Abstract
Fructansucrase enzymes polymerize the fructose moiety of sucrose into levan or inulin fructans, with β(2-6) and β(2-1) linkages, respectively. The probiotic bacterium Lactobacillus johnsonii strain NCC 533 possesses a single fructansucrase gene (open reading frame AAS08734) annotated as a putative levansucrase precursor. However, 13C nuclear magnetic resonance (NMR) analysis of the fructan product synthesized in situ revealed that this is of the inulin type. The ftf gene of L. johnsonii was cloned and expressed to elucidate its exact identity. The purified L. johnsonii protein was characterized as an inulosucrase enzyme, producing inulin from sucrose, as identified by 13C NMR analysis. Thin-layer chromatographic analysis of the reaction products showed that InuJ synthesized, besides the inulin polymer, a broad range of fructose oligosaccharides. Maximum InuJ enzyme activity was observed in a pH range of 4.5 to 7.0, decreasing sharply at pH 7.5. InuJ exhibited the highest enzyme activity at 55°C, with a drastic decrease at 60°C. Calcium ions were found to have an important effect on enzyme activity and stability. Kinetic analysis showed that the transfructosylation reaction of the InuJ enzyme does not obey Michaelis-Menten kinetics. The non-Michaelian behavior of InuJ may be attributed to the oligosaccharides that were initially formed in the reaction and which may act as better acceptors than the growing polymer chain. This is only the second example of the isolation and characterization of an inulosucrase enzyme and its inulin (oligosaccharide) product from a Lactobacillus strain. Furthermore, this is the first Lactobacillus strain shown to produce inulin polymer in situ.
Levansucrase and inulosucrase enzymes, collectively called fructansucrases (FSs) or fructosyltransferases (FTFs), polymerize the fructose moiety of their substrate sucrose into fructans which possess either levan or inulin structures with β(2-6) and β(2-1) linkages, respectively. Possible applications of inulin and its oligosaccharides have been reviewed earlier (19). Inulin-type fructans of a higher degree of polymerization are of particular interest due to their demonstrated pronounced in vitro prebiotic effects (45). In the food industry, inulin is used as a fat substitute and to provide texture and stability in several products, such as desserts, baked goods, and fermented dairy products, as well as infant formula (36). Inulin polymers also have a potential application as surfactants. Carbamoylated inulin has the ability to reduce interfacial tension, thus providing a biodegradable surface-active agent (40).
Inulosucrase enzymes (EC 2.4.1.9) are classified in glycoside hydrolase family GH68, along with other bacterial FS enzymes (http://www.cazy.org). Recently, a few mutagenesis studies focused on determining the structure-function relationship among FS enzymes have been published. For instance, modification of residues located at the −1 sugar-binding subsite of inulosucrase from Lactobacillus reuteri 121 strongly affected the size of the products synthesized (32). Mutagenesis of the Bacillus megaterium levansucrase Arg370 and Asn252 amino acids revealed that these residues are crucial for the polymer-versus-oligosaccharide product ratio of the enzyme (16). However, in spite of the availability of two high-resolution three-dimensional (3D) structures of the levansucrase proteins of Bacillus subtilis (24) and Gluconacetobacter diazotrophicus (23), also with sucrose bound in the active site, little is known about the structure-function relationships in these enzymes responsible for the specificity of the glycosidic linkage in the fructan products.
The type of linkages formed in fructans is most certainly based on differences in the identities and positions of specific amino acid residues present in the active sites of FS enzymes. All lactic acid bacteria FSs have a high level of amino acid sequence similarity (>60%), which does not allow straightforward discrimination between inulosucrase and levansucrase proteins (21, 47) by amino acid sequence alignments, as has been done for the identification of residues determining linkage specificity in glucansucrase enzymes (22). Furthermore, no structural information about inulosucrase enzymes is available and only a limited number of inulosucrase enzymes have been characterized. As reviewed in reference 47, levansucrase genes/enzymes from more than 17 species of gram-positive and gram-negative bacteria have been characterized whereas inulosucrase genes/enzymes have only been found in a few species of lactic acid bacteria, namely, Streptococcus mutans (37), Leuconostoc citreum CW28 (29), and L. reuteri 121 (51). An inulin-producing enzyme from Bacillus sp. has also been characterized, but the gene involved has not been identified (53).
The complete genome sequence (2.0 Mb in size) of the probiotic bacterium Lactobacillus johnsonii strain NCC 533 (formerly Lactobacillus acidophilus La1) has been published (35); it encodes a total of 1,821 proteins (5). L. johnsonii strain NCC 533 is a member of the acidophilus group of intestinal lactobacilli that have been extensively studied for their probiotic activities, pathogen inhibition, epithelial cell attachment, and immunomodulation (7, 12-14, 17, 20, 27). The food company Nestlé, which elucidated the genome sequence of the organism, uses it in a yogurt-like dairy product called LC1. According to the company, LC1 strengthens the body's natural defenses and keeps the bowels healthy (11). The published genome sequence of L. johnsonii NCC 533 contains an open reading frame, AAS08734 (GenBank accession no. AE017198), predicted to encode a levansucrase. Here we report a detailed molecular and biochemical characterization of this novel FS from the probiotic species L. johnsonii. We clearly show that L. johnsonii is capable of synthesizing an inulin polymer in situ. Furthermore, it uses open reading frame AAS08734 (GenBank accession no. AE017198), in fact encoding an inulosucrase, to synthesize this inulin.
MATERIALS AND METHODS
Amino acid sequence alignment of inulosucrase (InuJ) from L. johnsonii NCC 533 and phylogenetic tree construction.
Multiple amino acid sequence alignments of InuJ and known inulosucrases and levansucrases from other lactic acid bacteria were made with CLUSTAL W 1.74 (42). Characteristic features and the catalytic core of InuJ were deduced from these alignments with the aid of http://pfam.janelia.org (1). A phylogenetic tree of all known FSs of lactic acid bacteria was constructed with MEGA version 4 by the neighbor-joining method (41).
Bacterial strains and culturing conditions.
The L. johnsonii NCC 533 strain was obtained from the Nestlé Research Center, Lausanne, Switzerland. For genomic DNA isolation, the cells were cultivated anaerobically at 37°C in MRS medium containing 200 g liter−1 glucose. MRS with sucrose (200 g liter−1) was used for polysaccharide production by L. johnsonii. Escherichia coli TOP10 (Invitrogen) and BL21 Star (DE3) (Invitrogen) were used as hosts for cloning and expression, respectively. E. coli strains were grown at 37°C at 210 rpm in Luria-Bertani (LB) medium supplemented with 50 μg ml−1 ampicillin in order to maintain plasmid integrity. LB agar plates were made by adding 1.5% agar to the LB medium.
