Abstract
Chlorhexidine is a common-use antibacterial agent found in a range of personal-care products. We used rotating annular reactors to cultivate river biofilms under the influence of chlorhexidine or its molar equivalent in nutrients. Studies of the degradation of [14C]chlorhexidine demonstrated that no mineralization of the compound occurred. During studies with 100 μg liter−1 chlorhexidine, significant changes were observed in the protozoan and micrometazoan populations, the algal and cyanobacterial biomass, the bacterial biomass, and carbon utilization. Denaturing gradient gel electrophoresis (DGGE) in combination with statistical analyses showed that the communities developing under control and 100 μg liter−1 chlorhexidine were significantly different. At 10 μg liter−1 chlorhexidine, there was significantly increased algal and cyanobacterial biomass while the bacterial biomass was not significantly affected (P < 0.05). No significant effects on protozoan or metazoan grazing were detected at the 10-μg liter−1 chlorhexidine level. Fluorescent in situ hybridization indicated a significant reduction in the abundance of betaproteobacteria and gammaproteobacteria (P < 0.05). Archaeal cell counts were significantly reduced by both chlorhexidine and nutrient treatments. DGGE and statistical analyses indicated that 10 μg liter−1 chlorhexidine and molar equivalent nutrient treatments were significantly different from control communities. In contrast to community level observations, toxicological testing with a panel of cyanobacteria, algae, and protozoa indicated no detectable effects at 10, 50, and 100 μg liter−1 chlorhexidine. Thus, community level assessment indicated a risk of low levels of chlorhexidine in aquatic habitats while conventional approaches did not.
Chlorhexidine (1,1′-hexamethylene-bis[5-(p-chloro-phenyl)biguanide]; C22H30Cl2N102C6H12O7), first described in 1954, is a disinfectant with broad gram-positive and gram-negative antibacterial and antifungal activity and low mammalian toxicity (9, 36). Chlorhexidine is the active ingredient in many commercially available disinfectants, antiseptics, and oral-care products (41) and enters the environment primarily via sewage treatment plant effluents as a consequence of its significant usage in dental (15), medical (20), and veterinary applications (13). Its mode of action occurs via negatively charged groups on the cell surface, causing an irreversible loss of cytoplasmic constituents, as well as membrane damage and enzyme inhibition. At high concentrations (e.g., 0.5 to 1%), chlorhexidine causes extensive cell damage, coagulation of cytoplasmic constituents, and precipitation of proteins and nucleic acids (15). Chlorhexidine's biocidal activity is influenced by environmental factors, including pH, temperature, and the presence of interfering material (42). In medical and dental applications, effective application rates for chlorhexidine range from 0.5 to 1%, while in soaps and cleansers, it is used at a concentration of 2 to 4%. A number of studies have investigated the chlorhexidine susceptibilities of individual bacteria and multispecies biofilms of relevance to medical and dental applications (35).
There is a paucity of information concerning chlorhexidine's concentration in the environment and potential environmental effects, particularly on microorganisms. Kodama et al. (17) reported that domestic wastewater could contain up to 1.6 to 10.3 μg ml−1, depending upon the analytical method used. Although there is limited toxicological data for chlorhexidine, based on the values available, European regulators have classified chlorhexidine as environmentally harmful because it is highly toxic for water-dwelling organisms and is capable of causing harmful long-term effects in an aquatic environment, and evidence exists that it bioaccumulates. Chlorhexidine is a positively charged hydrophobic and lipophilic molecule that would be expected to interact with lipids and accumulate in the fatty tissues (e.g., lipids) of living organisms (5). This bioaccumulation has been confirmed in river biofilms by Dynes et al. (10). Soft X-ray transmission microscopy was used to demonstrate that chlorhexidine accumulated extensively in the lipids of both diatoms and bacteria in river biofilm communities (10). The U.S. Environmental Protection Agency considers chlorhexidine to be moderately toxic to fish and, based on a single study, highly toxic to aquatic invertebrates (2) at a minimum concentration of 32 μg liter−1. Based on these observations and the absence of relevant environmental-effects data, we carried out a series of microcosm experiments to assess the impacts of 10 and 100 μg liter−1 chlorhexidine on the development, diversity, and activity of river biofilm communities.
