Abstract
Cyanophycin [multi-l-arginyl-poly(l-aspartic acid) (CGP)] was, for the first time, produced in yeast. As yeasts are very important production organisms in biotechnology, it was determined if CGP can be produced in two different strains of Saccharomyces cerevisiae. The episomal vector systems pESC (with the galactose-inducible promoter GAL1) and pYEX-BX (with the copper ion-inducible promoter CUP1) were chosen to express the cyanophycin synthetase gene from the cyanobacterium Synechocystis sp. strain PCC 6308 (cphA6308) in yeast. Expression experiments with transgenic yeasts revealed that the use of the CUP1 promoter is much more efficient for CGP production than the GAL1 promoter. As observed by electrophoresis of isolated CGP in sodium dodecyl sulfate-polyacrylamide gels, the yeast strains produced two different types of polymer: the water-soluble and the water-insoluble CGP were observed as major and minor forms of the polymer, respectively. A maximum CGP content of 6.9% (wt/wt) was detected in the cells. High-performance liquid chromatography analysis showed that the isolated polymers consisted mainly of the two amino acids aspartic acid and arginine and that, in addition, a minor amount (2 mol%) of lysine was present. Growth of transgenic yeasts in the presence of 15 mM lysine resulted in an incorporation of up to 10 mol% of lysine into CGP. Anti-CGP antibodies generated against CGP isolated from Escherichia coli TOP10 harboring cphA6308 reacted with insoluble CGP but not with soluble CGP, if applied in Western or dot blots.
Cyanophycin, also referred to as multi-l-arginyl-poly(l-aspartic acid) or cyanophycin granule polypeptide (CGP), is a nonribosomally synthesized polypeptide consisting of a poly-(aspartic acid) backbone with arginine residues linked to the β-carboxyl group of each aspartate by the α-amino group (45). CGP is a polydispersed polymer; the molecular size distribution of CGP varies with the producing host strains (1, 15, 18, 20, 30, 37, 52). Due to its branched structure, CGP is not degradable by a wide range of proteinases (45). Biosynthesis of CGP from aspartate and arginine requires only one enzyme, cyanophycin synthetase, which is encoded by cphA (51). CGP is insoluble at neutral pH and under physiological ionic strength, but it is soluble at low (>3) or high (<9) pH. Ziegler et al. were the first to observe a water-soluble form of CGP after the heterologous expression of cphA from Desulfitobacterium hafniense strain DSM 10664 in Escherichia coli (52). A detailed study of the solubility behavior of CGP isolated from recombinant E. coli in inorganic salts has been carried out by Füser and Steinbüchel (16). It was shown that the occurrence of the soluble form was not dependent on the origin of cphA or on the host.
Recently, several putative applications for CGP and its derivatives have become available, indicating there is a need for its efficient biotechnological production (36, 42, 43). For the production of CGP at a technical scale, cyanobacteria were shown to be unsuitable due to their low cell densities and polymer contents (3.5% [wt/wt]) and slow growth and circumstantial growth conditions in a photobioreactor (18, 19). In contrast, much higher amounts of the polymer were produced with the heterotrophic bacterium Acinetobacter baylyi strain ADP1 (46% [wt/wt]) (14) and with recombinant strains of E. coli (24% [wt/wt] [15] and 34.5% [wt/wt] [21], respectively). Also, in the industrially relevant bacteria Pseudomonas putida, Ralstonia eutropha, and Corynebacterium glutamicum, considerable amounts of CGP could be produced after heterologous expression of cphA (3, 13, 48, 49). Recently, CGP production was also achieved for the first time in eukaryotic organisms. Transgenic tobacco plants accumulated up to 1.14% (wt/wt) and transgenic potato plants up to 0.24% (wt/wt) of water-soluble and water-insoluble CGP (37).
In the last century, yeasts have evolved into biotechnologically relevant production organisms for several products in industry. First, yeast systems that were developed for heterologous gene expression were based on Saccharomyces cerevisiae. This organism is traditionally used for large-scale production of baker's yeast and ethanol, with a considerable increase in the production of fuel ethanol in the last 3 decades (4, 6, 50). Additionally, this platform has been successfully applied to the production of valuable heterologous proteins on an industrial scale (26, 35), such as various FDA-approved pharmaceuticals, including insulin (34) and hepatitis B surface antigen (23). Yeast-based expression systems excel because of their available constitutive or strongly inducible promoters and their growth to high cell densities on inexpensive substrates. The range of today's established yeast expression systems includes S. cerevisiae, Kluyveromyces lactis, Pichia pastoris, Yarrowia lipolytica, Arxula adeninivorans, and Hansenula polymorpha (8, 17). Polymers such as human collagen and recombinant gelatin have also been produced successfully in several yeast strains (7, 12, 38, 47). The present study describes for the first time the expression of a cyanophycin synthetase gene and the successful production of CGP in yeast. These experiments emphasize the potential for biotechnological production of CGP in these industrially relevant organisms.
MATERIALS AND METHODS
Strains, media, and growth conditions.
All bacteria, yeast strains, and plasmids used in this study are listed in Table 1. E. coli XL1-Blue was used for plasmid maintenance and propagation and was grown in Luria-Bertani (LB) medium containing ampicillin (100 mg/liter) overnight at 37°C and agitated at 300 rpm. Yeast strains were cultivated in a rich YPD medium (1% [wt/vol] yeast extract, 2% [wt/vol] peptone, 2% [wt/vol] glucose) or in selective medium (0.67% [wt/vol] amino-acid-free Difco yeast nitrogen base containing 2% [wt/vol] galactose or glucose and supplemented as required with l-leucine [100 mg/liter], l-methionine [20 mg/liter], l-histidine [20 mg/liter], and l-tryptophan [20 mg/liter]). Solid medium contained 2% (wt/wt) agar for the growth of yeasts and E. coli. Yeasts were grown in 50-ml Erlenmeyer flasks for 12 to 48 h at 30°C and agitated at 300 rpm.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference or source |
|---|---|---|
| E. coli XL1-Blue | recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 λ−lac [F′ proAB lacIqlacZΔM15 Tn10 (Tcr)] | 11 |
| E. coli TOP10 | recA1 endA1 gyrA96 thi-1 hsdR17 (rK− mK+) supE44 relA1 ΔlacU169 | Invitrogen |
| E. coli BL21(DE3) | F′ ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| S. cerevisiae G175 | MATα ADE2 MET his3 leu2 ura3 trp1 TAG+ SE+ | Scandinavian Biotechnology Research (41) |
| S. cerevisiae BY4741 | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | 9 |
| pSK::cphAco | Apr; cphA from Synechocystis sp. strain PCC 6308 cloned as an HpaI fragment into pBluescript SK− (Stratagene) colinear to the lac mutant promoter | 1 |
| pET-23a::cphA6308 | Apr; cphA from Synechocystis sp. strain PCC 6308 cloned as an NdeI-XhoI fragment into pET-23a (Novagen) colinear to the T7 promoter | Steinle et al., unpublished data |
| pESC-URA | E. coli-S. cerevisiae shuttle vector; URA3 GAL1 GAL10 Apr | Stratagene |
| pYEX-BX | E. coli-S. cerevisiae shuttle vector; URA3 LEU2 CUP1 Apr | Clontech Laboratories |
| pESC-URA::cphA6308 | cphA from Synechocystis sp. strain PCC 6308 with an artificial Kozak initiation sequence cloned as a 2.6-kbp BamHI-SalI fragment into pESC-URA colinear to the GAL1 promoter | This study |
| pYEX-BX::cphA6308 | cphA from Synechocystis sp. strain PCC 6308 with an artificial Kozak initiation sequence cloned as a 2.6-kbp BamHI-SalI fragment into pESC-URA colinear to the CUP1 promoter | This study |
Apr, ampicillin resistance; Tcr, tetracycline resistance.
