Abstract
Renal cell carcinoma (RCC), the fifth leading malignant condition for men and tenth for women, accounts for 3% of all malignancies in Canada. It is a heterogeneous epithelial malignancy with different subtypes and varied tumour biology. Although most cases of RCC are sporadic, up to 4% of patients have an inherited predisposition for the disease. In this article, we review the current molecular genetics of the different subtypes in hereditary and sporadic RCC. Significant developments in understanding the underlying genetic basis of RCC over the last 2 decades are attributed to intensive research about rare inherited renal cancer syndromes and the identification of the genes responsible for them. Many of these genes are also found in sporadic RCC. Understanding the molecular mechanisms involved in the pathogenesis of RCC has aided the development of molecular-targeted drugs for this disease.
Renal cell carcinoma (RCC), the fifth leading malignant condition in men and the tenth in women, accounts for 3% of all malignancies in Canada.1 Though the mortality rate has been stable, the incidence of renal cancer has increased steadily for both sexes.1 In 2006, 4600 new cases were detected and resulted in 1500 deaths. The male to female ratio is roughly 2:1. Age-specific incidence shows a peak in early childhood, then follows the more usual pattern of a steep rise through adulthood.1
According to Heidelberg classification,2 malignant renal epithelial neoplasms are subclassified into common or conventional RCC, comprising 75% of renal cell neoplasms; papillary RCC (10%); chromophobe RCC (5%); collecting-duct carcinoma with medullary carcinoma of the kidney (1%); and RCC, unclassified (3%–5%).
Most cases of RCC are sporadic; up to 4% of patients have an inherited predisposition for this disease, including families with von Hippel-Lindau (VHL) disease, hereditary papillary renal cancer (HPRC), hereditary leiomyomatosis and renal cancer (HLRCC), and Birt-Hogg-Dubé (BHD) syndrome. The study of renal cancer genes and their molecular mechanisms is of utmost importance because it may help identify targets for therapy that will improve survival.
Hereditary kidney cancer
von Hippel-Lindau disease
The discovery of VHL disease occurred in 1894 when Collins reported a case of a bilateral vascular tumour in the retinas of 2 siblings.3 Since then, the constellation of lesions associated with this hereditary disease include hemangioblastoma of the central nervous system, visceral cysts or tumours, pheochromocytoma, endolymphatic sac tumours of the inner ear, epididymal and broad-ligament cystadenomas, and clear-cell RCC.4 VHL disease is a rare autosomal-dominant syndrome affecting approximately 1 in 35 000 people.5
The VHL gene, a tumour-suppressor gene,6,7 was mapped by linkage analysis to chromosome 3p258 and was isolated in 1993 with a positional cloning strategy.9 Its product, VHL protein (pVHL), consists of an α and β domain, and resides primarily in the cytoplasm, but can shuttle between the cytoplasm and the nucleus,10,11,12,13 This activity is important to its function in regulating transcription and production of growth factors.14
Through its α domain, pVHL forms a stable complex with Elongin subunits B and C,15,16,17 Cullin 2,18,19 and RING-box protein Rbx120 to form a ubiquitin E3 protein-ligase complex. pVHL serves as the substrate receptor of the complex that directly binds to specific proteins, such as hypoxia-inducible factor (HIF), and subsequently targets them for degradation through the ubiquitin proteolytic pathway.21
Hypoxia-inducible factor-1 (HIF-1) is a transcriptional activator that plays a crucial role in mediating cellular response to oxygen. HIF-1 is made up of a hypoxia-inducible subunit HIF-1α and a constitutively expressed subunit HIF-1β.22 Under adequate oxygenation, HIF-1α is hydroxylated by an oxygen-dependent prolyl hydroxylase in its 2 proline residues (Pro402,Pro564) located within the oxygen-dependent degradation domain in the cytoplasm,23,24 which then permits binding with the β domain of pVHL. This promotes the ubiquitination and destruction of HIF-1α. Also, hydroxylation by asparagine hydroxylase in the nucleus regulates its interaction with coactivator p300 and reduces the transcriptional activity of HIF-1.25
Patients with VHL disease were found to carry a germline mutated VHL allele and a wild-type VHL allele. Inactivation or loss of the wild-type VHL allele results in failure to bind with HIF-1α. Consequently, HIF-1α accumulates under normoxia, translocates from cytoplasm to nucleus and subsequently heterodimerizes with HIF-1β to enable binding to an HIF-responsive element26 and induce transcription of target genes leading to the production of GLUT-1 glucose transporter,27 platelet-derived growth factor β, carbonic anhydrase genes CA IX and CA XII,28,29 vascular endothelial growth factor (VEGF),30,31 and transforming growth factor α,32 thereby contributing to renal carcinogenesis.
pVHL has been involved in numerous cellular processes such as regulation of the extracellular matrix (ECM); cytoskeletal stability; and cell-cycle control and differentiation, other than regulation of HIF. Studies33,34 have shown that dysregulation of HIF does not completely explain the tumorigenesis in VHL-deleted cells,35,36 which suggests that pVHL may have HIF-independent tumour-suppressor functions.
A recent study37 has shown that pVHL in RCC cell lines is necessary for the normal organization of adherence and tight intercellular junctions, maintenance of cell polarity and control of paracellular permeability. Loss of VHL function leads to abnormalities in the deposition of extracellular fibronectin, a glycoprotein that interacts with integrins to bridge cells to the structural proteins of the ECM.38 A study39 reported that pVHL is required for adequate assembly of β1-integrin fibrillar adhesions and demonstrated that pVHL controls the strength of cell adhesion through this mechanism. pVHL also regulates the expression of tissue inhibitors of metalloproteinases and matrix metalloproteinase 2 and 9. Their dysregulation in RCC cells with loss of pVHL allows hepatocyte growth factor/scatter factor (HGF/SF)–dependent branching morphogenesis and invasion in vitro.40 Because epithelial-cell behaviour is clearly influenced by interaction with the ECM, disruption in the ECM may contribute to tumour invasion.
