Abstract
Using high-throughput gene-expression profiling technology, we can now gain a better understanding of the complex biology that is taking place in cancer cells. This complexity is largely dictated by the abnormal genetic makeup of the cancer cells. This abnormal genetic makeup can have profound effects on cellular activities such as cell growth, cell survival and other regulatory processes. Based on the pattern of gene expression, or molecular signatures of the tumours, we can distinguish or subclassify different types of cancers according to their cell of origin, behaviour, and the way they respond to therapeutic agents and radiation. These approaches will lead to better molecular subclassification of tumours, the basis of personalized medicine. We have, to date, done whole-genome microarray gene-expression profiling on several hundreds of kidney tumours. We adopt a combined bioinformatic approach, based on an integrative analysis of the gene-expression data. These data are used to identify both cytogenetic abnormalities and molecular pathways that are deregulated in renal cell carcinoma (RCC). For example, we have identified the deregulation of the VHL-hypoxia pathway in clear-cell RCC, as previously known, and the c-Myc pathway in aggressive papillary RCC. Besides the more common clear-cell, papillary and chromophobe RCCs, we are currently characterizing the molecular signatures of rarer forms of renal neoplasia such as carcinoma of the collecting ducts, mixed epithelial and stromal tumours, chromosome Xp11 translocations associated with papillary RCC, renal medullary carcinoma, mucinous tubular and spindle-cell carcinoma, and a group of unclassified tumours. Continued development and improvement in the field of molecular profiling will better characterize cancer and provide more accurate diagnosis, prognosis and prediction of drug response.
Approximately 34 000 new cases of renal cell carcinoma (RCC) are diagnosed each year in the United States; these cases account for 3% of all adult malignancies.1 About one-third of patients have metastatic disease, a median survival of 7–11 months and a 5-year survival of 10%. Every year about 13 000 patients die of RCC. Its incidence has been increasing, a phenomenon that wider use of imaging procedures cannot account for. RCC is a heterogeneous disease that is based on differences in morphology, genetic alterations and clinical behaviour. Several subtypes of RCC have been described: clear-cell, papillary, chromophobe, collecting-duct and unclassified (those that do not fit with any other diagnoses) RCC. Rarer types of renal neoplasia have been morphologically defined, including renal medullary RCC, Xp11 translocation carcinomas, carcinoma associated with neuroblastoma, and mucinous tubular and spindle-cell carcinoma. Benign renal tumours such as oncocytoma and papillary adenoma have also been well described.
Beginning with cytogenetic profiling decades ago, the diverse spectrum of renal pathology has generated tremendous interest in identifying their underlying molecular origin. Not surprisingly, distinct chromosomal aberrations have been correlated with some of the subtypes of renal tumours, for example, the loss of chromosome 3 in clear-cell RCC; the gain of chromosomes 7, 16 and 17 in papillary RCC; and the loss of chromosome Y in oncocytoma.2 These results were partially confirmed by more recent technology, for example, studies of the loss of heterozygosity that used microsatellite polymorphic markers and comparative genomic hybridization to confirm deletion of chromosome 3p in clear-cell RCC.3 Later on, identification of the genes responsible for hereditary RCC led to molecular studies of RCC. The genes identified in hereditary cases are thought to play an equal role in their sporadic counterparts. The best example is the VHL gene, the tumour-suppressor gene responsible for the autosomal-dominant cancer syndrome von Hippel-Lindau disease, which is characterized by clear-cell RCC, pheochromocytoma, retinal angioma and central nervous system hemangioblastoma. Mutations in VHL, loss of heterozygosity and imprinting were identified in about 70% of sporadic clear-cell RCCs, supporting an earlier hypothesis.4 However, studies of other hereditary RCC genes found either a small number of or no mutations in the sporadic tumours of subtypes, for example, the MET proto-oncogene for hereditary papillary RCC type 1,5,6 the BHD gene for Birt-Hogg-Dubé syndrome (characterized by a spectrum of RCC, but frequently chromophobe RCC and oncocytoma),7,8 and the fumarate hydratase FH gene for hereditary leiomyomatosis and papillary RCC type 2.9,10 It is, therefore, overly simplistic to associate each of these hereditary RCC genes with each distinct type of RCC.
