Abstract
The endospores formed by strains of type A Clostridium perfringens that produce the C. perfringens enterotoxin (CPE) are known to be more resistant to heat and cold than strains that do not produce this toxin. The high heat resistance of these spores allows them to survive the cooking process, leading to a large number of food-poisoning cases each year. The relative importance of factors contributing to the establishment of heat resistance in this species is currently unknown. The present study examines the spores formed by both CPE+ and CPE− strains for factors known to affect heat resistance in other species. We have found that the concentrations of DPA and metal ions, the size of the spore core, and the protoplast-to-sporoplast ratio are determining factors affecting heat resistance in these strains. While the overall thickness of the spore peptidoglycan was found to be consistent in all strains, the relative amounts of cortex and germ cell wall peptidoglycan also appear to play a role in the heat resistance of these strains.
The bacterium Clostridium perfringens is one of the most common causes of food-borne illness in the United States, leading to approximately 248,000 cases each year (24). Acute food poisoning (AFP) occurs when food is contaminated with C. perfringens spores possessing enough heat resistance to survive the cooking process (6). Rather than killing these spores, the heat stimulates dormant spores to germinate. If the food is not thoroughly refrigerated, the cells then propagate in the food. When this contaminated food is consumed, a large number of the cells survive the low pH of the stomach and begin sporulating in the small intestine. During the sporulation process, the C. perfringens enterotoxin (CPE) is produced (23). The cpe gene is usually found on the chromosome in strains that cause AFP, while strains that cause non-food-borne diarrhea usually carry the cpe gene on a plasmid (8).
Two primary bacterial factors contribute to C. perfringens AFP. The first requirement is that the contaminating strain produce endospores with a high degree of heat resistance. The second requirement is that the strain must contain and express the cpe gene. In an early report, Ando et al., (1) examined five strains of C. perfringens that produced highly heat-resistant endospores and compared them with five strains that produced spores with lower heat resistance. The study found that all five of the heat-resistant strains expressed the cpe gene, while the other strains did not. A recent study by Raju and Sarker (31) compared the heat resistance of CPE+ strain SM101 to that of a derived strain with a cpe knockout mutation and found that no change in heat resistance was produced by the mutation. It appears that although the expression of the cpe gene is associated with more heat-resistant strains, the gene or gene product itself does not confer heat resistance.
It has also been noted that the location of the cpe gene is associated with the heat resistance of the spores formed by strains of C. perfringens. Sarker et al., (33) compared the heat resistance of strains possessing a chromosomal cpe gene to the heat resistance of strains carrying a plasmid-borne cpe gene. They found that the spores formed by strains with a chromosomal cpe gene possessed a decimal reduction value (D value) that was, on average, 60-fold greater at 100°C than those of the other strains (33). Novak et al. (27) examined the endospores produced by these same strains in an attempt to determine if specific factors could be identified as contributing to this disparity in heat resistance. That study suggested that production of a smaller, potentially more dehydrated spore core was a major factor in determining heat resistance. More recently, Li and McClane (20) showed that strains with chromosomal cpe genes not only are more heat resistant, but also are more cold resistant in both the vegetative-cell and spore forms.
In this study, we examined the endospores of five CPE+ and four CPE− strains to determine what factors contribute to the disparity in heat resistance between these groups. The strains were assayed for spore heat resistance, protoplast water content, dipicolinic acid (DPA), and mineral concentrations. Spore structural dimensions were determined using transmission electron microscopy (TEM), and the spore cortex structure was determined using liquid chromatography (LC)-tandem mass spectrometry (MS-MS).
MATERIALS AND METHODS
Bacterial strains.
CPE+ strains, NCTC 8239, NCTC 8679, and NCTC 10240, are food-poisoning isolates, Hobbs serotypes 3, 6, and 13, respectively (12). SM101 is a derivative of NCTC 8798, Hobbs serotype 9, that is conducive to electroporation (43). Strain 3663 was obtained from the Norwegian Food Research Institute (18). A previously described duplex-PCR method (41) was used to demonstrate that all five CPE+ strains possessed chromosomal cpe genes, and the expression of CPE by these strains was previously verified by immunoblotting (16, 42). The CPE− strains FD-1 and T-65 were originally food isolates (11) but were verified to lack CPE (42). ATCC 3624 and strain 13 are clinical gangrene isolates (12, 36). PCR analysis (41) was used to verify the absence of the cpe gene in all four CPE− strains. Two-tailed Student's t tests were used to compare results for CPE+ versus CPE− strains. Linear regression analyses were used to detect significant relationships between D values and other parameters measured across all strains.
Spore preparation and determination of heat resistance parameters.
Spores for all strains were prepared in identical manners by growth at 37°C in Duncan and Strong sporulation medium with raffinose (DSSM) (10, 35). In order to increase the sporulation efficiency of the CPE− strains, 0.1% caffeine was added to sporulation media (32). This addition had no apparent effect on the sporulation rates of the CPE+ strains. The heat resistance of the spores of each strain was experimentally determined with and without the addition of caffeine, and no measurable difference was observed (data not shown). A variant of strain 13 that sporulates in the presence of phosphate (26) was isolated by growing the cells in DSSM for 72 h. The cells were concentrated, and vegetative cells were killed by the addition of 100 μg/ml lysozyme plus 200 μg/ml trypsin. The surviving spores were used to inoculate the next culture of DSSM, and the process was repeated. The surviving spores were saved as the variant strain 13V1. The variant was found to produce spores with heat resistance equal to that of the wild-type strain (data not shown). Spore heat resistance for all strains was assayed via determination of the D value for each strain at 90°C (1). Samples were removed directly from sporulating cultures 72 h postinoculation. The samples were heat treated at 70°C for 10 min to kill vegetative cells and then assayed immediately. Heat killing of each strain was followed for 4-log-unit reductions in CFU. Heat resistance assays were performed a minimum of five times on at least two separate cultures for each strain.
For additional assays, spores were purified 72 h after inoculation. Cultures were harvested by centrifugation at 10,000 × g at 4°C, and vegetative cells were lysed by the addition of 100 μg/ml lysozyme plus 200 μg/ml trypsin and incubation at 37°C for 4 h. Sodium dodecyl sulfate (SDS) was added to a concentration of 1% (wt/vol), and incubation was continued for an additional hour (27). Removal of SDS and cell debris was accomplished by washing the spore suspension five times with sterile deionized water at 50°C. The spores were verified to be greater than 90% free of vegetative cells by manual counts using phase-contrast microscopy.
Determination of spore core water content.