pVHL has been reported to bind directly to Jade-1,41 Chaperonin TRiC42 and a VHL-associated KRAB-A domain-containing protein (VHLak).43 Jade-1 is a short-lived kidney-enriched transcription factor that was found to suppress renal cancer growth by promoting apoptosis in experimental studies, whereas TRiC is essential for folding VHL into its native functional state. VHLak, on the other hand, functions as a negative regulator of HIF-1α transactivation. pVHL is also reported to ubiquitinate proteins such as the atypical protein kinase C isotypes PKCβ and PKCλ,44,45 and VHL-interacting deubiquitinating enzyme-1 and -2.46,47 Loss of pVHL function disrupts their interactions, which may contribute to tumorigenesis.
pVHL also binds to human RNA polymerase II seventh subunit (hsRPB7), facilitating its ubiquitination and proteasomal degradation. pVHL mutation enhances hsRPB7-induced VEGF expression.48 Recently, loss of pVHL was shown to impair p53-mediated cell-cycle arrest and apoptosis after DNA damage and to trigger the aberrant upregulation of HIF-α; this combination exerts a synergistic effect on the tumorigenesis of RCC.49 Loss of VHL also enhances cyclin D1 expression,50 whereas this loss downregulates the p27 gene in renal tumours;51 both result in cell-cycle progression.
Genotype-phenotype correlations in VHL disease suggest that pVHL has functions independent of its role in HIF regulation. The disease of families with VHL is subdivided, based on the absence (type I) or presence (type II) of pheochromocytoma. Patients with type II pheochromocytoma are further subdivided into types by level of risk (low [IIa] or high [IIb]) of RCC, or develop only pheochromocytoma or no RCC (IIc). Families with type I disease frequently harbour VHL-deletion or VHL-truncation mutations,52,53 whereas families with type II almost invariably harbour a VHL missense mutation. Patients with type I pheochromocytoma have no ability to regulate HIF, a condition that results in constitutive overexpression of HIF activity. Patients with type IIc disease retain their ability to bind and degrade HIF-α, linking the development of RCC to constitutive HIF upregulation.54,55
Hereditary papillary renal carcinoma
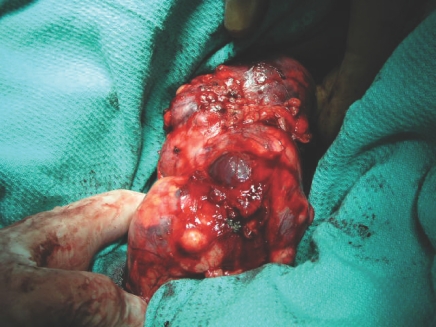
In 1994, Zbar et al56 reported an inherited form of papillary renal carcinoma with type 1 histologic features that affect people in their fourth to sixth decade of life. HPRC is an autosomal-dominant disease that is most often bilateral and multifocal (Fig. 1).56,57
Figure 1.
A case of a 47-year-old man with bilateral multifocal papillary renal cell carcinoma. Germline testing found an A-to-G mutation at nucleotide 3529 of the MET proto-oncogene.
MET, the gene linked to HPRC, is located at chromosome 7q31.1–34 in a 27-centimorgan interval between D7S496 and D7S1837, which encodes for a transmembrane receptor tyrosine kinase for MET.58
The MET-receptor tyrosine kinase is the prototypic member of a subfamily of growth factor receptors59–63 and binds to prototypic plasminogen-related growth factors, named HGF/SF.64,65 MET encodes a 150-kD amino acid precursor protein that undergoes glycosylation and subsequently cleaves into an α and a β chain.66
Activation of the signal transduction pathway in response to HGF/SF stimulation is mediated in part by autophosphorylation of specific tyrosine residues within the intracellular region of MET.
Phosphorylation of T1234 and T1235 located in the activation loop of the tyrosine-kinase domain activates the intrinsic kinase activity of the receptor,67,68 whereas phosphorylation of T1349 and T1356 in the C-terminal of MET activates a mutisubstrate-docking site69 that is responsible for signal transduction.70,71,72 HGF/SF stimulation and subsequent autophosphorylation results in the promotion of cell proliferation, inhibition of apoptosis, and increased motility and tubular-structure formation, termed “branching morphogenesis.”71
Although MET-HGF/SF signaling clearly mediates a variety of normal cellular processes, it has also been implicated in the generation and spread of tumours.73 MET is highly expressed in the kidney, and MET-HGF/SF signaling has been strongly implicated in the mediation of mitogenic74,75 and morphogenic differentiation76 in cultured kidney cells.
Several oncogenic forms of MET were discovered in tumour formation. One form is oncogenically activated through a missense germline mutation identified in HPRC.58 Trisomy 7 has also been reported in HPRC and is thought to play a role in the development of multiple renal tumours.77
The activation of c-MET in HPRC was found to possess constitutive kinase activity and malignant transforming ability.78,79 Jeffers and colleagues78 found that the transforming activity of c-MET was dependent on intracellular tyrosines at positions Y6 and Y10, in addition to Y8,9 and Y14,15, which influence a variety of MET-mediated responses, both in vitro (transformation, mitogenicity and invasion) and in vivo (tumorigenicity and metastasis).
A study80 has also shown that cell-surface interactions of c-MET in its β chain with extracellular signal-transduction molecules, such as plexin B1 and integrin α6β4, may enhance the invasiveness and metastatic potential of c-MET by inducing cytoskeletal changes.
In renal cancer, the MET and VHL signaling pathway is thought to intersect by means of pVHL-mediated regulation of HIF function. HIF stabilization through hypoxia or loss of VHL function results in overexpression of c-MET and forms a synergistic effect in inducing invasion.81
Hereditary leiomyomatosis and renal cancer
In 2001, Launonen and colleagues82 reported a disease complex of renal cancer associated in a studied kindred that included 11 family members with uterine leiomyoma and 7 members with a history of cutaneous nodules or leiomyoma. The family members affected were in their third to fourth decade of life and the histologic type of renal cancer was papillary type 2. This autosomal-dominant syndrome83 was later known as HLRCC. A small proportion of families with multiple cutaneous and uterine leiomyoma also have a cluster of renal collecting-duct cancer.84,85
The usual clinical presentation of HLRCC is the development of multiple leiomyoma in the skin and uterus. In 1 small series study,86 the frequency of papillary type 2 renal cancer was up to 62%. Often solitary but aggressive, this cancer has an early onset. The behaviour of these tumours seems contrary to that of HPRC tumours.
