Abstract
Molybdenum-independent nitrogenases were first described in the nitrogen-fixing bacterium Azotobacter vinelandii and have since been described in other diazotrophic bacteria. Previously, we reported the isolation of seven diazotrophs with Mo-independent nitrogenases from aquatic environments. In the present study, we extend these results to include diazotrophs isolated from wood chip mulch, soil, “paraffin dirt,” and sediments from mangrove swamps. Mo-deficient, N-free media under both aerobic and anaerobic conditions were used for the isolations. A total of 26 isolates were genetically and physiologically characterized. Their phylogenetic placement was determined using 16S rRNA gene sequence analysis. Most of the isolates are members of the gamma subdivision of the class Proteobacteria and appear to be specifically related to fluorescent pseudomonads and azotobacteria. Two other isolates, AN1 and LPF4, are closely related to Enterobacter spp. and Paenibacillus spp., respectively. PCR and/or Southern hybridization were used to detect the presence of nitrogenase genes in the isolates. PCR amplification of vnfG and anfG was used to detect the genetic potential for the expression of the vanadium-containing nitrogenase and the iron-only nitrogenase in the isolates. This study demonstrates that diazotrophs with Mo-independent nitrogenases can be readily isolated from diverse natural environments.
Azotobacter vinelandii, an aerobic, free-living, nitrogen-fixing soil bacterium, was the first diazotroph shown to have three distinct nitrogenases (6): the molybdenum (Mo)-containing nitrogenase (nitrogenase 1), the vanadium (V)-containing nitrogenase (nitrogenase 2), and the iron-only nitrogenase (nitrogenase 3).
Nitrogenase 1 is expressed when Mo is present. This enzyme complex consists of two components: dinitrogenase reductase 1 (Fe protein) and dinitrogenase 1 (Mo-Fe protein) (21, 22, 25). The structural genes that encode nitrogenase 1 subunits are organized as the nifHDK operon; the dinitrogenase reductase 1 is a homodimer whose subunit is encoded by nifH, while the α- and the β-subunits of dinitrogenase 1 are encoded by nifD and nifK, respectively (8).
Nitrogenase 2 is a V-containing enzyme complex that is synthesized under diazotrophic conditions lacking Mo but containing V (13, 23, 24, 28, 42-44). This enzyme complex consists of two components: dinitrogenase reductase 2 and dinitrogenase 2. The genes encoding nitrogenase 2 proteins are split between two operons: vnfHFd and vnfDGK. vnfH encodes dinitrogenase reductase 2 subunits, and vnfFd encodes a ferredoxin-like protein that is required for nitrogenase 2-dependent diazotrophic growth (42). The vnfDGK operon encodes the subunits for dinitrogenase 2. vnfD encodes the α-subunit, vnfK encodes the β-subunit, and vnfG encodes the small δ-subunit (28, 43, 44).
Nitrogenase 3 is made under Mo- and V-deficient conditions and does not appear to contain either Mo or V (10, 40). The components of this enzyme complex are dinitrogenase 3 and dinitrogenase reductase 3. The structural genes encoding nitrogenase 3 in A. vinelandii are located in the operon anfHDGKOR (27, 38). anfH encodes the subunits of dinitrogenase reductase 3, while anfD and anfK encode the α- and β-subunits of dinitrogenase 3, respectively, and anfG encodes the δ-subunit of dinitrogenase 3 (40). anfO and anfR are located immediately downstream of anfK and are cotranscribed with the anfDGK genes into one polycistronic mRNA (41). Both AnfO and AnfR are required for nitrogen fixation in the absence of both molybdenum and vanadium (38).
Over the years it has become clear that Mo-independent nitrogenases are present in a diverse group of diazotrophic microorganisms which include Clostridium pasteurianum (55), Rhodobacter capsulatus (46, 47), Anabaena variabilis (30, 50), Rhodospirillum rubrum (11, 32), Heliobacterium gestii (31), Azospirillum brasilense (9), Azotobacter salinestris (34), Azotobacter paspali (34), Azomonas macrocytogenes (34), Rhodopseudomonas palustris (39), and Methanosarcina acetivorans (19). Loveless et al. (35) demonstrated that diazotrophs with Mo-independent nitrogenases were easily isolated from aquatic environments using Mo-deficient, nitrogen-free media under aerobic conditions. In that study, seven isolates were shown to have Mo-independent nitrogenase systems. Analysis of the 16S rRNA gene sequences showed that these isolates fall into the gamma subdivision of the class Proteobacteria and seem to be specifically related to the fluorescent pseudomonads and azotobacteria. The ability to isolate diazotrophs with Mo-independent nitrogenases from these environments, including those that are known to have molybdenum concentrations sufficient for nitrogen fixation, suggests that factors other than molybdenum concentrations in the macroenvironment may be important in determining the presence of these organisms. For example, the concentration of molybdenum in a wastewater treatment plant was 90 nM, and in a salt marsh it was approximately 110 nM (unpublished observations).
In the present study, we demonstrate the widespread presence of Mo-independent nitrogenases in diazotrophs isolated from diverse environments by using Mo-deficient, nitrogen-free media under both aerobic and anaerobic conditions.
MATERIALS AND METHODS
Isolation of diazotrophic bacteria from natural environments.
Modified Burk medium (BM) containing 2% (wt/vol) glucose without any additions (−Mo, −N BM) (5) or supplemented with either 1 μM Na2MoO4, (+Mo, −N BM) or 1 μM V2O5 (−Mo, +V, −N BM) was used for growth and enrichment procedures with most of the isolates (35). All chemicals and reagents used were analytical grade and were ≥99.9% molybdenum free. To remove trace Mo contamination, glucose and phosphate buffer stock solutions for preparation of BM were extracted by the 8-hydroxyquinolone technique as described previously (5, 14). To remove trace metals, all glassware was base and acid washed as described by Benemann et al. (3). Enrichment was done under both aerobic and anaerobic conditions. Aerobic enrichment was accomplished by placing a spatula full of wood chip mulch, soil, or sediment in 10 ml of N-free, Mo-deficient BM (−Mo, −N BM) in a sterile 50-ml plastic centrifuge tube and incubating at 30°C with shaking until a noticeable increase in turbidity was observed. Approximately, six to seven transfers followed the initial enrichment by placing 100 μl of inoculum in 9.9 ml of −Mo, −N BM. Incubation conditions were as described above. A 100-μl aliquot from the sixth or seventh transfer was spread on −Mo, −N BM agar. Pure cultures were isolated from single colonies as previously described (35).
For anaerobic enrichment, the same samples (wood chip mulch, soil, or sediment) were placed in sterile 18- by 150-mm Bellco anaerobic culture tubes containing 9 ml of −Mo, −N BM. The tubes were sealed with a rubber stopper clamped with an aluminum ring. The sealed tubes were flushed with N2 and incubated at 30°C until growth was observed. Following the initial enrichment, the culture was transferred five to six times. Transfers were conducted as described above except that 10 ml of −Mo, −N BM was used and 100 μl of inoculum was transferred using a 1-ml syringe. After six transfers, 100 μl of inoculum was spread on −Mo, −N BM agar followed by incubation in a Coy anaerobic chamber (Coy, Ann Arbor, MI) with an anaerobic gas mixture of 10% H2, 5% CO2, and 85% N2 until growth was observed. Solid medium to be used for plates was poured inside the anaerobic chamber and allowed to equilibrate for 24 to 48 h prior to inoculation.