Purified spores were assayed for core density using an established density gradient sedimentation method (22). The density gradient material was Histodenz (Sigma), and the gradient range was 75% to 50% (wt/vol) Histodenz. Prior to core density determination, spore coats were permeabilized by incubation for 1 h at 37°C in 1% SDS, 8 M urea, 50 mM dithiothreitol, 50 mM Tris-HCl at pH 8.0, followed by four washes with 150 mM NaCl and a final wash in sterile deionized water (30). The effectiveness of the coat permeabilization procedure for each strain was demonstrated by a >3-log-unit reduction in CFU upon treatment of the coat-permeabilized samples with lysozyme, while lysozyme had no effect prior to permeabilization (data not shown). Permeabilized samples were also plated to verify that the loss in CFU was due to the lysozyme treatment rather than the permeabilization procedure. Each strain was assayed for spore core density a minimum of five times.
Electron microscopy measurements of spore cross sections.
Purified spores were fixed in 1.4% glutaraldehyde, embedded, and sectioned as described previously (17, 27). Transmission electron micrographs were analyzed using Scion Image, release alpha 4.0.3.2. (Scion Corporation, Frederick, MD). Spore measurements were performed on 50 to 100 cross sections at ×25,000 magnification. Spores that were obviously sectioned at an angle to either axis, apparent in pointed ends of the spore, were not measured. C. perfringens spores are ellipsoid, so the definition of an ellipsoid was used to divide the images into those showing sections through the short axis (length/width < 1.3, for width measurements) and those through the long axis (length/width ≥ 1.3, for length measurements). In both cases, noncentral cross sections that were located near the ends of the spores were defined as those that possessed a dimension >1 standard deviation (SD) from the mean and were excluded from calculations. The spore core (protoplast) volume was calculated by application of the formula for the volume of an ellipsoid: volume = (4/3) π (width/2)2 (length) (5). The protoplast-to-sporoplast (P/S) ratio was determined as previously described (5).
DPA and spore solute concentrations.
Purified spores were suspended to a known optical density at 600 nm (OD600), and DPA was extracted and quantified using an established colorimetric assay to determine the concentration of DPA per OD600 unit (14). Samples were also plated in triplicate to allow determination of the concentration of DPA per CFU. Spore core volume measurements were applied to this value to determine the concentration of DPA per μm3 spore core volume.
Core ion concentrations were determined using inductively coupled plasma (ICP) spectroscopy (38). All tubes, pipette tips, and glassware used in this procedure were first acid washed with 6 N Optima HCl (Fisher). Purified spores were incubated in 200 mM Tris-HCl (pH 8.0) for 20 min to release surface-associated ions. The spores were then washed three times in purified H2O, and samples were taken to determine the OD600. The spores were then suspended in 1 ml 6 N Optima HCl (Fisher) and heated at 100°C for 30 min. Samples were centrifuged for 15 min at 13,000 rpm, and the supernatant was diluted with fresh deionized H2O to a final volume of 5 ml. The samples were analyzed with an ICAP 61E simultaneous spectrometer equipped with a Thermo Elemental autosampler. The spectrometer was calibrated with 200 ml 1.2 N Optima HCl (Fisher). The samples were analyzed simultaneously for Ca2+, Fe2+, K+, Mg2+, Mn2+, and Na+. Spore core volume measurements were applied to determine cation concentrations per μm3 spore core volume.
Determination of spore cortex structure.
Purification of spore peptidoglycan (PG), digestion with muramidase, and separation of muropeptides by high-performance LC (HPLC) have been previously described (29). Following identification of muropeptide peaks, quantitation was performed using integrated peak areas as previously described (29). Muropeptides were collected from the separation of strain SM101 spore PG for analysis via LC-electrospray ionization-MS. The LC system was composed of an Agilent 1100 series stack containing a binary pump, AL8 autosampler, and vacuum degasser system connected in line to the mass spectrometer. The LC system was used to remove residual phosphate buffer from the original separation and to verify the presence of a single compound collected from the original HPLC fractionation. The LC gradient ran from 0 to 60% acetonitrile in 0.01% formic acid over a 20-min run at 200 μl/min on a HyPurity Aquastar column (2.1 mm by 150 mm; Thermo Electron Corporation).
The mass spectrometer was an ABI/MDS-Sciex 3200 QTrap with a Sciex TurboSpray ion source. For all muropeptides, the linear ion trap was employed in negative scan mode. The TurboSpray ion source had a voltage of −4,500 V at a temperature of 250°C. A declustering potential of −50 mV was used for all samples. For MS-MS fragmentations, collision energy was optimized for each individual muropeptide but in all cases was between −60 and −120 mV.
RESULTS
Heat resistance and spore core water content.
Cultures of each strain that had been allowed to sporulate for 72 h were heated in water at 90°C and plated to determine viability. This temperature was chosen to avoid difficulties in accurately determining D values in the range of 2 to <1 min at higher temperatures (1, 20, 27, 33). The D values at 90°C were computed for each strain (Table 1). As expected (1), the group of CPE+ strains demonstrated consistently higher heat resistance than the group of CPE− strains (P = 0.0026). Among the CPE+ strains, two were highly heat resistant, with D values of >45 min, while the other D values were clustered around 20 min. The CPE− strains had D values of ≤12 min, and the average D value of the CPE+ strains was more than five times greater than the average for the CPE− strains. Heat resistance assays were also performed on purified spores, with results indistinguishable from those using crude sporulated culture (data not shown).
TABLE 1.
C. perfringens spore heat resistance and water contenta
| Strain | CPEb | D value at 90°C (min) | Density (g/ml) | % Waterc |
|---|---|---|---|---|
| NCTC 8239 | + | 120.6 ± 4.5 | 1.329 ± 0.006 | 51.6 ± 2.4 |
| NCTC 8679 | + | 45.6 ± 9.4 | 1.333 ± 0.008 | 50.0 ± 3.2 |
| SM101 | + | 21.4 ± 0.8 | 1.308 ± 0.004 | 59.8 ± 1.6 |
| NCTC 10240 | + | 19.9 ± 2.4 | 1.308 ± 0.006 | 59.8 ± 2.4 |
| 3663 | + | 19.0 ± 2.1 | 1.306 ± 0.010 | 60.6 ± 3.9 |
| FD1 | − | 12.5 ± 4.6 | 1.307 ± 0.006 | 60.2 ± 2.4 |
| T-65 | − | 10.1 ± 1.6 | 1.316 ± 0.006 | 56.7 ± 2.4 |
| ATCC 3624 | − | 6.9 ± 0.7 | 1.322 ± 0.005 | 54.3 ± 2.0 |
| 13V1 | − | 5.5 ± 1.6 | 1.296 ± 0.006 | 64.6 ± 2.4 |
All values are averages ± SDs of at least three determinations using independent spore preparations.