Cloning of the inuJ gene.
Total genomic DNA was extracted from L. johnsonii NCC 533 and purified by the method described in reference 52 and modified as described in reference 26. DNA was amplified on a DNA thermal cycler PTC-200 (MJ Research) with high-fidelity DNA polymerase (Fermentas, Germany). L. johnsonii genomic DNA and primers FTF-Lj-F (5′-TATGTCAACCATGGATGTAAAACAAGTTGAAAAGAAAGAC-3′, containing an NcoI site [underlined]) and FTF-Lj-R (5′-TATGTCAAGGATCCTTAATGGTGATGGTGATGGTGTTGGTGTGGCTTCAA-3′, containing a BamHI site [underlined], a C-terminal His tag [italics], and stop codon [bold]), were used in a PCR to amplify the 5′- and 3′-truncated L. johnsonii FS (ftf) gene. This truncated ftf gene encodes amino acids 144 to 709 of the InuJ protein with a C-terminal His tag (designated InuJΔ144-709His). By using the NcoI and BamHI restriction sites, the inuJ amplicon was cloned into expression vector pET15b (Novagen). The resulting vector (pETInuJ) was transformed into E. coli BL21 Star (DE3) for expression studies. Correct construction of the plasmid was confirmed by nucleotide sequence analysis (GATC, Germany).
InuJ expression and purification.
E. coli BL21 harboring pETInuJ was grown overnight in 600 ml LB medium equally divided among three 1-liter Erlenmeyer flasks. The medium was supplemented with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) to induce protein expression. The cells were harvested by centrifugation at 3,500 × g for 15 min, and the pellet was resuspended in 20 ml binding buffer (20 mM Na2HPO4-NaH2PO4, pH 8.0) containing 5 mM β-mercaptoethanol and 4 mM imidazole. After sonication, the extract was centrifuged (20,000 × g for 20 min) and the protein present in the supernatant was purified to homogeneity by Ni affinity (Sigma) and anion-exchange chromatography (6-ml ResourceQ column; Amersham Pharmacia, Sweden). The purest enzyme fractions (as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis) possessing FS activity after anion-exchange chromatography were pooled, dialyzed overnight against sodium acetate buffer (25 mM, pH 5.4), and stored at 4°C for further studies.
Biochemical characterization of the recombinant inulosucrase.
All assays were performed at 55°C and pH 7.0 in Michaelis' barbital sodium acetate buffer (6) unless described otherwise. Purified enzyme (0.45 μg ml−1) was used for biochemical characterization and kinetic studies. One unit of InuJ enzyme activity is defined as the release of 1 μmol of monosaccharide per min from sucrose. Enzyme concentrations were determined with the Bradford reagent (Bio-Rad, Germany) with bovine serum albumin as the standard.
Effects of pH, temperature, CaCl2, and EDTA.
Michaelis' barbital sodium acetate buffer in a pH range of 4 to 8 was used to study the effect of pH on the activity of recombinant InuJΔ144-709His. Enzymatic incubations were performed with reaction mixtures containing 200 mM sodium acetate-sodium barbital buffer supplemented with 500 mM sucrose and 1 mM CaCl2. The activity of the enzyme (0.45 μg ml−1) was measured at 50°C. After preincubation of the assay mixture at the assay temperatures for 10 min, reactions were started by enzyme addition. Samples were taken every 3 min and used to determine the amounts of glucose and fructose released from sucrose (48). The amount of glucose formed reflects the total amount of sucrose utilized during the reaction (VG) (total activity). The amount of fructose (VF) formed is a measurement of hydrolytic activity. The transglycosylation activity was calculated by subtracting the amount of free fructose from glucose (VG − VF). The effect of temperature on the enzyme activity was studied in 200 mM sodium acetate-sodium barbital buffer, pH 7.0, supplemented with 500 mM sucrose and 1 mM CaCl2.
To study the effects of calcium ions on enzyme activity, the experiments were initially conducted with buffer without CaCl2 and all of the solutions used were prepared with MilliQ water. However, no differences in InuJ activity were observed in the presence or absence of Ca2+ ions. Therefore, further studies were conducted in the presence of gradually increasing concentrations of EDTA (from 0 to 800 μM) but without addition of CaCl2. Subsequently, the effect of Ca2+ ions (at concentrations of 0 to 800 μM) was studied in the presence of 600 μM EDTA in the reaction medium.
Kinetic parameters.
For the determination of kinetic parameters, activity assays were performed with 0.45 μg ml−1 protein and sucrose concentrations ranging from 5 to 1,000 mM. The Sigma Plot program (version 10.0) was used for curve fitting of the data by using either the standard Michaelis-Menten formula, y = (a × x)/(c + x), or the three-parameter Hill formula, y = (a × x)b/(cb + xb). In these formulas, y is the specific activity (units per milligram), x is the substrate concentration (millimolar sucrose), a is the maximum rate of metabolism (units per milligram), b is the Hill factor, and c is the Km or K50 (millimolar sucrose; Km in the case of Michaelis-Menten-type kinetics; K50 in the case of Hill-type kinetics).
Polysaccharide production and characterization.
The polysaccharide synthesized by L. johnsonii NCC 533 was produced by growing the strain anaerobically in 20 ml of MRS-sucrose medium for 7 days at 37°C. The culture was centrifuged at 4,000 × g for 10 min, and the supernatant was separated from the cells. The supernatant was run on a thin-layer chromatography (TLC) plate (Silica gel 60 F254; Merck, Darmstadt, Germany) overnight with 1-butanol-ethanol-water (5:5:3) as the mobile phase. The plates were air dried, sprayed with a urea developing solution specific for sugars containing fructose (44), and developed at 80°C. In situ L. johnsonii products were degraded by exo-inulinase of Aspergillus niger (Megazyme, Wicklow, Ireland). For this purpose, the exo-inulinase enzyme (final concentration, 10 U ml−1) was added to a mixture of the sample (the supernatant), 0.3 M phosphate buffer (pH 4.5), and MilliQ water (100 μl each). The reaction mixture was incubated at 40°C for 2 h, and 3 μl of this solution was analyzed with a TLC plate as described above.