Because tumorigenesis is a multistep, multigenic process, a global view of the gene-expression pattern and the deregulated pathways involved may offer a more accurate picture of RCC. Today, the use of high-throughput microarray gene-expression profiling in the molecular subclassification of tumours is prevalent, although it maybe years before it is fully appreciated and accepted by the clinical community. The main challenge is the conventional use of a single-gene, single-biomarker approach (e.g., prostate-specific antigen for prostate cancer), and the difficulty changing this and embracing the new concept of multiple genes or multiplex biomarkers, especially when the biological functions or relevance of these newly identified genes is not known. It is encouraging, however, that the Food and Drug Administration (FDA) has recently approved the first multiplex microarray prognostic tool based on a set of 70 genes (the MammaPrint test) for assessing the risk of distant metastasis for female patients with breast cancer. Barring the costs and regulatory requirements, microarray profiling should be considered a potential tool for molecular subclassification, and therefore better diagnosis, prognosis and prediction of drug response.
To determine the feasibility of the use of gene-expression information to develop improved tools for diagnosis, prognosis and prediction of drug response in RCC, we are leveraging the tremendous amount of microarray data that we have generated from 350 cases of primary renal tumours of varied morphology. These samples were obtained from multiple centres and have been associated with long-term clinical follow-up information. From the intersection of the gene-expression and clinical data, it is possible to identify either individual genes or sets of genes that can distinguish between the different RCC subtypes and prognostic classes. However, through careful analysis of the gene-expression profiling data, it is also possible to identify chromosomal abnormalities and abnormalities in signal-transduction pathways that can distinguish between the different RCC classes. In this paper, we demonstrate how the integration of the individual changes in gene expression, chromosomal abnormalities and signal-transduction abnormalities will allow the development of more powerful diagnostic and prognostic models of RCC.
Multiplexed markers or expression patterns detected by microarray gene-expression profiling
Gene-expression microarrays refer to the use of specific DNA sequences (the probes) arrayed onto a suitable surface such as nitrocellulose or glass.11 These probes cover the whole genome and therefore contain virtually all the genes in the human genome. The probes are allowed to hybridize with cDNA from a given sample that has been labelled with a detectable tag (such as fluorescent dye or radioactivity). After washing, the relative amount of cDNA that hybridizes to the DNA probe on the microarray can be measured and indicates the degree of gene (RNA) expression within the sample. An alternative technology builds short sequences of oligonucleotides (typically 20–60 base pairs long) onto the array surface. The sequence of these probes is preselected from genomic sequencing databases. Typically, when shorter sequences are used as probes, several nonoverlapping probe sequences that span the length of the gene are selected. This approach provides for multiple assessments of the expression of any given gene within a single array. In summary, the procedures allow detection of the expression of thousands of genes, forming a pattern. This pattern is then compared with that of either a matched normal or reference tissue serving as a control. This approach is based on multiple markers rather than a single marker. The combined value of these multiple markers outweighs the value of a single marker in the molecular subclassification of tumours.12 However, to those who are used to a single-marker assay and are not familiar with microarray profiling, this approach may be disappointing, even confusing, because no one single marker of a gene may predominate.