+, present; −, absent.
Water content was determined using the formula density = −0.00254x − 1.460 (22) and is expressed as a percentage of the wet mass.
Spores from each strain were centrifuged on Histodenz density gradients to determine the spore core wet densities (22) (Table 1). The spores with the two highest core densities were also those that possessed the highest D values. The strain with the lowest core density, strain 13V1, also had the lowest D value. For strains with intermediate D values, there was not a clear relationship between protoplast density and heat resistance.
Spore dimensions and core volume.
A large variation in the sizes of spores produced by various strains of C. perfringens has been previously noted (27). In order to accurately assay the spore core solute concentrations among our strains, it was necessary to determine the spore core unit volumes. Spores were fixed and examined by thin-section electron microscopy. As represented in Fig. 1, a great deal of structural variation was found in these images. Differences were apparent in the core area, cortex width, and coat structure. Several aspects of the spore dimensions were measured (Table 2). A twofold variation in spore core volume was observed among these strains. The three most heat-resistant strains, NCTC 8239, NCTC 8679, and SM101, possessed the smallest core diameters and corresponding core volumes, while FD-1 had a much larger diameter and a core volume over twice as large as those of the smallest strains. The average core volume of the CPE+ strains was calculated to be 0.38 ± 0.12 μm3, while the same value for our CPE− strains was 0.53 ± 0.12 μm3 (P = 0.053). The cortex PG thickness also varied, with the thickest cortex present in NCTC 8239, the most heat-resistant strain. The next-thickest PG layers were in the significantly larger spores of CPE− strains FD-1 and T-65. The thinnest PG was possessed by SM101. The spore coat thickness and structure differed dramatically between these strains, as shown in Fig. 1. The asymmetrical appearance of the coat in many strains made the average coat width difficult to calculate with accuracy (data not shown).
FIG. 1.
TEM images of C. perfringens spore cross sections. Representative images of spores produced by the designated strain are shown in each panel. The scale for each image is the same as that shown in the upper left panel. A, spore core; B, spore PG layer; C, coat layers.
TABLE 2.
C. perfringens spore dimensions and core volumes
| Strain | Core diama (μm) | Core lengtha (μm) | Core volb (μm3) | Cortex widtha (μm) | P/S ratioc |
|---|---|---|---|---|---|
| NCTC 8239 | 0.62 ± 0.06 | 0.72 ± 0.07 | 0.29 ± 0.05 | 0.13 | 0.36 ± 0.07 |
| NCTC 8679 | 0.64 ± 0.07 | 0.79 ± 0.08 | 0.34 ± 0.06 | 0.10 | 0.41 ± 0.08 |
| SM101 | 0.63 ± 0.06 | 0.75 ± 0.08 | 0.31 ± 0.05 | 0.09 | 0.49 ± 0.09 |
| NCTC 10240 | 0.68 ± 0.06 | 0.79 ± 0.07 | 0.38 ± 0.06 | 0.10 | 0.46 ± 0.08 |
| 3663 | 0.79 ± 0.07 | 0.89 ± 0.06 | 0.58 ± 0.08 | 0.10 | 0.48 ± 0.08 |
| FD1 | 0.83 ± 0.06 | 0.97 ± 0.08 | 0.70 ± 0.09 | 0.12 | 0.49 ± 0.08 |
| T65 | 0.72 ± 0.07 | 0.96 ± 0.05 | 0.52 ± 0.08 | 0.12 | 0.48 ± 0.08 |
| ATCC 3624 | 0.68 ± 0.05 | 0.89 ± 0.06 | 0.43 ± 0.05 | 0.11 | 0.45 ± 0.06 |
| 13V1 | 0.71 ± 0.07 | 0.87 ± 0.07 | 0.46 ± 0.07 | 0.11 | 0.45 ± 0.08 |
All values are averages of 50 to 120 measurements ± SDs. The SDs for the cortex measurements were found to be ≤0.02 in all instances.
Spore core volume (± SD) was calculated as follows: volume = (4/3) π (width/2)2 (length).
The P/S ratio (± SD) was calculated by dividing the calculated core volume by the volume of the core plus PG layer (5).
The P/S ratio (5) was determined from these measurements (Table 2). The protoplast was defined as the area of the spore within the inner membrane. The sporoplast was determined as the entire area within the outermost layer of PG (5). The lowest P/S ratios were found in the most heat-resistant strains, NCTC 8239 and 8679, with values of 0.36 and 0.41, respectively. The highest value, 0.49, was demonstrated by CPE− strain FD-1. Among the spore parameters measured, the P/S ratio exhibited the strongest relationship with the D value across the entire range of CPE+ and CPE− strains (Fig. 2) (P = 0.0025).
FIG. 2.
Heat resistance correlates with the P/S ratio in C. perfringens. The P/S ratio was calculated from measurements of TEM images as previously described (5). The line was drawn using the linear regression method and produced an r2 value of 0.77.
Spore solute contents.
The spore core DPA concentrations were determined and were found to differ among strains by as much as 20-fold (Table 3). While the average DPA concentration of the CPE+ strains was 65.4 fmol/μm3, more than twice the average for the CPE− strains, 26.7 fmol/μm3, linear regression analysis indicated there was no correlation between the DPA concentration and the D value. Spore monovalent and divalent cation contents were assayed (Table 3) with ICP spectroscopy. In all strains, the levels of Na+ and K+ were found to be below the limit of detection (data not shown). The Ca2+ concentration correlated strongly with DPA levels, with an r2 value of 0.99 (P < 0.0001). The average Ca2+, Fe2+, and Mg2+ concentrations were more than twofold higher in the CPE+ strains than in the CPE− strains, with the highest concentration of all the cations found in NCTC 8679. However, only Fe2+ showed a statistically significant difference between the two groups of strains (P = 0.03). A significant relationship (P = 0.0291) between the Fe2+ content and the D value was found across the full range of strains. Mn2+ concentrations were found to be very similar between the two groups, in the range of 3 to 10 fmol/μm3 (data not shown).
TABLE 3.