To produce fructan oligosaccharides and polymer, the purified recombinant inulosucrase (4.5 μg ml−1) was incubated with sucrose (600 mM) at 55°C in Michaelis' barbital sodium acetate buffer (pH 7.0) containing 1 mM CaCl2 and samples were taken at different time intervals. To characterize the oligosaccharide and polymer products formed, 1-μl aliquots from this reaction mixture (diluted four times) were run on TLC plates overnight as described above. The polymer was precipitated from the rest of the reaction mixture with 2 volumes of 96% cold ethanol and separated by centrifugation at 2,500 × g for 15 min. After being dissolved in MilliQ water, the polymer was precipitated (46). This process was repeated two more times, and the polymer was finally freeze-dried. For nuclear magnetic resonance (NMR) spectroscopy, samples were dissolved in 99.9 atom% D2O (Aldrich). One-dimensional 13C NMR spectra were recorded at 125 MHz on a 500-MHz Varian Inova NMR spectrometer at a probe temperature of 80°C. Chemical shifts are expressed in parts per million relative to the methyl group of internal acetone (δ = 31.07). Carbon spectra were recorded in 38K data sets, with a spectral width of 30.166 kHz. Prior to Fourier transformation, the time domain data were apodized with an exponential function corresponding to a 0.5-Hz line broadening.
High-pressure anion-exchange chromatography (HPAEC; Dionex, Sunnyvale, CA) was used to separate oligosaccharides produced by incubation of InuJ with sucrose for 48 h at 37°C as described above. Separation of oligosaccharides was achieved as described previously (32), with the following gradient for eluent A: 0 min, 100%; 10 min, 78%; 25 min, 60%; 80 min, 10%; 83 min, 0%; 91 min, 100%. Eluent A was 0.1 M sodium hydroxide, and eluent B was 0.1 M sodium hydroxide in 0.6 M sodium acetate. As the standard, a 1:1 mixture of Raftiline ST-Gel and Raftiline HP (Orafti, Tienen, Belgium) representing chicory inulin was used.
The molecular weight of the inulin was determined by high-performance size exclusion chromatography coupled on line with multiangle laser light scattering (MALLS) and differential refractive index detection (Schambeck SDF). A Dawn-F-DSP (Wyatt Technology, Santa Barbara, CA) He-Ne laser photometer (λ = 690 nm) equipped with a K5 flow cell and 18 detectors at angles ranging from 12.8 to 164.7° was used as a MALLS detector. Samples were filtered through a 0.45-μm filter (MILLEX), and the injection volume was 240 μl. NaNO3 (0.1 M) was used as the eluent at a flow rate of 1.0 ml min−1. Pullulan (pss-pulkitL; Polymer Standards Service) and dextran (T2000; Amersham Pharmacia Biotech, Uppsala, Sweden) samples with molecular masses ranging from 4 × 104 to 2 × 106 Da were used as standards.
RESULTS AND DISCUSSION
Amino acid sequence analysis of inulosucrase (InuJ) from L. johnsonii NCC 533.
The putative ftf gene of L. johnsonii consists of 2,394 bp and codes for a 797-amino-acid protein with a deduced molecular mass of 87.2 kDa. A putative signal peptidase cleavage site is present between amino acids 36 and 37 (http://www.cbs.dtu.dk/services/SignalP/). Following the N-terminal variable domain of 172 amino acids, a core region of 453 amino acids (residues 210 to 662) was identified (http://pfam.janelia.org) (9) that belongs to glycoside hydrolase family GH68 (http://www.cazy.org). A gram-positive cell wall-anchoring domain of 41 amino acids (amino acids 753 to 794) was detected at the C-terminal end of InuJ, following a C-terminal variable region of 89 amino acids. The C terminus of InuJ also contains a 17-fold repeat of the PXX motif and a hydrophobic stretch of 23 amino acids (residues 769 to 791), and the protein is terminated by six positively charged amino acids. This organization is similar to the protein-anchoring system reported for L. reuteri 121 inulosucrase (51), except that its LPQTG motif is replaced with an LPKAG motif in InuJ. The LPKAG motif also has been reported in an immunoglobulin light-chain-binding protein of a few strains of Peptostreptococcus magnus (18), where it is considered to be similar to the consensus sequence LPXTG, which is well conserved in gram-positive bacterial cell wall-associated proteins (10, 49, 51).
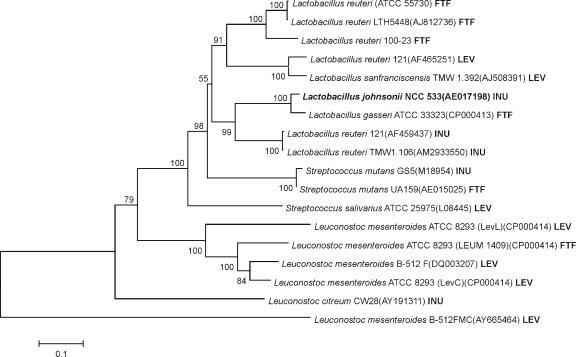
Alignments of the amino acid sequence of InuJ with FTF proteins of other lactic acid bacteria revealed the highest similarities to L. reuteri 121 inulosucrase (60% similarity) and L. gasseri FTF (82% similarity); InuJ clustered most closely with the latter in the phylogenetic tree (Fig. 1). The conserved amino acids reported to be involved in catalysis in FS enzymes were all present in the InuJ sequence. From the 3D structure of the B. subtilis levansucrase, residues D86, D247, and E342 have been identified as the catalytic nucleophile, the transition state stabilizer, and the general acid/base catalyst, respectively (24). In L. reuteri 121 inulosucrase, the importance of equivalent amino acids D272, D424, and E523, which correspond to D272, D425, and E524 in InuJ of L. johnsonii, have been proven by site-directed mutagenesis (34).
FIG. 1.
Unrooted phylogenetic tree of FTF proteins from lactic acid bacteria. Proteins: LEV, levansucrase β(2-6); INU, inulosucrase β(2-1); FTF, unknown linkage specificity. Alignments and dendrogram construction were done (with the complete amino acid sequences) by the neighbor-joining method with MEGA4. Bootstrap values (in percentages) are indicated at the branching points. The scale bar corresponds to a genetic distance of 0.1 substitution per position.
Cloning, expression, and purification of InuJ.
An N- and C-terminally truncated version of InuJ was constructed, aiming for high protein expression in E. coli. Previously, we reported that cloning and expression of the full-length ftf genes from L. reuteri 121 in E. coli led to expression problems. Only an ftf construct with a C-terminal truncation from the PXX amino acid residues onward (deleting the cell wall anchor) yielded good levels of protein expression (51). In addition, we made an N-terminal truncation in InuJ at a position previously shown not to affect the Lactobacillus sanfranciscensis levansucrase enzyme activity nor the composition of its polysaccharide products (43). The position of the truncation (at both the N- and C-terminal ends) was also based on the amino acid sequence of the B. subtilis SacB protein that has been crystallized (24), taking care not to delete conserved sequences.
