic Four-Way Junction DNA.
The four-way junction DNA was generated by annealing the following oligonucleotides (1, 5′-CCCTATAACCCCTGAGCGGAATTCCAGTCTGATAA; 2, 5′-GTAGTCGT GATAGGTGCAGGGGTTATAGGG; 3, 5′-AACAGTAGCTCTTATTCGAGCTCGC GCCCTATCACGACTA; 4, 5′-TTTATCAGACTGGAATTCCGCCGCGAGCTCGAATAAGAGCTACT-GT) as described (33). Annealing of oligonucleotides 1 and 6 (5′-TTATCAGACTGGAATTCCGCTCAGGGGTTATAGGG) and 3 and 5 (5′-GTAGTCGTGATAGGGCGCGAGCTCGAATAAGAGCTACTGT) generated two linear, double-stranded oligonucleotides that were used as control DNA probes to distinguish between sequence- and structure-specific DNA binding.
Electrophoretic Mobility-Shift Assay, DNaseI Footprint Experiments, and Determination of the Equilibrium Dissociation Constants.
Electrophoretic mobility-shift assays and DNaseI footprint experiments were performed essentially as previously described (20), with the concentration of LEF-HMG domain peptide specified in the figure legends. The dissociation constants for specific DNA binding were determined by performing saturation binding experiments as described (5).
Analysis of LEF-1-Mediated DNA Underwinding.
Oligonucleotides comprising one or two LEF-1-binding sites were ligated into the SalI site in plasmid Bluescript (Stratagene). Supercoiled plasmid DNA (200 ng) was nicked with DNaseI in the presence of ethidium bromide and incubated with varying amounts of purified LEF-HMG domain peptide for 30 min at 37°C. The DNA nicks were closed by adding T4 DNA ligase (34), and the topoisomer population was analyzed in 0.8% agarose gels in TBE buffer (90 mM Tris/64.6 mM boric acid/2.5 mM EDTA, pH 8.3) in the presence of different concentrations of chloroquine. The direction of the topological change was analyzed by determining the center of mass of the topoisomer population.
Circular Permutation Assay, Calculation of DNA Bending Parameters, and DNA Transfections.
The circularly permuted DNA fragments containing the wild-type and mutant LEF-1-binding sites were constructed and analyzed as described in ref. 6. Bend angles were estimated by determining the ratio of the mobility of the protein–DNA complex to the mobility of the unbound DNA in a mobility-shift assay (35). DNA transfections into T cells and determination of chloramphenicol acetyltransferase (CAT) activity were performed as described in ref. 30.
RESULTS
Binding of the HMG Domain of LEF-1 to Four-Way Junction DNA.
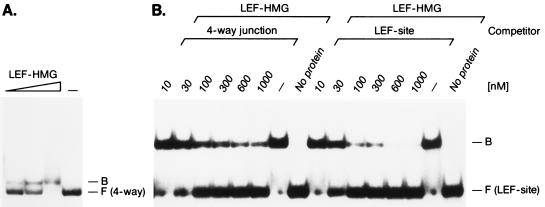
Several studies indicated that HMG domain proteins can recognize irregular DNA structures in a sequence-independent manner (36). In particular, the HMG domains of HMG-1 and human SRY were reported to recognize four-way junctions with higher affinity than linear duplex DNA that contains a binding site for SRY (7, 13, 18, 37). To examine whether the HMG domain of LEF-1 also shows a preference for binding to distorted DNA, we incubated purified LEF-HMG domain peptide with four-way junction DNA lacking a specific LEF-1-binding site. The HMG domain formed a complex with the four-way junction DNA probe in a concentration-dependent manner (Fig. 1A). In contrast, no binding of the LEF-HMG domain was observed with linear duplex oligonucleotides comprising the same nucleotide sequence as the four-way junction DNA (data not shown).
Figure 1.
Binding of the HMG domain of LEF-1 to four-way junction DNA. (A) Increasing amounts (3 ng, 10 ng, and 30 ng) of purified LEF-HMG domain peptide were incubated with radiolabeled four-way junction DNA and complex-formation-analyzed in electrophoretic mobility-shift assays. At the highest protein concentration of LEF-HMG domain a significant portion of the DNA probe formed protein/DNA aggregates that were retained in the well. The mobilities of the protein/DNA complexes (B) and the free probes (F) are indicated. (B) Analysis of the specificity of LEF-HMG domain binding to the four-way junction DNA in electrophoretic mobility-shift assays. Ten nanograms of purified HMG domain of LEF-1 was incubated with an oligonucleotide comprising a LEF-1-binding site in the presence of increasing amounts of unlabeled four-way junction DNA or specific DNA as competitor. The positions of the protein/DNA complexes (B) and the free probes (F) are indicated.