C. perfringens spore solute contents
| Strain | Contenta
|
|||
|---|---|---|---|---|
| DPA | Ca2+ | Fe2+ | Mg2+ | |
| NCTC 8239 | 49.7 ± 1.3 | 41.8 ± 5.0 | 1.8 ± 0.2 | 6.1 ± 2.3 |
| NCTC 8679 | 196.6 ± 3.0 | 130.6 ± 15.6 | 2.3 ± 0.1 | 18.8 ± 1.1 |
| SM101 | 18.9 ± 1.1 | 23.1 ± 1.9 | 0.9 ± 0.1 | 2.1 ± 1.0 |
| NCTC 10240 | 28.9 ± 1.0 | 22.5 ± 0.9 | 0.6 ± 0.1 | 4.3 ± 1.3 |
| 3663 | 33.1 ± 1.4 | 25.8 ± 1.1 | 0.8 ± 0.1 | 3.7 ± 0.9 |
| FD1 | 9.0 ± 0.9 | 10.5 ± 1.3 | 0.3 ± 0.1 | 1.2 ± 0.1 |
| T65 | 16.3 ± 0.8 | 14.1 ± 0.7 | 0.2 ± 0.1 | 2.0 ± 0.7 |
| ATCC 3624 | 64.2 ± 0.8 | 51.8 ± 6.4 | 0.7 ± 0.1 | 8.4 ± 1.4 |
| 13V1 | 17.2 ± 0.9 | 14.6 ± 1.4 | 0.5 ± 0.1 | 2.7 ± 0.9 |
All values are averages ± SDs of three measurements of independent spore preparations and are expressed in fmol/μm3.
Spore cortex structure.
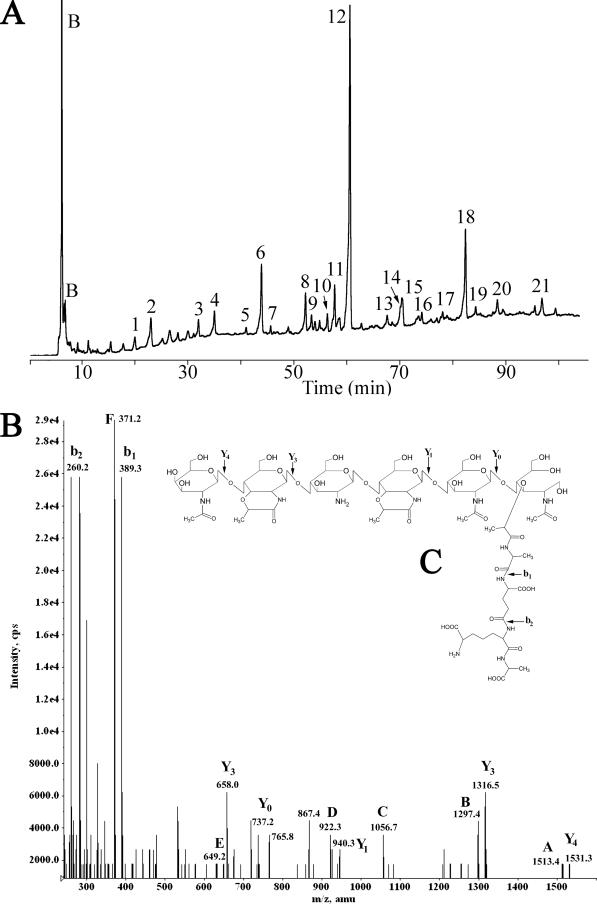
Spore PG was purified, muramidase digested, and analyzed using HPLC. Twenty-one muropeptides were collected from the SM101 sample (Fig. 3A) for structural analysis via electrospray ionization-MS-MS. The chromatograms for the other eight strains were very similar to that of SM101, producing a nearly identical series of peaks (data not shown). Eleven of the SM101 muropeptides had elution times closely matching those derived from Bacillus subtilis 168 endospores (reference 29 and data not shown), and MS analysis verified the coeluting peaks to be the same compounds. MS analysis (Fig. 3B and C and Table 4) revealed that each muropeptide ionized predominantly as a single proton loss (M-H+). Due to limitations of the instrument, compounds with masses of >1,700 were detected as the doubly charged ion (M-2H+). There was a lower frequency of Na+ adducts representing M + Na+-2H+ or M + 2Na+-3H+. MS-MS fragmentation was employed on all compounds to determine their structures (Table 5 and data not shown). In all MS-MS spectra, the saccharide chain fragmented on the nonreducing end of the glycosidic bond to produce primarily Y ions (Fig. 3B and C) (39). Stepwise assembly of these fragments from the nonreducing end allowed the characterization of the oligosaccharide chain (2). The fragmentation occurring within the peptide side chain followed the well-characterized pattern, with both b and y ions present in the same spectra, demonstrating fragmentation of each peptide bond (19). The spore PG peptide side chain structure was found to be Ala-Glu-diaminopimelic acid-Ala, the same side chain linkage observed in spores of Bacillus species and in Clostridium botulinum (3, 40). Unlike the cortexes of other endospores previously examined (3, 40), that of C. perfringens was found to lack N-acetylmuramic acid (NAM) residues possessing single alanine side chains. Dipeptide side chains were present, but at a level nearly 10-fold lower than the percentage of alanine side chains found in other species. Using the structures determined for the muropeptides, several structural parameters were calculated for the PG extracted from the spores of each strain (Table 6). Despite a higher percentage of tetrapeptide side chains, the C. perfringens strains demonstrated a degree of cross-linking lower than that of species previously examined (3). The CPE− strains all possess a significantly greater percentage of tripeptide side chains than the CPE+ strains, with averages of 9.4% and 5.5%, respectively (P = 0.0056). Approximately 10% of the N-acetylglucosamine present in the cortex was found to be deacetylated to glucosamine. This value was fairly consistent in all nine strains.
FIG. 3.
HPLC separation and MS analysis of C. perfringens SM101 spore muropeptides. (A) Spore PG was purified, digested with mutanolysin, reduced, and separated using a methanol gradient as previously described (29). Muropeptides were detected by absorbance at 206 nm and are numbered as in Table 4. Peaks labeled B are buffer components. (B) Fragmentation spectrum of muropeptide 18. Ion masses indicated by letters are those predicted in panel C and in Table 5. (C) Structure proposed for muropeptide 18. The arrows indicate fragmentation points to produce the masses indicated in panel B.
TABLE 4.