The truncated inuJ gene of 1,724 bp with a C-terminal His tag was successfully cloned into pET15b after amplification with primers FTF-Lj-F and FTF-Lj-R. This truncated gene encodes a recombinant FTF protein (InuJΔ144-709His) of 63 kDa that exhibits very high expression levels in E. coli, yielding about 25 mg of His-tagged, purified protein from a 1-liter culture.
Biochemical characterization of the InuJ enzyme. (i) Effects of pH, temperature, and CaCl2 on enzyme activity.
In order to define the best conditions for subsequent kinetic studies, the pH and temperature optima of InuJ, and the influence of Ca2+ ions on InuJ activity were determined. The effect of pH on enzyme activity was studied at 50°C (data not shown). The highest total inulosucrase activity was observed at pH 7.0, with a rapid decrease in activity at higher pH values. More than 85% of the activity was retained in the pH range of 4.5 to 6.0. The pH optima of several related enzymes have been reported previously, i.e., levansucrase of L. sanfranciscensis (optimum pH of 5.4) (43) and the FS enzymes of L. reuteri 121 (pH optima, 4.5 to 5.5) (49). Levansucrase (LevC) of L. mesenteroides exhibited the highest activity in the pH range of 6.5 to 7 (30). InuJ exhibited its maximum transglycosylation activity in a broad pH range of 4.5 to 7.0 (65 to 71%). Maximum hydrolytic activity was also observed in the same pH range, with a peak (29%) at pH 7.0.
The highest total InuJ enzyme activity was found at 55°C, with a drastic decrease at higher temperatures (data not shown). A large part (84%) of this total activity was coming from the transglycosylation activity. Maximum hydrolytic activity (28% of the total activity) was observed at 40°C. Relatively high optimum temperatures have also been reported for the FS enzymes from L. reuteri 121 (50°C) (50), L. sanfranciscensis (35 to 45°C) (43), and Bacillus sp. (60°C) (3). The increased transglycosylation activity observed at higher temperatures may be due to oligosaccharide formation (see below).
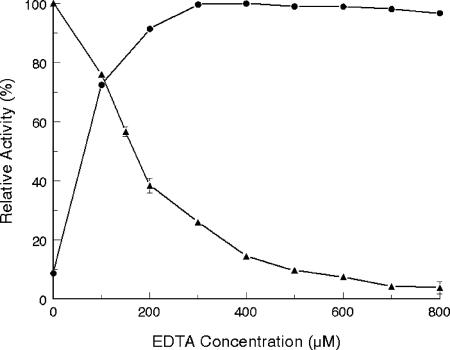
Initially, no differences in InuJ activity were observed in the presence or absence of Ca2+ ions. However, the InuJ enzyme activity decreased with increasing EDTA concentrations, with only 5% of the activity remaining at 700 μM EDTA. The enzyme activity was completely recovered upon the addition of 300 μM Ca2+ ions (Fig. 2). This result is in agreement with the proven essential role of calcium ions in FS activity (31). The latter study used site-directed mutagenesis to demonstrate that Asp520 in the inulosucrase of L. reuteri 121 plays an important role in Ca2+ binding. This residue is highly conserved in family GH68 proteins of gram-positive origin. In the 3D structure of B. subtilis levansucrase, the corresponding Asp339 residue is involved in calcium binding (24). In the case of InuJ, the equivalent amino acid residue is Asp521. The data show that the purified InuJ protein contained bound Ca2+ ions, which were scavenged when EDTA was added, thus decreasing enzyme activity. Calcium ions were also found to have a significant effect on enzyme stability. In the absence of EDTA, the enzyme retained 80% of its total activity after incubation at 55°C for 8 h, whereas it completely lost its activity within 3 h of incubation in the presence of 600 μM EDTA (data not shown).
FIG. 2.
Effects of EDTA (▴) and Ca2+ (at 600 μM EDTA) (•) on InuJ enzyme activity. The reaction was carried out in Michaelis' barbital sodium acetate buffer (pH 7.0) containing 500 mM sucrose. The reaction was started by the addition of 0.45 μg ml−1 enzyme in a 250-μl reaction mixture. The results are presented with the standard error of the mean indicated by vertical bars (n = 3).
(ii) Kinetic parameters.
Like many other lactobacilli, L. johnsonii NCC 533 grows optimally at 37°C, while its FS enzyme exhibited maximum activity at 55°C. Therefore, kinetic studies with InuJ were carried out at both temperatures (Table 1). The total transglycosylation and hydrolytic activity values of InuJ were determined in a reaction buffer with 500 mM sucrose. The ratio of transglycosylation to hydrolysis activities was almost 1:1 at 37°C but increased about 26-fold at 55°C. The total activity of InuJ at 37°C was about 2.5- and 23,000-fold higher than the activities of the C-terminally truncated inulosucrases from L. reuteri 121 (32) and L. citreum (28), respectively. The C-terminally truncated inulosucrase of L. citreum also displayed higher activity than the wild type, full-length, enzyme (28). The significantly higher activity of InuJ, compared to other inulosucrases, may be the result of its N- and C-terminal truncations. This remains to be studied in more detail.
TABLE 1.
FTF activities and comparison of the apparent kinetic constants for FTF activity of InuJΔ144-709His inulosucrase protein of L. johnsonii at different temperatures and at sucrose concentrations of 5 to 500 mMa
| Parameter | Valuec at temp of:
|
|
|---|---|---|
| 37°C | 55°C | |
| Total FTF activityb (U mg−1 of protein) | 365 | 719 |
| Transglycosylationb (U mg−1 of protein) | 187 | 692 |
| Hydrolysisb (U mg−1 of protein) | 178 | 27 |
| Transglycosylation/hydrolysis ratiob | 1.05 | 25.63 |
| k50/m, G (mM) | NDd | ND |
| kcat, G (s−1) | ND | ND |
| Mean Hill factor, G, ± SEM | 0.47 ± 0.06 | 0.76 ± 0.06 |
| Mean k50/m, F, ± SEM (mM) | 11.7 ± 1.9 | NTe |
| Mean kcat, F, ± SEM (s−1) | 176 ± 3.4 | NT |
| k50/m, G − F (mM) | ND | ND |
| kcat, G − F (s−1) | ND | ND |
| Mean Hill factor, G − F, ± SEM | 0.55 ± 0.10 | 0.97 ± 0.15 |
The kinetic constants are k50/m and kcat for (i) the formation of glucose (G; total enzyme activity), (ii) the formation of fructose (F; hydrolytic enzyme activity), and (iii) glucose minus fructose (G − F; transglycosylation enzyme activity).