Differentially expressed genes in different subtypes of renal neoplasia
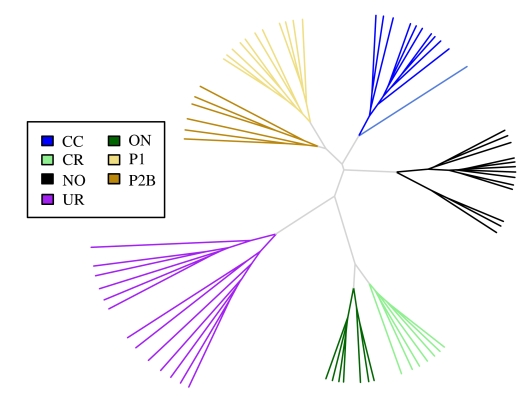
The role of gene-expression profiling involves the identification of differentially expressed genes — those that are either upregulated or downregulated in tumours when compared with normal renal tissue. For the majority of these genes, their function and role in tumorigenesis are unknown. In fact, some may be bystanders that play no role in tumorigenesis, but despite this, may serve as effective biomarkers because of their unique differential expression. On the other hand, some upregulated genes may serve as oncogenes or enhancers of cancer formation and progression. Similarly, some downregulated genes may normally function as tumour-suppressor genes. Both groups of genes may represent potential targets for therapies, based on the inhibition of the former and the activation of the latter. To date, several microarray studies13,14,15,16 have reported the ability of gene-expression patterns to distinguish between histological subtypes of RCC, such as conventional clear-cell RCCs, papillary type 1 and type 2 carcinomas, chromophobe carcinomas, oncocytomas and urothelial carcinoma of renal pelvis (Fig. 1). The close clustering of genes in each subtype points to different tumorigenic pathways, as seen in their histological features. In our previous study,17 16 unknown consecutive cases were subjected to gene-expression microarray analysis. The microarray diagnoses for 14 cases were consistent with those of pathologists; of the 2 remaining cases, one was a mixed oncocytoma and high-grade clear-cell RCC, and the other a chromophobe RCC whose histology mimicked that of papillary RCC.
Figure 1.
Distinction of the different subtypes of RCC by gene-expression profiling. Shown is an unrooted dendrogram of 68 renal gene-expression profiles derived from clear-cell (CC), chromophobe (CR), oncocytoma (ON), papillary type 1 (P1), papillary type 2b (P2b), and urothelial (UR) tumour samples. Also included are data derived from nondiseased renal tissue (NO). Hierarchical clustering is based on the log2-transformed gene-expression values from 8407 genes that have an overall interquartile range greater than 0.5. The dendrogram was produced with Euclidian distance and complete linkage clustering.
Currently, we are undertaking several studies to dissect the molecular signatures of rarer forms of renal tumours and the so-called unclassified RCC. The former include carcinoma of the collecting duct, renal medullary carcinoma, mucinous tubular and spindle-cell carcinoma, translocation carcinomas of the kidney, and mixed epithelial and stromal tumours. Completion of these studies will add to the repertoire of molecular signatures of renal neoplasia. New molecular entities may be found, especially in the group of unclassified RCC, a group of heterogenous tumours that do not fit into any of the current established histological subtypes.
From these studies, new diagnostic markers can be identified. To date, several studies have demonstrated the diagnostic value by immunohistochemical staining of these new markers on hundreds of cases. For example, by immunohistochemical staining, glutathione S-transferase α was found highly expressed in clear-cell RCC18 and α-methylacyl-CoA racemase in papillary RCC.19 Some of these markers will be especially useful in distinguishing histology with more subtle differences, such as the differences between type 1 and type 2 papillary RCC20 or between oncocytoma and chromophobe RCC (T.B.T., unpublished data, 2007).
Correlation with chromosomal changes
Before the advent of microarray technology, use of genetic tools such as genome-wide allelotyping were used to correlate chromosomal changes with clinical parameters such as histological subtypes.2,21–23 Recent studies24,25,26,27,28,29 have taken this process a step further by confirming that chromosomal abnormalities have dramatic effects on gene expression. For example, when a chromosome region is gained in a tumour cell, a large number of transcripts located in the amplified region have increased expression compared with cytogenetically normal (noncancerous) tissue.