Muropeptides produced from C. perfringens spore PG
| Peaka | Predicted structureb | (M-H)−d
|
|
|---|---|---|---|
| Calculated m/z | Observed m/z | ||
| 1 | MS-TP | 737.3 | 737.2 |
| 2 | DS-TP | 940.4 | 940.4 |
| 3 | TS-TP (+H2O) | 1,376.6 | 1,376.4 |
| 4 | TS-TP(no lactam) | 1,418.6 | 1,418.6 |
| 5 | TS(red)-TP(-acetyl) | 1,302.6 | 1,302.6 |
| 6 | TS(red)-TPe | 1,344.6 | 1,344.6 |
| 7 | DS-TriP-DS-TPc | 1,792.7 | 1,792.4 |
| 8 | TS-TP(-acetyl) | 1,316.6 | 1,316.4 |
| 9 | TS-TriP | 1,288.5 | 1,288.3 |
| 10 | TS-TP(-acetyl) | 1,316.6 | 1,316.5 |
| 11 | HS-TP(-acetyl+H2O) | 1,752.7 | 1,752.5 |
| 12 | TS-TP | 1,358.6 | 1,358.6 |
| 13 | TS-DP | 1,115.4 | 1,115.7 |
| 14 | OS-TP(-two acetyl) | 2,110.9 | 2,110.4 |
| 15 | HS(red)-TP(-acetyl) | 1,720.7 | 1,720.5 |
| 16 | DS-TP-TS-TP | 2,281.9 | 2,282.2 |
| 17 | OS-TP(-two acetyl) | 2,110.9 | 2,110.6 |
| 18 | HS-TP(-acetyl) | 1,734.7 | 1,734.6 |
| 19 | DS-TriP-TS-TP | 2,210.9 | 2,211.3 |
| 20 | TS-TP-TS-TP | 2,701.7 | 2,702.1 |
| 21 | HS-TP | 1,776.7 | 1,776.6 |
Peaks are numbered as in Fig. 3A.
MS, monosaccharide (NAM); DS, disaccharide; TS, tetrasaccharide; HS, hexasaccharide; OS, octasaccharide; DP, dipeptide (Ala + Glu); TriP, tripeptide (Ala + diaminopimelic acid [A2pm] + Glu); TP, tetrapeptide (Ala + A2pm + Glu + Ala).
Indicates a DS-TriP cross-linked to a DS-TP.
These masses correspond to the deprotonated molecule in negative ion mode. For m/z of >1,700, these were calculated from detection of the (M-2H)− ion.
Molecules containing reduced (red) muramic lactam.
TABLE 5.
Monosaccharide fragment analysis of muropeptide 18
| Fragmenta | Predicted structureb | Predicted m/z (M-H)− | Observed m/z (M-H)− |
|---|---|---|---|
| Y4 | PS-TP(-acetyl) z = 1 | 1,531.3 | 1,531.3 |
| z = 2c | 765.2 | 765.8 | |
| Y3 | TS-TP(-acetyl) z = 1 | 1,316.4 | 1,316.5 |
| z = 2c | 657.7 | 658.0 | |
| Y1 | DS-TP | 940.4 | 940.3 |
| Y0 | MS-TP | 736.2 | 737.2 |
| b1 | Glu+A2pm+Ala | 389.3 | 389.3 |
| b2 | Ala+A2pm | 260.2 | 260.2 |
| Products of multiple fragmentation and rearrangements of parent ion | |||
| A | PS-TP(-acetyl, -H2O)d | 1,513.3 | 1,513.4 |
| B | TS-TP(-acetyl, -H2O)d | 1,297.4 | 1,297.4 |
| C | TS-DP(-acetyl) | 1,056.4 | 1,056.7 |
| D | DS-TP(-H2O)d | 922.3 | 922.3 |
| E | MS-TriP | 649.3 | 649.2 |
| F | Glu+A2pm+Ala(-H2O)d | 371.1 | 371.2 |
Fragments present in Fig. 3B and predicted structures that support the proposed structure of compound 18 present in Fig. 3C.
PS, pentasaccharide. See Table 4, note b, for other abbreviations.
A doubly charged ion.
The loss of an H2O, apparently from a cyclization of the peptide side chain, appeared regularly in our fragmentation patterns of this and other previously well-defined muropeptides.
TABLE 6.
C. perfringens spore PG structural parametersa
| Strain | % Muramic acid withb:
|
||||||
|---|---|---|---|---|---|---|---|
| Lactam ring | No modificationc | TP | TriP | DP | Cross-link | Deacetylation of NAGd | |
| NCTC 8239 | 48.7 | 2.2 | 38.5 | 6.4 | 4.3 | 1.9 | 9.2 |
| NCTC 8679 | 50.0 | 1.2 | 38.6 | 6.2 | 4.2 | 1.8 | 9.6 |
| SM101 | 50.1 | 2.0 | 42.9 | 3.2 | 2.4 | 1.4 | 11.1 |
| NCTC 10240 | 49.6 | 1.8 | 41.1 | 4.1 | 3.6 | 1.8 | 9.0 |
| 3663 | 48.2 | 2.6 | 37.0 | 7.5 | 5.1 | 1.4 | 9.5 |
| FD1 | 47.9 | 2.5 | 37.0 | 11.4 | 1.6 | 2.0 | 9.5 |
| T65 | 48.3 | 2.7 | 37.7 | 7.9 | 3.9 | 1.8 | 9.0 |
| ATCC 3624 | 49.6 | 2.2 | 34.6 | 10.3 | 3.4 | 1.5 | 11.0 |
| 13V1 | 44.3 | 4.8 | 40.9 | 8.2 | 2.3 | 1.3 | 9.2 |
All values are averages of two analyses of independent spore preparations. The SD of each value was <5%.
See Table 4, note b, for abbreviations.
NAM present with no peptide side chain or lactam ring formation.
NAG, N-acetylglucosamine.
DISCUSSION
In this study, we selected five CPE+ strains and four CPE− strains of C. perfringens and evaluated the spores formed by these strains for factors known to affect heat resistance in other spore-forming species. While all of our CPE+ strains were more heat resistant than the CPE− strains, the division between the groups was not as great as observed previously. The D values at 90°C for our CPE+ strains ranged from 120 to 19 min, while those for the CPE− strains were between 13 and 5 min. Ando et al., (1) found D values at 95°C from 63 to 17 min for CPE+ and 3 to 1 min for CPE− strains. Sarker et al., (33) determined D values at 100°C of 124 to 30 min for strains with chromosomal cpe genes and 2 to 1 min for those with plasmid-borne cpe genes. The smaller difference between CPE+ and CPE− strain D values that we observed may have been due to the lower temperature we used. The broad range of D values across the strains reflects variation in the many factors that can affect heat resistance, as revealed by the measurements of spore structures and contents we obtained.
