Abstract
It has previously been reported that 1,N6-ethenoadenine (ɛA), deaminated adenine (hypoxanthine, Hx), and 7,8-dihydro-8-oxoguanine (8-oxoG), but not 3,N4-ethenocytosine (ɛC), are released from DNA in vitro by the DNA repair enzyme alkylpurine-DNA-N-glycosylase (APNG). To assess the potential contribution of APNG to the repair of each of these mutagenic lesions in vivo, we have used cell-free extracts of tissues from APNG-null mutant mice and wild-type controls. The ability of these extracts to cleave defined oligomers containing a single modified base was determined. The results showed that both testes and liver cells of these knockout mice completely lacked activity toward oligonucleotides containing ɛA and Hx, but retained wild-type levels of activity for ɛC and 8-oxoG. These findings indicate that (i) the previously identified ɛA-DNA glycosylase and Hx-DNA glycosylase activities are functions of APNG; (ii) the two structurally closely related mutagenic adducts ɛA and ɛC are repaired by separate gene products; and (iii) APNG does not contribute detectably to the repair of 8-oxoG.
There are numerous DNA repair enzymes that excise deleterious modified bases or a modifying group from DNA in mammalian cells (1–3). Efforts have been made to define the substrate range of most of these by using purified or partially purified or recombinant enzymes. However, such experiments can be ambiguous. In contrast, the recent use of targeted gene deletion in cells has provided more definitive information on the biological role of two base excision repair enzymes, hydroxymethyluracil-DNA glycosylase (4) and 3-methyladenine (m3A)-DNA glycosylase (alkylpurine-DNA-N-glycosylase; APNG) (5, 6). There are more examples of targeted deletion in mice of genes involved in nucleotide excision repair, generally lacking repair capacity for UV damage—e.g., XPA and XPC gene knockouts (7–9) as well as mismatch repair (10), O6-alkylguanine and O4-alkylthymine repair by O6-methylguanine methyltransferase (MGMT) (11).
In the case of hydroxymethyluracil-DNA glycosylase-deficient hamster cells, there was no phenotypic alteration (4). In an APNG-deficient mouse cell line, it was shown that two alkylating agents, N,N′-bis(2-chloroethyl)-N-nitrosourea (BCNU) and mitomycin C, induced chromosome damage and cell killing as a result of the lack of removal by m3A-DNA glycosylase of damage produced by these agents (5). Neither of these experiments was specifically directed toward the possible substrate range of enzymes, but both experiments resulted in new information on the role of a specific enzyme in maintaining genetic integrity against individual agents causing DNA damage.
A separate line of experimentation to assess the in vivo mutagenicity of various modified bases in differing repair backgrounds used defined synthetic oligonucleotides or globally modified vectors (12–15). Related to this are numerous in vitro experiments also using site-directed mutagenesis in which replication efficiency and changed base pairing could be quantitated [reviewed by Singer and Essigmann, (15)]. These data all yielded basic information on the potential biological role of modified bases in the absence of any repair capacity. By using the same oligonucleotides annealed to a complementary strand, but in the presence of single purified or cloned repair enzymes, it was possible to quantitate and identify which of known enzymes could repair an individual DNA adduct.
An early application of this in vitro system was used to study DNA glycosylase-mediated nicking activity toward chloroacetaldehyde adducts, particularly 1,N6-ethenoadenine (ɛA) and 3,N4-ethenocytosine (ɛC), which bound to partially purified HeLa extracts cleaving the oligonucleotide 5′ to the adduct (16, 17). It could be shown that both the purified “ɛA-binding protein” and cloned human m3A-DNA glycosylase (APNG) acted in the same manner on the same substrates—i.e., ɛA and m3A (18). ɛC was also released by partially purified HeLa cell-free extracts. Therefore it was concluded that it was a DNA glycosylase substrate (19). However, ɛC activity was later found to reside in a different glycosylase than APNG when an ɛC-DNA glycosylase activity was partially purified by column chromatography (20).
In addition, deamination resulting in adenine → hypoxanthine (Hx), which occurs naturally, was also repaired by the mammalian APNG protein (21). However, Hx was very poorly repaired by homologous clones from lower organisms, such as yeast and Escherichia coli (21). This species difference in repair was also true for ɛA (22).