To quantitate the relative binding affinities of the LEF-1-HMG domain for four-way junction DNA vs. the duplex TCR-1 oligonucleotide containing a LEF-1-binding site, we examined the formation of a LEF-HMG:TCR-1 complex in the presence of excess of unlabeled TCR-1 oligonucleotide or four-way junction DNA (Fig. 1B). Half-maximal competition was detected with the duplex TCR-1 DNA at 35 nM and with the four-way junction DNA at 70 nM. As a control, inefficient competition was observed with nonspecific duplex DNA (data not shown). Thus, the HMG domain of LEF-1 can also recognize irregular DNA structures, albeit with an approximately 2-fold lower affinity than a specific duplex binding site.
Specific Binding of the LEF-HMG Domain to Circular DNA Induces Negative Superhelical Turns.
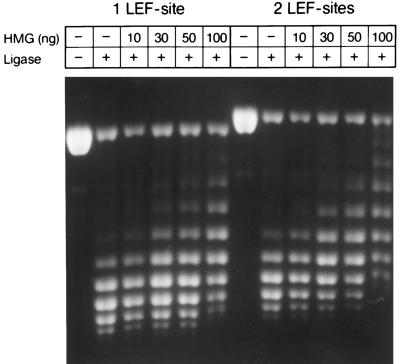
Previous experiments indicated that DNA binding by the HMG domain of LEF-1 induces a sharp bend of 120°–130° in the DNA helix (6, 12). The determination of DNA bending by circular permutation analysis, however, does not distinguish between a planar bend and a left-handed supercoil (38). Moreover, the severe distortion of the DNA double helix upon binding of the HMG domain of LEF-1 would be expected to induce negative superhelical turns (39). To address this possibility, we nicked plasmid DNA containing one or two LEF-1-binding sites with DNaseI and incubated the nicked DNA with increasing amounts of purified LEF-HMG domain peptide. Subsequently, we closed the DNA duplex with DNA ligase to preserve any changes in the topology of the DNA. Analysis of the topoisomers by agarose gel electrophoresis in the presence of chloroquine revealed that the addition of 30 ng of LEF-HMG domain shifted the topoisomer distribution (Fig. 2). A comparison of this analysis with the analysis of the topoisomer distribution in agarose gels in the absence of chloroquine indicated that the HMG domain of LEF-1 induces negative supercoils (Fig. 2; data not shown). Moreover, the topoisomer distribution changed with an increase in the number of the LEF-1-binding sites, indicating a site dependence of this effect. Although the data of this analysis are consistent with an unwinding of approximately half a helical turn per HMG domain, the modest sequence specificity of LEF-1 (5) and the lack of knowledge of the precise stoichiometry of the protein/DNA complex preclude a quantitative analysis of unwinding.
Figure 2.
Change in the linking number of DNA upon binding of the HMG domain of LEF-1. Five micrograms of singly nicked plasmid DNA containing either one or two LEF-1-binding sites was incubated with the indicated amounts of purified LEF-HMG domain peptide. The DNA nicks were closed by adding T4 DNA ligase, and the individual topoisomers were analyzed by agarose gel electrophoresis in the presence of 1 μg/ml chloroquine.
Detection of the Relative LEF-1 Bend Direction.