In B. subtilis, mutations or processes that decrease the spore core wet density by increasing the core water content lower the spores' resistance to wet heat (reviewed in reference 34). Within spores of a single strain prepared under varied conditions and within a set of isogenic strains, this relationship can approach linearity, but between strains and species, the results are less uniform, though a general trend is still clear (5). While our most heat-resistant strains, NCTC 8239 and NCTC 8679, possessed the highest core densities and our least heat-resistant strain, 13V1, had the lowest core density, several other strains varied from this trend. Most notably, the CPE− strains ATCC 3624 and T-65 possessed a high degree of dehydration and lower relative heat resistance. Overall, the average density of the CPE+ strains was very similar to that of the CPE− strains. These results indicate that spore core density is not always a good indicator of heat resistance in this species and that other factors can play significant roles in determining heat resistance.
A previous study found that the endospores formed by CPE+ C. perfringens strains demonstrated a large degree of variation in the core diameter and in the thickness of the cortex and coat layers (27). A similar analysis of our strains supported these findings. The three most heat-resistant strains possessed the smallest spore core diameters, and the thickest cortex belonged to the most heat-resistant strain. This may reflect the previous observation that across several sporulating species a decrease in the ratio of protoplast to sporoplast volume correlated with an increase in spore heat resistance (5). Among these factors, a low P/S ratio appears to be the most essential to the establishment of heat-resistant spores in C. perfringens.
In B. subtilis, it has been shown that decreasing the amount of DPA in a spore results in reduced heat resistance (28). While this relationship is apparent within a single strain that has been either depleted of DPA (7) or mutated to incorporate less DPA (28), this relationship has never been clearly demonstrated between strains or species. In all nine strains examined, DPA and Ca2+ concentrations correlated strongly with one another, with an r2 value of 0.99. This is not surprising, given the strong interaction between Ca2+ and the chelator DPA. NCTC 8679 was previously shown to possess extremely high levels of DPA (27), and our results verified this observation. While the average concentrations of DPA and Ca2+ among the CPE+ strains were more than twice as high as those among the CPE− strains, the great variability in concentrations within each group resulted in no correlation between these compounds and D values. This suggests that as long as a sufficient level of Ca2+ and DPA are present, other factors play greater roles in determining heat resistance.
Spore core Mg2+ and Fe2+ concentrations were also found to be higher in the CPE+ strains than in CPE− strains. The average Mg2+ concentration of the CPE+ strains was nearly twice that of the other strains, while the CPE+ strains had Fe2+ concentrations threefold higher. In C. botulinum, it was shown that sporulation in iron-deficient media led to production of spores with decreased heat resistance (37), though treatment of spores to incorporate dramatically higher levels of Fe2+ also led to a decrease in heat resistance (15). While a mechanism for this phenomenon is not clear, these studies suggested that Fe2+ levels in an endospore must remain within a relatively narrow range of concentrations, with both increases and decreases outside of this range negatively impacting heat resistance. Strain ATCC 3624 was found to possess a concentration of Fe2+ that was relatively low in comparison to those of DPA and the other solutes. Perhaps this helps to explain why the heat resistance of this CPE− strain is comparatively low, despite its relatively low P/S ratio and high core density.
The cortex PG structure was highly conserved among the nine strains tested and was slightly different from those of species previously examined (3, 29). The most notable difference from other species was the complete lack of single l-alanine side chains attached to NAM residues. The enzyme that cleaves peptide side chains to single l-Ala in B. subtilis is LytH (13), and we could find no gene in the sequenced C. perfringens genomes with a high degree of similarity to lytH. The result of failure to cleave peptides to single l-alanine residues is a nearly twofold increase in the number of tetrapeptide side chains in C. perfringens compared to other species previously studied (3, 29). Despite the increased number of peptide side chains available for cross-linking, the average percent cross-linking in C. perfringens spore PG is ∼1.7%; lower than that found in B. subtilis (2.4%) (4, 25) and Bacillus megaterium (2.2%) (2, 30).
Although many parameters of the cortex structure are very similar among these strains, the abundance of tripeptide side chains is significantly higher in the CPE− strains than in the CPE+ strains. In B. subtilis, tripeptide side chains are a characteristic of the germ cell wall rather than the cortex PG (4, 25). A high level of tripeptides in some strains may indicate that a larger percentage of the PG layer measured in our electron micrographs was germ cell wall as opposed to true cortex. This may help to explain the relative thickness of the cortex layers found in the CPE− strains that does not seem to contribute correspondingly to heat resistance.
A novel characteristic of the C. perfringens cortex structure is the high degree of de-N-acetylation present in the muropeptide structure. Nearly 10% of the glucosamine in the cortex was lacking an N-acetyl group compared to approximately 3% in B. megaterium (2). In the final stage of muropeptide preparation, the PG was digested with a muramidase. This enzyme cuts adjacent to NAM but does not act adjacent to muramic-δ-lactam residues. Thus, hexasaccharides result from the presence of two adjacent muramic-δ-lactam residues, and octasaccharides result from three adjacent muramic-δ-lactam residues. It is interesting that while up to 10% of the tetrasaccharides were deacetylated, the vast majority of the hexasaccharides lacked an acetyl group (data not shown) and 100% of the octasaccharides detected were deacetylated. This suggests that deacetylation of a glucosamine contributes to an increased likelihood of lactam ring formation on the adjacent NAM. The average degrees of deacetylation were nearly identical in the CPE+ and CPE− strains and did not appear to be a factor affecting heat resistance in these strains.
Table 7 presents a summary of the spore characteristics that appear to correlate with high heat resistance. The mean and SD for each factor was determined based on the values found for all nine strains. The two most heat-resistant strains, NCTC 8239 and 8679, exceeded the mean value in nearly every factor assessed in this study. Every CPE− strain fell more than 1 SD below the mean in at least one category, though the factor was not always the same one. It is interesting that strain FD-1 fell below the mean in every category yet was not the most heat-sensitive strain we assayed. Clearly, other factors contribute to the establishment of heat resistance in this strain.
TABLE 7.
Requirements for establishing a highly heat-resistant C. perfringens sporea
| Strain | High core densityb | Small core volc | Low P/S ratiod | High DPAe | High Fef | PG with low % TriPg | Total |
|---|---|---|---|---|---|---|---|
| NCTC 8239 | +1 | +1 | +1 | +1 | +1 | 0 | +5 |
| NCTC 8679 | +1 | 0 | +2 | +1 | +1 | 0 | +5 |
| SM101 | 0 | 0 | 0 | 0 | 0 | +1 | +1 |
| NCTC 10240 | 0 | 0 | 0 | 0 | 0 | +1 | +1 |
| 3663 | 0 | −1 | 0 | 0 | 0 | 0 | −1 |
| FD1 | 0 | −1 | 0 | −1 | 0 | −1 | −3 |
| T65 | 0 | 0 | 0 | 0 | −1 | 0 | −1 |
| ATCC 3624 | 0 | 0 | 0 | +1 | 0 | −1 | 0 |
| 13V1 | −1 | 0 | 0 | 0 | 0 | 0 | −1 |
Each value is based on the mean value and SD for all strains assayed. +1 indicates that the value for that particular strain was above 1 SD but less than 2 SDs above the mean; −1 indicates a value more than 1 SD but less than 2 SDs below the mean.