Growth experiments.
Growth of each strain was tested in −Mo, −N BM or −Mo, −N BM supplemented with 1 μM Na2MoO4, 1 μM V2O5, or 10 mM ammonium acetate. Each culture was grown overnight in the same medium in order to monitor growth. Inoculum culture (0.5 to 1.0 ml) was transferred to 30 ml of −Mo, −N BM in a sidearm flask (300 ml) and placed on a shaker at 30°C. Growth was monitored with a Klett-Summerson colorimeter equipped with a no. 66 filter (red). Generation times were calculated by log-linear regression.
Acetylene reduction assays.
To estimate nitrogenase activity, acetylene reduction assays were conducted on freshly grown cultures that had been starved for molybdenum by repeated transfers (at least three times) in molybdenum-deficient liquid media. Bellco sidearm flasks previously treated to remove metals were used to grow 30-ml cultures in modified BM. The media contained either no additive, 1 μΜ Na2MoO4, or 1 μM V2O5. To repress nitrogenase expression, ammonium acetate was added to the media at 1, 10, or 28 mM. A 10-ml volume of each culture grown to a density of 75 to 90 Klett units was then transferred to a 52- by 95-mm serum bottle and fitted with a rubber stopper. Two percent of the headspace gas was removed before injecting that same volume of acetylene into the bottle. A 3-ml volume was removed from the sample headspace and analyzed for ethylene and ethane using a Shimadzu GC-17 gas chromatograph after 0, 30, and 60 min of shaking at 200 rpm at 30°C. The gas chromatograph was fitted with a 1-ml sample loop on the injector port, a column of 50/80 Porapak N (182.88 cm by 3.175 mm), and a flame ionization detector. The temperatures of the injector, detector, and oven were 40°C, 200°C, and 35°C, respectively. Ethylene standards consisted of a mixture of ethylene and ethane, with various amounts of ethylene in increasing incremental amounts across seven tubes keeping ethane constant across all seven. For ethane standards, the amount of ethane was increased incrementally across seven tubes while the amount of ethylene remained constant. Incubation conditions were the same for both standards and samples.
Two isolates, AN1 and LPF4, failed to grow in Burk nitrogen-free liquid media. Therefore, in order to assay for acetylene reduction, the isolates were grown on slants prepared in anaerobic culture tubes. Strain LPF4 (Paenibacillus) was grown on modified Line's acetylene reduction medium (49) consisting of glucose (10 g), MgSO4·7H2O (0.5 g), C6H5FeO7·H20 (0.01 g), CaCl2·2H2O (0.07 g), K2HPO4 (2 g), KH2PO4 (0.5 g), thiamine (0.001 g), biotin (0.001 g), sodium thioglycolate (0.5 g), Jurgensen micronutrient solution (1 ml) (29), purified agar (15 g), and deionized water to a final volume of 1 liter. For AN1 (Enterobacter), the solid media consisted of −Mo, −N BM with 1.5% purified agar or the same medium supplemented with either 1 μΜ Na2MoO4, 1 μM V2O5, or 10 mM ammonium acetate. Slants were inoculated from an aerobic overnight culture grown in the same medium. The slants were incubated 24 to 48 h at 30°C in an anaerobic chamber or a Gas Pak anaerobic jar with an anaerobic system envelope generating H2 plus CO2. Once growth was observed, the culture tube was fitted with a rubber stopper and 0.5 ml of acetylene was added. After anaerobic incubation at 30°C for 24 h, 0.5-ml headspace samples were analyzed in duplicate as described above.
Determination of protein concentration.
Total cell protein concentration for the acetylene reduction assays was determined using a Pierce bicinchoninic acid protein assay kit (Rockford, IL), according to the protocol provided by the supplier.
DNA extraction.
DNA for all bacterial strains was prepared using the CTAB method (1).
PCR amplification and phylogenetic analysis of 16S rRNA genes.
The universal primers 515F and 1492R were used for 16S rRNA gene amplification (2). Amplification was accomplished using the protocol and reagents of the Qiagen Taq PCR core kit. A Bio-Rad iCycler thermal cycler or an Ericomp PowerBlock was used for the amplifications. The cycler settings were those described below for the anfG and vnfG amplifications. PCR products were sequenced at the biotechnology facilities of Iowa State University (Ames, IA).
A subalignment of 16S rRNA sequences from azotobacteria and diazotrophic Pseudomonas species was extracted from the Ribosomal Database Project (PubMed ID 17090583). The ‘Sequence Match’ function was used to identify the most similar sequence from a named organism for each of the isolates' 16S rRNA sequences; in the cases of AN1 and LPF4, the isolates were not azotobacteria, and so these relatives and a related well-known representative were included in the extracted alignment. The 16S rRNA sequences of the diazotrophic isolates were added to the alignment manually, and the alignment was refined manually. Trees were generated by the neighbor-joining method using Phylip v3.6 (18); these trees were not substantially different than those generated by maximum likelihood or unrooted parsimony.
PCR amplification of Mo-independent nitrogenase genes and phylogenetic analysis of AnfG and VnfG.
Primers for PCR amplification of vnfDGK and anfDGK DNA sequences were previously described (34). In addition, the 18-mer reverse primer K3r, 5′ GCAGTCGTACATCGGGTT 3′ (vnfK priming site positions 4312 to 4323 in reference to the Azotobacter vinelandii vnfDGK numbering [27, 28]), was used for the amplification of vnfG with forward primer D6f.
Amplification of vnfG and anfG was accomplished using the protocol and reagents of an Epicenter Fail Safe kit (Epicenter Biotechnologies, Madison, WI). The programmed temperature sequence for vnfG and anfG amplification was 94°C for hot start followed by 92°C for 1.5 min, 50°C for 1.5 min, and 72°C for 0.5 min. The temperature sequence was run for 30 cycles. The final product extension was conducted at 72°C for 7 min followed by a 4°C temperature hold. PCR products were isolated from a 0.4% (wt/vol) agarose gel using a GeneClean II kit (Bio 101, Inc., Vista, CA). PCR products were cloned using a Promega Easy vector kit and the associated protocols. Cloned PCR products were sequenced at the biotechnology facilities of Iowa State University (Ames, IA).
AnfG and VnfG sequences from the diazotrophic isolates and named bacterial species downloaded from the NCBI/GenBank database were added to the AnfG/VnfG protein family (PFAM 03139.13) alignment (PubMed ID 16381856), and nonconserved sequences at the N-termini were removed. Trees were generated by the neighbor-joining method using Phylip v3.6 (18); these trees were not substantially different than those generated by maximum likelihood or unrooted parsimony.
PCR amplification of nifH gene.
Primers for PCR amplification of the nifH gene were previously described (54).
Southern hybridization.