Activity values measured at a 500 mM sucrose concentration.
Enzyme assays were done with 0.45 μg ml−1 (final concentration) enzyme.
ND, kinetic parameters could not be determined because the enzyme was not saturated with sucrose, resulting in high standard errors with curve fits.
NT, kinetic parameters could not be determined due to high substrate inhibition.
The kinetic constants for the InuJ activities were determined with substrate concentrations ranging from 5 to 1,000 mM sucrose (Table 1). Even at higher substrate concentrations, the total (VG) and transfructosylation (VG minus VF) activities of the enzyme were not saturated by sucrose. Consequently, high standard errors were obtained with curve fits. Only the VF followed standard Michaelis-Menten kinetics at 37°C. The Km value of InuJ for hydrolysis was similar to that of inulosucrase from L. citreum CW28 (8) and L. reuteri 121 (50), while its kcat value was significantly higher (176 ± 3.4 s−1), indicating that this enzyme has a relatively higher efficiency. The InuJ hydrolysis reaction suffered from strong sucrose substrate inhibition at 55°C. However, the inhibition constant could not be determined because of high standard errors with the curve fit. A similar but lower substrate inhibition effect has been observed for the L. reuteri 121 Inu enzyme (50). The InuJ VG and VG minus VF activities did not obey Michaelis-Menten kinetics and the curve fit data with the three-parameter Hill equation could not be used to calculate the catalytic turnover rate (kcat). However, its Hill factor values were similar to those obtained for the L. reuteri 121 inulosucrase (50), except the value (0.76 ± 0.06) for total activity at 55°C. Hill-type kinetics is based on the assumption that there is more than one binding site present in the enzyme and/or that the enzyme exists in the multimeric form. A high Hill factor value reflects positive cooperativity, which indicates a positive interaction of the enzyme binding sites, and/or multimers, and vice versa. The Hill factor value for transfructosylation activity at 55°C was closer to 1, indicating that there were neither negative nor positive cooperativity effects. The non-Michaelian behavior of InuJ may be attributed to the oligosaccharides that were formed at an early stage of the reaction (Fig. 3), which may act as better acceptors than the growing polymer chain. Similar observations were made for the L. reuteri 121 inulosucrase (33).
FIG. 3.
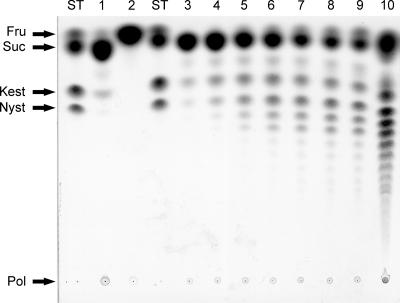
TLC analysis of polysaccharides produced by purified recombinant InuJ and growing cells of L. johnsonii NCC 533 after incubation with MRS-sucrose medium at 37°C for 7 days. Lane 1, in situ production of poly- and oligosaccharides by L. johnsonii cells; lane 2, in situ L. johnsonii products degraded by exo-inulinase of A. niger (TLC analysis of recombinant InuJ [0.45 μg ml−1 purified enzyme] products synthesized from sucrose [600 mM] after incubation at 55°C); lanes 3 to 10, samples taken after 5 min, 10 min, 20 min, 30 min, 40 min, 50 min, 60 min, and 17 h of incubation, respectively. ST, standard; Fru, fructose; Suc, sucrose; Kest, kestose; Nyst, nystose; Pol, polymer.
Production and identification of the polymer by InuJ.
Clear evidence was obtained for in situ production of poly- and oligosaccharides by L. johnsonii cells in MRS-sucrose medium (Fig. 3), although the levels remained relatively low under the experimental conditions used in the present studies. About 150 mg of polysaccharide product was obtained from 100 ml of L. johnsonii culture after 7 days of incubation with 20% sucrose in MRS-sucrose medium. The poly- and oligosaccharide material produced was degraded when inulinase was added (Fig. 3). Furthermore, 13C NMR analysis confirmed the polysaccharide to be an inulin (Table 2). The data thus indicate that L. johnsonii NCC 533 in situ produces inulin polymer and inulin oligosaccharides (Fig. 3), the first example of a probiotic strain that synthesizes prebiotic fructose oligosaccharides (FOS) and inulin in situ. It has already been reported that L. johnsonii possesses a cell surface-associated protein (GroEL) with activities for attachment to mucus and epithelial cells, in addition to other cellular functions (4), and this strain also has carbohydrate-binding properties in common with several enteropathogenic bacteria (27). In situ inulin and FOS production by L. johnsonii may further contribute to its probiotic properties. Sucrose is abundant in many fruits and is the most abundant carbon source in ungerminated cereal grains (38). The dietary sucrose that has escaped digestion in the upper gastrointestinal tract may serve as a substrate for inulin and FOS production by L. johnsonii cells in the colon.
TABLE 2.
Comparison of 13C NMR chemical shift values of fructans produced by lactic acid bacteria
| Carbon atom | Levan
|
Inulin
|
||||
|---|---|---|---|---|---|---|
| L. reuteri 121b | L. mesenteroidesa,c | S. mutans BHTd | L. citreume | L. reuteri 121a | L. johnsonii NCC 533 | |
| C-1 | 61.7 (59.6)a | 60.1 | 60.9 | 61.4 | 62.2 | 62.1 (62.2)a |
| C-2 | 105.0 (104.0)a | 104.3 | 103.2 | 103.60 | 104.2 | 104.1 (104.2)a |
| C-3 | 78.1 (76.0)a | 76.5 | 77.0 | 77.5 | 78.9 | 78.3 (78.4)a |
| C-4 | 76.6 (74.9)a | 75.4 | 74.3 | 74.9 | 75.6 | 75.6 (75.6)a |
| C-5 | 81.2 (80.0)a | 80.5 | 81.1 | 81.8 | 82.1 | 82.2 (82.2)a |
| C-6 | 64.3 (63.2)a | 63.6 | 62.2 | 62.7 | 63.1 | 63.2 (63.1)a |
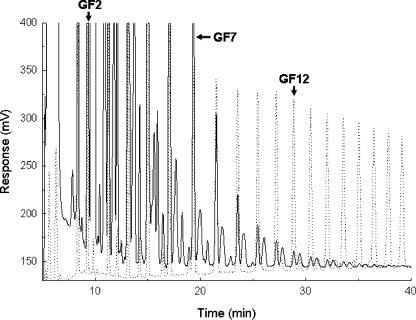
TLC analysis showed that pure recombinant InuJ synthesized a range of FOS in addition to inulin polymer (Fig. 3). Synthesis of FOS started within 5 min of incubation. The synthesis of a range of FOS has also been observed for Inu of L. reuteri 121 (33) and thus appears to be a typical property of inulosucrase enzymes. Oligosaccharides ranging from GF2 to GF15 were clearly detected by HPAEC, although synthesis of FOS with higher degrees of polymerization (>15) cannot be excluded, as is evident from the small peaks after 35 min of retention (Fig. 4). In addition, several unknown products, eluting next to the GFn oligosaccharides, were also present which were virtually undetectable in the FOS synthesized by L. reuteri 121 Inu (32).