Transfer of DNA.
Competent cells of E. coli were obtained using the CaCl2 procedure (40), and transformation of the cells was carried out according to the method described by Hanahan (22). Yeast transformation was performed by the lithium acetate procedure (28). Transformants were selected on minimal glucose medium lacking either uracil or leucine but supplemented for the other auxotrophic requirements of the respective yeast strain.
General DNA techniques.
Isolation of plasmids from E. coli was carried out using the alkaline lysis method described by Sambrook et al. (40). For DNA endonuclease digestion, standard ligation and agarose gel electrophoresis protocols were used (40). For the recovery of DNA fragments after electrophoresis, an Eppendorf Perfectprep gel clean-up kit was used by following the manufacturers instructions. Sequences of constructs were verified by capillary DNA sequencing (Universitätsklinikum, Münster, Germany). Isolation of total DNA from transgenic yeast was performed according to a method described by Kaiser et al. (29). For verification of the presence of cphA6308, PCRs with specific oligonucleotides (5′-AAAAGGATCCACTATGAAAATCCTCAAAACACAAACCC-3′ and 5′-TTTGTCGACCTATTCACTACTGAGATGATATTTCTCAATCATC-3′) as primers and with total DNA from transgenic yeasts as templates were carried out.
Cloning of cphA.
For cloning cphA6308 into the E. coli-yeast shuttle vectors pESC-URA and pYEX-BX (Table 1), PCR was done with Pfx DNA polymerase (Gibco BRL) according to the manufacturer's instructions, by using the oligonucleotides cphA-fw-BamHI (5′-AAAGGATCCACTATGAAAATCCTCAAAACACAAACCC-3′) as the sense and cphA-rw-SalI (5′-TTTGTCGACCTATTCACTACTGAGATGATATTTCTCAATCATC-3′) as the reverse primers. Thereby, an artificial Kozak site (underlined) upstream of the start codon, a BamHI restriction site in the upstream region and a SalI restriction site in the downstream region of cphA6308, were introduced. Plasmid pET-23a::cphA6308 (Table 1) was used as the template. Subsequently, the PCR products were cloned into the BamHI-SalI-treated E. coli-yeast shuttle vectors, yielding pESC-URA::cphA6308 and pYEX-BX::cphA6308, respectively.
Cell disruption and determination of CDM.
Yeast cells were harvested by using a bench centrifuge (5 min, 3,000 rpm, 4°C), and cell pellets were washed once with saline (0.9% [wt/vol] NaCl). For determination of the cell dry matter (CDM), pellets were lyophilized, and the cell mass was determined gravimetrically. For cell disruption, the cell pellet was resuspended in 1 ml buffer (20 mM Tris-HCl [pH 7.5]) per g of fresh or dry cell mass and disrupted for 5 min by a bead mill (type MM 301; Retsch, Haan, Germany). Soluble cell fractions were obtained by centrifugation of crude cell extracts (10 min, 13,000 rpm, 4°C).
Determination of protein concentrations.
Protein concentrations were determined using the methods described by Bradford (10) and Lowry et al. (32). Soluble cell fractions were used for the determination and were obtained as described above.
Cyanophycin synthetase assay.
The cyanophycin synthetase enzyme assay followed the procedure described by Aboulmagd et al. (1). Soluble cell fractions and crude cell extracts were used to determine the enzyme activity. Scintillation counting was carried out using a model LS 6500 scintillation counter (Beckman Instruments GmbH, München, Germany).
Isolation of CGP.
For the isolation of CGP, yeast cells were disrupted as described above. The crude cell extract obtained after cell disruption was centrifuged for 10 min at 13,000 rpm at 4°C. The supernatant was used for the isolation of water-soluble CGP by applying a modified method described by Ziegler et al. (52), using heat treatment, proteinase K digestion, and precipitation with 3 volumes of ethanol. After samples were subjected to proteinase K digestion, they were applied to Vivaspin 20 concentrators (Vivascience AG, Hannover, Germany), with a 10-kDa membrane to remove low-molecular-weight substances. CGP was subsequently precipitated with ethanol and washed once with acetone. Water-insoluble CGP was isolated from the cell debris by resuspending the sample in 0.1 M HCl. After the suspension was centrifuged (15 min, 13,000 rpm), the supernatant was neutralized by adding NaOH. After another centrifugation step, the polymer was obtained from the pellet and washed twice with demineralized water and lyophilized to determine the dry weight. Isolation of CGP from E. coli BL21(DE3)(pET-23a::cphA6308) was performed as described by Frey et al. (15).
Characterization of CGP.
The amino acid constituents of the water-soluble and water-insoluble CGP isolated from the transgenic yeasts G175(pESC-URA::cphA6308), G175(pYEX-BX::cphA6308), and BY4741(pYEX-BX::cphA6308) were determined by high-performance liquid chromatography (HPLC) (1). Calibration was done with samples from an amino acid reference kit (Kollektion AS-10 from Serva Feinbiochemica, Heidelberg, Germany).
Electrophoretic methods.
Analysis of polymers by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed in 11.5% (wt/vol) acrylamide gels according to the method described by Laemmli (31). Prestained standard molecular weight proteins were purchased from Fermentas; the applied mixture contained β-galactosidase from E. coli (117 kDa), bovine serum albumin from bovine plasma (90 kDa), ovalbumin from chicken egg white (49 kDa), carbonic anhydrase from bovine erythrocytes (35 kDa), β-lactoglobulin from bovine milk (26 kDa), and lysozyme from chicken egg white (19 kDa). Proteins and CGP-like material were stained with Serva Blue R. Protein concentrations were determined as described by Hartree (24).
Immunological methods.
For the preparation of anti-CGP antiserum, cells of E. coli TOP10(pSK::cphAco) (Table 1) were grown in LB medium under ampicillin selection (100 μg/ml). After the cells were harvested by centrifugation for 15 min at 4°C, they were washed once with 0.9% (wt/vol) NaCl and disrupted by using a French pressure cell with 1,000 mPa (Amicon, Silver Spring, MD). Subsequently, CGP was isolated according to the method described by Simon (44). Purified CGP (100 mg/ml) was used for the generation of custom polyclonal antibodies in rabbits by Eurogentec (Seraing, Belgium). The immunoglobulin G (IgG) fraction was purified by affinity chromatography using protein A-Sepharose CL-4B (27). To purify anti-CGP-specific IgG, CGP was blotted onto a Hybond-P membrane (Amersham Biosciences). The membrane was incubated in 2.5% (wt/vol) skim milk in TBS buffer (8% [wt/vol] NaCl; 2% [vol/vol] Tris/HCl [pH 7.6]) for 1 h. After the membrane was washed three times for 10 min each in TBS, 400 μl of the IgG solution from protein A chromatography was added. After a 3-h incubation, the membrane was washed again, and anti-CGP-specific IgGs were eluted with 1 ml of elution buffer (5 mM glycine [pH 3], 0.5 M NaCl, 0.05% [vol/vol] Tween 20), neutralized with 1 M potassium phosphate buffer (pH 8.0), and stored at −20°C. For immunological detection of CGP, proteins and CGP were transferred from gels onto Hybond-P membranes as described by the manufacturer (Amersham Biosciences). Immunological analysis was performed as described in reference 25 with slight modifications, employing anti-CGP-specific IgGs (diluted 1:100 in TBS buffer). Dot blot experiments were performed as described by the manufacturer of the polyvinylidene difluoride membrane (GE Healthcare, Buckinghamshire, United Kingdom). IgGs were visualized on immunoblots by using anti-rabbit IgG-alkaline phosphatase conjugates (Sigma-Aldrich), converting 5-bromo-4-chloro-3-indolyl-phosphate dipotassium nitrotetrazolium blue chloride (Sigma-Aldrich) into a dark insoluble product.