In a genome-wide linkage analysis with 370 microsatellite markers, the predisposition locus of HLRC maps to chromosome 1q42–44,82 which is caused by a germline mutation in fumarate hydratase (FH) in the majority of screened patients. Most of the germline mutations are missense mutations, but small deletions, insertions and nonsense mutations have been reported.87 Molecular analysis has shown that the wild-type of the FH allele is lost in HLRCC-associated tumours, indicating that FH acts as tumour-suppressor gene.88
FH catalyzes the hydration of fumarate to malate as part of the tricarboxylic acid cycle in the mitochondrial matrix. Loss of FH activity during progressive catalysis in the mitochondria during the Krebs cycle disrupts the process, resulting in increased fumarate levels and an increased concentration of its precursor, succinate.
Although the mechanism of tumorigenesis in HLRCC remains unclear, 2 notable proposed theories may explain its tumorigenesis: pseudohypoxic drive and defective apoptosis caused by mitochondrial dysfunction or structural changes. Further research will further elucidate these mechanisms.
Birt-Hogg-Dubé syndrome
BHD syndrome was first reported in 1977 in a large kindred who had small papular lesions that originated in the hair follicles of the head and neck, appearing during the third and fourth decades of life.89 This rare autosomal-dominant syndrome is characterized by hair follicle hamartomas, pulmonary cyst, spontaneous pneumothorax and renal cell tumours.90
The BHD gene was localized to chromosome 17p11.2,91,92 which encodes folliculin,90 a protein currently of unknown function. But a recent a study93 suggested that folliculin and its interacting partner FNIP1 may be involved in the energy or nutrient pathway regulated by 5′AMP-activated protein kinase and mammalian target of rapamycin, a protein kinase that regulates cell growth, proliferation, motility and transcription.
Renal cancer, which may develop in 15%–30% of patients with BHD,94 is usually multiple and bilateral.95 Renal tumours in BHD usually demonstrate more than 1 coexisting histologic type, such as chromophobe (34%), oncocytoma (5%), clear-cell (9%) and papillary (1.5%) RCC, but the largest subset (65%) is a hybrid comprising oncocytoma and chromophobe RCC.96
In a study by Schmidt and colleagues,97 affected members inherited an insertion or deletion of a cytosine in a C8 tract in exon 11, which represents a hypermutable hotspot for mutation in the BHD gene. Significantly fewer renal tumours were observed in patients with the C-deletion than those with the C-insertion mutation.
On the other hand, Vocke and colleagues98 reported that somatic mutations in the second copy of BHD were distributed across the entire gene. The majority resulted in frameshifts that are predicted to truncate the BHD protein while the loss of heterozygosity at the BHD locus was detected in a minority of additional tumours. These results support a role for BHD as a tumour-suppressor gene that predisposes a person to develop renal tumours when both copies are inactivated.
Sporadic kidney cancer
Clear-cell (conventional) RCC
Clear-cell RCC accounts for approximately 75% of all sporadic forms of RCC. The origin of this cell type is thought to be from proximal tubules.94 From a study9 of families with VHL and linkage analysis, the VHL gene responsible for clear-cell RCC was identified on a small locus on the short arm of chromosome 3. This gene was found to act as a loss-of-function tumour-suppressor gene and was mutated or underwent methylation in a high proportion of tumours from patients with sporadic forms of clear-cell RCC.99 The frequency of this gene inactivation has been reported in up to 60%–70% of these patients.100
Similar to the pathway found in familial disease, one allele is mutated and the other is inactivated.99 The difference in sporadic RCC is that an additional step is required for the development of cancer: both genetic changes must occur independently because the VHL gene has no inherited pre-existent abnormality. The downstream effects are identical; loss of inhibition of the VHL gene on HIF-1α leads to its accumulation, resulting in increased transcription of the target genes. The hypervascularity of the clear-cell RCC and other VHL-mediated tumours has been attributed to this overexpression.101
Unlike the familial form, the resulting phenotype is commonly a solitary tumour that occurs later in life. No mutations of the VHL gene have been found in sporadic RCC outside of the clear-cell variant.102 To date, no phenotypic patterns have been associated with specific tumour mutations.
Papillary RCC
Papillary RCC accounts for 15% of sporadic tumours. The cells originate in the proximal tubule.103 The 2 distinct histological subtypes are classified as type 1 and 2 papillary RCC.104 type1, which is rare, is associated with MET gene mutations, whereas type 2 has none of these mutations and is most commonly encountered in sporadic forms of RCC and more recently in HLRCC.105
Papillary RCC, which is morphologically and cytogenetically different from clear-cell RCC, is found 5–8 times more frequently in males than in females, and more often in the elderly.103,106 In hereditary papillary RCC, as discussed earlier, the gene responsible is MET on chromosome 7, which encodes for receptor tyrosine kinase that is normally activated by HGF/SF.