DNA fragments containing A. vinelandii nifD (0.8-kb nifD insert of pTMR18 [7]), vnfD (1.4-kb vnfD insert of pVDSJ1 [26]), and anfD (1.08-kb anfD insert of pPJD3A2 [41]) were used as hybridization probes. Probes were labeled with digoxigenin (Boehringer Mannheim), and the Southern blot procedure was performed as described by the supplier. Hybridization temperatures ranged from 50 to 60°C.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the nucleotide sequences of the nitrogen fixation genes are as follows: BR2anfG, EF694542; BR3anfG, EF694543; BR5anfG, EF694544; BR6anfG, EF694545; BR7anfG, EF694546; BR8anfG, EF694547; BR10anfG, EF694548; DP6anfG, EF694549; DP7anfG, EF694550; LPF4anfG, EF694551; MU4anfG, EF694552; MU5anfG, EF694553; MU11anfG, EF694554; MU12anfG, EF694555; MU13anfG, EF694556; NC2anfG, EF694557; PRM1anfG, EF694558; MU7anfG, EF694559; ARB2vnfG, EF694560; ARB3vnfG, EF694561; BR2vnfG, EF694562; BR3vnfG, EF694563; BR5vnfG, EF694564; BR6vnfG, EF694565; BR7vnfG, EF694566; BR8vnfG, EF694567; BR10vnfG, EF694568; DP6vnfG, EF694569; DP7vnfG, EF694570; LPF4vnfG, EF694571; MB4vnfG, EF694572; MU1vnfG, EF694573; MU2vnfG, EF694574; MU4vnfG, EF694575; MU6vnfG, EF694576; MU7vnfG, EF694577; MU11vnfG, EF694578; MU12vnfG, EF694579; MU13vnfG, EF694580; NC2vnfG, EF694581; PB3vnfG, EF694582; and AN1anfHDGK, EF694583. The GenBank accession numbers for the 16S rRNA gene sequences are as follows: ARB2, AY588643.1; ARB3, AY590431.1; NC2, AY590432.1; MU2, AY590433.1; MU4, AY590434.1; MU5, AY590435.1; MU7, AY590436.1; BR7, AY590437.1; BR3, AY590438.1; BR10, AY590439.1; BR5, AY590440.1; MU11, AY590441.1; MB4, AY590442.1; MU6, AY590443.1; MU1, AY590444.1; DP7, AY590445.1; MU13, AY590446.1; MU12, AY590447.1; PRM1, AY590448.1; BR8, AY590449.1; PB3, AY590450.1; BR2, AY590451.1; DP6, AY590452.1; BR6, AY590453.1; LPF4, AY590454.1; and AN1, AY590455.1.
RESULTS AND DISCUSSION
Isolation of environmental isolates.
A total of 26 diazotrophic strains were isolated from soil samples from Brazil, Puerto Rico, and the United States using Mo-deficient, N-free medium (Table 1). Group 1 consists of 19 isolates where genes for nitrogenases 1, 2, and 3 were detected. Group 2 consists of 6 isolates where genes for nitrogenases 1 and 2 were detected. Group 3 consists of only 1 isolate where genes for nitrogenases 1 and 3 were detected. Modified BM was used for growth and enrichment procedures for most of the isolates (5). Strains AN1 and LPF4 were isolated using anaerobic enrichment conditions.
TABLE 1.
Isolates containing Mo-independent nitrogenases and their environmental sources
| Group and isolatea | Environmental source | Enrichmentb |
|---|---|---|
| Group I | ||
| BR2 | Soil, Foz do Iguazu, Brazil | −Mo, −N BM |
| BR3 | Soil, Foz do Iguazu, Brazil | −Mo, −N BM |
| BR5 | Soil, Foz do Iguazu, Brazil | −Mo, +V, −N BM |
| BR6 | Soil, Foz do Iguazu, Brazil | −Mo, −N BM |
| BR7 | Soil, Foz do Iguazu, Brazil | −Mo, −N BM |
| BR8 | Soil, Foz do Iguazu, Brazil | −Mo, −N BM |
| BR10 | Soil, Foz do Iguazu, Brazil | −Mo, −N BM |
| DP6 | Wastewater treatment plant, Durham, NC | −Mo, +V, −N BM |
| DP7 | Wastewater treatment plant, Durham, NC | −Mo, +V, −N BM |
| LPF4 | “Paraffin dirt,” Avery Island, LA | −Mo, −N BM (anaerobic) |
| MU4 | Wood chip mulch, Durham, NC | −Mo, +V, −N BM |
| MU5 | Wood chip mulch, Durham, NC | −Mo, +V, −N BM |
| MU6 | Wood chip mulch, Durham, NC | −Mo, +V, −N BM |
| MU7 | Wood chip mulch, Durham, NC | −Mo, +V, −N BM |
| MU11 | Wood chip mulch, Durham, NC | −Mo, −N BM |
| MU12 | Wood chip mulch, Durham, NC | −Mo, −N BM |
| MU13 | Wood chip mulch, Durham, NC | −Mo, −N BM |
| NC2 | Creek sediment, Raleigh, NC | −Mo, −N BM |
| PRM1 | Mangrove sediment, Boqueron, Puerto Rico | −Mo, −N BM |
| Group II | ||
| ARB2 | Wood chip mulch, Durham, NC | −Mo, −N BM |
| ARB3 | Wood chip mulch, Durham, NC | −Mo, −N BM |
| MB4 | Mangrove sediment, Boqueron, Puerto Rico | −Mo, −N BM |
| MU1 | Wood chip mulch, Durham, NC | +Mo, −N BM |
| MU2 | Wood chip mulch, Durham, NC | +Mo, −N BM |
| PB3 | Soil, Raleigh, NC | −Mo, −N BM |
| Group III | ||
| AN1 | Wood chip mulch, Durham, NC | −Mo, −N BM (anaerobic) |
Group I, genes for nitrogenases 1, 2, and 3 were detected; group II, genes for nitrogenases 1 and 2 were detected; and group III, genes for nitrogenases 1 and 3 were detected.
“−Mo, −N BM,” N-free, Mo-deficient Burk medium; “−Mo, +V, −N BM,” N-free, Mo-deficient, +V Burk medium; and “+Mo, −N BM,” N-free, +Mo Burk medium.
The ability of diazotrophs with Mo-independent nitrogenases to grow in a wide variety of environments, including those known to have sufficient Mo concentrations for Mo-dependent nitrogen fixation, might be determined by other environmental factors (e.g., temperature) in addition to the concentration of Mo in the macroenvironment (4, 36). As such, we were able to isolate strains with Mo-independent nitrogenases using enrichment media containing 1 μM Na2MoO4 (strains MU1 and MU2). Another possible environmental factor that may be involved in the requirement of Mo-independent nitrogenases is temperature. Several studies suggest that the V-containing nitrogenase may function more efficiently at low temperatures than does the Mo-containing nitrogenase (37, 51).
Previously, we suggested that diazotrophic growth of A. vinelandii on a solid medium surface might generate Mo-depleted microzones due to the organism's powerful Mo uptake system. This could lead to a competitive advantage by generating ecological niches that exclude diazotrophs lacking Mo-independent nitrogenases. Consistent with this hypothesis are the results of competition experiments where wild-type A. vinelandii had an advantage over a mutant lacking Mo-independent nitrogenase 3 on Mo-sufficient solid media but not in liquid media (36). If this hypothesis holds for other diazotrophs with Mo-independent nitrogenases, then it is reasonable to expect to find these diazotrophs in macroenvironments that have sufficient Mo concentrations for Mo-dependent nitrogen fixation. The fact that diazotrophs with Mo-independent nitrogenases were isolated from many different soil environments suggests that this is probably the case, although we did not measure molybdenum concentrations in the samples used in this study.