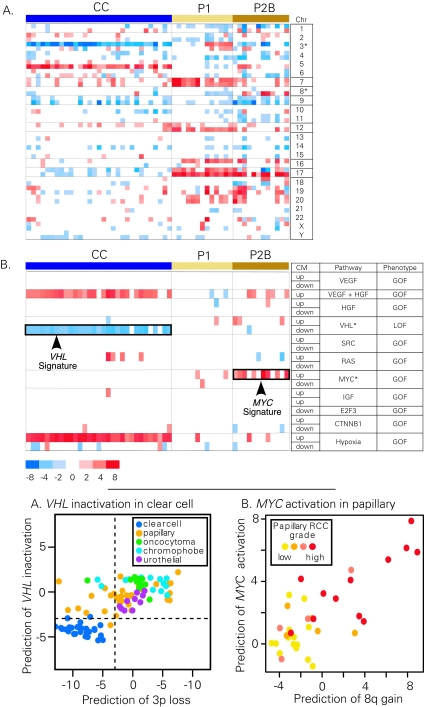
Logically then, based on regional expression biases, cytogenetic abnormalities may be indirectly deduced from gene-expression profiling data. By using such gene-expression values, investigators can develop 2 data sets: individual gene-expression values and an approximation of the underlying sample karyotype. These, in turn, enable correlation studies with histological subtypes, based on gene-expression profiles, as well as on chromosomal changes. One method used for predicting chromosomal aberrations from the microarray data has been termed “comparative genomics microarray analysis,” which has been successfully applied to the studies of kidney cancer (Fig. 2A).30
Figure 2.
Fig. 2A shows the identification of chromosomal abnormalities and signaling pathway abnormalities from gene-expression data. Gene-expression profiles derived from clear-cell (CC), papillary type 1 (P1) and type 2b (P2b) RCC samples were compared with gene-expression profiles derived from nondiseased kidney tumour tissue. Predictions of chromosomal abnormalities were made with the comparative genomics microarray analysis method.30 Fig. 2B shows predictions of pathway abnormalities made with the parametric gene-set enrichment analysis method.31 For pathway analysis, the signature component labelled up indicates the set of genes that show increased expression relative to control cells for each pathway. Likewise, the signature component labelled dn indicates the set of genes that show decreased expression relative to control cells for each pathway. The labels GOF and LOF indicate whether the signature components are associated with a gain of function or a loss of function of each pathway, respectively. Gene sets corresponding to the following pathways were analyzed: VEGF, HGF, VHL, avian sarcoma viral oncogene homologue (SRC), rat sarcoma viral oncogene homologue (RAS), MYC, insulin-like growth factor 1 (IGF), E2F transcription factor 3 (E2F3), β-catenin (CTNNB1), and hypoxia-related genes (Hypoxia). The resulting summary statistic (t statistic) is plotted for each gene set. Red indicates that a significant number of genes in the set have increased expression in the tumour samples relative to the normal kidney, whereas blue indicates that a significant number of genes in each set have decreased expression. Only the most significant data are displayed (p < 0.005).
Detection of oncogenic pathway in RCC
Identification of oncogenic signatures in tumour samples is a useful tool for unravelling the mechanisms of tumour development and potentially could be used to guide treatment choices.32 The majority of clear-cell RCCs have a loss of chromosome 3p and inactivation of the VHL gene located on 3p25.33 As a proof-of-concept of the oncogenic-signature approach, we wanted to determine whether sets of genes whose expression is reported to be modified by VHL in cell-line studies34 could also be detected as misregulated in the gene-expression profiles of clear-cell RCCs. Identification of a downregulated VHL signature in clear-cell RCC would validate the use of literature-curated gene sets to identify misregulated signal-transduction pathways in RCC. Gene-expression data from clear-cell and papillary RCCs were used for analysis of gene-set enrichment (Fig. 2B).35 Although the molecular-signature map of RCC can be a rich source of information, most notably, a significant number of genes included in a gene set reflective of the activation state of the VHL pathway are downregulated in all classes of clear-cell tumours, but not in papillary or chromophobe RCC (Fig. 2B, VHL dn). Specifically, most of the genes downregulated by VHL loss of function in the cell-line model are also significantly downregulated in the clear-cell RCC tumour samples. This is consistent with the high frequency of VHL pathway mutations in clear-cell RCC, but not other RCC subtypes. Interestingly, a number of genes reflective of a synergistic hepatocyte growth factor and vascular endothelial growth factor (HGF + VEGF) activation state36 are increased in clear-cell RCC, and may reflect the sensitivity of RCC to VEGF-pathway inhibitors. Using pathway analysis, we also identified a gene signature indicative of c-Myc activation in high-grade papillary RCC.31 Our subsequent molecular work31 demonstrated that amplification of chromosome 8q, the region where the c-Myc gene resides, contributes to c-Myc overexpression and subsequent pathway activation in these tumours. Overall, the success of the gene-set enrichment analysis in identifying both previously reported and novel molecular pathways misregulated in clear-cell and papillary RCC, supports the idea that this type of analysis helps identify signaling pathways that are disrupted in RCC.