An activating mutation to this gene in HPRC allows its transformation from a proto-oncogene into an oncogene, which becomes autonomous and results in eventual unregulated cellular proliferation through signal-transduction pathways.107 Chromosome 7 is then duplicated, increasing its level of expression and producing greater numbers of protein copies. However, this activated mutation of the MET gene is found in only 13% of cases in its sporadic counterpart.58,108
Changes that were mostly found in sporadic papillary tumours were 2- or 3-fold gains of wild-type chromosome 7.109 It is postulated that these gains may be sufficient to result in tumour formation without an activating mutation in the MET gene.109 These gains are found in up to 80% of sporadic papillary forms of RCC, along with duplications of chromosome 17 or loss of the Y chromosome.110
A few cases of sporadic forms of papillary RCC have been associated with translocations involving chromosome X, in which it has been fused to the TFE3 transcription gene.111 This transcription-encoding gene is related to the proto-oncogene myc; this dysregulated fusion may be the basis for RCC in this subtype.112,113 These tumours often affect children and young adults. However, the genes responsible for the majority of cases of sporadic papillary RCC are yet to be elucidated.113
Chromophobe, oncocytoma and collecting-duct RCC
The incidence of chromophobe, oncocytoma and collecting-duct tumours is 4%, 4% and less than 1%, respectively.103 No specific genes have been characterized as essential to the development of sporadic forms of these tumours.
The BHD gene is implicated in the rare familial autosomal-dominant condition, BHD syndrome. BHD mutations can occur rarely in sporadic RCC.114,115 Further understanding of the role of alterations in the BHD gene remains a focus of research study today.
Molecular profiling in sporadic forms of RCC
RCC is a heterogeneous epithelial malignancy with different subtypes and tumour biology. Development of novel molecular methods may one day enhance diagnosis before treatment, which may, in turn, increase detection of incidental renal masses and the existence of its benign counterpart. Incorporation of new molecular markers into current staging systems can revolutionize the staging of RCC and provide more accurate prognostication.116
Molecular markers will play an important role in moving from nonspecific treatments to targeting specific therapy in local, locally advanced and metastatic RCC. Methods based on the emergence of gene arrays have made it possible to investigate the expression of large numbers of genes together. An example of this form of treatment is targeting the proteins in the hypoxia-inducible pathway in clear-cell RCC.
Conclusion
Significant developments in understanding the underlying genetic basis of RCC have occurred over the last 2 decades. These advances have been attributed to intensive research into rare inherited renal cancer syndromes and identification of the genes responsible for these syndromes. Many of the genes involved are also found in sporadic RCC.
Understanding the molecular mechanisms involved in the pathogenesis of RCC has aided the development of molecular-targeted drugs for this disease. Experimental drugs with promising results are currently targeting the components of the VEGF (angiogenesis) and transforming-growth-factor-α (growth-factor) pathway of advanced RCC. During the next decade, many more clinical trials will clarify the effectiveness of this strategy. Urologists need to ensure that their understanding of the molecular events in the development of RCC remains current because future management of RCC may belong to a different paradigm.
Footnotes
This article has been peer reviewed.
Competing interests: None declared for Drs. Lim and Ko. Dr. Pautler has received educational grants from Pfizer and Bayer.
References
- 1.Maclaughlin J, Dryer D, Logan H, et al. Canadian cancer society/National Cancer Institute of Canada: Canadian cancer statistics 2006. Toronto: Canada; 2006. [Google Scholar]
- 2.Kovacs G, Akhtar M, Beckwith BJ, et al. Heidelberg classification of renal cell tumours. J Pathol 1997;183:131-3. [DOI] [PubMed] [Google Scholar]
- 3.Collins E. Intra-ocular growths (two cases, brother and sister, with peculiar vascular new growth, probably retinal, affecting both eyes). Trans Ophthalmol Soc U K 1894;14: 1419. [Google Scholar]
- 4.Maher ER, Kaelin WG. von Hippel-Lindau disease. Medicine 1997;76:381-91. [DOI] [PubMed] [Google Scholar]
- 5.Kim WY, Kaelin WG. Role of VHL gene mutation in human cancer. J Clin Oncol 2004; 22: 4991-5004. [DOI] [PubMed] [Google Scholar]
- 6.Iliopoulos O, Kibel A, Gray S, et al. Tumor suppression by the human von Hippel-Lindau gene product. Nat Med 1995;1:822-6. [DOI] [PubMed] [Google Scholar]
- 7.Chen F, Kishida T, Duh FM, et al. Suppression of growth of renal carcinoma cells by the von Hippel-Lindau tumor suppressor gene. Cancer Res 1995;55:4804-7. [PubMed] [Google Scholar]
- 8.Seizinger BR, Rouleau GA, Ozelius LJ, et al. Von Hippel-Lindau disease maps to the region of chromosome 3 associated with renal cell carcinoma. Nature 1988; 332: 2689. [DOI] [PubMed] [Google Scholar]
- 9.Latif F, Tory K, Gnarra J, et al. Identification of von Hippel-Lindau disease tumor suppressor gene. Science 1993;260:1317-20. [DOI] [PubMed] [Google Scholar]
- 10.Ye Y, Vasavada S, Kuzmin I, et al. Subcellular localization of the von Hippel-Lindau disease gene product is cell cycle-dependent. Int J Cancer 1998;78:62-9. [DOI] [PubMed] [Google Scholar]