MATERIALS AND METHODS
Microcosm operation.
The experimental setup and reactor design for biofilm development has been described in detail (21, 24). Natural river water (South Saskatchewan River, Saskatoon, SK, Canada) was used as an inoculum and as a source of carbon and nutrients. The nutrients and antimicrobial were added directly to the individual reactors using a peristaltic pump. Nutrient levels were assessed as described by Chénier et al. (7). River temperatures varied between 0.5 and 4°C over the course of the experiment; the reactors were maintained at a constant 21 ± 2°C. The water was pumped through the reactors at a rate of 500 ml per day (1 reactor volume) by using a multichannel peristaltic pump (Watson Marlow, Wilmington, MA). Treatments included the addition of chlorhexidine at 10 or 100 μg liter−1 and the molar equivalent in carbon (glucose) and nitrogen (ammonium chloride). Chlorhexidine has a molecular weight of 505.5, and chlorhexidine dihydrochloride has a solubility of 0.6 g liter−1 in water. The chlorhexidine concentrations used were selected to be several orders of magnitude below medical-industrial application levels, which are 0.2 to 1.0%, to bracket the minimum toxicity level of 32 μg liter−1 reported for Daphnia magna by the U.S. Environmental Protection Agency (2) and to reflect levels detected in wastewater (17). In addition, control reactors that received river water alone were operated. Biofilms were grown under treatment and control conditions in bioreactors for a period of 8 weeks, at which time coupons were removed for immediate analysis (confocal laser scanning microscopy [CLSM], microscopic, isotopic, and carbon utilization assays), fixed for in situ hybridization, or frozen at −80°C and stored for subsequent DNA extraction and analysis (denaturing gradient gel electrophoresis [DGGE]).
CLSM and image analysis.
Examination of all stained and control materials was carried out with an MRC 1024 Bio-Rad confocal laser scanning microscope (previously Bio-Rad, Hemel Hempstead, United Kingdom; now Zeiss, Jena, Germany) attached to a Microphot SA microscope (Nikon, Tokyo, Japan). Slides from each of the replicate reactors were cut into 1-cm2 pieces and mounted in small petri dishes using Dow Corning no. 3140 acid-free silicone coating (WPI, Inc., Sarasota, FL) and then stained and analyzed according to the following procedures. For observation, the following water-immersible lenses were used: 63×, 0.9 numerical aperture (NA) (Zeiss, Jena, Germany) and 40×, 0.55 NA, and 10×, 0.35 NA (Nikon, Japan). The biofilms were observed by using a double-labeling procedure and three-channel fluorescence detection; bacteria were stained with the fluorescent nucleic acid stain Syto 9 (excitation wavelength, 488; bandwidth of filter set, 522/32), a lectin probe (Triticum vulgaris-tetramethyl rhodamine isothiocyanate) (excitation wavelength, 568; bandwidth of filter set, 605/32) was used to visualize exopolymer, and autofluorescence (excitation wavelength, 647; bandwidth of filter set, 680/32) was used to detect algal and cyanobacterial cells (31). Digital image analysis of the CLSM optical thin sections in each of the three channels was used to determine such parameters as biofilm depth, bacterial cell area (biomass), exopolymer biomass, cyanobacterial biomass, and total photosynthetic biomass at various depths. Image analyses were performed by using NIH Image version 1.61 (http://rsb.info.nih.gov/nih-image/) with macros written for semiautomated quantification, as described by Manz et al. (26). In addition, three-color red-green-blue projections of the biofilms were computed.
Exopolymer analyses.
The lectins of Arachis hypogaea, Canavalia ensiformis, Glycine max, T. vulgaris, and Ulex europaeus were used alone or in combination for in situ analyses of polymer composition, as described by Neu et al. (32). Image analyses and calculations of lectin binding volumes were carried out using the equations of Neu et al. (32).