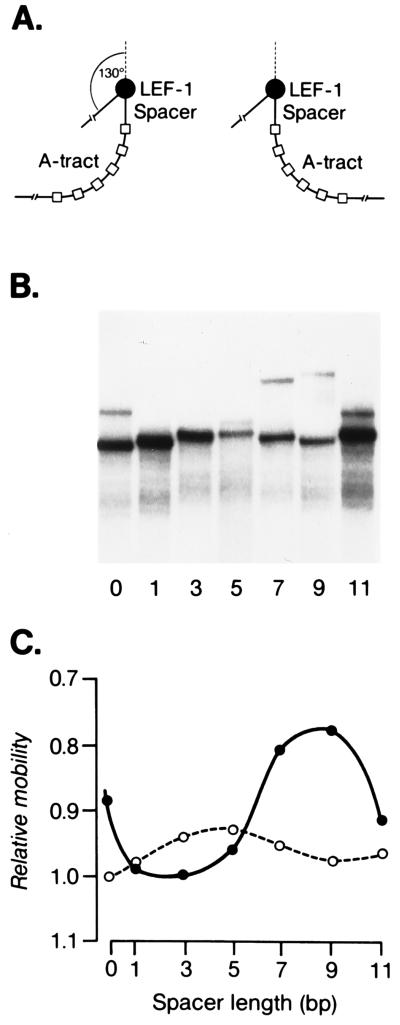
The HMG domain of LEF-1 was shown to substitute for the function of IHF in a site-specific recombination assay (6). Recombination was found to be dependent on a specific position and orientation of the LEF-1-binding site relative to other factor-binding sites, suggesting a precise directionality of the LEF-induced DNA bend. To determine the direction of the HMG domain-induced DNA bend, we extended our previous analysis by performing a phasing experiment (40, 41). Toward this end, we used DNA probes in which the position of the LEF-1-binding site was changed in increments of 1 or 2 nt relative to an intrinsic DNA bend of A tracts (Fig. 3). The LEF-HMG:DNA complex formed with the DNA probe containing a spacer of 3 nt migrated with the fastest mobility. In this phasing relationship, the HMG-domain-induced DNA bend and the intrinsic DNA bend are in an opposite orientation and counteract each other. By contrast, the protein:DNA complex formed with a DNA probe containing a spacer with 9 nt migrated with the slowest mobility, suggesting that the DNA bends are in the same direction. The free DNA probes also varied with a periodicity of approximately 10 bp, suggesting that the DNA upstream of the A tracts DNA contains an intrinsic but modest bend. Assuming a periodicity of 10.5 bp per helical turn of DNA and based on the bending at the center of A tracts toward the minor groove of the DNA, these data suggest that the LEF-induced DNA bend is directed toward the major groove of the DNA helix. This directionality of the LEF-induced helix is consistent with the solution structure of the LEF-HMG:DNA complex (12).
Figure 3.
Phasing analysis of DNA bending by the HMG domain of LEF-1. (A) Schematic structures of the protein/DNA complexes in which the protein-induced DNA bend is in phase with the intrinsic DNA bend (spacer length of 9) or out of phase (spacer length of 3). The filled circle indicates the LEF-1-binding site, which is separated by DNA of variable lengths (70–81 nt) from six A tracts. (B) Electrophoretic mobility-shift assay of purified LEF-HMG domain peptide (3 ng) bound to phased DNA probes. The numbers below indicate nucleotides inserted between the LEF-1-binding site and the A tract DNA. (C) Relative mobilities of protein/DNA complexes as a function of the spacer length (solid line). The broken line shows the variation in the mobilities of the free DNA probes.
Effects of Nucleotide Mutations on DNA Bending.
To gain insight into the relationship between DNA binding and DNA bending, we searched for mutations in the LEF-1-binding site that change the bending angle without affecting the binding affinity. In saturation binding experiments, the equilibrium dissociation constants for the interaction of the LEF-HMG domain with the mutant binding site probes TCR-8 and TCR-15 were increased markedly (Table 1). By contrast, three other mutant binding sites (probes TCR-11, TCR-12, and TCR-13) were recognized by the LEF-HMG domain with an affinity that was only 2- to 3-fold higher than that of the TCR-1 probe, which represents the wild-type LEF-1-binding site (5).
Table 1.
Schematic structure of wild-type and mutant LEF-1-binding sites
| Binding site | Nucleotide sequence | Kd [M] | Bending angle |
|---|---|---|---|
| 6084 | |||
| TCR-1 | 5′ GTAGGGCACCCTTTGAAGCTCTCCC | 1 × 10−9 | 130° |
| TCR-8 | 5′ ---------------TTT------- | 1 × 10−8 | 90° |
| TCR-11 | 5′ -----------A---TTΔ------- | 3 × 10−9 | 85° |
| TCR-12 | 5′ -----------------T------- | 2 × 10−9 | 90° |
| TCR-13 | 5′ -----------A------------- | 2 × 10−9 | 90° |
| TCR-15 | 5′ -----------A---TT-------- | 2 × 10−8 | 85° |
The determined equilibrium dissociation constants for binding of the HMG domain of LEF-1 to the different LEF-1-binding sites and the estimated angles of HMG-domain-induced DNA bending are indicated.