- 11.Lee S, Chen DY, Humphrey JS, et al. Nuclear/cytoplasmic localization of the von Hippel-Lindau tumor suppressor gene product is determined by cell density. Proc Natl Acad Sci U S A 1996;93:1770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee S, Neumann M, Stearman R, et al. Transcription-dependent nuclear-cytoplasmic trafficking is required for the function of the von Hippel-Lindau tumor suppressor protein. Mol Cell Biol 1999;19:1486-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duan DR, Humphrey JS, Chen DY, et al. Characterization of the VHL tumor suppressor gene product: localization, complex formation, and the effect of natural inactivating mutations. Proc Natl Acad Sci U S A 1995;92:6459-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groulx I, Lee S. Oxygen-dependent ubiquitination and degradation of hypoxia-inducible factor requires nuclear-cytoplasmic trafficking of the von Hippel-Lindau tumor suppressor protein. Mol Cell Biol 2002;22:5319-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duan DR, Pause A, Burgess WH, et al. Inhibition of transcription elongation by the VHL tumor suppressor protein. Science 1995;269:1402-6. [DOI] [PubMed] [Google Scholar]
- 16.Kibel A, Iliopoulos O, DeCaprio JA, et al. Binding of the von Hippel-Lindau tumor suppressor protein to Elongin B and C. Science 1995;269:1444-6. [DOI] [PubMed] [Google Scholar]
- 17.Takagi Y, Pause A, Conaway RC, et al. Identification of Elongin C sequences required for interaction with the von Hippel-Lindau tumor suppressor protein. J Biol Chem 1997; 272: 27444-9. [DOI] [PubMed] [Google Scholar]
- 18.Pause A, Lee S, Worrell RA, et al. The von Hippel-Lindau tumor-suppressor gene product forms a stable complex with human CUL-2, a member of the Cdc53 family of proteins. Proc Natl Acad Sci U S A 1997;94:2156-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lonergan KM, Iliopoulos O, Ohh M, et al. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol Cell Biol 1998;18:732-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamura T, Koepp DM, Conrad MN, et al. Rbx1, a component of the VHL tumor suppressor complex and SCF ubiquitin ligase. Science 1999;284:657-61. [DOI] [PubMed] [Google Scholar]
- 21.Kamura T, Sato S, Iwai K, et al. Activation of HIF1alpha ubiquitination by a reconstituted von Hippel-Lindau (VHL) tumor suppressor complex. Proc Natl Acad Sci U S A 2000;97:10430-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang GL, Jiang B-H, Rue EA, et al. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci U S A 1995;92:5510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Srinivas V, Zhang LP, Zhu XH, et al. Characterization of an oxygen/redox-dependent degradation domain of hypoxia-inducible factor alpha (HIF-alpha) proteins. Biochem Biophys Res Commun 1999;260:557-61. [DOI] [PubMed] [Google Scholar]
- 24.Masson N, William C, Maxwell PH, et al. Independent function of two destruction domains in hypoxia-inducible factor-alpha chains activated by prolyl hydroxylation. EMBOJ 2001;20:5197-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lando D, Peet DJ, Whelan DA, et al. Asparagine hydroxylation of the HIF transactivation domain: a hypoxic switch. Science 2002;295:858-61. [DOI] [PubMed] [Google Scholar]
- 26.Semenza GL, Nejfelt MK, Chi SM, et al. Hypoxia-inducible nuclear factors bind to an enhancer element located 3′ to the human erythropoietin gene. Proc Natl Acad Sci U S A 1991;88:5680-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen C, Pore N, Behrooz A, et al. Regulation of glut 1 mRNA by hypoxia-inducible factor-1.Interaction between H-ras and hypoxia. J Biol Chem 2001;276:9519-25. [DOI] [PubMed] [Google Scholar]
- 28.Ivanov SV, Kuzmin I, Wei MH, et al. Down-regulation of transmembrane carbonic anhydrase in renal cell carcinoma cell lines by wild-type von Hippel-Lindau transgene. Proc Natl Acad Sci U S A 1998;95:12596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tureci O, Sahin U, Vollmar E, et al. Human carbonic anhydrase XII: cDNA cloning expression and chromosomal localization of a carbonic anhydrase gene that is overexpressed in some renal cell cancers. Proc Natl Acad Sci U S A 1998;95:7608-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mukhopadhyay D, Knebelmann B, Cohen HT, et al. The von Hippel-Lindau tumor suppressor gene product interacts with Sp1 to repress vascular endothelial growth factor promoter activity. Mol Cell Biol 1997;17:5629-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gnarra JR, Zhou S, Merrill MJ, et al. Post-transcriptional regulation of vascular endothelial growth factor mRNA by the product of the VHL tumor suppressor gene. Proc Natl Acad Sci U S A 1996;93:10589-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Knebelmann B, Ananth S, Cohen HT, et al. Transforming growth factor α is a target for the von Hippel-Lindau tumor suppressor. Cancer Res 1998;58:226-31. [PubMed] [Google Scholar]
- 33.Mack FA, Rathmell WK, Arsham AM, et al. Loss of pVHL is sufficient to cause HIF dysregulation in primary cells but does not promote tumor growth. Cancer Cell 2003;3: 7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maranchie JK, Vasselli JR, Riss J, et al. The contribution of VHL substrate binding and HIF-1 alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell 2002;1: 24755. [DOI] [PubMed] [Google Scholar]
- 35.Jiang Y, Zhang W, Kondo K, et al. Gene expression profiling in a renal cell carcinoma cell line: dissecting VHL and hypoxia-dependent pathways. Mol Cancer Res 2003;1: 45362. [PubMed] [Google Scholar]