Phylogenetic analysis.
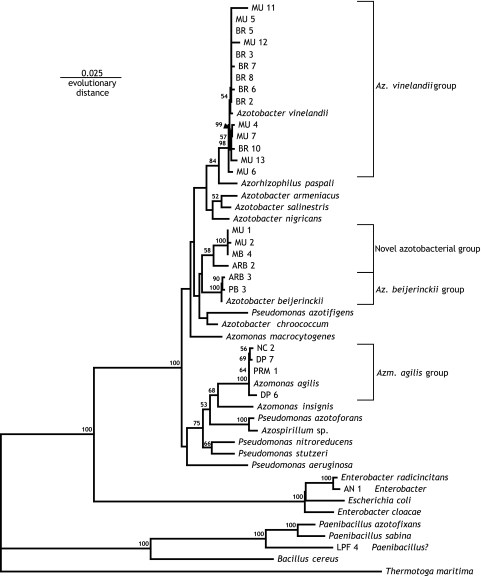
Phylogenetic analysis of 16S rRNA sequences places all but two of the diazotrophic isolates among the gammaproteobacteria, and more specifically as members of the Pseudomonas/Azotobacteria “fluorescent pseudomonad” clade (Fig. 1). The genus Pseudomonas is ubiquitous in soil, and its members are capable of growing on diverse carbon sources and under diverse environmental conditions (12). Pseudomonas is phenotypically similar to the genera Azotobacter and Azomonas, and the phylogenetic relationship between these organisms has not been well defined (52, 53) (PubMed ID 15133068). Some diazotrophic strains of Pseudomonas stutzeri, a nonfluorescent pseudomonad, have been described, and for this reason the azotobacteria have been placed in this group (53). This study as well as previous work (35) show that Pseudomonas/Azotobacteria is the predominant group isolated using −Mo, −N BM, a medium commonly used for growth and maintenance of Azotobacter species. Approximately half of the isolates (MU11, MU5, BR5, MU12, BR3, BR7, BR8, BR6, BR2, MU4, MU7, BR10, MU13, and MU6) obtained in this study are very closely related to the well-studied Azotobacter vinelandii; at least in terms of rRNA similarity, these might be considered strains of this species. Likewise, two isolates (ARB3 and PB3) are more similar to strains of Azotobacter beijerinckii, and four isolates (NC2, DP6, DP7, and PRM1) are more similar to strains of Azomonas agilis. An additional four isolates (MU1, MU2, MU4, and ARB2) form a clade that, although clearly falling among the azotobacteria, was not specifically affiliated with any characterized species.
FIG. 1.
16S rRNA phylogenetic analysis of the environmental isolates and azotobacteria/diazotrophic Pseudomonas species. A subalignment of 16S rRNA sequences from azotobacteria and diazotrophic Pseudomonas species was extracted from the Ribosomal Database Project (PubMed ID 17090583). The ‘Sequence Match’ function was used to identify the most similar sequence from a named organism for each of the isolated 16S rRNA sequences; in the cases of AN1 and LPF4, these were not azotobacteria, and so these relatives and a related well-known representative were included in the extracted alignment. The 16S rRNA sequences of the diazotrophic isolates were added to the alignment manually. Trees were generated by the neighbor-joining method using Phylip v3.6.
Isolate AN1 also is a member of the gammaproteobacteria, but it is most closely related to species of the genus Enterobacter, and specifically to E. radicincitans, a known diazotrophic and plant growth-promoting species (PubMed ID 15900968). Nitrogen-fixing enterics have also been found as common inhabitants of soils (33), plant material (16), decaying wood (48), and pulp and paper mill effluents (20). However, alternative nitrogenases have not previously been observed in this group.
The only nonproteobacterial strain was LPF4 isolated from “paraffin dirt” (Avery Island, LA). LPF4 is a member of the Firmicutes (low G+C gram-positive bacteria) and is most closely related to species of the genus Paenibacillus, although this affiliation is more distant than that of the other isolates and is not specific to any particular species of the genus. Diazotrophic Paenibacillus species (e.g., P. azotofixans) are common soil inhabitants, but alternative nitrogenases have not previously been identified in this group (15, 45, 49, 53).
Nitrogenase genes.
The presence of the different nitrogenase genes, nifH, nifD, vnfD, vnfG, anfD, and anfG, was detected using Southern blot hybridization and/or PCR amplification (Table 2). In each of the isolates, except AN1, LPF4, and MB4, we were able to detect genes that encode the Mo-nitrogenase using Southern blot hybridization. Detection of the Mo-nitrogenase nifH gene on AN1, LPF4, and MB4 was done using PCR. In most of the isolates, the molecular characterization of the Mo-independent nitrogenases to detect the δ-subunit (vnfG/anfG) of the V-nitrogenase and iron-only nitrogenase was done using PCR. Southern blot hybridization was used to characterize several isolates for the α-subunit (vnfD/anfD) of the V-nitrogenase and iron-only nitrogenase (respectively).
TABLE 2.
Detection of nitrogenase genes in the environmental isolates using PCR and Southern hybridization
| Isolate | DNA fragment size(s) (kb)a
|
||
|---|---|---|---|
| nifD/nifH | vnfD/vnfG | anfD/anfG | |
| AN1 | + | ND | 3.1 |
| ARB2 | 15 | 4.4, 5.0 | ND |
| ARB3 | 12 | 2.3, 4.3 | ND |
| BR2 | 1.4 | + | + |
| BR3 | 1.5 | + | + |
| BR5 | 1.4 | + | + |
| BR6 | 1.5 | + | + |
| BR7 | 1.4 | + | + |
| BR8 | 1.5 | + | + |
| BR10 | + | + | + |
| DP6 | 9 | + | + |
| DP7 | 8 | + | + |
| LPF4 | + | + | + |
| MB4 | + | 1.7, 4.5 | ND |
| MU1 | >10 | 5.0, 4.5 | ND |
| MU2 | >10 | 5.0, 4.5 | ND |
| MU4 | 4.4, 1.4 | 4.9, 3.0, 1.3 | 6.2, 3.4 |
| MU5 | 1.4 | 3.0, 1.3 | 3.4 |
| MU6 | 1.5 | + | ND |
| MU7 | 1.4 | 3.0, 1.3 | 3.4 |
| MU11 | 1.4 | + | + |
| MU12 | 1.4 | + | + |
| MU13 | 1.7 | + | + |
| NC2 | 7.1 | + | + |
| PB3 | >10 | 2.2 | ND |
| PRM1 | 7.8 | 2 | >10 |
The vnfG PCR products obtained with the D6f/K1r primer set were 1,700 bp in size, and those obtained with D6f/K3r were 1,213 bp in size. The anfG PCR product generated by D7f/K2r was approximately 760 bp. A comparison of the predicted amino acid sequences for VnfG and AnfG indicates a high degree of identity among the gene products for most of the isolates examined.