Another ubiquitous endogenously produced mutagenic modified base in mammals is 7,8-dihydro-8-oxoguanine (8-oxoG), which was also reported as a substrate for APNG (23). It is now known that human and mouse cells contain a DNA glycosylase, termed 8-oxoG-DNA glycosylase, which was recently cloned (24–29).
More definite proof of each of these enzyme specificities became feasible when a base excision repair gene, the APNG gene, was deleted in a knockout (ko) mouse (30) and tissue extracts were used to measure glycosylase activity toward these four substrates. This paper presents biochemical evidence obtained by using extracts from genetically deficient mice to prove that both ɛA and Hx are substrates for APNG, but ɛC and 8-oxoG are not.
MATERIALS AND METHODS
Materials.
[γ-32P]ATP (specific activity 6,000 Ci/mmol; 1 Ci = 37 GBq) was purchased from Amersham. T4 polynucleotide kinase was purchased from United States Biochemical. Uracil-DNA glycosylase (UDG) was obtained from GIBCO/BRL. The major human apurinic site (AP) endonuclease (HAP1) was a gift from I. D. Hickson (University of Oxford). Phosphocellulose P11 was from Whatman. Sep-Pak columns were from Waters.
Generation of APNG ko Mice.
The generation and initial characterization of our APNG ko mice will be described in detail elsewhere. Brief details are included here for clarity. The murine APNG gene consists of four exons and is situated in the α-globin locus on chromosome 11 (31). The targeting vector was designed to disrupt the expression of APNG gene, while leaving intact DNase-hypersensitive sites, both 5′ and internal to the APNG gene, that may be involved in the regulation of other genes in the locus. Thus, by using standard gene targeting procedures, a 2.5-kb XhoI–BamHI fragment encompassing the APNG promoter sequences and exons 1 and 2 was deleted and replaced by a neo expression cassette. Correctly targeted embryonic stem cells (129/Ola) were identified by PCR and Southern blotting and microinjected into C57BL/6 mouse blastocytes. APNG ko mice were then obtained by crossing APNG(+/−) offspring of the resulting chimeras.
Preparation of Cell-Free Extracts.
Cell-free extracts were prepared by sonication of macerated testes and liver tissue in ice-cold buffer (50 mM Tris⋅HCl, pH 8.3/1 mM EDTA/3 mM DTT) containing 2 μg/ml leupeptin, followed by the addition of phenylmethylsulfonyl fluoride (PMSF) to 0.5 mM. Proteins precipitating between 30% and 60% saturated ammonium sulfate were recovered by centrifugation and frozen at −80°C until required.
The above ammonium sulfate precipitates were dissolved in a buffer containing 25 mM Hepes–KOH at pH 7.8, 0.5 mM EDTA, 0.125 mM PMSF, 3 mM 2-mecaptoethanol, and 10% (vol/vol) glycerol and then used in enzyme assays.
Preparation of Murine APNG.
Full-length cDNA was obtained from mouse (BALB/c) testis by reverse transcription–PCR, inserted into pUC 8.0 on EcoRI ends, and confirmed by DNA sequencing. After incubation overnight at 37°C, transformed E. coli DH5α cells (GIBCO/BRL) from 4 liters of culture were recovered by centrifugation and sonicated for 30 sec in buffer A (20 mM Tris⋅HCl, pH 8.0/1 mM EDTA/1 mM DTT/5% glycerol) containing 2 μg/ml leupeptin and 0.5 mM PMSF. Cell debris was removed by centrifugation, and proteins precipitating between 30% and 55% saturated ammonium sulfate were purified further by phosphocellulose P11 chromatography, essentially as described by Roy et al. (32). Briefly, the ammonium sulfate precipitate was dissolved in buffer A containing 0.1 M NaCl and dialyzed against the same at 4°C overnight. The dialysate was then loaded on to a preequilibrated P11 column, and the enzyme was eluted with 0.3 M NaCl in buffer A. Fractions were assayed for APNG activity by the method of Bjelland and Seeberg (33), using N-[3H]methyl-N-nitrosourea-treated calf thymus DNA as substrate, and active fractions were pooled. The resulting partially purified material was essentially nuclease free under the conditions of the assays described in this article.
Oligonucleotide Substrates.