Protozoan and micrometazoan enumeration.
Protozoa and micrometazoa were enumerated in accordance with the method of Packroff et al. (33). Samples were removed from the reactors on a weekly basis, and the numbers of protozoa and micrometazoa were manually counted on replicate 2-cm2 subsamples using phase-contrast microscopy.
Carbon utilization assays.
Carbon utilization assays were carried out for biofilm samples using commercial Eco-plates (Biolog, Hayward, CA) (19, 24). Biofilm coupons were scraped using a sterile silicon rubber spatula to remove the biofilm and sonicated in a Bransonic 5120 water bath sonifier (Branson Ultrasonics, Danbury, CT) for 5 min to disperse the cells, and appropriate dilutions (10−4) were inoculated (150 μl) into all 96 wells of the Biolog microtiter plates and incubated at 23 ± 3°C with atmospheric oxygen. The plates were read using a standard microtiter plate reader each day until a stable result was obtained (7 days).
Toxicity testing.
Chlorhexidine was added at 0, 10, 50, and 100 μg liter−1 to mineral salts growth media in 24-well microtiter plates. A panel of algae (Scenedesmus quadricauda, Selenastrum sp., Ulothrix sp., Ankistrodesmus falcatus, Oscillatoria tenius, Synedra sp., and Thalassiosira sp.) and cyanobacteria (Anabaena sp., Glaucocystis nostochinea, Lyngbya sp., Microcystis aeruginosa, and Nostoc sp.) were inoculated into the media and incubated at 23 ± 3°C under continuous illumination. The following protozoa and metazoa were also exposed to the same levels of chlorhexidine in microtiter plates: Euplotes sp., Dileptus sp., Blepharisma sp., Stentor sp., Spirostomum sp., Euglena sp., Paramecium sp., and rotifera. The cultures were assessed by eye and by microscopic examination for growth, activity, and movement relative to control cultures over a 14-day period. Incubations were carried out in triplicate for all concentrations.
Stable-isotope analyses.
For stable-isotope analyses, approximately 1 mg each of dried biofilm material, reagent grade chlorhexidine, and nutrients was weighed into 4- by 9-mm tin cups. Samples were analyzed for stable-isotope ratios of carbon using a Micromass Isoprime EA CF-IRMS (GV Instruments, Manchester, United Kingdom) at the National Hydrology Research Centre, Saskatoon, Saskatchewan, Canada, using standard techniques (44). Isotope ratios are expressed in delta (δ) notation as parts per thousand (δ13C) differences from the Pee Dee Bee reference standard as follows: δ13C (‰) = [(Rsample − Rstandard)/Rstandard] × 1,000, where R denotes the 13C/12C ratio. All results are reported relative to the Pee Dee Bee limestone standard for δ13C. International standards and an internal working protein reference material (δ13C = −12.6 ± 0.2) were used to verify sample reproducibility. The results are expressed as mean ± standard error.
Radioisotope analyses.
After the 8-week growth period in the bioreactors, fresh biofilm samples on polycarbonate strips were used in microcosms to assess the impacts of treatments on bacterial activity. The polycarbonate strips were aseptically cut (2 cm2), and each piece with its associated biofilm was transferred into a 20-ml sealable glass vial with 10 ml of water from the corresponding bioreactors. Chlorhexidine mineralization results were assessed as the percentage of 14CO2 produced from the uniformly labeled ring [14C]chlorhexidine (specific activity, 0.1 mCi mmol−1 added at 100 μg liter−1 and 36,000 dpm) as measured by liquid scintillation spectrometry (Tri-Carb 2100TR; Packard Instruments, Downers Grove, IL). Thymidine incorporation was carried out following the standard protocol of Robarts and Wicks (39). All negative controls were killed with formaldehyde at 4% final concentration.
Molecular analyses. (i) Total-community DNA extraction.