The molecular characterization of the nitrogenase genes either by Southern blotting and/or PCR (Table 2) clearly supports our 16S phylogenetic analysis (Fig. 1). In all of the isolates within the Azotobacter vinelandii group and the Azomonas agilis group, except for isolate MU6, genes for the three nitrogenases were detected. The presence of all three nitrogenases in Azotobacter vinelandii and Azomonas agilis had been previously reported and well characterized (4, 17). In isolate MU6 of the A. vinelandii group, although a nitrogenase 3 gene was not detected, it is possible that an Fe-only nitrogenase is present, based on diazotrophic growth and the acetylene reduction assay as discussed below. In the Azotobacter beijerinckii group, which includes strains ARB3 and PB3, only the genes for nitrogenases 1 and 2 were detected. These results are consistent with those reported by Fallik et al. for Azotobacter beijerinckii (17). In strains MU1, MU2, MB4, and ARB2, which are included in the novel azotobacteria group of the 16S phylogenetic tree (Fig. 1), only nitrogenase 1 and 2 genes were detected (Table 2).
For PCR amplification of anf genes in AN1, degenerate primers D2f/D5r (34) were used. The only product detected was anfD. Similar results were obtained with Southern blot hybridizations using a probe containing A. vinelandii anfD (1.08-kb anfD insert of pPJD3A2 [41]). Blast sequence comparison of the anfD sequence shows that it is 85% identical to that of Azotobacter vinelandii.
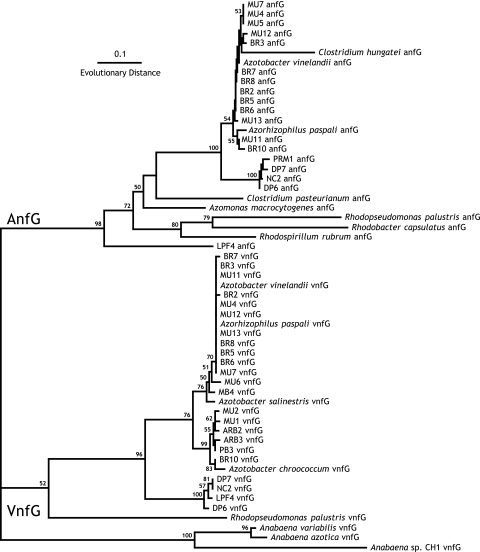
Phylogenetic analysis of vnfG and anfG.
All of the PCR amplification products obtained using either anfDGK or vnfDGK PCR primers contained sequences encoding proteins related to AnfG or VnfG, respectively (Fig. 2), and these correspond to the nutritional requirements of the organisms for Mo-independent growth that are presented below. The relationships between the anfG and vnfG sequences of characterized species do not correspond to the inferred relationships between these organisms in the 16S rRNA phylogenetic tree, as seen previously (34). This suggests that anf and vnf operons have been frequently transferred horizontally. However, the relationships between the anfG and vnfG sequences of the newly isolated diazotrophs are generally consistent with the phylogenetic placement of the organisms on the basis of 16S rRNA sequence. anfG and vnfG sequences from isolates of the A. vinelandii group are both also closely related to anfG and/or vnfG of A. vinelandii, with the single exception of the vnfG sequence from isolate BR10, which is more closely related to the distinct vnfG of Azotobacter chroococcum. All of the anfG and vnfG sequences from the A. beijerinckii and “novel azotobacteria” groups form clusters in both the anfG and vnfG subtrees; in the case of the vnfG sequences, these clusters are very similar to that of A. chroococcum; this may reflect a relationship between these organisms that is not resolved in the 16S rRNA phylogeny. The single exception is the vnfG of strain MB4, which is related to those of the large A. vinelandii group. The anfG and vnfG sequences from the A. agilis group form clusters distinct from any characterized anfG or vnfG sequence, consistent with the 16S phylogenetic tree. The isolate LPF4 (Paenibacillus) anfG sequence forms a novel, deep branch in the anfG portion of the tree, as might be expected, but its vnfG sequence falls among those of the A. agilis group. Taken together, these relationships support the vertical evolution of the alternative nitrogenase operons generally, interspersed with an occasional horizontal transfer.
FIG. 2.
Phylogeny of the AnfG and VnfG sequences. AnfG and VnfG sequences from the environmental isolates and named bacterial species downloaded from the NCBI/GenBank database were added to the AnfG/VnfG protein family (PFAM 03139.13) alignment (PubMed ID 16381856), and nonconserved sequences at the N-termini were removed. Trees were generated by the neighbor-joining method using Phylip v3.6.
Diazotrophic growth and acetylene reduction.
Table 3 summarizes the diazotrophic growth and whole-cell nitrogenase activity of representative isolates selected according to their placement in the 16S phylogenetic tree (Fig. 1). Nitrogenase activity as measured by acetylene reduction followed the usual pattern, where it was highest during growth in the presence of Mo and lowest under Mo-deficient conditions. As mentioned previously, most of the isolates in this study are closely related to Azotobacter vinelandii. All of the strains tested within this clade—BR5, BR6, BR7, BR8, BR10, MU4, MU5, MU6, MU11, and MU13—grew diazotrophically in the presence of Mo or V, or in the absence of these metals. When grown on +Mo, −N BM, all isolates reduced acetylene to ethylene and no ethane was produced. When grown on −Mo, +V, −N BM and −Mo, −N BM, acetylene was reduced to both ethylene and ethane. These results suggest that all of these isolates utilize nitrogenases 1, 2, or 3. All isolates from the Azomonas agilis clade—NC2, DP6, DP7, and PRM1—reduced acetylene to ethylene; however, ethane production was not determined when grown on −Mo, +V, −N BM and −Mo, −N BM.
TABLE 3.