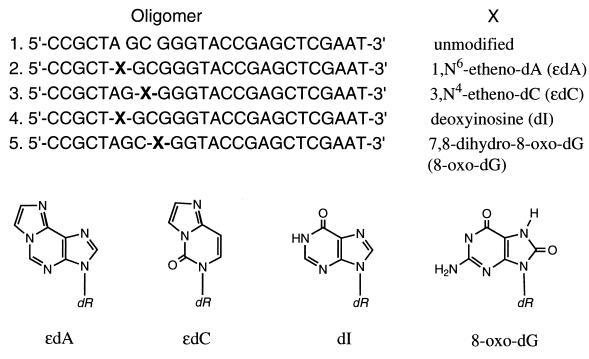
The four sequences of oligonucleotides used as substrates for examining repair enzymatic activities in the cell-free extracts from wild-type (wt) and ko mice are listed in Fig. 1. The phosphoramidite technique was used for synthesis. The phosphoramidite of X in sequences 2, 4, and 5 was obtained from Glen Research (Sterling, VA). The phosphoramidite of ɛdC (sequence 3) was synthesized as discussed previously (19). Each of these oligomers was annealed to a universal complementary 25-mer strand, 5′-ATTCGAGCTCGGTACCCGCTAGCGG-3′.
Figure 1.
Structure of the modified nucleosides used and their positions in a defined 25-mer oligonucleotide. The unmodified oligomer is in sequence 1. The substitutions are shown as X (sequences 2–5).
The deoxyuridine (dU)-containing 25-mer oligomer was obtained by placing the dU at position 8 from the 5′ side in sequence 1. The double-stranded 25-mer oligonucleotide containing a single AP site was prepared by treating the same dU-containing oligomer with UDG (0.002 unit) for 15 min at 37°C.
DNA Nicking Assay.
The [γ-32P]ATP end-labeling of oligomers and DNA nicking assay in this work were performed essentially as described by Rydberg et al. (16, 17). Briefly, the nicking reaction was carried out in a total volume of 10 μl in 25 mM Hepes–KOH, pH 7.8/0.5 mM EDTA/0.5 mM DTT containing 0.5 mg of poly(dI-dC), 0.5 mM spermidine, and 10% glycerol for 1 hr at 37°C. Quantitation of the extent of cleavage was by densitometry of autoradiograms after electrophoresis in denaturing 12% acrylamide gels. Size markers were synthesized, purified, and co-electrophoresed to determine the size of the 5′ 32P-labeled cleavage product (16).
RESULTS
Cleavage of ɛA, Hx, and ɛC by Testes Extracts from wt and ko Mice.
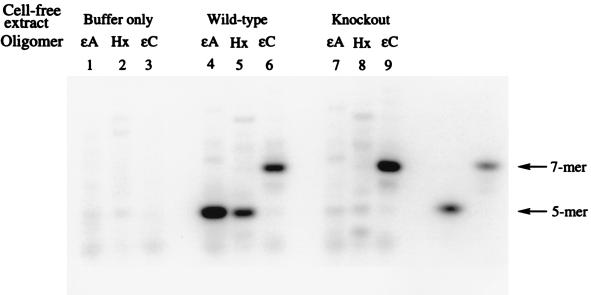
A comparison of nature and extent of glycosylase activity in wt and ko mice showed that all three modified bases were cleaved from defined 25-mer oligonucleotides by the wt cell-free extracts (Fig. 2, lanes 4–6). However, the best substrate is ɛA, while both Hx and ɛC are cleaved about 50% less efficiently in the wt crude extracts. When the ko mouse testes extracts were used, only ɛC was cleaved (Fig. 2, lane 9), indicating that glycosylase activity toward ɛA and Hx was absent as a result of deletion of the APNG gene.
Figure 2.
Autoradiogram of denaturing 12% polyacrylamide gel comparing extent of nicking by mouse testes cell-free extracts of ɛA, Hx, and ɛC in control samples (lanes 1–3), wt mice (lanes 4–6) and ko mice (lanes 7–9). The positions of 5′ cleavage are shown on the far right, as indicated by arrows. The oligomers were allowed to react with 30 μg of protein for 1 h at 37°C in a standard nicking assay.
Quantitation of the same enzymatic activities in testes extracts by gel scanning confirmed that the activities are protein concentration dependent (Fig. 3), and even at the highest protein level used, there was no detectable ɛA or Hx incised by the ko mouse preparation (Fig. 3 Right). The activity toward ɛC was unchanged from the data for the wt extract. That is, there is a similar extent of cleavage in both wt and ko extracts.