For each treatment bioreactor, a frozen (−80°C) polycarbonate strip was aseptically cut (2 cm2) and transferred into a 50-ml polypropylene tube (Falcon; Becton Dickinson, Franklin Lanes, NJ). Bacterial cells from the frozen biofilm samples were removed from the polycarbonate strip with a sterile metal scraper, and total DNA was extracted by using the FastDNA spin kit for soil (Bio101 Systems Qbiogene, Carlsbad, CA) according to the manufacturer's instructions.
(ii) PCR amplification.
The gene encoding eubacterial 16S rRNA was also amplified to perform DGGE. PCR amplification was conducted in a 25-μl reaction mixture containing 1 μl of DNA template, 10 pmol of each appropriate primer as described by Muyzer et al. and Muyzer and Ramsing (28, 29), 1.25 U Taq DNA polymerase (New England Biolabs, Ipswich, MA) 1× PCR buffer, 2.5 mM MgCl2, and 200 μM deoxynucleoside triphosphate. A touchdown PCR program using the PTC-200 thermocycler (MJ Research, Inc., Waltham, MA) consisted of an initial denaturation step of 94°C for 5 min, followed by 10 cycles of denaturation at 94°C for 1 min, annealing at 66°C (decreasing in each cycle by 1°C) for 1 min, and an elongation step of 72°C for 1 min. Following these steps, another 20 cycles of 95°C for 1 min, annealing at 56°C for 1 min, and elongation at 72°C for 1 min, with a final elongation step of 72°C for 7 min, were performed. The appropriately sized PCR product was verified by electrophoresis on a 1.5% (wt/vol) agarose gel in 1.0× Tris-acetate-EDTA buffer (40 mM Tris, 20 mM acetic acid, and 1 mM EDTA) for 1.0 h at 100 V. The gels were stained using ethidium bromide and documented using the AlphaImager 3300 gel documentation and image analysis system (Alpha Innotech Corp., San Leandro, CA).
(iii) DGGE analysis.
After the specificities and sizes of the amplified products were checked on agarose gels, the PCR product was separated by DGGE (28, 29) using an Ingeny phorU2 system (Ingeny, Leiden, The Netherlands). Aliquots (20 μl) of the PCR product were mixed with 4 μl of loading-dye buffer and resolved on a 6% (wt/vol) polyacrylamide gel in 1.0× Tris-acetate-EDTA buffer, using denaturing gradients from 45 to 65% (100% denaturant contains 7 M urea and 40% deionized formamide). DGGE was carried out at 40 V for 10 min and then 100 V for 18 h at 60°C. After electrophoresis, the gel was stained with Sybr green I (1:10,000 dilution; Molecular Probes, Eugene, OR) for 15 min with gentle agitation and photographed using the AlphaImager 3300 gel documentation and image analysis system (Alpha Innotech Corp., San Leandro, CA).
(iv) FISH.
Biofilm fixation was done following the protocol of Manz et al. (26). The probes used were as follows (probe, target organisms, percent formamide, NaCl concentration [mM], and source): EUB338, Bacteria, 20, 250 (1); BET42a, Betaproteobacteria, 35, 88 (25); GAM42a, Gammaproteobacteria, 35, 88 (25); and ARC915, Archaea, 20, 250 (43). Oligonucleotide probes (Interactiva, Berlin, Germany) conjugated to Oregon green were stored in TE buffer (10 mM Tris, 1 mM EDTA, pH 7.5) at −20°C. Working solutions were adjusted to 50 ng DNA per ml. Prewarmed hybridization buffer (0.9 M NaCl, 20 mM Tris-HCl [pH 7.2], 0.01% sodium dodecyl sulfate) and a probe-specific formamide concentration were mixed with the fluorescently labeled oligonucleotide (1 ng ml−1 hybridization buffer) and applied to the fixed biofilm material. The slides were placed in humid chambers and incubated for 90 min at 46°C. After this, the hybridization buffer was drawn off with tissue placed at the edges of the slides. Subsequently, the slides were transferred to 50 ml prewarmed washing buffer (20 mM Tris-HCl, 0.01% sodium dodecyl sulfate, NaCl) and incubated at 48°C for 20 min. For microscopic analysis, slides were carefully rinsed with distilled water, air dried, and mounted in antifading medium (Slow Fade; Molecular Probes Inc., Eugene OR). All hybridization and washing steps were performed in the dark. Autofluorescence signals were eliminated from the images by two means: (i) all probes were conjugated to green fluorescent reporters (excitation wavelength, 488; bandwidth of filter set, 522/32) and (ii) autofluorescence signals in the red (excitation wavelength, 568; bandwidth of filter set, 605/32) and far-red (excitation wavelength, 647; bandwidth of filter set, 680/32) channels were subtracted from the green channel during image collection. These two procedures eliminated algal and cyanobacterial signals from the fluorescent in situ hybridization (FISH) analyses. Cyanobacteria are therefore not included in the results of FISH analyses. However, this approach precluded dual staining with EUB338 and other probes, requiring that total eubacterial counts and specific probe counts be determined on separate samples. For digital analyses, images were individually thresholded based on their signal-to-noise ratios, and the percent area of hybridized or Sytox-stained cells was determined using NIH Image vl.64.