- 36.Wykoff CC, Pugh CW, Maxwell PH, et al. Identification of novel hypoxia dependent and independent target genes of the von Hippel-Lindau (VHL) tumour suppressor by mRNA differential expression profiling. Oncogene 2000;19:6297-305. [DOI] [PubMed] [Google Scholar]
- 37.Calzada MJ, Esteban MA, Feijoo-Cuaresma M, et al. von Hippel-Lindau tumor suppressor protein regulates the assembly of intercellular junctions in renal cancer cells through hypoxia-inducible factor-independent mechanisms. Cancer Res 2006;66: 155360. [DOI] [PubMed] [Google Scholar]
- 38.Ohh M, Yauch R, Lonergan M, et al. The von Hippel–Lindau tumor suppressor protein is required for proper assembly of an extracellular fibronectin matrix. Mol Cell 1998; 1: 959-68. [DOI] [PubMed] [Google Scholar]
- 39.Esteban-Barragan MA, Avila P, Alvarez-Tejado M, et al. Role of the von Hippel-Lindau tumor suppressor gene in the formation of beta1-integrin fibrillar adhesions. Cancer Res 2002; 62:2929-36. [PubMed] [Google Scholar]
- 40.Koochekpour S, Jeffers M, Wang PH, et al. The von Hippel-Lindau tumor suppressor gene inhibits hepatocyte growth factor/scatter factor-induced invasion and branching morphogenesis in renal carcinoma cells. Mol Cell Biol 1999;19:5902-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou MI, Wang H, Ross JJ, et al. The von Hippel-Lindau tumor suppressor stabilizes novel plant homeodomain protein Jade-1. J Biol Chem 2002;277:39887-98. [DOI] [PubMed] [Google Scholar]
- 42.Feldman DE, Spiess C, Howard DE, et al. Tumorigenic mutations in VHL disrupt folding in vivo by interfering with chaperonin binding. Mol Cell 2003;12:1213-24. [DOI] [PubMed] [Google Scholar]
- 43.Li Z, Wang D, Na X, et al. The VHL protein recruits a novel KRAB-A domain protein to repress HIF-1alpha transcriptional activity. EMBO J 2003;22:1857-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Okuda H, Saitoh K, Hirai S, et al. The von Hippel-Lindau tumor suppressor protein mediates ubiquitination of activated atypical protein kinase C. J Biol Chem 2001;276: 436117. [DOI] [PubMed] [Google Scholar]
- 45.Okuda H, Hirai S, Takaki Y, et al. Direct interaction of the beta-domain of VHL tumor suppressor protein with the regulatory domain of atypical PKC isotypes. Biochem Biophys Res Commun 1999;263:491-7. [DOI] [PubMed] [Google Scholar]
- 46.Li Z, Wang D, Na X, et al. Identification of a deubiquitinating enzyme subfamily as substrates of the von Hippel-Lindau tumor suppressor. Biochem Biophys Res Commun 2002; 294: 700-9. [DOI] [PubMed] [Google Scholar]
- 47.Li Z, Na X, Wang D, et al. Ubiquitination of a novel deubiquitinating enzyme requires direct binding to von Hippel-Lindau tumor suppressor protein. J Biol Chem 2002; 277: 4656-62. [DOI] [PubMed] [Google Scholar]
- 48.Na X, Duan HO, Messing E, et al. Identification of the RNA polymerase II subunit hsRPB7 as a novel target of the von Hippel-Lindau protein. EMBO J 2003;22:4249-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Roe JS, Kim H, Lee SM, et al. p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol Cell 2006;22:395-405. [DOI] [PubMed] [Google Scholar]
- 50.Bindra RS, Vasselli JR, Stearman R, et al. VHL mediated hypoxia regulation of cyclin D1 in renal cancer cells. Cancer Res 2002;62:3014-9. [PubMed] [Google Scholar]
- 51.Osipov V, Keating JT, Faul PN, et al. Expression of p27 and VHL in renal tumors. Appl Immunohistochem Mol Morphol 2002;10:344-50. [DOI] [PubMed] [Google Scholar]
- 52.Zbar B, Kishida T, Chen F, et al. Germline mutations in the von Hippel-Lindau disease (VHL) gene in families from North America, Europe, and Japan. Hum Mutat 1996;8: 34857. [DOI] [PubMed] [Google Scholar]
- 53.Chen F, Kishida T, Yao M, et al. Germline mutations in the von Hippel-Lindau disease tumor suppressor gene: correlations with phenotype. Hum Mutat 1995;5:66-75. [DOI] [PubMed] [Google Scholar]
- 54.Hoffman MA, Ohh M, Yang H, et al. von Hippel-Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum Mol Genet 2001; 10: 1019-27. [DOI] [PubMed] [Google Scholar]
- 55.Clifford SC, Cockman ME, Smallwood A, et al. Contrasting effects on HIF1a regulation by disease-causing pVHL mutations correlate with patterns of tumorigenesis in von Hippel-Lindau disease. Hum Mol Genet 2001;10:1029-38. [DOI] [PubMed] [Google Scholar]
- 56.Zbar B, Tory K, Merino M, et al. Hereditary papillary renal carcinoma. J Urol 1994; 151: 561-6. [DOI] [PubMed] [Google Scholar]
- 57.Zbar B, Glenn G, Lubensky I, et al. Hereditary papillary renal carcinoma clinical studies in 10 families. J Urol 1995;153:907-12. [PubMed] [Google Scholar]
- 58.Schmidt L, Duh FM, Chen F, et al. Germline and somatic mutations in the tyrosine kinase domain of the Met proto-oncogene in papillary renal carcinomas. Nat Genet 1997; 16:68-73. [DOI] [PubMed] [Google Scholar]
- 59.Brinkmann V, Foroutan H, Sachs M, et al. Hepatocyte growth factor/scatter factor induces a variety of tissue-specific morphogenic programs in epithelial cells. J Cell Biol 1995; 131:1573-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeffers M, Rao MS, Rulong S, et al. Hepatocyte growth factor/scatter factor-Met signaling induces proliferation, migration, and morphogenesis of pancreatic oval cells. Cell Growth Differ 1996;7:1805-13. [PubMed] [Google Scholar]
- 61.Medico E, Mongiovi AM, Huff J, et al. The tyrosine kinase receptors Ron and Sea control “scattering” and morphogenesis of liver progenitor cells in vitro. Mol Biol Cell 1996; 7:495-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Montesano R, Schaller G, Orci L. Induction of epithelial tubular morphogenesis in vitro by fibroblast-derived soluble factors. Cell 1991;66:697-711. [DOI] [PubMed] [Google Scholar]