Figure 3.
Quantitation of the 32P-labeled bands from PAGE of the same three oligomers in Fig. 2 as a function of protein concentration of testes extracts (0–30 μg), under the same reaction conditions as in Fig. 2. The percent of oligomer cleaved is expressed as % nicking efficiency. Note that in the ko mice (Right) there was no detectable ɛA or Hx cleavage above background.
Cleavage of 8-oxoG by Testes Extracts from wt and ko Mice.
In Fig. 4 Left, gel electrophoresis of an 8-oxoG-containing oligomer allowed to react with increasing amounts of wt and ko testes extract indicated that the cleavage occurred in an enzyme concentration-dependent manner. This was supported by densitometry of the gel as shown in Fig. 4 Right, in which the extent of cleavage was approximately 40% of the oligonucleotide at the highest protein concentration used and there was no difference between wt and ko mice.
Figure 4.
(Left) PAGE of activities of wt and ko mouse testes extract toward 8-oxoG after 1-h incubation, which produces a 5′ 8-mer fragment as indicated by the arrow. Lane 1 is a control with buffer alone. Lanes 2–5 and 6–9 are with increasing amounts of testes protein (5, 10, 15, and 30 μg). (Right) Quantitation of the autoradiogram in Left. Note that there is no detectable difference between the wt and ko mouse extracts.
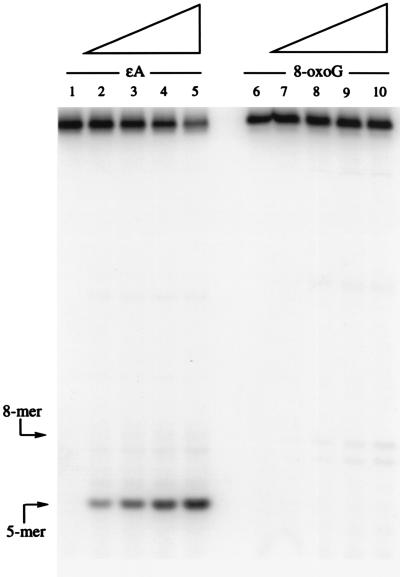
Cleavage of ɛA and 8-oxoG by Cloned Mouse APNG.
In this experiment a cloned partially purified APNG gene product was used in a standard nicking assay (see Materials and Methods) to compare enzyme activity toward ɛA and 8-oxoG. In Fig. 5 Left it can be seen that the expected 5-mer (see Fig. 1) from the ɛA-containing oligonucleotide was cleaved to a high extent as a function of protein concentration. In contrast, in Fig. 5 Right, the same protocol used with the 8-oxoG-containing oligonucleotide showed no detectable 8-mer, which would be the 5′ cleavage product from this oligomer (see Fig. 1). It should be noted that a 5′ AP endonuclease, the major human AP endonuclease (HAP1), was present to produce a uniform 5′ cleavage of the AP site resulting from APNG activity.
Figure 5.
Autoradiogram of partially purified murine APNG-treated ɛA oligomer (Fig. 1, sequence 2) (Left) and 8-oxoG oligomer (Fig. 1, sequence 5) (Right) after treatment with increasing amounts of the recombinant protein, from 2.5 to 15 ng, shown by the slope of the triangles above. Lanes 1 and 6 are controls with no added protein. The reaction conditions were the same as in Fig. 2 except that HAP1 at 0.5 μg/ml was added to cleave the AP site on the 5′ side. The use of HAP1 alone under the same conditions does not cause any cleavage of either oligomer (data not shown). The positions of the size markers are indicated by arrows.
Use of Liver Cell-Free Extracts from wt and ko Mice.
The liver cell-free extracts, from both wt and ko mice, contained less enzyme activity toward all four adducts. This was in agreement with the levels of mRNA expression in these tissues (data not shown). However, the substrate specificity in both wt and ko liver was the same as that of testes. In ko mice both ɛA and Hx cleavage was absent, whereas ɛC and 8-oxoG cleavage occurred (data not shown).
Other Repair Activities Tested in wt and ko Mice.