Experimental design and statistical analyses.
The experimental design consisted of controls (untreated), nutrient controls to which the molar equivalent of the chlorhexidine treatment was added as carbon (glucose) and nitrogen (ammonium chloride), and chlorhexidine treatment at 10 or 100 μg liter−1. Each treatment had three identical replicate reactors randomly assigned to it on the reactor bench (replications). Furthermore, each analysis was done on subsamples of randomly selected biofilm coupons from among the 12 identical coupons in each replicate reactor. The CLSM imaging was done at five random locations at five positions on transects across the 1-cm2 piece of the biofilm coupon. Subsampling for other analyses (protozoan counts, thymidine incorporation, and carbon utilization analyses) was also carried out using randomly selected subsamples from among the 12 identical coupons in each replicate reactor. Analysis of variance was used to detect significant differences among sample means at a P value of <0.05. Analyses were carried out using the commercial package MiniTab (State College, PA). Band detection, matching, and processing of DGGE gels were completed with GelCompare II software 4.6 (Applied Maths, Kotrijk, Belgium). Fingerprint data were processed by generating a band-matching table (4). The binary data were exported and compared by principal-component analysis (PCA) with PRIMER v6 software (PrimerE, Ltd., Lutton, United Kingdom). Statistical analyses of PCA scores generated from the first two axes were run using an analysis of similarity (ANOSIM) with PRIMER v6 software (8). The inclusion of DGGE ladders allowed GelCompare II to normalize the positions of bands in all of the lanes under examination.
RESULTS AND DISCUSSION
Effects on river biofilm community composition and architecture.
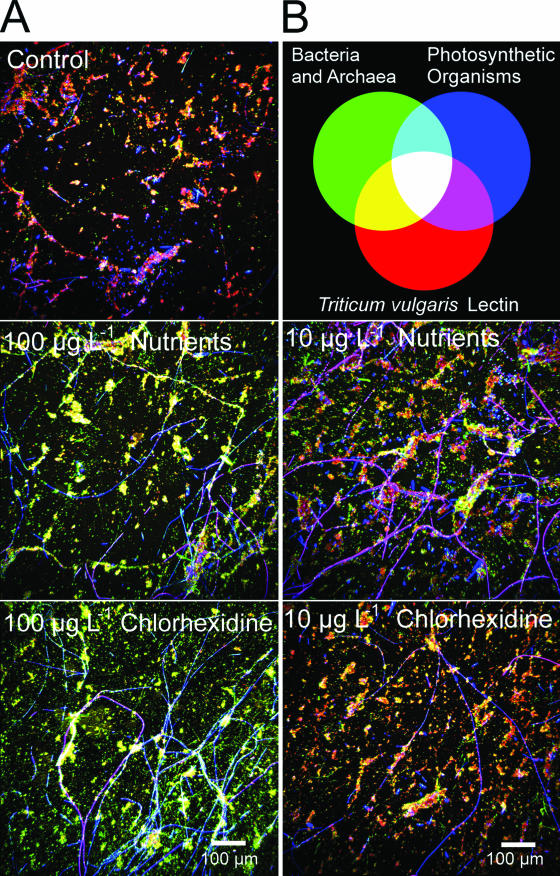
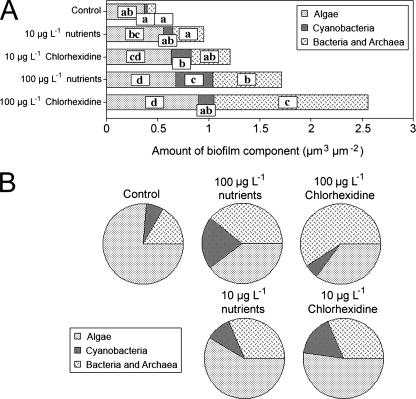
As noted by Porsbring et al. (34), the use of community-level testing provides a relatively high degree of realism and reliability in assessing the potential ecological effects of contaminants. Biofilm architecture and community composition have been demonstrated to be highly sensitive to environmental stress, with changes in biofilm thickness, cellular distribution, and community structure at the level of major functional groupings, e.g., cyanobacteria, algae, and bacteria (6, 22, 23, 24). Detailed comparisons of the river biofilms cultivated under control conditions and with chlorhexidine indicated marked changes with treatment. The effects of the treatments on the community structure and biofilm architecture are evident in the representative (images were selected as representative based on their similarity to the mean values determined for the treatment) confocal