Prognosis by gene-expression profiling
In our previous study,37 for example, the availability of 29 clear-cell RCC frozen-tissue specimens with up to 12 years of follow-up information provided a striking opportunity to understand the molecular mechanisms that underlie tumorigenesis and prognosis. By using 21 632 cDNA microarrays, we demonstrated a significant distinction in gene-expression profiles between those for patients with a relatively nonaggressive form of the disease (100% survival after 5 years, with the majority [88%] having no clinical evidence of metastasis v. patients with a relatively aggressive form of the disease [average survival time 25.4 mo; 0% 5-yr survival]). About 40 genes accurately distinguished the 2 forms of the disease. The results suggested that 2 molecularly distinct forms of clear-cell RCC exist and that the integration of expression-profile data with clinical parameters could enhance the diagnosis and prognosis of clear-cell RCC. More recently, a study of 177 clear-cell RCC tumours38 elicited a set of genes whose expression of mRNA correlated significantly with length of survival after surgery, even after controlling for conventional clinical parameters. Notably, overlapping genes were found in both studies,37,38 further confirming the underlying molecular difference between nonaggressive and aggressive tumours and their clinical behaviour. This finding has significant clinical implications for intervention strategies. With the recent approval of sorafenib and sunitinib, which are pleiotropic-kinase inhibitors, a trial of adjuvant therapy based on microarray analysis can be initiated. Similar to our clear-cell study,37 we recently reported a set of prognostic genes that distinguished between classes of papillary RCC.20 The first class, with excellent survival, corresponded to 3 histologic subtypes of RCC: type 1, low-grade type 2, and mixed type 1 and low-grade type 2. The second class, with poor survival, corresponded to high-grade type 2 tumours. These prognostic gene profiles can be incorporated in trials of adjuvant therapy.
Conclusion
With the advent of high-throughput technology, the form of future diagnostic and prognostic tools is likely to be multiplexed, consisting of multiple rather than single markers. With the recent FDA approval of the prognostic set of 70 genes for breast cancer, we expect to see the use of similar devices for other types of cancer, including RCC. The promising results obtained so far for RCC certainly point to the worthiness of prospective multicentred trials for its use clinically. The use of fresh-frozen tumours for the extraction of high-quality RNA and other products, such as DNA and protein for microarray analysis, and other molecular studies, will, we hope, become commonplace. Moreover, there is a clear need for microarray gene-expression profiling to be done on paraffin-embedded blocks because they are more easily procured and have a more clearly defined morphology than fresh-frozen tumours. Alternatively, the multigene method of reverse transcriptase polymerase chain reaction can be adopted to study RNA extracted from paraffin-embedded blocks. However, this approach involves tremendous effort to optimize the conditions for the primers for the reverse transcriptase polymerase chain reaction.39 High-throughput technologies have clear clinical implications, and their clinical application will have a direct impact on patient management.
Acknowledgments
Acknowledgements: We acknowledge The Gerber Foundation, Hauenstein Foundation, Fischer Family Trust, the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor, the Schregardus Family Foundation, and Amway Japan. We thank the Cooperative Human Tissue Network of the National Cancer Institute for providing tumour samples for this research. We thank Sabrina Noyes for manuscript preparation and submission.
Footnotes
This article has been peer reviewed.
Competing interests: None declared.