Diazotrophic growth and whole-cell nitrogenase activity in liquid media
| Isolate | Addition to −Mo, −N BM | Generation time (h) | Nitrogenase activity ± SD (n)a
|
|
|---|---|---|---|---|
| nmoles C2H4·mg protein−1·30 min−1 | nmoles C2H6·mg protein−1·30 min−1 | |||
| A. vinelandii CA | NH4+ (28 mM) | 2.83 | − | − |
| Na2MoO4 (1 μΜ) | 3.49 | 2,542 ± 27 (2) | − | |
| V2O5 (1 μM) | 5.20 | 255 ± 28 (2) | 6 ± 1 (2) | |
| None | 5.92 | 66 ± 1 (2) | 5 ± 1 (2) | |
| ARB3 | NH4+ (10 mM) | 2.4 | − | − |
| Na2MoO4 (1 μΜ) | 1.8 | 672 ± 52 (2) | − (2) | |
| V2O5 (1 μM) | 3 | 100 | 6 | |
| None | 24 | 24 | 4 | |
| BR5 | NH4+ (10 mM) | 3.0 | 8 | − |
| Na2MoO4 (1 μΜ) | 3.1 | 1,328 | − | |
| V2O5 (1 μM) | 4.5 | 31 | 2 | |
| None | 5.3 | 15 | 3 | |
| BR6 | NH4+ (10 mM) | 2.3 | 7 ± 1.0 (2) | − (2) |
| Na2MoO4 (1 μΜ) | 2.7 | 1,277 | − (2) | |
| V2O5 (1 μM) | 8.6 | 40 ± 4 (2) | 3 ± 0.2 (2) | |
| None | 8.4 | 27 ± 15 (2) | 4 ± 1 (2) | |
| BR7 | NH4+ (10 mM) | 4 | − | − |
| Na2MoO4 (1 μΜ) | 4.3 | 1,293 ± 302 (2) | − (2) | |
| V2O5 (1μM) | 3.7 | 65 ± 29 (3) | 4 ± 0.4 (3) | |
| None | 4.9 | 30 ± 11 (3) | 4 ± 1 (3) | |
| BR8 | NH4+ (10 mM) | 2.5 | 36 ± 4 (2) | NDb |
| Na2MoO4 (1 μΜ) | 2.4 | 1,369 ± 55 (2) | ND | |
| V2O5 (1 μM) | 3.5 | 794 | 16 | |
| None | 3.5 | 89 ± 10 (2) | 5 ± 2 (2) | |
| BR10 | NH4+ (10 mM) | 1.1 | − | ND |
| Na2MoO4 (1 μΜ) | 2.3 | 1,616 ± 97 (2) | ND | |
| V2O5 (1 μM) | 2.5 | 781 | 25 | |
| None | ND | 320 | 14 | |
| DP6 | NH4+ (10 mM) | 4.7 | − | ND |
| Na2MoO4 (1 μΜ) | 3.6 | 1,324 ± 13 (2) | ND | |
| V2O5 (1 μM) | 3.7 | 136 ± 7 (2) | ND | |
| None | 4.0 | 67 ± 7 (2) | ND | |
| DP7 | NH4+ (10 mM) | 4.0 | − | ND |
| Na2MoO4 (1 μΜ) | 3.4 | 836 ± 134 (2) | ND | |
| V2O5 (1 μM) | 3.8 | 61 ± 36 (2) | ND | |
| None | 3.7 | 42 ± 7 (2) | ND | |
| MB4 | NH4+ (10 mM) | 4.8 | − | − |
| Na2MoO4 (1 μΜ) | 4.6 | 900 ± 319 (3) | − | |
| V2O5 (1 μM) | 4 | 52 ± 7 (2) | 5 ± 1 (2) | |
| None | 20.4 | 18 ± 2 | − | |
| MU4 | NH4+ (10 mM) | 3.7 | 42 ± 34 (2) | ND |
| Na2MoO4 (1 μΜ) | 4.8 | 481 ± 19 (2) | ND | |
| V2O5 (1 μM) | 6.1 | 352 ± 89 (2) | 10 ± 2 (2) | |
| None | 7.0 | 234 | 8 | |
| MU5 | NH4+ (10 mM) | 4.1 | 50 ± 10 (2) | ND |
| Na2MoO4 (1 μΜ) | 4.7 | 310 ± 9 (2) | ND | |
| V2O5 (1 μM) | 7.4 | 45 ± 2 (2) | ND | |
| None | 9.4 | 138 ± 6 (2) | ND | |
| MU6 | NH4+ (10 mM) | 1.2 | − (3) | − (3) |
| Na2MoO4 (1 μΜ) | 1.5 | 1,513 ± 636 (3) | − (3) | |
| V2O5 (1 μM) | 2.7 | 84 ± 33 (2) | 5 ± 2 (2) | |
| None | 2.4 | 24 | 4 | |
| MU11 | NH4+ (10 mM) | 2.5 | − | − |
| Na2MoO4 (1 μΜ) | 2.6 | 1,355 | − | |
| V2O5 (1 μM) | 3.1 | 92 ± 19 (2) | 4 ± 1 (2) | |
| None | 5.4 | 18 ± 9 (2) | 3 ± 1 (2) | |
| MU13 | NH4+ (10 mM) | 2.6 | − | − |
| Na2MoO4 (1 μΜ) | 2.9 | 865 | − | |
| V2O5 (1 μM) | 4.3 | 32 | 3 | |
| None | 4.8 | 16 | 3 | |
| NC2 | NH4+ (10 mM) | 1.9 | − | ND |
| Na2MoO4 (1 μΜ) | 2.3 | 2,841 ± 17 (2) | ND | |
| V2O5 (1 μM) | 2.7 | 431 ± 3 (2) | ND | |
| None | 3.1 | 123 ± 1 (2) | ND | |
| PB3 | NH4+ (10 mM) | 2.4 | − | ND |
| Na2MoO4 (1 μΜ) | 2.3 | 1,142 ± 34 (2) | ND | |
| V2O5 (1 μM) | 3.5 | 1,575 | 30 | |
| None | 11.1 | 693 | 43 | |
| PRM1 | NH4+ (10 mM) | 2.5 | 85 ± 10 (2) | ND |
| Na2MoO4 (1 μΜ) | 2.5 | 1,877 ± 338 (2) | ND | |
| V2O5 (1 μM) | 3.2 | 656 ± 39 (2) | ND | |
| None | 2.9 | 196 ± 18 (2) | ND | |
−, measured activity was no greater than activity of the blank (−Mo, −N BM); n, number of acetylene reduction assays on different days.
ND, not determined.
Isolates ARB3, PB3, and MB4 grew diazotrophically in the presence of V and in the absence of Mo and V. However, generation times when grown on −Mo, −N BM were 11.1 h for PB3, 20.4 h for MB4, and 24 h for ARB3. Southern blot hybridization and PCR amplification suggested that only nitrogenase 1 and 2 genes are present in these isolates (Table 2). Thus, it is possible that these strains are using dinitrogenase 2 containing an Fe-only cofactor when cells are grown under Mo- and V-deficient conditions. An analogous situation has been observed with a mutant of A. vinelandii lacking the structural genes for dinitrogenases 1 and 2 where FeMoCo was incorporated in dinitrogenase 3 to form an active nitrogenase complex (40). Further studies with isolates ARB3, MB4, and PB3 will be needed to test the cofactor substitution hypothesis.
All isolates grew with generation times of 1.8 to 4 h in +Mo, −N BM. Isolates BR10, MU4, MU5, MU11, and NC2 follow a growth rate pattern (NH4+ > Mo > V > −Mo) similar to that for A. vinelandii CA and that described for strains isolated in a previous study (6). Interestingly, this pattern is altered for other isolates; for example, ARB3, BR7, BR8, DP6, DP7, MB4, and PRM1 grow as fast or faster under at least one nitrogen-fixing condition than they do in the presence of 10 mM ammonium acetate.
BR10 grows diazotrophically in the presence of Mo or V. However, growth was not detected for BR10 in the absence of Mo and V even though anfG was detected using PCR. This strain also did not exhibit nitrogenase activity under Mo-deficient conditions. Thus, it is unclear as to whether BR10 lacks a full complement of genes for nitrogenase 3 expression or whether other conditions were not met during our attempts to grow it under diazotrophic conditions in the absence of Mo and V.
In most of the isolates, nitrogenase activity was greatly reduced or absent in the presence of 10 mM NH4+.
Two isolates, AN1 (closely related to Enterobacter) and LPF4 (closely related to Paenibacillus) failed to grow in liquid, nitrogen-free BM. Therefore, these isolates were grown on slants (BM or Line's acetylene reduction medium) for acetylene reduction studies. The finding that these isolates grew on N-free agar medium in the absence of Mo and V but not in liquid medium could be attributable to the presence of Mo and V (or other micronutrients) in the purified agar. Loveless et al. (35) previously described similar diazotrophic growth for isolates SM1, SM3, and WB3. Strains AN1 and LPF4 exhibited nitrogenase activity when cultured under diazotrophic conditions in the presence and absence of Mo, with the highest values obtained in the presence of Mo (data not shown). Nitrogenase activity was repressed by 10 mM NH4+. The acetylene reduction assays are consistent with the genetic characterization of these isolates and suggest the presence of at least one of the Mo-independent nitrogenases.