FIG. 4.
HPAEC analysis of the FOS products synthesized by purified recombinant InuJΔ144-709His. Solid line, FOS produced by InuJ; dotted line, inulin standards.
The main signals in the 13C NMR spectrum of the fructan synthesized by InuJ corresponds to a fructose polymer with β(2-1)-linked fructose units, typical for the structure found in inulin (Fig. 5). The NMR data also indicate that the inulin product is linear, only one signal is present at C-2 (2), or the degree of branching is too low to be detected.
FIG. 5.
13C NMR spectrum of inulin produced by purified recombinant InuJΔ144-709His recorded in D2O at 80°C. Chemical shifts are given in parts per million relative to the signal (δ = 31.07) of the acetone internal reference.
Therefore, the L. johnsonii ftf gene encodes an inulosucrase enzyme (Table 2). The ftf gene present on the genome of L. johnsonii NCC 533 has been annotated (http://www.ncbi.nlm.nih.gov/entrez/viewer.fcgi?db=nucleotide&val=AAS08734.1) to encode a levansucrase precursor. Our data clearly show that this ftf actually encodes an inulosucrase enzyme.
As determined by high-performance size exclusion chromatography-MALLS, the average molecular mass of the inulin produced by recombinant InuJ of L. johnsonii was determined as 4 × 107 Da, which is comparable to the molecular masses, 6 × 107 to 9 × 107 Da and 1 × 107 Da, reported for the inulins produced by S. mutans GS-5 (15) and L. reuteri 121 (51), respectively. The polymer also showed a remarkably small polydispersity index (Mw/Mn; where Mw and Mn denote, in grams per mole, the average molar mass by weight and the average molar mass by number, respectively) of 1.05 to 1.17, indicating that there is not much variation in the Mw of polymer chains and they all contain approximately the same number of monosaccharide units.
Conclusions.
Our results unequivocally show that the putative levansucrase gene present in the genome of L. johnsonii NCC 533 actually encodes an inulosucrase enzyme that is responsible for the synthesis of an inulin-type fructan. With our truncation strategy for improved E. coli protein expression, we were able to obtain high yields of pure inulosucrase protein. The InuJ catalytic properties, Ca2+ dependence, and FOS synthesis largely resemble those of the Inu enzyme of L. reuteri 121.
This is only the second Lactobacillus inulosucrase which has been purified and characterized in detail, the first having been reported in L. reuteri 121 (51). The available FS amino acid sequence information does not yet allow straightforward identification of inulosucrase or levansucrase enzymes. Characterization of more inulosucrase sequences, and/or crystallographic information for an inulosucrase protein, may provide this information in the future. The isolation and characterization of the inulosucrase gene/enzyme that we have reported here may be an important step in this direction.
Acknowledgments
The work presented here is part of the Ph.D. study of Munir A. Anwar, who obtained a scholarship from the Higher Education Commission (HEC) of Pakistan for this purpose.
We thank the Nestlé Research Centre, Lausanne, Switzerland, for providing the L. johnsonii NCC 533 strain and Pieter van der Meulen and Klaas Dijkstra (Department of Biophysical Chemistry, University of Groningen) for their excellent technical support in NMR spectroscopy. We also thank Albert Woortman and Peter Sanders (Netherlands Organization for Applied Scientific Research, TNO) for molecular weight determination and FOS analysis by HPAEC. We are grateful to Rachel van der Kaaij and Sander van Leeuwen for their help in the construction of the phylogenetic tree and the interpretation of NMR spectra, respectively.
Footnotes
Published ahead of print on 11 April 2008.
REFERENCES
- 1.Bateman, A., L. Coin, R. Durbin, R. D. Finn, V. Hollich, S. Griffiths-Jones, A. Khanna, M. Marshall, S. Moxon, E. L. Sonnhammer, D. J. Studholme, C. Yeats, and S. R. Eddy. 2004. The Pfam protein families database. Nucleic Acids Res. 32:D138-D141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baumgartner, S., T. G. Dax, W. Praznik, and H. Falk. 2000. Characterisation of the high-molecular weight fructan isolated from garlic (Allium sativum L.). Carbohydr. Res. 328:177-183. [DOI] [PubMed] [Google Scholar]
- 3.Ben Ammar, Y., T. Matsubara, K. Ito, M. Iizuka, T. Limpaseni, P. Pongsawasdi, and N. Minamiura. 2002. Characterization of a thermostable levansucrase from Bacillus sp. TH4-2 capable of producing high molecular weight levan at high temperature. J. Biotechnol. 99:111-119. [DOI] [PubMed] [Google Scholar]
- 4.Bergonzelli, G. E., D. Granato, R. D. Pridmore, L. F. Marvin-Guy, D. Donnicola, and I. E. Corthesy-Theulaz. 2006. GroEL of Lactobacillus johnsonii La1 (NCC 533) is cell surface associated: potential role in interactions with the host and the gastric pathogen Helicobacter pylori. Infect. Immun. 74:425-434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boekhorst, J., R. J. Siezen, M. C. Zwahlen, D. Vilanova, R. D. Pridmore, A. Mercenier, M. Kleerebezem, W. M. de Vos, H. Brussow, and F. Desiere. 2004. The complete genomes of Lactobacillus plantarum and Lactobacillus johnsonii reveal extensive differences in chromosome organization and gene content. Microbiology 150:3601-3611. [DOI] [PubMed] [Google Scholar]
- 6.Chipperfield, A. R., and D. M. Taylor. 1970. Binding of plutonium to glycoproteins in vitro. Radiat. Res. 43:393-402. [PubMed] [Google Scholar]
- 7.Cruchet, S., M. C. Obregon, G. Salazar, E. Diaz, and M. Gotteland. 2003. Effect of the ingestion of a dietary product containing Lactobacillus johnsonii La1 on Helicobacter pylori colonization in children. Nutrition 19:716-721. [DOI] [PubMed] [Google Scholar]