References
- 1.Eble JN, International Agency for Research on Cancer, World Health Organization, editors. Pathology and genetics of tumours of the urinary system and male genital organs. Lyon: IARC Press; 2004. [Google Scholar]
- 2.Kovacs G, Akhtar M, Beckwith BJ, et al. The Heidelberg classification of renal cell tumours. J Pathol 1997;183:131-3. [DOI] [PubMed] [Google Scholar]
- 3.Alimov A, Kost-Alimova M, Liu J, et al. Combined LOH/CGH analysis proves the existence of interstitial 3p deletions in renal cell carcinoma. Oncogene 2000;19: 13929. [DOI] [PubMed] [Google Scholar]
- 4.Kenck C, Wilhelm M, Bugert P, et al. Mutation of the VHL gene is associated exclusively with the development of non-papillary renal cell carcinomas. J Pathol 1996;179:157-61. [DOI] [PubMed] [Google Scholar]
- 5.Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat Genet 1997;16:68-73. [DOI] [PubMed] [Google Scholar]
- 6.Schmidt L, Junker K, Nakaigawa N, et al. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene 1999;18:2343-50. [DOI] [PubMed] [Google Scholar]
- 7.Khoo SK, Kahnoski K, Sugimura J, et al. Inactivation of BHD in sporadic renal tumors. Cancer Res 2003;63:4583-7. [PubMed] [Google Scholar]
- 8.Pavlovich CP, Walther MM, Eyler RA, et al. Renal tumors in the Birt-Hogg-Dube syndrome. Am J Surg Pathol 2002;26:1542-52. [DOI] [PubMed] [Google Scholar]
- 9.Tomlinson IP, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 2002;30:406-10. [DOI] [PubMed] [Google Scholar]
- 10.Toro JR, Nickerson ML, Wei MH, et al. Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America. Am J Hum Genet 2003;73:95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Quackenbush J. Microarray analysis and tumor classification. N Engl J Med 2006;354:2463-72. [DOI] [PubMed] [Google Scholar]
- 12.Rhodes DR, Chinnaiyan AM. Integrative analysis of the cancer transcriptome. Nat Genet 2005;37:S31-7. [DOI] [PubMed] [Google Scholar]
- 13.Boer JM, Huber WK, Sultmann H, et al. Identification and classification of differentially expressed genes in renal cell carcinoma by expression profiling on a global human 31,500-element cDNA array. Genome Res 2001;11:1861-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Higgins JP, Shinghal R, Gill H, et al. Gene expression patterns in renal cell carcinoma assessed by complementary DNA microarray. Am J Pathol 2003;162:925-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takahashi M, Yang XJ, Sugimura J, et al. Molecular subclassification of kidney tumors and the discovery of new diagnostic markers. Oncogene 2003;22:6810-8. [DOI] [PubMed] [Google Scholar]
- 16.Young AN, Amin AB, Moreno CS, et al. Expression profiling of renal epithelial neoplasms: a method for tumor classification and discovery of diagnostic molecular markers. Am J Pathol 2001;158:1639-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang XJ, Sugimura J, Schafernak KT, et al. Classification of renal neoplasms based on molecular signatures. J Urol 2006;175:2302-6. [DOI] [PubMed] [Google Scholar]
- 18.Chuang ST, Chu P, Sugimura J, et al. Overexpression of glutathione s-transferase alpha in clear cell renal cell carcinoma. Am J Clin Pathol 2005;123:421-9. [DOI] [PubMed] [Google Scholar]
- 19.Tretiakova MS, Sahoo S, Takahashi M, et al. Expression of alpha-methylacyl-CoA racemase in papillary renal cell carcinoma. Am J Surg Pathol 2004;28:69-76. [DOI] [PubMed] [Google Scholar]
- 20.Yang XJ, Tan MH, Kim HL, et al. A molecular classification of papillary renal cell carcinoma. Cancer Res 2005;65:5628-37. [DOI] [PubMed] [Google Scholar]
- 21.Gettman MT, Blute ML, Spotts B, et al. Pathologic staging of renal cell carcinoma: significance of tumor classification with the 1997 TNM staging system. Cancer 2001;91:354-61. [DOI] [PubMed] [Google Scholar]