Conclusion.
Twenty-six environmental isolates and the nitrogenases detected in them are summarized in Table 1. Of these, 19 isolates possessed the three nitrogenases, 6 isolates contained nitrogenases 1 and 2, and only 1 isolate, AN1, contained nitrogenases 1 and 3.
We were able to identify the presence of Mo-independent nitrogenases in strains closely related to Enterobacter spp. and Paenibacillus spp. as well as the fluorescent pseudomonads. The 16S phylogenetic analysis is consistent with a previous study and shows that the majority of isolates are closely related to the fluorescent pseudomonads and azotobacteria.
Lastly, the ability to isolate these types of diazotrophs directly from different geographical areas should expand our knowledge of bacteria that express Mo-independent nitrogenases.
Acknowledgments
This work was supported by the USDA Agricultural Research Service and the North Carolina Agricultural Research Service. D.A.B. was supported by a MARC Faculty Predoctoral Fellowship from the National Institutes of Health.
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidmann, and J. A. Smith. 1987. Preparation of genomic DNA from bacteria. In K. Struhl (ed.), Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 2.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone National Park hot spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benemann, J. R., C. E. McKenna, R. F. Lie, T. G. Traylor, and M. D. Kamen. 1972. The vanadium effect in nitrogen fixation by azotobacter. Biochim. Biophys. Acta 264:25-38. [DOI] [PubMed] [Google Scholar]
- 4.Bishop, P. E. 1993. Three genetically distinct nitrogenase systems in Azotobacter vinelandii, p. 301-324. In L. L. Barton and B. C. Hemming (ed.), Iron chelation in plants and soil microorganisms. Academic Press, Inc., New York, NY.
- 5.Bishop, P. E., M. E. Hawkins, and R. R. Eady. 1986. Nitrogen fixation in molybdenum-deficient continuous culture by a strain of Azotobacter vinelandii carrying a deletion of the structural genes for nitrogenase (nifHDK). Biochem. J. 238:437-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bishop, P. E., and R. Premakumar. 1992. Alternative nitrogen fixation systems, p. 736-762. In G. Stacey, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman and Hall, New York, NY.
- 7.Bishop, P. E., T. M. Rizzo, and K. F. Bott. 1985. Molecular cloning of nif DNA from Azotobacter vinelandii. J. Bacteriol. 162:21-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brigle, K. E., W. E. Newton, and D. R. Dean. 1985. Complete nucleotide sequence of the Azotobacter vinelandii nitrogenase structural gene cluster. Gene 37:37-44. [DOI] [PubMed] [Google Scholar]
- 9.Chakraborty, B., and K. R. Samadar. 1995. Evidence for the occurrence of an alternative nitrogenase system in Azospirillum brasilense. FEMS Microbiol. Lett. 127:127-131. [Google Scholar]
- 10.Chisnell, J. R., R. Premakumar, and P. E. Bishop. 1988. Purification of a second alternative nitrogenase from a nifHDK deletion strain of Azotobacter vinelandii. J. Bacteriol. 170:27-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Davis, R., L. Lehman, R. Petrovich, V. K. Shah, G. P. Roberts, and P. W. Ludden. 1996. Purification and characterization of the alternative nitrogenase from the photosynthetic bacterium Rhodospirillum rubrum. J. Bacteriol. 178:1445-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doudoroff, M., and N. J. Palleroni. 1974. Part 7. Gram negative aerobic rods and cocci, p. 217. In R. E. Buchanan and N. E. Gibbons (ed.), Bergey's manual of determinative bacteriology, 8th ed. The Williams and Wilkins Co., Baltimore, MD.
- 13.Eady, R. R., T. H. Richardson, R. W. Miller, M. Hawkins, and D. J. Lowe. 1988. The vanadium nitrogenase of Azotobacter chroococcum. Purification and properties of the Fe protein. Biochem. J. 256:189-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eady, R. R., and R. L. Robson. 1984. Characteristics of N2 fixation in Mo-limited batch and continuous cultures of Azotobacter vinelandii. Biochem. J. 224:853-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elo, S., I. Suominen, P. Kampfer, J. Juhanoja, M. Salkinoja-Salonen, and K. Haahtela. 2001. Paenibacillus borealis sp. nov., a nitrogen-fixing species isolated from spruce forest humus in Finland. Int. J. Syst. Evol. Microbiol. 51:535-545. [DOI] [PubMed] [Google Scholar]
- 16.Evans, H. J., N. E. Campbell, and S. Hill. 1972. Asymbiotic nitrogen-fixing bacteria from the surfaces of nodules and roots of legumes. Can. J. Microbiol. 18:13-21. [DOI] [PubMed] [Google Scholar]
- 17.Fallik, E., Y. K. Chan, and R. L. Robson. 1991. Detection of alternative nitrogenases in aerobic gram-negative nitrogen-fixing bacteria. J. Bacteriol. 173:365-371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Felsenstein, J. 2005. PHYLIP (Phylogeny Inference Package), version 3.6. Distributed by the author. Department of Genome Sciences, University of Washington, Seattle.
- 19.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of Methanosarcina acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gauthier, F., J. D. Neufeld, B. T. Driscoll, and F. S. Archibald. 2000. Coliform bacteria and nitrogen fixation in pulp and paper mill effluent treatment systems. Appl. Environ. Microbiol. 66:5155-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgiadis, M. M., H. Komiya, P. Chakrabarti, D. Woo, J. J. Kornuc, and D. C. Rees. 1992. Crystallographic structure of the nitrogenase iron protein from Azotobacter vinelandii. Science 257:1653-1659. [DOI] [PubMed] [Google Scholar]
- 22.Gillum, W. O., L. E. Mortenson, J. S. Chen, and R. H. Holm. 1977. Quantitative extrusions of the Fe4S4 cores of the active sites of ferredoxins and the hydrogenase of Clostridium pasteurianum. J. Am. Chem. Soc. 99:584-595. [DOI] [PubMed] [Google Scholar]
- 23.Hales, B. J., E. E. Case, J. E. Morningstar, M. F. Dzeda, and L. A. Mauterer. 1986. Isolation of a new vanadium-containing nitrogenase from Azotobacter vinelandii. Biochemistry 25:7251-7255. [DOI] [PubMed] [Google Scholar]
- 24.Hales, B. J., D. J. Langosch, and E. E. Case. 1986. Isolation and characterization of a second nitrogenase Fe-protein from Azotobacter vinelandii. J. Biol. Chem. 261:15301-15306. [PubMed] [Google Scholar]
- 25.Hausinger, R. P., and J. B. Howard. 1983. Thiol reactivity of the nitrogenase Fe-protein from Azotobacter vinelandii. J. Biol. Chem. 258:13486-13492. [PubMed] [Google Scholar]