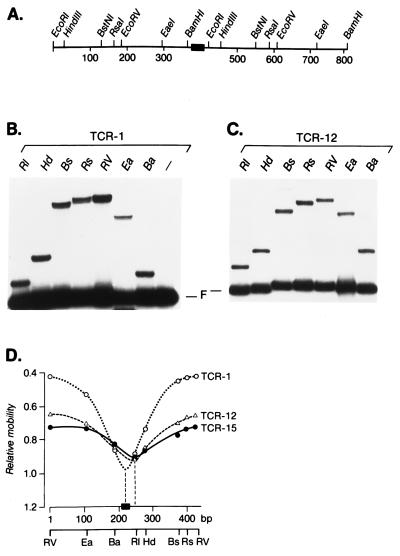
The extent of DNA bending by LEF-1 at these mutant sites was examined by circular permutation assays (Fig. 4). We generated sets of circularly permuted DNA probes with wild-type and mutant LEF-1-binding sites and determined the rates of migration of the protein:DNA complexes in electrophoretic mobility-shift assays (Fig. 4 B and C). Calculation of the relative rates of migration of the protein:DNA complexes as a function of the relative positions of the LEF-1-binding site within the DNA probes indicated that several mutant DNA probes have an altered DNA bend as compared with the wild-type probe (Fig. 4 B and C). The graphic representation of the electrophoretic mobility-shift data obtained with the mutant LEF-1-binding sites showed a significant 40°–45° decrease in the bend angle and a shift of the bend center by approximately 30 nucleotides toward the 3′ end of the DNA probes (Fig. 4D and Table 1). Taken together, these data indicate that various mutations in the LEF-1-binding site alter the DNA bending angle and the bend center. Notably, the bend angle did not correlate strictly with changes in the affinity of DNA binding by LEF-1. For example, the mutant sites TCR-8 and TCR-15 showed a decrease in both the binding affinity and the bend angle (Table 1). Mutant sites TCR-12 and TCR-13, however, are recognized by the LEF-HMG domain with almost wild-type affinity but show the same reduction in the bend angle as compared with the low-affinity sites.
Figure 4.
Circular permutation analysis of the DNA flexure induced by binding of the HMG domain of LEF-1. (A) Structure of the DNA fragment used to generate circularly permuted probes. The filled box shows the position of the wild-type or mutant LEF-1-binding site. (B) Electrophoretic mobility-shift assay of binding of the LEF-HMG domain to circularly permuted DNA probes. Five nanograms of purified HMG domain peptide of LEF-1 was incubated with circularly permuted DNA probes containing the TCR-1 (wild-type) binding site and electrophoresed in polyacrylamide gel. The positions of protein/DNA complexes and the free probes (F) are shown. (C) Five nanograms of purified HMG domain peptide of LEF-1 was incubated with circularly permuted DNA probes containing the TCR-12 (mutant) binding site and electrophoresed in polyacrylamide gel. (D) Determination of the center of bending of the different binding sites. The relative mobilities of the protein/DNA complexes as shown in B and C (and data not shown) were plotted as a function of the position of the wild-type or mutant LEF-1-binding sites relative to the probe ends. The filled box represents the wild-type or mutant LEF-1-binding site, and the numbers below indicate the distance in base pairs (bp) from the left-hand EcoRV (RV) site to the right-hand RV site. The bend centers were estimated by extrapolating the linear portions of the curves to a position on the DNA fragment.
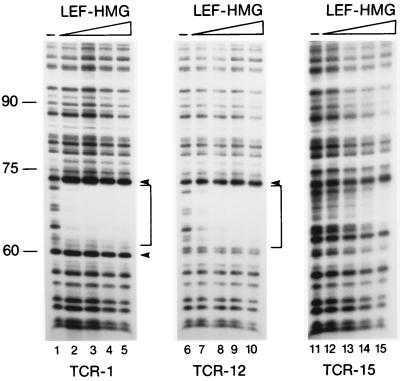
The observed shift in the bend center of the mutant LEF-1-binding sites raised the possibility that the introduced mutations revealed an intrinsic bend of the DNA or generated a cryptic binding site for LEF-1. To examine the binding of LEF-1 to the wild-type and the mutant sites, we performed a quantitative DNaseI footprint assay (Fig. 5). This experiment revealed a similar level of binding of the LEF-1-HMG domain peptide to the wild-type TCR-1 and mutant TCR-12 probes and a reduced level of binding to the mutant TCR-15 probe. Moreover, the position and extent of the DNaseI footprints obtained with the mutant TCR-12 and TCR-15 probes were identical to that of the wild-type TCR-1 probe, suggesting that the shift in the bend center in the mutant LEF-1-binding sites may reflect differences in the protein/DNA interaction rather than binding of LEF-1 to a cryptic downstream sequence. The resolution of the DNaseI footprint analysis, however, may not be sufficient to visualize subtle changes in DNA binding by LEF-1.
Figure 5.
In vitro DNaseI footprinting analysis of the HMG domain of LEF-1 bound to different LEF-1-binding sites. Increasing amounts (0 ng, 3 ng, 30 ng, and 100 ng) of purified LEF-HMG domain peptide were incubated with 5′ end-labeled DNA fragments containing the wild-type (TCR-1) or mutant (TCR-12 and TCR-15) LEF-1-binding site. Brackets indicate the region protected from DNaseI digestion.
Contribution of LEF-Induced DNA Bending to Regulation of TCRα Enhancer Function.