Isolation of RNA and RT-PCR.
For isolation of RNA, yeast cells were grown in 5 ml of selective medium under induced conditions. Therefore, cells harboring the vector pESC-URA::cphA6308 were grown in minimal medium with 2% (wt/vol) galactose as the inducer for the GAL1 promoter and the carbon source, and cells harboring pYEX-BX::cphA6308 were grown in minimal medium containing 0.1 mM CuSO4 as the inducer for the CUP1 promoter and 2% (wt/vol) glucose as the carbon source. Cells were harvested (3 min, 5,000 rpm) and broken by treatment in a bead mill (type MM 301; Retsch, Haan, Germany). RNA was isolated by using an RNeasy mini-kit (Qiagen, Hilden, Germany) as described by the manufacturer. After RNA was isolated, the remaining DNA was hydrolyzed by DNaseI (Roche Diagnostics, Mannheim, Germany) during an incubation of 45 min at 37°C. To determine if the cphA gene was transcribed in the transgenic yeast cells, reverse transcription-PCR (RT-PCR) was performed as described by the manufacturer (OneStep RT-PCR kit; Qiagen, Hilden, Germany), using cphA-specific oligonucleotides (5′-GCCATCGCTGATGTCGGTGG-3′ and 5′-CGATGGCAATACCCCCGGTAC-3′) as the primers and 0.5 ng RNA as the template. DNA controls were carried out to exclude any DNA contamination.
Transmission electron microscopy (TEM) studies.
Cells were fixed with 2.5% (vol/vol) glutaraldehyde in 0.1 M phosphate-buffered saline (PBS) (pH 7.3) for 45 min. After the cells were washed three times with PBS for 20 min each, they were postfixed in 1% (wt/vol) osmium tetroxide in 0.1 M PBS (pH 7.3) and washed once with PBS for 20 min. Then, the water was removed by using a graded water/ethanol series (30, 50, 70, 90, and 96%, and absolute ethanol) in which each step lasted about 15 min. To obtain thin sections, the samples were embedded in Spurr resin without propylene oxide (46). Sections with a thickness of 70 to 80 nm were cut with an ultramicrotome (Leica Mikroskopie und Systeme, Wetzlar, Germany) by using a diamond knife and were placed on a 200-mesh copper grid. Subsequently, the sections were stained with saturated uranyl acetate solution for 30 min and with a lead citrate solution according to the method described by Reynolds (39) for 3 min. Imaging was performed with an H-500 model TEM (Hitachi, Tokyo, Japan) in the brightfield mode at an acceleration voltage of 80 kV at room temperature.
RESULTS
Generation of transgenic yeasts.
The cyanophycin synthetase gene carried by Synechocystis sp. strain PCC 6308 (cphA6308) was amplified by PCR and ligated into two different E. coli-yeast shuttle vectors to compare the suitabilities of two different inducible promoters for the expression of CphA6308 and the establishment of CGP biosynthesis in S. cerevisiae. In vector pESC, cphA6308 is under the control of a galactose-inducible promoter (GAL1), whereas in vector pYEX-BX, cphA6308 is controlled through the CUP1 promoter, which is induced by the addition of 0.1 mM copper ions. Whereas vector pYEX-BX::cphA6308 mediated uracil and also leucine prototrophy, vector pESC-URA mediated only uracil prototrophy. S. cerevisiae strains G175 and BY4741 (Table 1) were transformed with both generated plasmids (Table 1), yielding transgenic yeast strains G175(pESC-URA::cphA6308), BY4741(pESC-URA::cphA6308), G175(pYEX-BX::cphA6308), and BY4741(pYEX-BX::cphA6308). In addition, the vectors without insertions were also transformed into both yeast strains. When total DNA was isolated from cells of the resulting clones, and when PCR was carried out with cphA-specific primers, all the obtained transformants, except for those transformed with only the vectors, showed specific PCR products, verifying the presence of cphA (data not shown).
Transcriptional analysis of transgenic yeast for cphA.
For expression experiments, cells of transgenic yeasts harboring the plasmid pESC-URA::cphA6308 were grown in minimal medium containing 2% (wt/vol) galactose as the sole carbon source and as the inducer for the GAL1 promoter. In contrast, cells of the transgenic yeasts harboring plasmid pYEX-BX::cphA6308 were grown in minimal medium containing 2% (wt/vol) glucose as the carbon source and 0.1 mM CuSO4 as the inducer for the CUP1 promoter. To determine whether cphA6308 is transcribed in the transgenic yeast cells, RNA was isolated, and RT-PCR was performed. All the obtained transgenic yeasts that were transformed with cphA-containing plasmids showed cphA-specific PCR products, in contrast to that of the DNA control, indicating that cphA is transcribed in these cells (data not shown). Negative controls (strains G175 and BY4741 harboring only the vectors) were also analyzed; they did not show cphA-specific PCR products.
Analysis of CphA enzyme activity in transgenic yeasts.
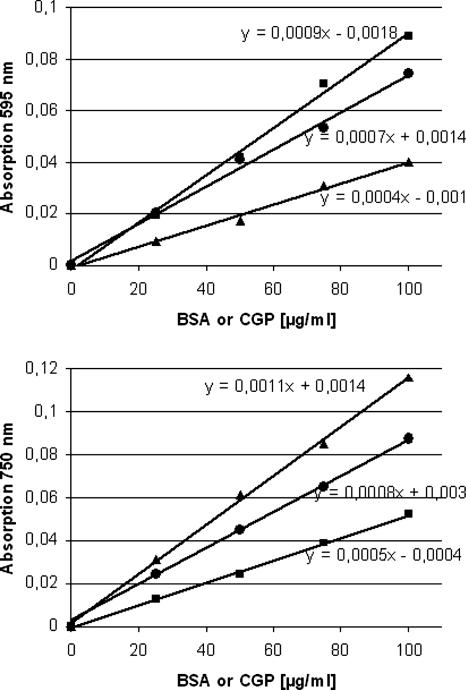
CphA enzyme activity was measured in soluble cell fractions obtained from S. cerevisiae strains G175 and BY4741 harboring pESC-URA::cphA6308, pYEX-BX::cphA6308, or the respective vector controls by employing a radiometric assay. The disintegrations per minute (dpm) were determined to be below 75 dpm for vector controls and the strains BY4741 and G175 harboring pESC-URA::cphA6308, indicating the absence of significant CphA enzyme activity and that pESC-URA::cphA6308 did not confer considerable CphA activity on the strains. In contrast, measurements in soluble cell fractions of strains BY4741 or G175 harboring pYEX-BX::cphA6308 gave 756 or 931 dpm, respectively. Soluble cell fractions from E. coli strain BL21(DE3) harboring pET-23a::cphA6308 were used as positive controls and gave 9,420 dpm. These data clearly demonstrated that pYEX-BX::cphA6308 conferred CphA enzyme activity on both of the investigated yeast strains. Unfortunately, it was not possible to calculate real specific enzyme activities because protein concentrations could not be measured accurately due to the presence of CGP. The presence of CGP in protein samples affected the total protein concentration when it was determined by the Bradford method (10) or that of Lowry et al. (32). CGP gave higher protein values if determined with the Bradford reagent and quencher values if determined with the Lowry reagent, as revealed by standard curves (Fig. 1). Therefore, the real protein values could only be estimated and had to be carefully considered. After the isolation of soluble CGP, the polymer was solubilized in buffer, and the “protein” concentration of this solution was determined according to the Bradford method; the value obtained was subtracted from the protein concentration determined for the respective soluble cell fraction. The values obtained were used to calculate specific enzyme activities, which were 2.00, 0.91, and 0.76 U/mg protein for E. coli BL21(DE3)(pET-23a::cphA6308), S. cerevisiae G175(pYEX-BX::cphA6308), and S. cerevisiae BY4741(pYEX-BX::cphA6308), respectively. Specific activities for vector controls and yeasts harboring pESC-URA::cphA6308 were below 0.02 U/mg protein.