- 22.Moch H, Mihatsch MJ. Genetic progression of renal cell carcinoma. Virchows Arch 2002;441:320-7. [DOI] [PubMed] [Google Scholar]
- 23.Moch H, Presti JC Jr, Sauter G, et al. Genetic aberrations detected by comparative genomic hybridization are associated with clinical outcome in renal cell carcinoma. Cancer Res 1996;56:27-30. [PubMed] [Google Scholar]
- 24. Crawley JJ, Furge KA. Identification of frequent cytogenetic aberrations in hepatocellular carcinoma using gene expression data. Genome Biol 2002; Epub 2002 Nov 25;3: RESEARCH0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haddad R, Furge KA, Miller J, et al. Genomic profiling and cDNA microarray analysis of human colon adenocarcinoma and associated peritoneal metastasis reveals consistent cytogenetic and transcriptional aberrations associated with progression of multiple metastases. Appl Genomics Proteomics 2002;1:123-34. [Google Scholar]
- 26.Harding MA, Arden KC, Gildea JW, et al. Functional genomic comparison of lineage-related human bladder cancer cell lines with differing tumorigenic and metastatic potentials by spectral karyotyping, comparative genomic hybridization, and a novel method of positional expression profiling. Cancer Res 2002;62:6981-9. [PubMed] [Google Scholar]
- 27.Hughes TR, Roberts CJ, Dai H, et al. Widespread aneuploidy revealed by DNA microarray expression profiling. Nat Genet 2000;25:333-7. [DOI] [PubMed] [Google Scholar]
- 28.Phillips JL, Hayward SW, Wang Y, et al. The consequences of chromosomal aneuploidy on gene expression profiles in a cell line model for prostate carcinogenesis. Cancer Res 2001;61:8143-9. [PubMed] [Google Scholar]
- 29.Pollack JR, Sorlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A 2002;99:12963-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Furge KA, Lucas KA, Takahashi M, et al. Robust classification of renal cell carcinoma based on gene expression data and predicted cytogenetic profiles. Cancer Res 2004; 64: 4117-21. [DOI] [PubMed] [Google Scholar]
- 31.Furge KA, Chen J, Koeman J, et al. Detection of DNA copy number changes and oncogenic signaling abnormalities from gene expression data reveals MYC activation in high-grade papillary renal cell carcinoma. Cancer Res 2007;67:3171-6. [DOI] [PubMed] [Google Scholar]
- 32.Bild AH, Yao G, Chang JT, et al. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature 2005;439:353-7. [DOI] [PubMed] [Google Scholar]
- 33.Kenck C, Wilhelm M, Bugert P, et al. Mutation of the VHL gene is associated exclusively with the development of non-papillary renal cell carcinomas. J Pathol 1996; 179: 157-61. [DOI] [PubMed] [Google Scholar]
- 34.Staller P, Sulitkova J, Lisztwan J, et al. Chemokine receptor CXCR4 downregulated by von Hippel-Lindau tumour suppressor pVHL. Nature 2003;425:307-11. [DOI] [PubMed] [Google Scholar]
- 35.Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 2005;102:15545-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gerritsen ME, Tomlinson JE, Zlot C, et al. Using gene expression profiling to identify the molecular basis of the synergistic actions of hepatocyte growth factor and vascular endothelial growth factor in human endothelial cells. Br J Pharmacol 2003;140:595610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takahashi M, Rhodes DR, Furge KA, et al. Gene expression profiling of clear cell renal cell carcinoma: gene identification and prognostic classification. Proc Natl Acad Sci US A 2001;98:9754-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhao H, Ljungberg B, Grankvist K, et al. Gene expression profiling predicts survival in conventional renal cell carcinoma. PLoS Med 2006;3:e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paik S, Shak S, Tang G, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med 2004;351:2817-26. [DOI] [PubMed] [Google Scholar]