- 63.Santoro MM, Collesi C, Grisendi S, et al. Constitutive activation of the RON gene promotes invasive growth but not transformation. Mol Cell Biol 1996;16:7072-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stoker M, Perryman M. An epithelial scatter factor released by embryo fibroblasts. J Cell Sci 1985;77:209-23. [DOI] [PubMed] [Google Scholar]
- 65.Bottaro DP, Rubin JS, Faletto DL, et al. Identification of the hepatocyte growth factor receptor as the c-met proto-oncogene product. Science 1991;251:802-4. [DOI] [PubMed] [Google Scholar]
- 66.Faletto DL, Tsarfaty I, Kmiecik TE, et al. Evidence for non-covalent clusters of the c-met proto-oncogene product. Oncogene 1992;7:1149-57. [PubMed] [Google Scholar]
- 67.Naldini L, Vigna E, Ferracini R, et al. The tyrosine kinase encoded by the MET proto-oncogene is activated by autophosphorylation. Mol Cell Biol 1991;11:1793-803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rodrigues GA, Park M. Autophosphorylation modulates the kinase activity and oncogenic potential of the Met receptor tyrosine kinase. Oncogene 1994;9:2019-27. [PubMed] [Google Scholar]
- 69.Ponzetto C, Bardelli A, Zhen Z, et al. A multifunctional docking site mediates signaling and transformation by the hepatocyte growth factor/scatter factor receptor family. Cell 1994;77:261-71. [DOI] [PubMed] [Google Scholar]
- 70.Komada M, Kitamura N. The cell dissociation and motility triggered by scatter factor/hepatocyte growth factor are mediated through the cytoplasmic domain of the c-Met receptor. Oncogene 1993;8:2381-90. [PubMed] [Google Scholar]
- 71.Weidner KM, Sachs M, Birchmeier W. The met receptor tyrosine kinase transduces motility, proliferation and morphogenic signals of scatter factor/hepatocyte growth factor in epithelial cells. J Cell Biol 1993;121:145-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhu H, Naujokas MA, Park M. Receptor chimeras indicate that the met tyrosine kinase mediates the motility and morphogenic responses of hepatocyte growth/scatter factor. Cell Growth Differ 1994;5:359-66. [PubMed] [Google Scholar]
- 73.Jeffers M, Rong S, Vande Woude GF. Hepatocyte growth factor/scatter factor-Met signaling in tumorigenicity and invasion/metastasis. J Mol Med 1996;74:505-13. [DOI] [PubMed] [Google Scholar]
- 74.Igawa T, Kanda S, Kanetake H. Hepatocyte growth factor is a potent mitogen for cultured rabbit renal tubular epithelial cells. Biochem Biophys Res Commun 1991; 174: 831-8. [DOI] [PubMed] [Google Scholar]
- 75.Kan M, Zhang G, Zarnegar R, et al. Hepatocyte growth factor/hepatopoietin A stimulates the growth of rat kidney proximal tubule epithelial cells (RPTE), rat nonparenchymal liver cells, human melanoma cells, mouse keratinocytes and stimulates anchorage-independent growth of SV-40 transformed RPTE. Biochem Biophys Res Commun 1991; 174: 331-7. [DOI] [PubMed] [Google Scholar]
- 76.Montesano R, Matsumoto K, Nakamura T, et al. Identification of a fibroblast-derived epithelial morphogen as hepatocyte growth factor. Cell 1991;67:901-8. [DOI] [PubMed] [Google Scholar]
- 77.Zhuang Z, Park W, Pack S, et al. Trisomy 7-harbouring non-random duplication of the mutant met allele in hereditary papillary renal carcinomas. Nat Genet 1998;20:669. [DOI] [PubMed] [Google Scholar]
- 78.Jeffers M, Koochekpour S, Fiscella M, et al. Signaling requirements for oncogenic forms of the Met tyrosine kinase receptor. Oncogene 1998;17:2691-700. [DOI] [PubMed] [Google Scholar]
- 79.Jeffers M, Schmidt L, Nakaigawa N, et al. Activating mutations for the Met tyrosine receptor in human cancer. Proc Natl Acad Sci U S A 1997;94:11445-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Giordano S, Corso S, Conrotto P, et al. The semaphorin 4D receptor controls invasive growth by coupling with Met. Nat Cell Biol 2002;4:720-4. [DOI] [PubMed] [Google Scholar]
- 81.Pennacchietti S, Michieli P, Galluzzo M, et al. Hypoxia promotes invasive growth by transcriptional activation of met protooncogene. Cancer Cell 2003;3:347-61. [DOI] [PubMed] [Google Scholar]
- 82.Launonen V, Vierimaa O, Kiuru M, et al. Inherited susceptibility to uterine leiomyomas and renal cell cancer. Proc Natl Acad Sci U S A 2001;98:3387-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tomlinson IP, Alam NA, Rowan AJ, et al. Germline mutations in FH predispose to dominantly inherited uterine fibroids, skin leiomyomata and papillary renal cell cancer. Nat Genet 2002;30:406-10. [DOI] [PubMed] [Google Scholar]
- 84.Alam NA, Rowan AJ, Wortham NC, et al. Genetic and functional analyses of FH mutations in multiple cutaneous and uterine leiomyomatosis, hereditary leiomyomatosis and renal cancer, and fumarate hydratase deficiency. Hum Mol Genet 2003;12:124152. [DOI] [PubMed] [Google Scholar]
- 85.Toro JR, Nickerson ML, Wei MH, et al. Mutations in the fumarate hydratase gene cause hereditary leiomyomatosis and renal cell cancer in families in North America. Am J Hum Genet 2003;73:95-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wei MH, Toure O, Glenn GM, et al. Novel mutation in FH and expansion of the spectrum of phenotypes expressed in families with hereditary leiomyomatosis and renal cell cancer. J Med Genet 2006;43:18-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Alam NA, Olpin S, Leigh IM. Fumarate hydratase mutations and predisposition to cutaneous leiomyomas, uterine leiomyomas and renal cancer. Br J Dermatol 2005; 153:11-7. [DOI] [PubMed] [Google Scholar]