FIG. 1.
Standard curves for protein analysis employing BSA (▴), soluble CGP (▪), and a mixture of BSA and CGP (•). The substances were applied in concentrations of 0 to 100 μg/ml and were solubilized in water. (A) Determination according to the Bradford method (10). (B) Determination according to the method described by Lowry et al. (32). Linear equations are given in the graphs.
Analysis of cell extracts of transgenic yeasts for the presence of CGP.
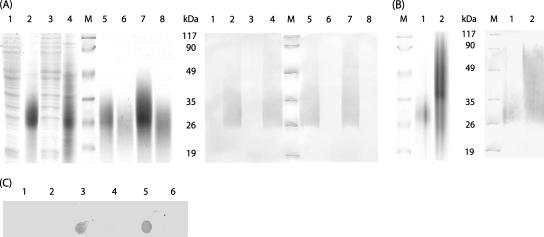
To determine whether the obtained transgenic yeast strains are able to synthesize CGP, we isolated the polymer by different methods and analyzed it by SDS-PAGE. For this procedure, cells were disrupted, and the crude extracts obtained were separated through centrifugation into a soluble cell fraction and the cell debris. To detect the water-soluble form of CGP, the soluble cell fractions were incubated with proteinase K. To detect the water-insoluble CGP, the cell debris fractions were incubated in 0.1 M HCl and centrifuged, and the resulting supernatant was then transferred to a new tube. Aliquots of both obtained solutions were then analyzed by SDS-PAGE (Fig. 2A). The gels clearly indicated that most transgenic yeast strains were producing considerable amounts of water-insoluble and even higher amounts of water-soluble CGP. Only analysis of the transgenic S. cerevisiae strain BY4741 harboring the plasmid pESC-URA::cphA6308 did not reproducibly show any detectable amount of CGP (data not shown). Presumably, the vector pESC-URA was not suitable for the production of significant amounts of CGP in strain BY4741. SDS-PAGE revealed for the polydispersed soluble CGP a molecular mass distribution from 20 to 35 kDa, whereas for the polydispersed insoluble CGP, a slightly higher molecular mass range from about 26 to 45 kDa (Fig. 2A) was found.
FIG. 2.
Analysis and detection of CGP isolated from cells of recombinant strains of S. cerevisiae and E. coli and from cyanobacteria. (A) Yeast cell extracts were analyzed by SDS-PAGE (left) and Western blotting (right). Acidic extracts were obtained by resuspension of cell debris in 0.1 M HCl and centrifugation. Proteinase K fractions were obtained by digestion of soluble cell fractions with proteinase K. In each lane, 10 to 20 μg CGP was applied. Lanes 1 to 4 represent crude extracts from yeast: 1, BY4741 harboring pYEX-BX; 2, BY4741 harboring pYEX-BX::cphA6308; 3, G175 harboring pYEX-BX: 4, G175 harboring pYEX-BX::cphA6308. Lanes 5 to 8 represent soluble and insoluble CGP from yeast: 5, acidic extract from BY4741 harboring pYEX-BX::cphA6308; 6, proteinase K fraction from BY4741 harboring pYEX-BX::cphA6308: 7, acidic extract from G175 harboring pYEX-BX::cphA6308; 8, proteinase K fraction from G175 harboring pYEX-BX::cphA6308. (B) CGP isolated from E. coli BL21(DE3)(pET-23a::cphA6308) and from Synechocystis sp. strain PCC 6308 was analyzed by SDS-PAGE (left) and by immunological detection in Western blotting (right) using anti-CGP IgGs. In each lane, 10 μg CGP was applied. M, protein marker; lane 1, CGP from recombinant E. coli; lane 2, CGP from Synechocystis sp. (C) Acidic fractions and proteinase K fractions from S. cerevisiae analyzed by dot blot employing anti-CGP IgGs. Lanes 1, 3 and 5, acidic fractions; 2, 4 and 6, proteinase K fractions. Lanes: 1 and 2, G175 harboring pYEX-BX; 3 and 4, G175 harboring pYEX-BX::cphA6308; 5 and 6, BY4741 harboring pYEX-BX::cphA6308. CGP (30 μg) was applied for cphA-harboring strains.
Detection of CGP by anti-CGP IgGs antibodies.
To have an additional tool for analysis of CGP and CGP-harboring cells, polyclonal antibodies were raised against CGP isolated and purified from a recombinant strain of E. coli expressing the cyanophycin synthetase from Synechocystis sp. strain PCC 6308, as described in Materials and Methods. Specific immunoreactions (Fig. 2A and B) occurring with Western blots from SDS-PAGE gels, in which 10 μg of CGP solubilized in HCl either from E. coli or Synechocystis sp. PCC 6308 cells was separated, indicated the functionality of these antibodies (Fig. 2B). When different amounts (1, 3, 6, 30, or 60 μg) of CGP isolated from E. coli BL21(DE3)(pET-23a::cphA6308) were applied to an SDS-PAGE gel and blotted, 6 μg of CGP was the minimal detectable amount in the Western blots (data not shown).
Subsequently, the anti-CGP antibodies were applied to crude extracts prepared from transgenic yeasts cells and also to purified CGP isolated from these yeast cells (Fig. 2A). In the applied crude extracts, only CGP and no distinct protein molecules gave an immunoreaction, indicating that anti-CGP IgGs bound specifically to CGP molecules and not to other proteins (Fig. 2A). A strong immunoreaction occurred with insoluble CGP isolated from yeast as it was also observed for CGP isolated from the cells of E. coli or Synechocystis sp. Surprisingly, no immunoreaction at all occurred with the soluble CGP isolated from yeast (Fig. 2A). Due to the solubility behavior of this CGP form, blotting was also carried out by applying three membranes instead of one, to exclude the possibility that soluble CGP passed the first membrane during blotting and was therefore not detectable. However, no immunoreaction occurred on any of the three membranes (data not shown). In addition, dot blot experiments were carried out to confirm the observation that the anti-CPG antibodies used do not react with soluble CGP (Fig. 2C). Obviously, the results obtained in this experiment were the same as those in the Western blotting analysis; therefore, it was concluded that an immunoreaction occurred with insoluble CGP but not with soluble CGP (Fig. 2A and C) and that these antibodies can be used to discriminate between the two forms.
Determination of the amino acid composition.