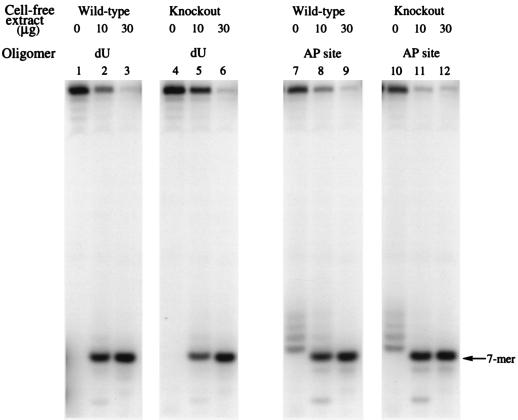
In Fig. 6 it is shown that normal levels of UDG and 5′ AP endonuclease were present in ko mice studied by using the same 25-mer sequence with dU at position 8 or the same dU-containing oligomer pretreated with UDG, indicating that the ko mouse construct did not affect the enzyme involved in the next step of base excision repair, i.e., AP endonuclease cleavage of the AP site.
Figure 6.
Activity of both mouse extracts against dU (lanes 1–6) and the AP site (lanes 7–12). The AP site was obtained by treating the dU-containing oligomer with purified UDG. The AP site was then created and incubated with testes cell-free extracts from both wt and ko mice. Lanes 7 and 10 illustrate the lability of the AP site under experimental conditions in the absence of any added enzyme. For this reason the purified HAP1 was added to the reaction mixture in Fig. 5.
DISCUSSION
Assigning a specific repair enzyme function to particular DNA adducts has been difficult when using bacterial and mammalian cells for testing because there are many repair enzymes present. Isolation, protein purification, and molecular cloning techniques have helped to define the enzyme and its substrate, in vitro, but not necessarily unambiguously, because it appeared that more than one pathway may exist for the repair of particular DNA lesions (3). In addition, it has not always been possible to achieve the absolute purity of some enzyme preparations. A more definite proof of substrate specificity and in vivo function can be achieved by using a genetic approach that involves deletion of specific genes coding for DNA repair proteins in cells or animals.
To our knowledge, this is the first instance of the deletion of an enzyme of the base excision repair pathway being used to study its substrate specificity. Thus, the initial finding that two DNA glycosylases, separated by their chromatographic properties, are required for the repair of ɛA and ɛC in human cells (20) has been confirmed in the mouse model. Additionally, we have presented compelling evidence that APNG is the principal glycosylase involved in the repair of ɛA and Hx, corroborating the biochemical evidence reported by this (18) and other (21, 22) groups. However, for 8-oxoG, another biologically important, endogenously formed promutagenic lesion (34–36), no evidence for the involvement of APNG in its repair was found in this study, in contrast to a previous report in which both partially purified recombinant mouse and human APNG were used (23). Recently a number of groups have reported the cloning of a human and mouse 8-oxoG-DNA glycosylase, the functional homologue of the E. coli fpg gene (24–29), and this is likely to be the principal route for the repair of this DNA lesion.
Therefore, when a ko mouse lacking APNG activity was obtained (30), it was feasible to use cell-free tissue extracts to evaluate the effects on each of these modified bases, site-specifically placed in defined oligonucleotides, by using a nicking assay (16) and quantitation by densitometry of the 5′ 32P end-labeled fragment. From these experiments, it was then clear that ɛA and ɛC were substrates for separate DNA glycosylases (Figs. 2 and 3). This work also confirmed that ɛA and Hx were both substrates for APNG (Fig. 3).
In addition, we find that both wt and ko mice remove 8-oxoG efficiently and to the same extent (Fig. 4), indicating that deletion of the APNG gene does not affect 8-oxoG removal, in accordance with the discovery of a separate 8-oxoG-DNA glycosylase gene in mammalian cells (24–29). In the work reported here, we have evidence only that APNG does not play a measurable role in repair of 8-oxoG under the conditions of our assay system. We have, however, been able to show that UDG and AP endonuclease activities were not affected by APNG gene deletion.
The technique used here in conjunction with in vitro repair studies of oligonucleotides with site-specifically placed adducts can provide an unambiguous means to facilitate designation of the substrate specificity of a given enzyme.