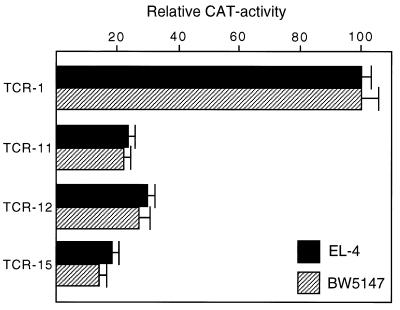
The observation that some mutations decreased the angle of LEF-induced DNA bending, without significantly changing the equilibrium dissociation constant for protein:DNA interaction, raised the question as to the effect of these mutations on the activity of the TCRα enhancer. We transfected T cells with reporter gene constructs in which the wild-type or various mutant TCRα enhancers have been linked to the minimal fos promoter and the CAT gene as indicator. A reproducible 4-fold reduction in CAT activity was observed with the TCR-11 and TCR-12 enhancer fragments carrying mutated LEF-1 binding sites (Fig. 6). Taking into account a 2-fold lower binding affinity of the TCR-12 site for LEF-1 and the partial change in DNA bending, this observed decrease in enhancer activity indicates that the protein-induced DNA bend contributes at least by a factor of two to enhancer function. Consistent with previous studies in which we analyzed other point mutations in the LEF-1-binding site (20, 28), the function of the mutant TCR-15 enhancer that reduces LEF-1 binding by a factor of 20 was impaired approximately 5-fold (Fig. 6 and Table 1).
Figure 6.
DNA bending participates in the regulation of the TCRα enhancer. Reporter plasmids consisting of the minimal TCRα enhancer containing either the wild-type (TCR-1) or mutant (TCR-11, TCR-12, TCR-15) LEF-1-binding sites were linked to the minimal fos promoter/CAT gene. The different plasmid DNAs were transfected into the T cell lines EL-4 and BW5147, and CAT activity was determined after 48 hr. CAT activities are indicated relative to 100% obtained with the wild-type TCRα enhancer construct.
DISCUSSION
In this study, we further examine the structural and functional properties of LEF-1-induced DNA bending. We show by a helical phasing analysis that the protein-induced DNA bend is directed toward the major groove of the DNA helix. In this protein:DNA complex partial intercalation of aromatic residues into the minor groove and wrapping of a basic C-terminal tail around the major groove bent the DNA helix by 120° (12). The structure of the LEF-HMG:DNA complex is reminiscent of those of the HMG domain protein SRY (11) and the unrelated TATA-binding protein TBP (42, 43). We also observed in the helical phasing analysis that the migration of the free DNA probes varied with a periodicity of approximately 10 bp. This anomalous migration of the free DNA probes can, in principle, be accounted for by a structural distortion in the sequences upstream of the A tract DNA or in the LEF-1-binding site. An intrinsic DNA bend of 20° was detected in the binding site of the bacterial catabolic activator protein (CAP), which induces a sharp bend in the DNA helix (44). Together, these observations raise the possibility that this structural distortion, which might be transient in solution, contributes to DNA binding by the HMG domain of LEF-1. Consistent with this possibility, the HMG domain of LEF-1 efficiently recognizes four-way junction DNA, which contains stably distorted non-B-DNA (37). However, the relative binding affinity of LEF-1 to its cognate binding site in linear B-DNA is higher than that detected with the four-way junction DNA lacking the LEF-1-binding site. Although SRY, another sequence-specific HMG domain protein, was reported to bind four-way junction DNA more efficiently than to linear DNA, it is likely that this study did not use the optimal binding site for SRY (6, 7, 10, 22). In contrast, nonspecific HMG domain proteins, which include HMG-1, strongly prefer four-way junction DNA (33). Thus, the structure-specific recognition of DNA is an intrinsic property of this class of DNA-binding proteins and might reflect the structural fold of the HMG domain (11, 12, 24, 45).
DNA binding of LEF-1 to circular DNA is accompanied by a significant unwinding of the DNA helix and by negative supercoiling. This is reminiscent of the supercoiling of circular DNA that has been observed with the HMG proteins HMG-1 and mtTF (8, 46). Based on the functional role of LEF-1 in the regulation of TCRα enhancer activity, protein-induced unwinding of DNA may contribute to the function of LEF-1 in the context of chromatin. Studies with in vitro-assembled nucleosomes indicated that LEF-1 can stimulate the activity of the HIV-1 and TCRα enhancers on chromatin templates (47, 48). The potential of LEF-1 to unwind the DNA helix may affect the assembly or stability of a nucleosome (49). Alternatively, LEF-induced unwinding of DNA may play a role in initiation of transcription by facilitating the binding of other DNA-bending proteins, such as TBP (50), or by facilitating DNA strand separation. Consistent with this possibility, binding of the HMG domain of LEF-1 to a site immediately upstream of the minimal fos promoter increases in vitro transcription 3-fold (K.G., unpublished observations). Moreover, the function of LEF-1 in stimulating TCRα enhancer-mediated transcription in vitro is dependent on DNA topology. LEF-1 activity was observed on circular DNA molecules but was significantly impaired by relaxing the DNA with topoisomerase I (51).