As observed by SDS-PAGE (Fig. 2A), the transgenic yeasts produced two different types of CGP which are significantly distinguishable by their solubility behavior and their reaction with the anti-CGP antibodies. Both types were isolated by different procedures, as described in Materials and Methods. HPLC analysis of the two CGP types revealed that both types of CGP isolated from the same cells exhibited the same amino acid compositions. CGP isolated from either the BY4741 or the G175 strain, respectively, consisted mainly of aspartic acid or arginine, which occurred at molar fractions of 52 or 46%, respectively. Lysine was detected at a maximum fraction of only 2 mol%.
Microscopy analysis of transgenic yeast.
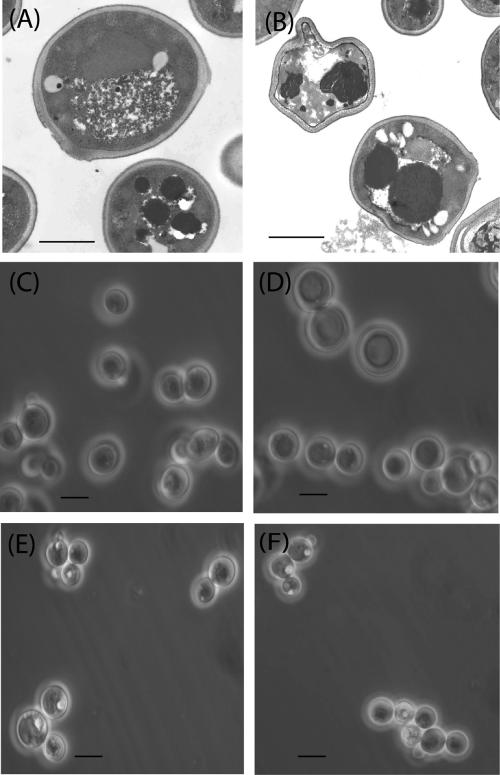
For the visualization of CGP granules in cells of S. cerevisiae strain G175 harboring pESC-URA::cphA6308, phase-contrast light microscopy and electron microscopy were applied (Fig. 3B and D). S. cerevisiae strain G175 harboring pESC-URA was analyzed for comparison in parallel (Fig. 3A and C). Both strains were grown in minimal media containing 2% (wt/vol) galactose as the sole carbon source for 48 h at 30°C. From cells of cphA-carrying strains, water-soluble and water-insoluble forms of CGP were isolated in amounts similar to those described above (soluble CGP amounted to about 2% of the cell dry mass; insoluble CGP amounted to <0.1% of the cell dry mass). Since both strains showed weak light-scattering granules by light microscopy (Fig. 3C and D), electron microscopy views should have elucidated where the CGP was deposited in the cells. The micrographs revealed black areas in all investigated cells (Fig. 3A and B). Cells of strain G175 harboring pYEX-BX::cphA6308 showed stronger light-scattering inclusions in several cells (Fig. 3E and F).
FIG. 3.
Microscopy examinations of transgenic S. cerevisiae cells. TEM (A and B) and light microscopy (C, D, E, and F) pictures of S. cerevisiae G175 harboring pESC-URA (A and C) and of S. cerevisiae G175 harboring pESC-URA::cphA6308 (B and D) are shown. Cells were cultivated in minimal medium containing 2% (wt/vol) galactose and harvested after 48 h. Thin sections were prepared, and electron micrographs were obtained for TEM as described in Materials and Methods. (E) A light microscopy picture of S. cerevisiae G175 harboring pYEX-BX::cphA6308 grown in minimal medium containing 2% (wt/vol) glucose and 0.1 mM CuSO4 and harvested after 48 h is shown. (F) A light microscopy picture of S. cerevisiae G175 harboring pYEX-BX::cphA6308 is shown; cells were cultivated in minimal medium containing 2% (wt/vol) glucose and 0.4 mM CuSO4 and harvested after 48 h. Bars A and B, 2 μm; bars C to F, 4 μm.
Cultivation experiments.
Without varying the cultivation conditions, the total maximal CGP contents of 1.1%, 6.9%, and 5.9% (wt/wt) were measured for S. cerevisiae strain G175(pESC-URA::cphA6308), G175(pYEX-BX::cphA6308), and BY4741(pYEX-BX::cphA6308), respectively (Table 2). The CDM averages were 81, 85, and 46 mg for the respective strains. CDM averages of strains harboring the vectors without inserts were 85, 106, and 81 mg for S. cerevisiae strain G175(pESC-URA), G175(pYEX-BX), and BY4741(pYEX-BX), respectively (Table 2). To enhance the cell densities and the amounts of polymer produced in the transgenic yeasts and to investigate the compositions of the accumulated CGP in detail, several cultivation experiments with differing conditions were performed (Table 2). Glycerol was added to the cells of S. cerevisiae strain G175(pESC-URA::cphA6308) to increase the cell density. Strains B4741 and G175 harboring pYEX-BX::cphA6308 were cultivated in the presence of different CuSO4 concentrations ranging from 0.0 to 0.4 mM. Furthermore, all cphA-carrying cells were cultivated in the presence of the CGP constituents aspartic acid, arginine, and lysine. In addition, all transgenic yeasts were cultivated in complex medium (YPD) instead of minimal medium. All CDM values obtained and the resulting CGP contents are listed in Table 2. Obviously, cells reproducibly lost their plasmids when they were grown in complex medium, since CGP contents were drastically decreased to a maximal 0.3% insoluble CGP. Interestingly, no soluble CGP was synthesized if CGP constituents were added to the medium, but the content of insoluble CGP and the cell dry weight amount increased for all strains. The addition of glycerol in cultures of strain G175 harboring pESC-URA::cphA6308 did not result in higher CDM values or polymer contents of the cells. This was also observed for strain G175 and strain BY4741 harboring pYEX-BX::cphA6308 when they were grown in the presence of two- or fourfold higher concentrations of CuSO4. However, HPLC analysis of CGP isolated from cells grown in the presence of 15 mM lysine revealed that this polymer consisted of up to 10 mol% of lysine.
TABLE 2.