micrographs shown in Fig. 1. Images for the experiment indicated a replacement of algal/cyanobacterial species with an enhancement of the abundance of filamentous taxa. At 100 μg liter−1, chlorhexidine had a significant impact on the appearance of the biofilm community at the ×10 scale of observation relative to the control biofilms (Fig. 1). Digital image analyses of the biofilm communities indicated shifts in the algal and bacterial components (Fig. 2A and B). The 10-μg liter−1 molar equivalent nutrient and chlorhexidine treatments resulted in significant increases in the algal and cyanobacterial biomass. However, no significant (P < 0.05) increase in the bacterial biomass was detected. The increases were in keeping with the increased nutrient level, although chlorhexidine resulted in greater increases. Typically there was a differential effect of nutrients versus the addition of chlorhexidine. For example, although both treatments tended to increase the overall biomass, chlorhexidine had a greater effect. This may be explained by its suppression of grazer populations versus the positive effects of nutrient additions (see below).
FIG. 1.
Representative CLSM photomicrographs of control, chlorhexidine-treated, and nutrient control-treated river biofilm communities. The color wheel shows bacteria (green), T. vulgaris-tetramethyl rhodamine isothiocyanate lectin binding polymer (red), and photosynthetic biomass (blue/magenta).
FIG. 2.
(A and B) Results of image analyses of confocal laser micrographs illustrating the effects of the chlorhexidine and nutrient treatments on the relative abundances of algae, cyanobacteria, and bacteria in the river biofilms, by treatment. The parameters indicated by different letters are significantly different (P < 0.05). (B) Proportional illustration of the impacts of treatments on the relative abundances of algae, cyanobacteria, and bacteria in the river biofilms.
The apparent lack of published results of toxicological assessments of chlorhexidine prevents comparison to established values. To provide a set of comparative values, we assessed the impacts of 0, 10, and 100 μg liter−1 of chlorhexidine on a selection of algal and cyanobacterial species, and no effects were observed at the highest concentration of 100 μg liter−1. No effects on growth were reported for the following algae and cyanobacteria in our tests: S. quadricauda, Selenastrum sp., Ulothrix sp., A. falcatus, O. tenius, Synedra sp., Thalassiosira sp., and the cyanobacteria Anabaena sp., G. nostochinea, Lyngbya sp., M. aeruginosa, and Nostoc sp. (data not shown). These findings were not in keeping with our in situ microscale evaluations showing significant impacts on the composition of the complex community, particularly at 100 μg liter−1.