The mutational analysis of the LEF-1-binding site identified several nucleotides in the core recognition sequence that affect the extent of protein-induced DNA bending. Most of the mutations that decreased the bend angle also reduced the efficiency of DNA binding by LEF-1 (up to 20-fold for TCR-15). A similar correlation has been reported previously for the CAP protein (52). However, we identified some mutations in the LEF-1-binding site that reduced the bend angle but were recognized by LEF-1 with an efficiency that was only 2-fold lower than that observed with the wild-type binding site (TCR-11, TCR-12, and TCR-13). These results indicate that the DNA-binding affinity of LEF-1 does not strictly correlate with the extent of the protein-induced DNA bend.
A common feature of the mutant binding sites is the observed shift in the apparent bend centers to the 3′ site of the recognition sequence. A shift of the bend center, although less pronounced, was also observed in a circular permutation analysis of a mutant binding site for the bacterial CAP protein (53). The structural basis for the shift in the bend center is unclear. The DNaseI footprint analysis suggests that LEF-1 does not recognize cryptic downstream binding sites. Instead, the presence of a natural bend in the DNA could bias the position of the LEF-1 bend more clearly in the case of the small bends at the mutant binding sites than in the large bend at the wild-type binding site.
The identification of high-affinity binding sites that showed a reduced bend angle allowed us to examine the contribution of DNA bending to the activation of the TCRα enhancer. In these experiments, we observed a correlation between the LEF-HMG domain-induced DNA bend angle and the transcriptional potential of the mutant binding sites in the context of the TCRα enhancer. The decrease of the bend angle from 130° to 90° for the high-affinity TCR-11 and TCR-12 mutant sites correlated with a 4-fold reduction of enhancer activity in T cells. A more pronounced decrease in enhancer activity was observed with a low-affinity TCR-15 mutant site. Taking into account the 2-fold decrease in the binding affinity of LEF-1 for the mutant TCR-11 and -12 enhancers, the effect of the altered bend angle on the enhancer activity is 2-fold. Although this effect is modest, the point mutations in the TCR-11 and -12 enhancers did not abrogate bending but rather reduced the bend angle. Moreover, an interaction of LEF-1 with other enhancer-binding proteins may obscure the effects of a decrease in the bend angle. Recent studies have shown that the function of LEF-1 in the regulation of the TCRα enhancer is augmented by an interaction with a protein termed ALY that also associates with the TCRα enhancer-binding protein AML-1 (32). Additional support for a role of protein-induced DNA bending in the regulation of the TCRα enhancer was provided by experiments in which LEF-1 was replaced by the HMG domain of LEF-1 or SRY (28, 29). These experiments indicated that the HMG domain is sufficient to mediate part of the activation of TCRα enhancer. However, DNA bending may not be an obligatory function of LEF-1. Recent experiments demonstrated that LEF-1 can associate with β-catenin, which is a component of the Wnt signaling pathway (54, 55). In association with β-catenin, LEF-1 can function as a conventional transcriptional activator that stimulates transcription from a synthetic enhancer containing multimerized LEF-1 binding sites (56). In contrast, in vivo experiments in Drosophila suggest that LEF-1, in association with the β-catenin orthologue armadillo, also regulates gene expression in a context-dependent manner (57). Thus, the architectural function of LEF-1 may be a general feature of this protein.
Acknowledgments
We thank H. Nash for providing pBEND1. This work was supported by the Howard Hughes Medical Institute.