The mean density for all strains assayed was 1.314 ± 0.012 g/ml.
The mean core volume was 0.44 ± 0.13 μm3.
The mean protoplast/sporoplast ratio was 0.45 ± 0.4.
Due to the outlying DPA content of NCTC 8679 (>2.5 SDs above the mean), the value was excluded for calculating the mean DPA and Fe content. The mean value for DPA in the remaining strains was 29.7 ± 18.8 fmol/μm3.
The mean value for Fe2+ was 0.9 ± 0.7 fmol/μm3.
The mean value for % TriP was 7.23 ± 2.6.
A persistent question remaining in the field is what is the selective advantage to maintaining chromosomal cpe genes and the ability to produce highly heat-resistant spores. Our first thought is that high heat resistance and accompanying cold resistance (20) are simply measures of long-term stability at ambient temperatures that C. perfringens might encounter in the environment (soil and aquatic reservoirs). We consider that the broad range of C. perfringens strains may reflect three separable populations. A population that stably inhabits environmental samples, as opposed to gut flora, is the CPE− strains that are the cause of many gas gangrene infections. If these strains occupy a stable niche in the soil, then CPE expression or long-term spore stability may not be selected for. This is consistent with a recent report that the majority of soil isolates are CPE− (21). The CPE+ isolates found in that study had plasmid-borne cpe genes and were thus not likely reservoirs for contaminants leading to AFP.
Another population is human gut flora isolates, which can be further divided into two classes: ≥95% of gut isolates are CPE−, and the remainder are CPE+ (9). This uneven ratio may be determined by negative selection against CPE by the host immune system and a selective advantage in competition for the small population that maintains CPE. During any transfer between the gut floras of individual hosts, long-term spore stability would provide a selective advantage for the smaller number of CPE+ spores. This could therefore drive the coinheritance of chromosomal cpe and high heat resistance. These strains, though rare among the total C. perfringens population, would be frequently encountered in cases of AFP simply due to their ability to survive cooking and refrigeration temperatures.
We have identified several spore properties that appear to play important roles in determining C. perfringens spore heat resistance. Further insight into the relative importance of these factors may be obtained by genetically altering a strain to modify these properties within a consistent genetic background.
Acknowledgments
This project was supported by the National Research Initiative of the USDA Cooperative State Research, Education and Extension Service, grant number 2004-04069.
We thank Katie Sucre and Jessica McElligott for technical assistance and the Soil Testing Laboratory at Virginia Tech for ICP spectroscopy analysis.
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Ando, Y., T. Tsuzuki, H. Sunagawa, and S. Oka. 1985. Heat resistance, spore germination, and enterotoxigenicity of Clostridium perfringens. Microbiol. Immunol. 29:317-326. [DOI] [PubMed] [Google Scholar]
- 2.Atrih, A., G. Bacher, R. Korner, G. Allmaier, and S. J. Foster. 1999. Structural analysis of Bacillus megaterium KM spore peptidoglycan and its dynamics during germination. Microbiology 145:1033-1041. [DOI] [PubMed] [Google Scholar]
- 3.Atrih, A., and S. J. Foster. 2001. Analysis of the role of bacterial endospore cortex structure in resistance properties and demonstration of its conservation amongst species. J. Appl. Microbiol. 91:364-372. [DOI] [PubMed] [Google Scholar]
- 4.Atrih, A., and S. J. Foster. 1999. The role of peptidoglycan structure and structural dynamics during endospore dormancy and germination. Antonie van Leeuwenhoek 75:299-307. [DOI] [PubMed] [Google Scholar]
- 5.Beaman, T. C., J. T. Greenamyre, T. R. Corner, H. S. Pankratz, and P. Gerhardt. 1982. Bacterial spore heat resistance correlated with water content, wet density, and protoplast/sporoplast volume ratio. J. Bacteriol. 150:870-877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borriello, S. P. 1995. Clostridial disease of the gut. Clin. Infect. Dis. 20(Suppl. 2):S242-S250. [DOI] [PubMed] [Google Scholar]
- 7.Church, B. D., and H. Halvorson. 1959. Dependence of the heat resistance of bacterial endospores on their dipicolinic acid content. Nature 183:124-125. [DOI] [PubMed] [Google Scholar]
- 8.Collie, R. E., and B. A. McClane. 1998. Evidence that the enterotoxin gene can be episomal in Clostridium perfringens isolates associated with non-food-borne human gastrointestinal diseases. J. Clin. Microbiol. 36:30-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daube, G., P. Simon, B. Limbourg, C. Manteca, J. Mainil, and A. Kaeckenbeeck. 1996. Hybridization of 2,659 Clostridium perfringens isolates with gene probes for seven toxins (alpha, beta, epsilon, iota, theta, mu, and enterotoxin) and for sialidase. Am. J. Vet. Res. 57:496-501. [PubMed] [Google Scholar]
- 10.Duncan, C. L., and D. H. Strong. 1968. Improved medium for sporulation of Clostridium perfringens. Appl. Microbiol. 16:82-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duncan, C. L., H. Sugiyama, and D. H. Strong. 1968. Rabbit ileal loop response to strains of Clostridium perfringens. J. Bacteriol. 95:1560-1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hall, H. E., R. Angelotti, K. H. Lewis, and M. J. Foter. 1963. Characteristics of Clostridium perfringens strains associated with food and food-borne disease. J. Bacteriol. 85:1094-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horsburgh, G. J., A. Atrih, and S. J. Foster. 2003. Characterization of LytH, a differentiation-associated peptidoglycan hydrolase of Bacillus subtilis involved in endospore cortex maturation. J. Bacteriol. 185:3813-3820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janssen, F. W., A. J. Lund, and L. E. Anderson. 1958. Colorimetric assay for dipicolinic acid in bacterial spores. Science 127:26-27. [DOI] [PubMed] [Google Scholar]
- 15.Kihm, D. J., M. T. Hutton, J. H. Hanlin, and E. A. Johnson. 1990. Influence of transition metals added during sporulation on heat resistance of Clostridium botulinum 113B spores. Appl. Environ Microbiol. 56:681-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kokai-Kun, J. F., J. G. Songer, J. R. Czeczulin, F. Chen, and B. A. McClane. 1994. Comparison of Western immunoblots and gene detection assays for identification of potentially enterotoxigenic isolates of Clostridium perfringens. J. Clin. Microbiol. 32:2533-2539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kozuka, S., and K. Tochikubo. 1983. Triple fixation of Bacillus subtilis dormant spores. J. Bacteriol. 156:409-413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Labbe, R. G. 1981. Enterotoxin formation by Clostridium perfringens type A in a defined medium. Appl. Environ Microbiol. 41:315-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Le Blanc, J. C., J. W. Hager, A. M. Ilisiu, C. Hunter, F. Zhong, and I. Chu. 2003. Unique scanning capabilities of a new hybrid linear ion trap mass spectrometer (Q. TRAP) used for high sensitivity proteomics applications. Proteomics 3:859-869. [DOI] [PubMed] [Google Scholar]
- 20.Li, J., and B. A. McClane. 2006. Further comparison of temperature effects on growth and survival of Clostridium perfringens type A isolates carrying a chromosomal or plasmid-borne enterotoxin gene. Appl. Environ Microbiol. 72:4561-4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, J., S. Sayeed, and B. A. McClane. 2007. Prevalence of enterotoxigenic Clostridium perfringens isolates in Pittsburgh (Pennsylvania) area soils and home kitchens. Appl. Environ Microbiol. 73:7218-7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsay, J. A., T. C. Beaman, and P. Gerhardt. 1985. Protoplast water content of bacterial spores determined by buoyant density sedimentation. J. Bacteriol. 163:735-737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McClane, B. A. 2005. Clostridial enterotoxins, p. 385-406. In P. Durre (ed.), Handbook on clostridia. Taylor & Francis Group, Boca Raton, FL.