Acknowledgments
We thank Dr. A. Chenna of Lawrence Berkeley National Laboratory for preparation of the oligonucleotides used in this work. We also thank R. J. Weeks, M. A. Willington, J. A. Rafferty, and B. Deans for their invaluable help in the generation of the APNG ko mice and K. J. Mynett for his excellent technical assistance during part of this work at the Paterson Institute for Cancer Research. This work was supported by National Institutes of Health Grants CA 47723, CA 72079, and ES 07363 (to B.S.) and was administered by the Lawrence Berkeley National Laboratory under Department of Energy Contract DE-AC03-76SF00098, and it was supported by a Cancer Research Campaign Project Grant (to G.P.M. and R.H.E.).
ABBREVIATIONS
- APNG
alkylpurine-DNA-N-glycosylase
- UDG
uracil-DNA glycosylase
- ɛA
1,N6-ethenoadenine
- ɛC
3,N4-ethenocytosine
- Hx
hypoxanthine
- dI
deoxyinosine
- 8-oxoG
7,8-dihydro-8-oxoguanine
- m3A
3-methyladenine
- wt
wild-type
- ko
knockout
- AP
apurinic site
- HAP1
human AP endonuclease 1
References
- 1.Sancar A. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. [DOI] [PubMed] [Google Scholar]
- 2.Friedberg E, Walker G C, Seide W. DNA Repair and Mutagenesis. Washington, DC: Am. Soc. Microbiol.; 1995. [Google Scholar]
- 3.Singer B, Hang B. Chem Res Toxicol. 1997;10:713–732. doi: 10.1021/tx970011e. [DOI] [PubMed] [Google Scholar]
- 4.Mi L J, Mahl E, Chaung W, Boorstein R J. Mutat Res. 1997;374:287–295. doi: 10.1016/s0027-5107(96)00247-3. [DOI] [PubMed] [Google Scholar]
- 5.Engelward B P, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson L. EMBO J. 1996;15:945–952. [PMC free article] [PubMed] [Google Scholar]
- 6.Sidorkina O, Saparbaev M, Laval J. Mutagenesis. 1997;12:23–28. doi: 10.1093/mutage/12.1.23. [DOI] [PubMed] [Google Scholar]
- 7.Sands A T, Abuin A, Sanchez A, Conti C J, Bradley A. Nature (London) 1995;377:162–165. doi: 10.1038/377162a0. [DOI] [PubMed] [Google Scholar]
- 8.de Vries A, van Oostrom C T M, Hofhuis F M A, Dortant P M, Berg R J, de Gruijl F R, Wester P W, van Kreijl C F, Capel P J A, van Steeg H, Verbeek S J. Nature (London) 1995;377:169–173. doi: 10.1038/377169a0. [DOI] [PubMed] [Google Scholar]
- 9.Nakane H, Takeuchi S, Yuba S, Saijo M, Nakatsu Y, Murai H, Nakatsuru Y, Ishikawa T, Hirota S, Kitamura Y, Kato Y, Tsunoda Y, Miyauchi H, Horio T, Tokunaga T, Matsunaga T, Nikaido O, Nishimune Y, Okada Y, Tanaka K. Nature (London) 1995;377:165–169. doi: 10.1038/377165a0. [DOI] [PubMed] [Google Scholar]
- 10.Reitmair A H, Schmits R, Ewel A, Bapat B, Redston M, Mitri A, Waterhouse P, Mittrucker H-W, Wakeham A, Liu B, Thomason A, Griesser H, Gallinger S, Ballhausen W G, Fishel R, Mak T W. Nat Genet. 1995;11:64–70. doi: 10.1038/ng0995-64. [DOI] [PubMed] [Google Scholar]
- 11.Sakumi K, Shiraishi A, Shimizu S, Tsuzuki T, Ishikawa T, Sekiguchi M. Cancer Res. 1997;57:2415–2418. [PubMed] [Google Scholar]
- 12.Dosanjh M K, Singer B, Essigmann J M. Biochemistry. 1991;30:7027–7033. doi: 10.1021/bi00242a031. [DOI] [PubMed] [Google Scholar]
- 13.Jacobsen J S, Humayun M Z. Biochemistry. 1990;29:496–504. doi: 10.1021/bi00454a025. [DOI] [PubMed] [Google Scholar]
- 14.Moriya M, Zhang W, Johnson F, Grollman A P. Proc Natl Acad Sci USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singer B, Essigmann J M. Carcinogenesis. 1991;12:949–955. doi: 10.1093/carcin/12.6.949. [DOI] [PubMed] [Google Scholar]