- 26.Jacobitz, S., and P. E. Bishop. 1992. Regulation of nitrogenase-2 in Azotobacter vinelandii by ammonium, molybdenum, and vanadium. J. Bacteriol. 174:3884-3888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joerger, R. D., M. R. Jacobson, R. Premakumar, E. D. Wolfinger, and P. E. Bishop. 1989. Nucleotide sequence and mutational analysis of the structural genes (anfHDGK) for the second alternative nitrogenase from Azotobacter vinelandii. J. Bacteriol. 171:1075-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joerger, R. D., T. M. Loveless, R. N. Pau, L. A. Mitchenall, B. H. Simon, and P. E. Bishop. 1990. Nucleotide sequences and mutational analysis of the structural genes for nitrogenase 2 of Azotobacter vinelandii. J. Bacteriol. 172:3400-3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jurgensen, M. F., and C. B. Davey. 1971. Nonsymbiotic nitrogen-fixing microorganisms in forest and tundra soils. Plant Soil 34:341-356. [Google Scholar]
- 30.Kentemich, T., G. Danneberg, B. Hundeshagen, and H. Bothe. 1988. Evidence for the occurrence of the alternative, vanadium-containing nitrogenase in the cyanobacterium Anabaena variabilis. FEMS Microbiol. Lett. 51:19-24. [Google Scholar]
- 31.Kimble, L. K., and M. T. Madigan. 1992. Evidence for an alternative nitrogenase in Heliobacterium gestii. FEMS Microbiol. Lett. 100:255-260. [Google Scholar]
- 32.Lehman, L. J., and G. P. Roberts. 1991. Identification of an alternative nitrogenase system in Rhodospirillum rubrum. J. Bacteriol. 173:5705-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Line, M. A., and M. W. Loutit. 1971. Non-symbiotic nitrogen-fixing organisms from New Zealand tussock-grassland soils. J. Gen. Microbiol. 66:309-318. [Google Scholar]
- 34.Loveless, T. M., and P. E. Bishop. 1999. Identification of genes unique to Mo-independent nitrogenase systems in diverse diazotrophs. Can. J. Microbiol. 45:312-317. [PubMed] [Google Scholar]
- 35.Loveless, T. M., J. R. Saah, and P. E. Bishop. 1999. Isolation of nitrogen-fixing bacteria containing molybdenum-independent nitrogenases from natural environments. Appl. Environ. Microbiol. 65:4223-4226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maynard, R. H., R. Premakumar, and P. E. Bishop. 1994. Mo-independent nitrogenase 3 is advantageous for diazotrophic growth of Azotobacter vinelandii on solid medium containing molybdenum. J. Bacteriol. 176:5583-5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Miller, R. W., and R. R. Eady. 1988. Molybdenum and vanadium nitrogenases of Azotobacter chroococcum. Low temperature favours N2 reduction by vanadium nitrogenase. Biochem. J. 256:429-432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mylona, P. V., R. Premakumar, R. N. Pau, and P. E. Bishop. 1996. Characteristics of orf1 and orf2 in the anfHDGK genomic region encoding nitrogenase 3 of Azotobacter vinelandii. J. Bacteriol. 178:204-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Oda, Y., S. K. Samanta, F. E. Rey, L. Wu, X. Liu, T. Yan, J. Zhou, and C. S. Harwood. 2005. Functional genomic analysis of three nitrogenase isozymes in the photosynthetic bacterium Rhodopseudomonas palustris. J. Bacteriol. 187:7784-7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pau, R. N., M. E. Eldridge, D. J. Lowe, L. A. Mitchenall, and R. R. Eady. 1993. Molybdenum-independent nitrogenases of Azotobacter vinelandii: a functional species of alternative nitrogenase-3 isolated from a molybdenum-tolerant strain contains an iron-molybdenum cofactor. Biochem. J. 293:101-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Premakumar, R., M. R. Jacobson, T. M. Loveless, and P. E. Bishop. 1992. Characterization of transcripts expressed from nitrogenase-3 structural genes of Azotobacter vinelandii. Can. J. Microbiol. 38:929-936. [DOI] [PubMed] [Google Scholar]
- 42.Raina, R., U. K. Bageshwar, and H. K. Das. 1993. The ORF encoding a putative ferredoxin-like protein downstream of the vnfH gene in Azotobacter vinelandii is involved in the vanadium-dependent alternative pathway of nitrogen fixation. Mol. Gen. Genet. 236:459-462. [DOI] [PubMed] [Google Scholar]
- 43.Robson, R. L., R. R. Eady, T. H. Richardson, R. W. Miller, M. Hawkins, and J. R. Postgate. 1986. The alternative nitrogenase of Azotobacter chroococcum is a vanadium enzyme. Nature (London) 322:388-390. [Google Scholar]
- 44.Robson, R. L., P. R. Woodley, R. N. Pau, and R. R. Eady. 1989. Structural genes for the vanadium nitrogenase from Azotobacter chroococcum. EMBO J. 8:1217-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rosado, A. S., G. F. Duarte, L. Seldin, and J. D. Van Elsas. 1998. Genetic diversity of nifH gene sequences in Paenibacillus azotofixans strains and soil samples analyzed by denaturing gradient gel electrophoresis of PCR-amplified gene fragments. Appl. Environ. Microbiol. 64:2770-2779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schneider, K., A. Muller, U. Schramm, and W. Klipp. 1991. Demonstration of a molybdenum- and vanadium-independent nitrogenase in a nifHDK-deletion mutant of Rhodobacter capsulatus. Eur. J. Biochem. 195:653-661. [DOI] [PubMed] [Google Scholar]
- 47.Schuddekopf, K., S. Hennecke, U. Liese, M. Kutsche, and W. Klipp. 1993. Characterization of anf genes specific for the alternative nitrogenase and identification of nif genes required for both nitrogenases in Rhodobacter capsulatus. Mol. Microbiol. 8:673-684. [DOI] [PubMed] [Google Scholar]
- 48.Seidler, R. J., P. E. Aho, P. N. Raju, and H. J. Evans. 1972. Nitrogen fixation by bacterial isolates from decay in living whole fir trees, Abies concolor (Gold and Glend) Lindl. J. Gen. Microbiol. 73:413-416. [Google Scholar]
- 49.Seldin, L., J. D. Van Elsas, and E. G. C. Penido. 1983. Bacillus nitrogen fixers from Brazilian soils. Plant Soil 70:243-255. [Google Scholar]
- 50.Thiel, T. 1993. Characterization of genes for an alternative nitrogenase in the cyanobacterium Anabaena variabilis. J. Bacteriol. 175:6276-6286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walmsley, J., and C. Kennedy. 1991. Temperature-dependent regulation by molybdenum and vanadium of expression of the structural genes encoding three nitrogenases in Azotobacter vinelandii. Appl. Environ. Microbiol. 57:622-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Woese, C. R., W. G. Weisburg, B. J. Paster, C. M. Hahn, R. S. Tanner, N. R. Krieg, H. P. Koops, H. Harms, and E. Stackenbrandt. 1984. The phylogeny of purple bacteria: the beta subdivision. Syst. Appl. Microbiol. 5:327-336. [DOI] [PubMed] [Google Scholar]
- 53.Young, J. P. W. 1992. Phylogenetic classification of nitrogen-fixing organisms, p. 43-86. In G. Stacy, R. H. Burris, and H. J. Evans (ed.), Biological nitrogen fixation. Chapman and Hall, New York, NY.
- 54.Zehr, J. P., and L. A. McReynolds. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl. Environ. Microbiol. 55:2522-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zinoni, F., R. M. Robson, and R. L. Robson. 1993. Organization of potential alternative nitrogenase genes from Clostridium pasteurianum. Biochim. Biophys. Acta 1174:83-86. [DOI] [PubMed] [Google Scholar]