- 24.Mead, P. S., L. Slutsker, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:841-842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meador-Parton, J., and D. L. Popham. 2000. Structural analysis of Bacillus subtilis spore peptidoglycan during sporulation. J. Bacteriol. 182:4491-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Myers, G. S., D. A. Rasko, J. K. Cheung, J. Ravel, R. Seshadri, R. T. DeBoy, Q. Ren, J. Varga, M. M. Awad, L. M. Brinkac, S. C. Daugherty, D. H. Haft, R. J. Dodson, R. Madupu, W. C. Nelson, M. J. Rosovitz, S. A. Sullivan, H. Khouri, G. I. Dimitrov, K. L. Watkins, S. Mulligan, J. Benton, D. Radune, D. J. Fisher, H. S. Atkins, T. Hiscox, B. H. Jost, S. J. Billington, J. G. Songer, B. A. McClane, R. W. Titball, J. I. Rood, S. B. Melville, and I. T. Paulsen. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031-1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novak, J. S., V. K. Juneja, and B. A. McClane. 2003. An ultrastructural comparison of spores from various strains of Clostridium perfringens and correlations with heat resistance parameters. Int. J. Food Microbiol. 86:239-247. [DOI] [PubMed] [Google Scholar]
- 28.Paidhungat, M., B. Setlow, A. Driks, and P. Setlow. 2000. Characterization of spores of Bacillus subtilis which lack dipicolinic acid. J. Bacteriol. 182:5505-5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Popham, D. L., J. Helin, C. E. Costello, and P. Setlow. 1996. Analysis of the peptidoglycan structure of Bacillus subtilis endospores. J. Bacteriol. 178:6451-6458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Popham, D. L., B. Illades-Aguiar, and P. Setlow. 1995. The Bacillus subtilis dacB gene, encoding penicillin-binding protein 5*, is part of a three-gene operon required for proper spore cortex synthesis and spore core dehydration. J. Bacteriol. 177:4721-4729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raju, D., and M. R. Sarker. 2005. Comparison of the levels of heat resistance of wild-type, cpe knockout, and cpe plasmid-cured Clostridium perfringens type A strains. Appl. Environ Microbiol. 71:7618-7620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sacks, L. E., and P. A. Thompson. 1978. Clear, defined medium for the sporulation of Clostridium perfringens. Appl. Environ Microbiol. 35:405-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sarker, M. R., R. P. Shivers, S. G. Sparks, V. K. Juneja, and B. A. McClane. 2000. Comparative experiments to examine the effects of heating on vegetative cells and spores of Clostridium perfringens isolates carrying plasmid genes versus chromosomal enterotoxin genes. Appl. Environ Microbiol. 66:3234-3240. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Setlow, P. 2006. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J. Appl. Microbiol. 101:514-525. [DOI] [PubMed] [Google Scholar]
- 35.Shih, N. J., and R. G. Labbe. 1996. Sporulation-promoting ability of Clostridium perfringens culture fluids. Appl. Environ Microbiol. 62:1441-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu, T., K. Ohtani, H. Hirakawa, K. Ohshima, A. Yamashita, T. Shiba, N. Ogasawara, M. Hattori, S. Kuhara, and H. Hayashi. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. USA 99:996-1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sugiyama, H. 1951. Studies on factors affecting the heat resistance of spores of Clostridium botulinum. J. Bacteriol. 62:81-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swerdlow, B. M., B. Setlow, and P. Setlow. 1981. Levels of H+ and other monovalent cations in dormant and germinating spores of Bacillus megaterium. J. Bacteriol. 148:20-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tang, H., Y. Mechref, and M. V. Novotny. 2005. Automated interpretation of MS/MS spectra of oligosaccharides. Bioinformatics 21(Suppl. 1):i431-i439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warth, A. D., and J. L. Strominger. 1969. Structure of the peptidoglycan of bacterial spores: occurrence of the lactam of muramic acid. Proc. Natl. Acad. Sci. USA 64:528-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wen, Q., K. Miyamoto, and B. A. McClane. 2003. Development of a duplex PCR genotyping assay for distinguishing Clostridium perfringens type A isolates carrying chromosomal enterotoxin (cpe) genes from those carrying plasmid-borne enterotoxin (cpe) genes. J. Clin. Microbiol. 41:1494-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wnek, A. P., R. J. Strouse, and B. A. McClane. 1985. Production and characterization of monoclonal antibodies against Clostridium perfringens type A enterotoxin. Infect. Immun. 50:442-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao, Y., and S. B. Melville. 1998. Identification and characterization of sporulation-dependent promoters upstream of the enterotoxin gene (cpe) of Clostridium perfringens. J. Bacteriol. 180:136-142. [DOI] [PMC free article] [PubMed] [Google Scholar]