Cell dry weights and CGP contents determined from cultivations under different conditionsa
| Strain(plasmid) | Growth condition variable | CDM (mg) | % CGP content (mean ± SD)
|
|
|---|---|---|---|---|
| Insoluble | Soluble | |||
| G175(pESC-URA) | 85 | - | - | |
| G175(pYEX-BX) | Plus 0.1 mM CuSO4 | 106 | - | - |
| BY4741(pYEX-BX) | Plus 0.1 mM CuSO4 | 81 | - | - |
| G175(pESC-URA::cphA6308) | 81 | 0.1 ± 0.1 | 1 ± 1.0 | |
| Plus glycerol 3% (wt/wt) | 82 | 0.1 ± 0.1 | 1 ± 1.0 | |
| YPD medium | 249 | 0.1 ± 0.1 | - | |
| Plus 10 mM Asp, 15 mM Arg | 99 | 0.3 ± 0.1 | - | |
| Plus 15 mM Arg | 98 | 1.2 ± 0.2 | - | |
| Plus 15 mM Lys | 97 | 1.4 ± 0.2 | - | |
| G175(pYEX-BX::cphA6308) | Without CuSO4 | 88 | 1.3 | 3.1 |
| Plus 0.1 mM CuSO4 | 85 | 2.5 ± 1.0 | 3.7 ± 0.2 | |
| Plus 0.2 mM CuSO4 | 85 | 2.2 ± 0.2 | 4.7 ± 0.2 | |
| Plus 0.4 mM CuSO4 | 83 | 1.5 ± 0.2 | 4.2 ± 0.3 | |
| Plus 0.1 mM CuSO4; YPD medium | 288 | 0.1 ± 0.1 | - | |
| Plus 0.1 mM CuSO4, 10 mM Asp, and 15 mM Arg | 129 | 2.7 ± 0.2 | - | |
| Plus 0.1 mM CuSO4 and 15 mM Arg | 121 | 3.1 ± 0.2 | - | |
| Plus 0.1 mM CuSO4 and 15 mM Lys | 83 | 2.2 ± 0.2 | - | |
| BY4741(pYEX-BX::cphA6308) | Without CuSO4 | 61 | 0.7 | 3.0 |
| Plus 0.1 mM CuSO4 | 46 | 2.0 ± 1.0 | 3.9 ± 0.2 | |
| Plus 0.2 mM CuSO4 | 39 | 0.6 ± 0.2 | 3.9 ± 0.3 | |
| Plus 0.4 mM CuSO4 | 37 | 0.6 ± 0.1 | 4.2 ± 0.1 | |
| Plus 0.1 mM CuSO4; YPD medium | 227 | 0.3 ± 0.1 | - | |
| Plus 0.1 mM CuSO4, 10 mM Asp, and 15 mM Arg | 78 | 3.9 ± 0.4 | - | |
| Plus 0.1 mM CuSO4 and 15 mM Arg | 75 | 3.3 ± 0.4 | - | |
| Plus 0.1 mM CuSO4 and 15 mM Lys | 71 | 3.2 ± 0.4 | - | |
Cell dry weights and CGP contents are the results of cultivating S. cerevisiae strains BY4741 and G175 harboring vector pESC-URA or pYEX-BX or pESC-URA::cphA6308 and pYEX-BX::cphA6308, respectively, under different conditions. All strains were grown at 30°C for 48 h. Strains harboring pESC-URA and pESC-URA::cphA6308 were grown in minimal medium containing 2% (wt/vol) galactose except where YPD medium is listed. Strains harboring pYEX-BX::cphA6308 were grown in minimal medium containing 2% (wt/vol) glucose except where YPD medium is listed. Cell dry matter (CDM) amounts were determined as described in Materials and Methods. Insoluble and soluble CGP samples were isolated as described in Materials and Methods. Most cultivations were performed at least three times. Values given are means ± standard deviations (SD). -, CGP could not be isolated.
DISCUSSION
This study describes for the first time the synthesis of CGP in recombinant yeast and, besides the recently described synthesis of CGP in plants (37), the only investigation of the production of this polymer in eukaryotic organisms. It was shown that both of the investigated S. cerevisiae strains were able to produce the polymer in considerable amounts. However, the two applied vectors containing different promoters were not equally suitable. The vector pYEX-BX::cphA6308 conferred synthesis of CGP to both of the S. cerevisiae strains, whereas strain BY4741 showed significant CGP accumulation only if it harbored the plasmid pYEX-BX::cphA6308 but not if it harbored pESC-URA::cphA6308. The applied vectors differ not only in their promoters but also in their auxotrophy markers. Vector pESC-URA mediates only uracil prototrophy, whereas vector pYEX-BX mediates uracil and leucine (leu2-d) prototrophy. However, the leu2-d gene present in pYEX-BX is degenerated, resulting in enhanced replication and an increase in the copy number of the whole vector, to provide enough leucine for the viability of the cells (33). The increased replication of the whole vector should result in an increased transcription of cphA. Cells of S. cerevisiae strain BY4741 and strain G175 harboring the vector pYEX-BX::cphA6308 exhibited higher CphA enzyme activity than cells harboring vector pESC-URA::cphA6308. This observation also correlated with the higher polymer yields in pYEX-BX::cphA6308-harboring strains.
Immunological analyses showed that the generated anti-CGP IgGs were highly specific to CGP isolated from various organisms (Fig. 2). However, no immunoreaction occurred with water-soluble CGP synthesized by transgenic yeast (Fig. 2A). This observation was surprising as the chemical structure of this form of CGP has been reported to be identical to the water-insoluble form (16, 52); also, our analysis did not reveal any differences (data not shown). Presumably, relevant groups or regions in the CGP molecule that are recognized by the antibodies are disguised, resulting in an inability of the IgGs to bind to the soluble form. Eventually these disguised residues could also be the cause for the solubility of CGP, which is not fully elucidated yet (16). About 6 μg of CGP could be detected with Western blotting. However, this was not the real detection limit as CGP did not form a distinct band on the gel but was instead dispersed over a wide area on the gel due to its polydispersity. Nevertheless, the antibodies generated provide a suitable tool for the detection of insoluble CGP from different organisms. The molecular mass distributions of CGP was between 20 and 35 kDa for the soluble CGP and between 26 and 45 kDa for the insoluble CGP and was similar to distributions observed with recombinant bacteria and plants (3, 15, 37). In contrast, in cyanobacteria, the apparent molecular masses were much higher, ranging up to 130 kDa (18).
An interesting aspect was the amino acid composition of the isolated CGP. CphA6308 has been described as having a broad substrate range in vitro (2) and incorporates up to 10 mol% of lysine, replacing arginine in the side chain of CGP, when it is expressed in E. coli (30). In contrast, CGP isolated from the natural host Synechocystis sp. strain PCC 6308 is composed of aspartic acid and arginine only (5). Without a variation in the medium composition, S. cerevisiae produced CGP with a maximum fraction of only 2 mol% of lysine. However, an increase in the amount of lysine, up to 10 mol%, was detected when 15 mM of lysine was added to the media; this composition correlated with that observed by Krehenbrink et al. (30).
Light microscopy investigations of cells harboring pESC-URA::cphA6308 did not reveal the presence of CGP granules as they did with A. baylyi strain ADP1 or with recombinant E. coli strains (14, 15). Also, electron microscopy views did not visualize CGP granules in the yeast cells, which usually appear as black areas in the cells (14). However, such areas were widespread in the yeast strains harboring cphA and also in the negative control. As the investigated yeast strains produced mainly soluble CGP and less than 0.1% of the insoluble form, which occurs as granules, CGP could not be visualized by this method in yeast. Only S. cerevisiae strain G175 harboring pYEX-BX::cphA6308 showed, to a larger extent, light-scattering inclusions which might be CGP granules (Fig. 3E and F).
Determination of the cell densities of transgenic yeasts harboring cphA6308 yielded lower values than the negative controls without cphA, thereby indicating that the synthesis and accumulation of CGP resulted in slower growth. Such an inhibition due to CGP biosynthesis was recently reported in transgenic plants, too (37). However, a total CGP content of almost 7%, which was obtained without having varied the cultivation conditions, is still high in comparison to that of CGP-producing plants or some recombinant strains of C. glutamicum and P. putida (3, 37). Through slight modifications of the cultivation conditions, the CDM average was increased 1.5-fold for strains harboring pYEX-BX::cphA6308, and interestingly, soluble CGP was no longer produced when the CGP constituents aspartate and arginine were added to the medium. This observation could be useful for the directed production of one form of CGP.