- 8.Elena Oritz-Soto, M. A., V. Olivares-Illana, and A. Lopez-Munguia. 2004. Biochemical properties of inulosucrase from Leuconostoc citreum CW28 used for inulin synthesis. Biocatal. Biotransform. 22:275-281. [Google Scholar]
- 9.Finn, R. D., J. Mistry, B. Schuster-Bockler, S. Griffiths-Jones, V. Hollich, T. Lassmann, S. Moxon, M. Marshall, A. Khanna, R. Durbin, S. R. Eddy, E. L. Sonnhammer, and A. Bateman. 2006. Pfam: clans, web tools and services. Nucleic Acids Res. 34:D247-D251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fischetti, V. A., V. Pancholi, and O. Schneewind. 1990. Conservation of a hexapeptide sequence in the anchor region of surface proteins from gram-positive cocci. Mol. Microbiol. 4:1603-1605. [DOI] [PubMed] [Google Scholar]
- 11.Fukushima, Y. 2007. Probiotics and natural defence function of the host. Biosci. Microflora 26:1-10. [Google Scholar]
- 12.Gotteland, M., L. Poliak, S. Cruchet, and O. Brunser. 2005. Effect of regular ingestion of Saccharomyces boulardii plus inulin or Lactobacillus acidophilus LB in children colonized by Helicobacter pylori. Acta Paediatr. 94:1747-1751. [DOI] [PubMed] [Google Scholar]
- 13.Granato, D., G. E. Bergonzelli, R. D. Pridmore, L. Marvin, M. Rouvet, and I. E. Corthesy-Theulaz. 2004. Cell surface-associated elongation factor Tu mediates the attachment of Lactobacillus johnsonii NCC533 (La1) to human intestinal cells and mucins. Infect. Immun. 72:2160-2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Granato, D., F. Perotti, I. Masserey, M. Rouvet, M. Golliard, A. Servin, and D. Brassart. 1999. Cell surface-associated lipoteichoic acid acts as an adhesion factor for attachment of Lactobacillus johnsonii La1 to human enterocyte-like Caco-2 cells. Appl. Environ. Microbiol. 65:1071-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyer, A. G., B. Schroeer, S. Radosta, D. Wolff, S. Czapla, and J. Springer. 1998. Structure of the enzymatically synthesized fructan inulin. Carbohydr. Res. 313:165-174. [DOI] [PubMed] [Google Scholar]
- 16.Homann, A., R. Biedendieck, S. Gotze, D. Jahn, and J. Seibel. 2007. Insights into polymer versus oligosaccharide synthesis: mutagenesis and mechanistic studies of a novel levansucrase from Bacillus megaterium. Biochem. J. 407:189-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ibnou-Zekri, N., S. Blum, E. J. Schiffrin, and T. Von der Weid. 2003. Divergent patterns of colonization and immune response elicited from two intestinal Lactobacillus strains that display similar properties in vitro. Infect. Immun. 71:428-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kastern, W., U. Sjobring, and L. Bjorck. 1992. Structure of peptostreptococcal protein L and identification of a repeated immunoglobulin light chain-binding domain. J. Biol. Chem. 267:12820-12825. [PubMed] [Google Scholar]
- 19.Kaur, N., and A. K. Gupta. 2002. Applications of inulin and oligofructose in health and nutrition. J. Biosci. 27:703-714. [DOI] [PubMed] [Google Scholar]
- 20.Kirjavainen, P. V., A. C. Ouwehand, E. Isolauri, and S. J. Salminen. 1998. The ability of probiotic bacteria to bind to human intestinal mucus. FEMS Microbiol. Lett. 167:185-189. [DOI] [PubMed] [Google Scholar]
- 21.Korakli, M., and R. F. Vogel. 2006. Structure/function relationship of homopolysaccharide producing glycansucrases and therapeutic potential of their synthesised glycans. Appl. Microbiol. Biotechnol. 71:790-803. [DOI] [PubMed] [Google Scholar]
- 22.Kralj, S., I. G. van Geel-Schutten, E. J. Faber, M. J. Van der Maarel, and L. Dijkhuizen. 2005. Rational transformation of Lactobacillus reuteri 121 reuteransucrase into a dextransucrase. Biochemistry 44:9206-9216. [DOI] [PubMed] [Google Scholar]
- 23.Martínez-Fleites, C., M. Ortiz-Lombardia, T. Pons, N. Tarbouriech, E. J. Taylor, J. G. Arrieta, L. Hernandez, and G. J. Davies. 2005. Crystal structure of levansucrase from the gram-negative bacterium Gluconacetobacter diazotrophicus. Biochem. J. 390:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng, G., and K. Futterer. 2003. Structural framework of fructosyl transfer in Bacillus subtilis levansucrase. Nat. Struct. Biol. 10:935-941. [DOI] [PubMed] [Google Scholar]
- 25.Morales-Arrieta, S., M. E. Rodriguez, L. Segovia, A. Lopez-Munguia, and C. Olvera-Carranza. 2006. Identification and functional characterization of levS, a gene encoding for a levansucrase from Leuconostoc mesenteroides NRRL B-512 F. Gene 376:59-67. [DOI] [PubMed] [Google Scholar]
- 26.Nagy, I., G. Schoofs, F. Compernolle, P. Proost, J. Vanderleyden, and R. De Mot. 1995. Degradation of the thiocarbamate herbicide EPTC (S-ethyl dipropylcarbamothioate) and biosafening by Rhodococcus sp. strain NI86/21 involve an inducible cytochrome P-450 system and aldehyde dehydrogenase. J. Bacteriol. 177:676-687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neeser, J. R., D. Granato, M. Rouvet, A. Servin, S. Teneberg, and K. A. Karlsson. 2000. Lactobacillus johnsonii La1 shares carbohydrate-binding specificities with several enteropathogenic bacteria. Glycobiology 10:1193-1199. [DOI] [PubMed] [Google Scholar]
- 28.Olivares-Illana, V., A. Lopez-Munguia, and C. Olvera. 2003. Molecular characterization of inulosucrase from Leuconostoc citreum: a fructosyltransferase within a glucosyltransferase. J. Bacteriol. 185:3606-3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olivares-Illana, V., C. Wacher-Rodarte, S. Le Borgne, and A. López-Munguía. 2002. Characterization of a cell-associated inulosucrase from a novel source: a Leuconostoc citreum strain isolated from Pozol, a fermented corn beverage from Mayan origin. J. Ind. Microbiol. Biotechnol. 28:112-117. [DOI] [PubMed] [Google Scholar]