ABBREVIATIONS
- LEF-1
lymphoid enhancer-binding factor 1
- HMG
high mobility group
- TCRα
T cell receptor α
- IHF
integration host factor
- CAT
chloramphenicol acetyltransferase
References
- 1.Echols H. Science. 1986;233:1050–1056. doi: 10.1126/science.2943018. [DOI] [PubMed] [Google Scholar]
- 2.Nash H. Trends Biochem Sci. 1990;15:222–227. doi: 10.1016/0968-0004(90)90034-9. [DOI] [PubMed] [Google Scholar]
- 3.Grosschedl R, Giese K, Pagel J. Trends Genet. 1994;10:94–100. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 4.Tjian R, Maniatis T. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 5.Giese K, Amsterdam A, Grosschedl R. Genes Dev. 1991;5:2567–2578. doi: 10.1101/gad.5.12b.2567. [DOI] [PubMed] [Google Scholar]
- 6.Giese K, Cox J, Grosschedl R. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- 7.Ferrari S, Harley V, Pontiggia A, Goodfellow P, Lovell-Badge R, Bianchi M. EMBO J. 1992;11:4497–4506. doi: 10.1002/j.1460-2075.1992.tb05551.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fischer R P, Lisowsky T, Parisi M A, Clayton D A. J Biol Chem. 1992;267:3358–3367. [PubMed] [Google Scholar]
- 9.Van de Wetering M, Clevers H. EMBO J. 1992;11:3039–3044. doi: 10.1002/j.1460-2075.1992.tb05374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giese K, Pagel J, Grosschedl R. Proc Natl Acad Sci USA. 1994;91:3368–3372. doi: 10.1073/pnas.91.8.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werner M H, Gronenborn A M, Clore G M. Science. 1996;271:778–784. doi: 10.1126/science.271.5250.778. [DOI] [PubMed] [Google Scholar]
- 12.Love J J, Case D A, Giese K, Grosschedl R, Wright P E. Nature (London) 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 13.Pil P M, Lippard S J. Science. 1992;256:234–237. doi: 10.1126/science.1566071. [DOI] [PubMed] [Google Scholar]
- 14.Laudet V, Stehelen D, Clevers H. Nucleic Acids Res. 1993;21:2493–2501. doi: 10.1093/nar/21.10.2493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bustin M, Lehn D A, Landsman D. Biochim Biophys Acta. 1990;1049:231–243. doi: 10.1016/0167-4781(90)90092-g. [DOI] [PubMed] [Google Scholar]
- 16.Jantzen H-M, Admon A, Bell P B, Tjian R. Nature (London) 1990;344:830–836. doi: 10.1038/344830a0. [DOI] [PubMed] [Google Scholar]
- 17.Kuhn A, Voit R, Stefanovsky V, Evers R, Bianchi M, Grummt I. EMBO J. 1994;5:416–424. doi: 10.1002/j.1460-2075.1994.tb06276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bianchi M, Beltrame M, Paonessa G. Science. 1989;243:1056–1059. doi: 10.1126/science.2922595. [DOI] [PubMed] [Google Scholar]
- 19.Sinclair A M, Berta P, Palmer M S, Hawkins J R, Griffiths B L, Smith D J, Foster J W, Frischauf A-M, Lovell-Badge R, Goodfellow P. Nature (London) 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 20.Travis A, Amsterdam A, Belanger C, Grosschedl R. Genes Dev. 1991;5:880–894. doi: 10.1101/gad.5.5.880. [DOI] [PubMed] [Google Scholar]
- 21.Waterman M, Fischer W H, Jones K A. Genes Dev. 1991;5:656–669. doi: 10.1101/gad.5.4.656. [DOI] [PubMed] [Google Scholar]
- 22.Harley V R, Jackson D I, Hextall P J, Hawkins J R, Berkovitz G D, Sockanathan S, Lovell-Badge R, Goodfellow P. Science. 1992;255:453–456. doi: 10.1126/science.1734522. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka I, Appelt K, Dijk J, White S W, Wilson K S. Nature (London) 1984;310:376–381. doi: 10.1038/310376a0. [DOI] [PubMed] [Google Scholar]
- 24.Read C M, Cary P D, Crane-Robinson C, Driscoll P C, Norman D G. Nucleic Acids Res. 1993;21:3427–3436. doi: 10.1093/nar/21.15.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonnefoy E, Takahashi M, Rouviere-Yaniv J. J Mol Biol. 1994;128:116–129. doi: 10.1006/jmbi.1994.1563. [DOI] [PubMed] [Google Scholar]
- 26.Hoover T R, Santero E, Porter S, Kustu S. Cell. 1990;63:11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- 27.Segall A M, Goodman S D, Nash H A. EMBO J. 1994;13:4536–4548. doi: 10.1002/j.1460-2075.1994.tb06775.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giese K, Kingsley C, Kirchner J, Grosschedl R. Genes Dev. 1995;9:995–1008. doi: 10.1101/gad.9.8.995. [DOI] [PubMed] [Google Scholar]