These experiments concerning the production of CGP in S. cerevisiae clearly indicated that CGP synthesis can be conferred to yeast and that these microorganisms are therefore putative candidates for the biotechnical production of CGP in the future. However, the CGP contents of the cells need to be enhanced. This could be achieved, for example, by using stronger induction systems, by engineering the metabolism of yeasts, and by varying the cultivation conditions. A chromosomal integration of cphA in yeast would probably be especially advantageous, thereby allowing the use of technical, low-cost media instead of specific minimal media. In addition, the use of other yeasts such as P. pastoris and H. polymorpha (17), which have become increasingly interesting for biotechnical purposes because they exhibit high levels of productivity and because they can be grown to high cell densities, could be applied in the future.
Acknowledgments
We thank Ursula Malkus (Institut für Medizinische Physik und Biophysik) for expert preparation of the electron microscopic specimen. We thank Roland Klassen and Friedhelm Meinhardt (Institut für Molekulare Mikrobiologie und Biotechnologie) for providing S. cerevisiae strain BY4741 and vectors pESC-URA and pYEX-BX. We thank M. Gustavsson, E. Wiberg, P. Stolt, and S. Stymne (Scandinavian Biotechnology Research, Alnarp, Sweden) for providing S. cerevisiae strain G175. We also thank Kay M. Frey, a member of our laboratory, for providing the cyanobacterial CGP.
This project was supported by a grant (EOSLT02034) from SenterNovem (Utrecht, The Netherlands).
Footnotes
Published ahead of print on 11 April 2008.
REFERENCES
- 1.Aboulmagd, E., F. B. Oppermann-Sanio, and A. Steinbüchel. 2000. Molecular characterization of the cyanophycin synthetase from Synechocystis sp. strain PCC6308. Arch. Microbiol. 174:297-306. [DOI] [PubMed] [Google Scholar]
- 2.Aboulmagd, E., F. B. Oppermann-Sanio, and A. Steinbüchel. 2001. Purification of Synechocystis sp. strain PCC6308 cyanophycin synthetase and its characterization with respect to substrate and primer specificity. Appl. Microbiol. Biotechnol. 67:2176-2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aboulmagd, E., I. Voss, F. B. Oppermann-Sanio, and A. Steinbüchel. 2001. Heterologous expression of cyanophycin synthetase and cyanophycin synthesis in the industrial relevant bacteria Corynebacterium glutamicum and Ralstonia eutropha and in Pseudomonas putida. Biomacromolecules 2:1338-1342. [DOI] [PubMed] [Google Scholar]
- 4.Alfenore, S., X. Cameleyre, L. Benbadis, C. Bideaux, J.-L. Uribelarrea, G. Goma, C. Molina-Jouve, and S. E. Guillouet. 2004. Aeration strategy: a need for very high ethanol performance in Saccharomyces cerevisiae fed-batch process. Appl. Microbiol. Biotechnol. 63:537-542. [DOI] [PubMed] [Google Scholar]
- 5.Allen, M. M., and P. J. Weathers. 1980. Structure and composition of cyanophycin granules in the cyanobacterium Aphanocapsa 6308. J. Bacteriol. 141:959-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Attfield, P. V. 1997. Stress tolerance: the key to effective strains of industrial baker's yeast. Nat. Biotechnol. 15:1351-1357. [DOI] [PubMed] [Google Scholar]
- 7.Báez, J., D. Olsen, and J. W. Polarek. 2005. Recombinant microbial systems for the production of human collagen and gelatin. Appl. Microbiol. Biotechnol. 69:245-252. [DOI] [PubMed] [Google Scholar]
- 8.Böer, E., G. Steinborn, A. Matros, H. P. Mock, G. Gellissen, and G. Kunze. 2007. Production of interleukin-6 in Arxula adeninivorans, Hansenula polymorpha and Saccharomyces cerevisiae by applying the wide-range yeast vector (CoMed) system to simultaneous comparative assessment. FEMS Yeast Res. 7:1181-1187. [DOI] [PubMed] [Google Scholar]
- 9.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14:115-132. [DOI] [PubMed] [Google Scholar]
- 10.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 11.Bullock, W. O., J. M. Fernandez, and J. M. Stuart. 1987. XL1-Blue: a high efficiency plasmid transforming recA Escherichia coli strain with β-galactosidase selection. BioTechniques 5:376-379. [Google Scholar]
- 12.de Bruin, E. C., M. W. T. Werten, C. Laane, and F. A. de Wolf. 2002. Endogenous prolyl 4-hydroxylation in Hansenula polymorpha and its use for the production of hydroxylated recombinant gelatin. FEMS Yeast Res. 1:291-298. [DOI] [PubMed] [Google Scholar]
- 13.Diniz, C. S., I. Voss, and A. Steinbüchel. 2006. Optimization of cyanophycin production in recombinant strains of Pseudomonas putida and Ralstonia eutropha employing elementary mode analysis and statistical experimental design. Biotechnol. Bioeng. 93:698-717. [DOI] [PubMed] [Google Scholar]
- 14.Elbahloul, Y., M. Krehenbrink, R. Reichelt, and A. Steinbüchel. 2005. Physiological conditions conducive to high cyanophycin content in biomass of Acinetobacter calcoaceticus strain ADP1. Appl. Environ. Microbiol. 71:858-866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frey, K. M., F. B. Oppermann-Sanio, H. Schmidt, and A. Steinbüchel. 2002. Technical-scale production of cyanophycin with recombinant strains of Escherichia coli. Appl. Environ. Microbiol. 68:3377-3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Füser, G., and A. Steinbüchel. 2005. Investigations on the solubility behavior of cyanophycin. Solubility of cyanophycin in solutions of simple inorganic salts. Biomacromolecules 6:1367-1374. [DOI] [PubMed] [Google Scholar]
- 17.Gellissen, G., G. Kunze, C. Gaillardin, J. M. Cregg, E. Berardi, M. Veenhuis, and I. van der Klei. 2005. New yeast expression platforms based on methylotrophic Hansenula polymorpha and Pichia pastoris and on dimorphic Arxula adeninivorans and Yarrowia lipolytica: a comparison. FEMS Yeast Res. 5:1079-1096. [DOI] [PubMed] [Google Scholar]
- 18.Hai, T., F. B. Oppermann-Sanio, and A. Steinbüchel. 1999. Purification and characterization of cyanophycin and cyanophycin synthetase from the thermophilic Synechococcus sp. MA19. FEMS Microbiol. Lett. 181:229-236. [DOI] [PubMed] [Google Scholar]
- 19.Hai, T., H. Ahlers, H. Gorenflo, and A. Steinbüchel. 2000. Axenic cultivation of anoxygenic phototrophic bacteria, cyanobacteria, and microalgae in a new closed tubular glass photobioreactor. Appl. Microbiol. Biotechnol. 53:383-389. [DOI] [PubMed] [Google Scholar]
- 20.Hai, T., F. B. Oppermann-Sanio, and A. Steinbüchel. 2002. Molecular characterization of a thermostable cyanophycin synthetase from the thermophilic cyanobacterium Synechococcus sp. MA19 and in vitro synthesis of cyanophycin and related polyamides. Appl. Environ. Microbiol. 68:93-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hai, T., K. M. Frey, and A. Steinbüchel. 2006. Activation of cyanophycin synthetase of Nostoc ellipsosporum strain NE1 by truncation at the carboxy-terminal region. Appl. Microbiol. Biotechnol. 72:7652-7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 23.Harford, N., T. Cabezon, B. Colau, A. M. Delisse, T. Rutgers, and M. De Wilde. 1987. Construction and characterization of a Saccharomyces cerevisiae strain (RIT4376) expressing hepatitis B surface antigen. Postgrad. Med. J. 63:65-70. [PubMed] [Google Scholar]