In other instances (22), there have also been clear effects of a pharmaceutical or personal-care product on the photosynthetic biomass in a community context. The proportional analyses presented in Fig. 2 further illustrate the general shift to a more bacterium-dominated community, particularly at 100 μg liter−1 chlorhexidine or its molar equivalent in nutrients. These shifts have significant ecological implications for community carbon and energy flow as a consequence of the close coupling of algae and bacterial activity. Effects on the algal and protozoan-metazoan groups can result in a broad range of effects on biofilm development, given their critical roles as “ecosystem engineers” (3, 16). Ordinarily, studies have indicated a positive relationship between phototrophic and heterotrophic organisms (11, 12, 14). Changes in the ratio of phototrophs to heterotrophs result in shifts in both the nutrient-processing capacity and the natural food web structure of river communities. Wilson et al. (46), examining the effects of antimicrobials, noted substantial changes in community composition for algae in river biofilms exposed to <1-μg liter−1 concentrations of agents such as triclosan. An additional concern is that although biomass increases similar to those resulting from a nutrient addition may occur, underlying changes in the species composition may have negative effects on functional diversity in the community. That such changes occur in this instance is supported by the detection of changes in functional-activity levels, as indicated by carbon utilization analyses (see below).
Community activity.
Carbon utilization assays indicated that the 100-μg liter−1 chlorhexidine treatments had a strong and significant negative effect relative to control values, as well as to the addition of chlorhexidine's molar equivalent as nutrients in all categories (Fig. 3A). In contrast, the 10-μg liter−1 nutrient equivalent and chlorhexidine treatments had significant positive effects, particularly on the utilization of carboxylic acids, amino acids, and amines (Fig. 3B). The South Saskatchewan River is carbon and nutrient limited (24, 30), and as a consequence, it has typically been highly responsive to additions of potential nutrient sources. Thus, the response to nutrients was in keeping with previous observations (30).
FIG. 3.
(A and B) Differential displays of the carbon utilization assays of control biofilms and those growing with 10 μg liter−1 (B) and 100 μg liter−1 (A) chlorhexidine treatments and their respective nutrient controls. The circled data points are significantly different from their respective control values (P ≤ 0.05); those in solid black are significantly different from their respective treatments, either nutrient or chlorhexidine (P < 0.05).
Thymidine incorporation provided another measure of community metabolic potential. Although subject to bias, thymidine incorporation is a generally accepted measure of the activity of the bacterial component of microbial communities (38). In this case, only the 10-μg liter−1 molar equivalent nutrient treatments resulted in a statistically significant change in thymidine incorporation, with a 50% decrease. Similarly, thickness or depth has often been used as a measure of response to treatment in biofilm research and has proven responsive to a variety of stresses, including nutrients, oxygen, metals, and pharmaceuticals (6, 22, 23, 24, 30). In the current study, however, none of the treatments resulted in a significant change in average biofilm thickness relative to control biofilms. Thickness measurements are often used as an integrator of many effects, as indicated above; however, as can be seen in the changes in biomass observed in the biofilms (Fig. 2), it may mask important changes in biofilm architecture. Indeed, it is not unusual for biofilm thickness to be maximal when biofilm density is minimal; this is due in particular to the distribution of pores and channels, which may arise from growth patterns or grazing activity, both of which may be invoked to explain the observed patterns in this study. Thus, the effects of treatments on thymidine incorporation were in keeping with the observed effects on bacterial biomass, biofilm thickness, and other measures that detected a minimal impact of chlorhexidine on these relatively gross measures of the biofilm community. It is useful to consider that from a community analysis perspective, these measures tend to have a bias toward a small fraction of the community, as in carbon utilization assays (40), or lack the resolution, i.e., thickness and thymidine incorporation, to detect subtle but highly relevant changes. Therefore, these measures may have limited utility in assessing community level responses to environmental stress.