- 30.Olvera, C., S. Centeno-Leija, and A. Lopez-Munguia. 2007. Structural and functional features of fructansucrases present in Leuconostoc mesenteroides ATCC 8293. Antonie van Leeuwenhoek 92:11-20. [DOI] [PubMed] [Google Scholar]
- 31.Ozimek, L. K., G. J. Euverink, M. J. Van der Maarel, and L. Dijkhuizen. 2005. Mutational analysis of the role of calcium ions in the Lactobacillus reuteri strain 121 fructosyltransferase (levansucrase and inulosucrase) enzymes. FEBS Lett. 579:1124-1128. [DOI] [PubMed] [Google Scholar]
- 32.Ozimek, L. K., S. Kralj, T. Kaper, M. J. Van der Maarel, and L. Dijkhuizen. 2006. Single amino acid residue changes in subsite −1 of inulosucrase from Lactobacillus reuteri 121 strongly influence the size of products synthesized. FEBS J. 273:4104-4113. [DOI] [PubMed] [Google Scholar]
- 33.Ozimek, L. K., S. Kralj, M. J. Van der Maarel, and L. Dijkhuizen. 2006. The levansucrase and inulosucrase enzymes of Lactobacillus reuteri 121 catalyse processive and non-processive transglycosylation reactions. Microbiology 152:1187-1196. [DOI] [PubMed] [Google Scholar]
- 34.Ozimek, L. K., S. A. van Hijum, G. A. van Koningsveld, M. J. Van der Maarel, G. H. Van Geel-Schutten, and L. Dijkhuizen. 2004. Site-directed mutagenesis study of the three catalytic residues of the fructosyltransferases of Lactobacillus reuteri 121. FEBS Lett. 560:131-133. [DOI] [PubMed] [Google Scholar]
- 35.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roberfroid, M. B. 2000. Prebiotics and probiotics: are they functional foods? Am. J. Clin. Nutr. 71:1682S-1687S. [DOI] [PubMed] [Google Scholar]
- 37.Rosell, K. G., and D. Birkhed. 1974. An inulin-like fructan produced by Streptococcus mutans strain JC2. Acta Chem. Scand. B 28:589. [DOI] [PubMed] [Google Scholar]
- 38.Schwab, C., J. Walter, G. W. Tannock, R. F. Vogel, and M. G. Ganzle. 2007. Sucrose utilization and impact of sucrose on glycosyltransferase expression in Lactobacillus reuteri. Syst. Appl. Microbiol. 30:433-443. [DOI] [PubMed] [Google Scholar]
- 39.Shimamura, A., K. Tsuboi, T. Nagase, M. Ito, H. Tsumori, and H. Mukasa. 1987. Structural determination of d-fructans from Streptococcus mutans, serotype b, c, e, and f strains, by 13C-n.m.r. spectroscopy. Carbohydr. Res. 165:150-154. [DOI] [PubMed] [Google Scholar]
- 40.Stevens, C. V., A. Meriggi, M. Peristeropoulou, P. P. Christov, K. Booten, B. Levecke, A. Vandamme, N. Pittevils, and T. F. Tadros. 2001. Polymeric surfactants based on inulin, a polysaccharide extracted from chicory. 1. Synthesis and interfacial properties. Biomacromolecules 2:1256-1259. [DOI] [PubMed] [Google Scholar]
- 41.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 42.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tieking, M., M. A. Ehrmann, R. F. Vogel, and M. G. Ganzle. 2005. Molecular and functional characterization of a levansucrase from the sourdough isolate Lactobacillus sanfranciscensis TMW 1.392. Appl. Microbiol. Biotechnol. 66:655-663. [DOI] [PubMed] [Google Scholar]
- 44.Trujillo, L. E., R. Gomez, A. Banguela, M. Soto, J. G. Arrieta, and L. Hernández. 2004. Catalytical properties of N-glycosylated Gluconacetobacter diazotrophicus levansucrase produced in yeast. Electron. J. Biotechnol. 7:116-123. [Google Scholar]
- 45.van de Wiele, T., N. Boon, S. Possemiers, H. Jacobs, and W. Verstraete. 2007. Inulin-type fructans of longer degree of polymerization exert more pronounced in vitro prebiotic effects. J. Appl. Microbiol. 102:452-460. [DOI] [PubMed] [Google Scholar]
- 46.Van Geel-Schutten, G. H., E. J. Faber, E. Smit, K. Bonting, M. R. Smith, B. Ten Brink, J. P. Kamerling, J. F. Vliegenthart, and L. Dijkhuizen. 1999. Biochemical and structural characterization of the glucan and fructan exopolysaccharides synthesized by the Lactobacillus reuteri wild-type strain and by mutant strains. Appl. Environ. Microbiol. 65:3008-3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Van Hijum, S. A. F. T., S. Kralj, L. K. Ozimek, L. Dijkhuizen, and I. G. van Geel-Schutten. 2006. Structure-function relationships of glucansucrase and fructansucrase enzymes from lactic acid bacteria. Microbiol. Mol. Biol. Rev. 70:157-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Van Hijum, S. A. F. T., K. Bonting, M. J. E. C. Van der Maarel, and L. Dijkhuizen. 2001. Purification of a novel fructosyltransferase from Lactobacillus reuteri strain 121 and characterization of the levan produced. FEMS Microbiol. Lett. 205:323-328. [DOI] [PubMed] [Google Scholar]
- 49.Van Hijum, S. A. F. T., E. Szalowska, M. J. Van der Maarel, and L. Dijkhuizen. 2004. Biochemical and molecular characterization of a levansucrase from Lactobacillus reuteri. Microbiology 150:621-630. [DOI] [PubMed] [Google Scholar]
- 50.Van Hijum, S. A. F. T., M. J. Van der Maarel, and L. Dijkhuizen. 2003. Kinetic properties of an inulosucrase from Lactobacillus reuteri 121. FEBS Lett. 534:207-210. [DOI] [PubMed] [Google Scholar]
- 51.Van Hijum, S. A. F. T., G. H. Van Geel-Schutten, H. Rahaoui, M. J. Van der Maarel, and L. Dijkhuizen. 2002. Characterization of a novel fructosyltransferase from Lactobacillus reuteri that synthesizes high-molecular-weight inulin and inulin oligosaccharides. Appl. Environ. Microbiol. 68:4390-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Verhasselt, P., F. Poncelet, K. Vits, A. Van Gool, and J. Van Der Leyden. 1989. Cloning and expression of a Clostridium acetobutylicum α-amylase gene in Escherichia coli. FEMS Microbiol. Lett. 50:135-140. [DOI] [PubMed] [Google Scholar]
- 53.Wada, T., M. Ohguchi, and Y. Iwai. 2003. A novel enzyme of Bacillus sp. 217C-11 that produces inulin from sucrose. Biosci. Biotechnol. Biochem. 67:1327-1334. [DOI] [PubMed] [Google Scholar]