- 24.Hartree, E. F. 1972. Determination of protein: a modification of the Lowry method that gives a linear photometric response. Anal. Biochem. 48:422-427. [DOI] [PubMed] [Google Scholar]
- 25.Hein, S., T. Hai, and A. Steinbüchel. 1998. Synechocystis sp. PCC6803 possesses a two-component polyhydroxyalkanoic acid synthase similar to that of anoxygenic purple sulfur bacteria. Arch. Microbiol. 170:162-170. [DOI] [PubMed] [Google Scholar]
- 26.Hensing, M. C. M., R. J. Rouwenhorst, J. J. Heijnen, J. P. van Dijken, and J. T. Pronk. 1995. Physiological and technological aspects of large-scale heterologous-protein production with yeast. Antonie van Leeuwenhoek 67:261-279. [DOI] [PubMed] [Google Scholar]
- 27.Hjelm, H., K. Hjelm, and J. Sjöquist. 1972. Protein A from Staphylococcus aureus. Its isolation by affinity chromatography and its use as an immunosorbent for isolation of immunoglobulins. FEBS Lett. 28:73-76. [DOI] [PubMed] [Google Scholar]
- 28.Ito, H., Y. Fukura, K. Murata, and A. Timura. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 15:163-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kaiser, C., S. Michaelis, and A. Mitchell. 1994. Methods in yeast genetics: a Cold Spring Harbor Laboratory course manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Krehenbrink, M., F. B. Oppermann-Sanio, and A. Steinbüchel. 2002. Evaluation of non-cyanobacterial genome sequences for occurrence of genes encoding proteins homologous to cyanophycin synthetase and cloning of an active cyanophycin synthetase from Acinetobacter sp. strain DSM 587. Arch. Microbiol. 177:371-380. [DOI] [PubMed] [Google Scholar]
- 31.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 32.Lowry, O. H., N. J. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 33.Macreadie, I. G., O. Horaitis, A. J. Verkuylen, and K. W. Savin. 1991. Improved shuttle vectors for cloning and high-level Cu2+-mediated expression of foreign genes in yeast. Gene 104:107-111. [DOI] [PubMed] [Google Scholar]
- 34.Melmer, G. 2005. Biopharmaceuticals and the industrial environment, p. 361-383. In G. Gellissen (ed), Production of recombinant proteins: novel microbial and eukaryotic expression systems. Wiley-VCH, Weinheim, Germany.
- 35.Mendoza-Vega, O., J. Sabatié, and S. W. Brown. 1994. Industrial production of heterologous proteins by fed-batch cultures of the yeast Saccharomyces cerevisiae. FEMS Microbiol. Rev. 15:369-410. [DOI] [PubMed] [Google Scholar]
- 36.Mooibroek, H., N. Oosterhuis, M. Giuseppin, M. Toonen, H. Franssen, E. Scott, J. Sanders, and A. Steinbüchel. 2007. Assessment of technological options and economical feasibility for cyanophycin biopolymer and high-value amino acid production. Appl. Microbiol. Biotechnol. 77:257-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neumann, K., D. P. Stephan, K. Ziegler, M. Hühns, I. Broer, W. Lockau, and E. K. Pistorius. 2005. Production of cyanophycin, a suitable source for the biodegradable polymer polyaspartate, in transgenic plants. Plant Biotechnol. J. 3:249-258. [DOI] [PubMed] [Google Scholar]
- 38.Nokelainen, M., H. Tu, A. Vuorela, H. Notbohm, K. I. Kivirikko, and J. Myllyharju. 2001. High-level production of human type I collagen in the yeast Pichia pastoris. Yeast 18:797-806. [DOI] [PubMed] [Google Scholar]
- 39.Reynolds, E. S. 1963. The use of lead citrate at high pH as an electron opaque stain in electron microscopy. J. Cell Biol. 17:208-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 41.Sandager, L., M. H. Gustavsson, U. Ståhl, A. Dahlqvist, E. Wiberg, A. Banas, M. Lenmann, H. Ronne, and S. Stymne. 2002. Storage lipid synthesis is non-essential in yeast. J. Biol. Chem. 277:6478-6482. [DOI] [PubMed] [Google Scholar]
- 42.Sanders, J., E. Scott, R. Weusthuis, and H. Mooibroek. 2007. Bio-refinery as the bio-inspired process to bulk chemicals. Macromol. Biosci. 7:105-117. [DOI] [PubMed] [Google Scholar]
- 43.Scott, E., F. Peter, and J. Sanders. 2007. Biomass in the manufacture of industrial products: the use of proteins and amino acids. Appl. Microbiol. Biotechnol. 75:751-762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon, R. D. 1976. The biosynthesis of multi-l-arginyl-poly(l-aspartic acid) in the filamentous cyanobacterium Anabaena cylindrica. Biochim. Biophys. Acta 422:407-418. [DOI] [PubMed] [Google Scholar]
- 45.Simon, R. D., and P. Weathers. 1976. Determination of the structure of the novel polypeptide containing aspartic acid and arginine which is found in cyanobacteria. Biochim. Biophys. Acta 420:165-176. [DOI] [PubMed] [Google Scholar]
- 46.Spurr, A. R. 1969. A low-viscosity epoxy resin embedding medium for electron microscopy. J. Ultrastruct. Res. 26:31-43. [DOI] [PubMed] [Google Scholar]
- 47.Vaughn, P. R., M. Galanis, K. M. Richards, T. A. Tebb, J. A. M. Ramshaw, and J. A. Werkmeister. 1998. Production of recombinant hydroxylated human type III collagen fragment in Saccharomyces cerevisiae. DNA Cell Biol. 17:511-518. [DOI] [PubMed] [Google Scholar]
- 48.Voss, I., S. Cardoso Diniz, E. Aboulmagd, and A. Steinbüchel. 2004. Identification of the Anabaena sp. strain PCC 7120 cyanophycin synthetase as suitable enzyme for production of cyanophycin in Gram-negative bacteria like Pseudomonas putida and Ralstonia eutropha. Biomacromolecules 5:1588-1595. [DOI] [PubMed] [Google Scholar]
- 49.Voss, I., and A. Steinbüchel. 2006. Application of a KDPG-aldolase gene-dependent addiction system for enhanced production of cyanophycin in Ralstonia eutropha strain H16. Metab. Eng. 8:66-78. [DOI] [PubMed] [Google Scholar]
- 50.Wisselink, H. W., M. J. Toirkens, M. del Rosario Franco Berriel, A. A. Winkler, J. P. van Dijken, J. T. Pronk, and A. J. A. van Maris. 2007. Engineering of Saccharomyces cerevisiae for efficient anaerobic alcoholic fermentation of l-arabinose. Appl. Environ. Microbiol. 73:4881-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ziegler, K., A. Diener, C. Herpin, R. Richter, R. Deutzmann, and W. Lockau. 1998. Molecular characterization of cyanophycin synthetase, the enzyme catalyzing the biosynthesis of the cyanobacterial reserve material multi-l-arginyl-poly-l-aspartate (cyanophycin). Eur. J. Biochem. 254:154-159. [DOI] [PubMed] [Google Scholar]
- 52.Ziegler, K., R. Deutzmann, and W. Lockau. 2002. Cyanophycin synthetase-like enzymes of non-cyanobacterial Eubacteria: characterization of the polymer produced by a recombinant synthetase of Desulfitobacterium hafniense. Z. Naturforsch. 57:522-529. [DOI] [PubMed] [Google Scholar]