- 29.Van de Wetering M, Castrop J, Korinek V, Clevers H. Mol Cell Biol. 1996;16:745–752. doi: 10.1128/mcb.16.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giese K, Grosschedl R. EMBO J. 1993;12:4667–4676. doi: 10.1002/j.1460-2075.1993.tb06155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carlson P, Waterman M, Jones K. Genes Dev. 1993;7:2418–2430. doi: 10.1101/gad.7.12a.2418. [DOI] [PubMed] [Google Scholar]
- 32.Bruhn L, Munnerlyn A, Grosschedl R. Genes Dev. 1997;11:640–653. doi: 10.1101/gad.11.5.640. [DOI] [PubMed] [Google Scholar]
- 33.Bianchi M. EMBO J. 1988;7:843–849. doi: 10.1002/j.1460-2075.1988.tb02883.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klippel A, Kanaar R, Kahmann R, Cozzarelli N R. EMBO J. 1993;12:1047–1057. doi: 10.1002/j.1460-2075.1993.tb05746.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson J F, Landy A. Nucleic Acids Res. 1988;16:9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lilley D M J. Trends Genet. 1988;4:111–114. doi: 10.1016/0168-9525(88)90099-6. [DOI] [PubMed] [Google Scholar]
- 37.Lilley D M J. Nature (London) 1992;357:282–283. doi: 10.1038/357282a0. [DOI] [PubMed] [Google Scholar]
- 38.Cozzarelli N R, Boles T C, White J H. In: DNA Topology and Its Biological Effects. Cozzarelli N R, Wang J C, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1990. pp. 139–184. [Google Scholar]
- 39.Travers A. Nature (London) 1989;341:184–185. doi: 10.1038/341184a0. [DOI] [PubMed] [Google Scholar]
- 40.Salvo J J, Grindley N D F. Nucleic Acids Res. 1987;15:9771–9779. doi: 10.1093/nar/15.23.9771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zinkel S S, Crothers D M. Nature (London) 1987;328:178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]
- 42.Kim Y, Geiger J, Kahn S, Sigler P. Nature (London) 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 43.Kim J, Nikolov D, Burley S. Nature (London) 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 44.Kahn J D, Crothers D M. Proc Natl Acad Sci USA. 1992;92:6343–6346. doi: 10.1073/pnas.89.14.6343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weir H M, Kraulis P J, Hill C S, Raine A R, Laue E D, Thomas J J. EMBO J. 1993;12:1311–1319. doi: 10.1002/j.1460-2075.1993.tb05776.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bazett-Jones D, Leblanc B, Herfort M, Moss T. Science. 1994;264:1134–1137. doi: 10.1126/science.8178172. [DOI] [PubMed] [Google Scholar]
- 47.Sheridan P L, Sheline C T, Cannon K, Voz M L, Pazin M J, Kadonaga J T, Jones K A. Genes Dev. 1995;9:2090–2104. doi: 10.1101/gad.9.17.2090. [DOI] [PubMed] [Google Scholar]
- 48.Mayall T P, Sheridan P L, Montminy M R, Jones K A. Genes Dev. 1997;11:887–899. doi: 10.1101/gad.11.7.887. [DOI] [PubMed] [Google Scholar]
- 49.White J H, Cozzarelli N R, Bauer W R. Science. 1988;241:323–327. doi: 10.1126/science.3388041. [DOI] [PubMed] [Google Scholar]
- 50.Parvin J A, McCormick R J, Sharp P A, Fisher D E. Nature (London) 1995;373:724–727. doi: 10.1038/373724a0. [DOI] [PubMed] [Google Scholar]
- 51.Bagga R, Emerson B M. Genes Dev. 1997;11:629–639. doi: 10.1101/gad.11.5.629. [DOI] [PubMed] [Google Scholar]
- 52.Gartenberg M R, Crothers D M. Nature (London) 1988;333:824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- 53.Liu-Johnson H-N, Gartenberg M R, Crothers D M. Cell. 1986;47:995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- 54.Behrens J, von Kies J P, Kühl M, Bruhn L, Wedlich D, Grosschedl R, Birchmeier W. Nature (London) 1996;382:638–642. doi: 10.1038/382638a0. [DOI] [PubMed] [Google Scholar]
- 55.Molenaar M, van de Wetering M, Oosterwegel M, Peterson-Maduro J, Godsave S, Korinek V, Roose J, Destrée O, Clevers H. Cell. 1996;86:391–399. doi: 10.1016/s0092-8674(00)80112-9. [DOI] [PubMed] [Google Scholar]
- 56.van de Wetering M, Cavallo R, Rooijes D, van Beest M, van Es J, Loureiro J, Ypma A, Hursh D, Jones T, Bejsovek A, Peifer M, Mortin M, Clevers H. Cell. 1997;88:789–799. doi: 10.1016/s0092-8674(00)81925-x. [DOI] [PubMed] [Google Scholar]
- 57.Riese J, Yu X, Munnerlyn A, Eresh E, Hsu S-C, Grosschedl R, Bienz M. Cell. 1997;88:777–787. doi: 10.1016/s0092-8674(00)81924-8. [DOI] [PubMed] [Google Scholar]