- 16.Rydberg B, Dosanjh M K, Singer B. Proc Natl Acad Sci USA. 1991;88:6839–6842. doi: 10.1073/pnas.88.15.6839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rydberg B, Qiu Z H, Dosanjh M K, Singer B. Cancer Res. 1992;52:1377–1379. [PubMed] [Google Scholar]
- 18.Singer B, Antoccia A, Basu A K, Dosanjh M K, Fraenkel-Conrat H, Gallagher P E, Kuśmierek J T, Qiu Z H, Rydberg B. Proc Natl Acad Sci USA. 1992;89:9386–9390. doi: 10.1073/pnas.89.20.9386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dosanjh M K, Chenna A, Kim E, Fraenkel-Conrat H, Samson L, Singer B. Proc Natl Acad Sci USA. 1994;91:1024–1028. doi: 10.1073/pnas.91.3.1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hang B, Chenna A, Rao S, Singer B. Carcinogenesis. 1996;17:155–157. doi: 10.1093/carcin/17.1.155. [DOI] [PubMed] [Google Scholar]
- 21.Saparbaev M, Laval J. Proc Natl Acad Sci USA. 1994;91:5873–5877. doi: 10.1073/pnas.91.13.5873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saparbaev M, Kleibl K, Laval J. Nucleic Acids Res. 1995;23:3750–3755. doi: 10.1093/nar/23.18.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bessho T, Roy R, Yamamoto K, Kasai H, Nishimura S, Tano K, Mitra S. Proc Natl Acad Sci USA. 1993;90:8901–8904. doi: 10.1073/pnas.90.19.8901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu R Z, Nash H M, Verdine G L. Curr Biol. 1997;7:397–407. doi: 10.1016/s0960-9822(06)00187-4. [DOI] [PubMed] [Google Scholar]
- 25.Rosenquist T A, Zharkov D O, Grollman A P. Proc Natl Acad Sci USA. 1997;94:7429–7434. doi: 10.1073/pnas.94.14.7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roldan-Arjona T, Wei Y F, Carter K C, Klungland A, Anselmino C, Wang R P, Augustus M, Lindahl T. Proc Natl Acad Sci USA. 1997;94:8016–8020. doi: 10.1073/pnas.94.15.8016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radicella J P, Dherin C, Desmaze C, Fox M S, Boiteux S. Proc Natl Acad Sci USA. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arai K, Morishita K, Shinmura K, Kohno T, Kim S R, Nohmi T, Taniwaki M, Ohwada S, Yokota J. Oncogene. 1997;14:2857–2861. doi: 10.1038/sj.onc.1201139. [DOI] [PubMed] [Google Scholar]
- 29.Aburatani H, Hippo Y, Ishida T, Takashima R, Matsuba C, Kodama T, Takao M, Yasui A, Yamamoto K, Asano M, Fukasawa K, Yoshinari T, Inoue H, Ohtsuka E, Nishimura S. Cancer Res. 1997;57:2151–2156. [PubMed] [Google Scholar]
- 30.Elder R H, Weeks R J, Willington M A, Cooper D P, Rafferty J A, Deans B, Hendry J H, Margison G P. Proc Am Assoc Cancer Res. 1997;38:360. [Google Scholar]
- 31.Kielman M F, Smits R, Bernini L F. Mamm Genome. 1995;6:499–504. doi: 10.1007/BF00356165. [DOI] [PubMed] [Google Scholar]
- 32.Roy R, Brooks C, Mitra S. Biochemistry. 1994;33:15131–15140. doi: 10.1021/bi00254a024. [DOI] [PubMed] [Google Scholar]
- 33.Bjelland S, Seeberg E. Nucleic Acids Res. 1987;15:2787–2801. doi: 10.1093/nar/15.7.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ames B N. Science. 1983;221:1256–1264. doi: 10.1126/science.6351251. [DOI] [PubMed] [Google Scholar]
- 35.Moriya M, Ou C, Bodepudi V, Johnson F, Takeshita M, Grollman A P. Mutat Res. 1991;254:281–288. doi: 10.1016/0921-8777(91)90067-y. [DOI] [PubMed] [Google Scholar]
- 36.Shibutani S, Takeshita M, Grollman A P. Nature (London) 1991;349:431–434. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]