- 88.Kiuru M, Launonen V, Hietala M, et al. Familial cutaneous leiomyomatosis is a two hit condition associated with renal cell cancer of characteristic histopathology. Am J Pathol 2001;159:825-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Birt AR, Hogg GR, Dubé J. Hereditary multiple fibrofolliculomas with trichodiscomas and acrochordons. Arch Dermatol 1977;113:1674-7. [PubMed] [Google Scholar]
- 90.Nickerson ML, Warren MB, Toro J, et al. Mutations in a novel gene lead to kidney tumors, lung wall defects, and benign tumors of the hair follicle in patients with the Birt-Hogg-Dubé syndrome. Cancer Cell 2002;2:157-64. [DOI] [PubMed] [Google Scholar]
- 91.Schmidt LS, Warren MB, Nickerson ML, et al. Birt-Hogg-Dubé syndrome, a genodermatosis associated with spontaneous pneumothorax and kidney neoplasia, maps to chromosome 17p11.2. Am J Hum Genet 2001;69:876-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khoo SK, Bradley M, Wong FK, et al. Birt-Hogg-Dubé syndrome: mapping of a novel hereditary neoplasia gene to chromosome 17p12-q11.2. Oncogene 2001;20:523942. [DOI] [PubMed] [Google Scholar]
- 93.Baba M, Hong SB, Sharma N, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci U S A 2006;103:15552-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Linehan WM, Walther MM, Zbar B, et al. The genetic basis of cancer of the kidney. J Urol 2003;170:2163-72. [DOI] [PubMed] [Google Scholar]
- 95.Roth JS, Rabinowitz AD, Benson M, et al. Bilateral renal cell carcinoma in the Birt-Hogg-Dubé syndrome. J Am Acad Dermatol 1993;29:1055-6. [DOI] [PubMed] [Google Scholar]
- 96.Pavlovich CP, Hewitt S, Walther MM, et al. Renal tumors in the Birt-Hogg-Dubé syndrome. Am J Surg Pathol 2002;26:1542-52. [DOI] [PubMed] [Google Scholar]
- 97.Schmidt LS, Nickerson ML, Warren MB, et al. Germline BHD-mutation spectrum and phenotype analysis of a large cohort of families with Birt-Hogg-Dubé syndrome. Am J Hum Genet 2005;76:1023-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vocke CD, Yang Y, Pavlovich CP, et al. High frequency of somatic frameshift BHD gene mutations in Birt-Hogg-Dubé-associated renal tumors. J Natl Cancer Inst 2005; 97: 931-5. [DOI] [PubMed] [Google Scholar]
- 99.Herman JG, Latif F, Weng Y, et al. Silencing of the VHL tumor-suppressor gene by DNA methylation in renal carcinoma. Proc Natl Acad Sci U S A 1994;91:9700-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Iliopoulos O. Molecular biology of renal cell cancer and the identification of therapeutic targets. J Clin Oncol 2006;24:5593-600. [DOI] [PubMed] [Google Scholar]
- 101.Sufan RI, Jewett MA, Ohh M. The role of von Hippel-Lindau tumor suppressor protein and hypoxia in renal clear cell carcinoma. Am J Physiol Renal Physiol 2004;287:F16. [DOI] [PubMed] [Google Scholar]
- 102.Gnarra JR, Tory K, Weng Y, et al. Mutations of the VHL tumour suppressor gene in renal carcinoma. Nat Genet 1994;7:85-90. [DOI] [PubMed] [Google Scholar]
- 103.Amin MB, Tamboli J, Javidan H, et al. Prognostic impact of histologic subtyping of adult renal epithelial neoplasms: an experience of 405 cases. Am J Surg Pathol 2002;26: 28191. [DOI] [PubMed] [Google Scholar]
- 104.Delahunt B, Eble JN. Papillary renal cell carcinoma: a clinicopathologic and immunohistochemical study of 105 tumors. Mod Pathol 1997;10:537-44. [PubMed] [Google Scholar]
- 105.Lubensky IA., Schmidt L, Zhuang Z, et al. Hereditary and sporadic papillary renal carcinomas with c-MET mutations share a distinct morphological phenotype. Am J Pathol 1999; 155:517-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kovacs G. The value of molecular genetic analysis in the diagnosis and prognosis of renal cell tumours. World J Urol 1994;12:64-8. [DOI] [PubMed] [Google Scholar]
- 107.Phillips JL, Pavlovich CP, Walther M, et al. The genetic basis of renal epithelial tumors: advances in research and its impact on prognosis and therapy. Curr Opin Urol 2001; 11: 463-9. [DOI] [PubMed] [Google Scholar]
- 108.Schmidt L, Junker K, Nakaigawa N, et al. Novel mutations of the MET proto-oncogene in papillary renal carcinomas. Oncogene 1999;18:2343-50. [DOI] [PubMed] [Google Scholar]
- 109.Glukhova L, Lavialle C, Fauvet D, et al. Mapping of the 7q31 subregion common to the small chromosome 7 derivatives from two sporadic papillary renal cell carcinomas: increased copy number and overexpression of the MET proto-oncogene. Oncogene 2000; 19:754-61. [DOI] [PubMed] [Google Scholar]
- 110.Zambrano NR, Lubensky IA, Merino MJ, et al. Histopathology and molecular genetics of renal tumors toward unification of a classification system. J Urol 1999;162: 124658. [PubMed] [Google Scholar]
- 111.Shipley JM, Birdsall S, Clark J, et al. Mapping the X chromosome breakpoint in two papillary renal cell carcinoma cell lines with a t(X;1)(p11.2;q21.2) and the first report of a female case. Cytogenet Cell Genet 1995;71:280-4. [DOI] [PubMed] [Google Scholar]
- 112.Cohen HT, McGovern FJ. Renal cell carcinoma. N Engl J Med 2005;353:2477-90. [DOI] [PubMed] [Google Scholar]
- 113.Linehan WM, Zbar B. Focus on kidney cancer. Cancer Cell 2004;6:223-8. [DOI] [PubMed] [Google Scholar]
- 114.Da Silva NF, Gentle D, Hesson LB, et al. Analysis of the Birt-Hogg-Dubé (BHD) tumour suppressor gene in sporadic renal cell carcinoma and colorectal cancer. J Med Genet 2003; 40:820-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Khoo SK., Kahnoski K, Sugimura J, et al. Inactivation of BHD in sporadic renal tumors. Cancer Res 2003;63:4583-7. [PubMed] [Google Scholar]
- 116.Lam JS, Leppert JT, Figlin RA, et al. Role of molecular markers in the diagnosis and therapy of renal cell carcinoma. Urology 2005;66:1-9. [DOI] [PubMed] [Google Scholar]