Stable-isotope analysis may be used to assess potential incorporation of added carbon sources and as a global indicator of changes in community composition. We have demonstrated that pharmaceuticals may significantly shift community structure, resulting in a parallel shift in the δ13C signature; this has been the case when, for example, cyanobacteria have been eliminated from the community (22). Analyses of chlorhexidine indicated that its δ13C value was −22.8, while that of the added glucose carbon was −10. Analyses of community carbon indicated that there was a significant change in both the 10-μg liter−1 (−29.7 chlorhexidine; −29.8 nutrients) and 100-μg liter−1 (−30.1 chlorhexidine; −30.1 nutrients) treatments relative to control biofilms (−28.3) (P < 0.05). The general values reported are in keeping with those obtained in similar studies for river biofilms (22). Given the shift to a more negative signature, there is no indication that chlorhexidine, with a δ13C value of −22.8, was substantially incorporated into community carbon, which has a δ13C value of −29 to −30. However, the sensitivity of the method is an issue, given the proportion of added chlorhexidine carbon to total available carbon in the river biofilm community. The lack of difference between nutrient and chlorhexidine treatments at all levels would be consistent with relatively subtle changes in community structure that may not be reflected in these global values but might be captured in compound-specific analyses examining DNA, lipid, or protein.
Effects on community composition.
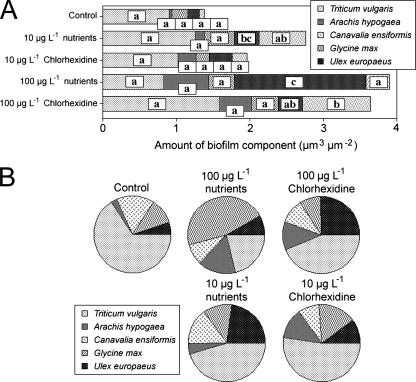
Altered community structure, particularly in bacterial functional groups, may be indicated by in situ lectin binding analyses (22, 23, 30). In this case, relatively few significant changes in lectin binding patterns were observed. The 10-μg liter−1 molar equivalent of nutrients resulted in an increase in G. max binding, as did the 100-μg liter−1 nutrient equivalent (Fig. 4). In the chlorhexidine treatments, there was a significant increase only in the binding of U. europaeus lectin at 100 μg liter−1. No other significant effects were observed. However, the contrast in glycoconjugate composition between the 100-μg liter−1 nutrient equivalent and the 100-μg liter−1 chlorhexidine treatments is indicative of differences in the community composition and thus denotes a contrast in the effects of the nutrients versus chlorhexidine. The change in the relative abundances of the five lectins is evident in Fig. 4B; the similarity of effects between the 10-μg liter−1 treatments and the contrasts at 100-μg liter−1 molar equivalent of nutrients or chlorhexidine are particularly apparent. Changes in lectin binding patterns and glycoconjugate nature are consistent with underlying changes in the population structure of the community, including changes in the algal, cyanobacterial, and bacterial components, which all contribute to the biofilm exopolymeric matrix (22, 30).
FIG. 4.
(A) Effects of chlorhexidine and nutrient treatments on the composition of river biofilm exopolymer, as determined by in situ lectin binding analyses. Parameters indicated by different letters are significantly different (P < 0.05). (B) Impacts of treatments on the relative abundances of lectin binding sites in the river biofilms. A panel of five lectins was used to assess the nature of the glycoconjugates present in the biofilms.
The results obtained for fluorescent lectin binding studies were in keeping with subtle changes in the underlying community species composition that were not evident in the gross measures of biofilm mass and thymidine incorporation. Fluorescent lectin binding also indicated that there was a differential effect of antimicrobial and nutrient additions in the 10- and 100-μg liter−1 treatments.
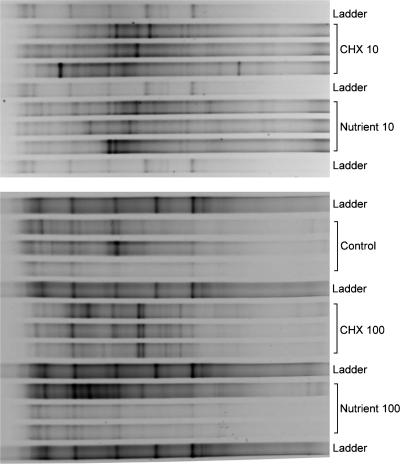
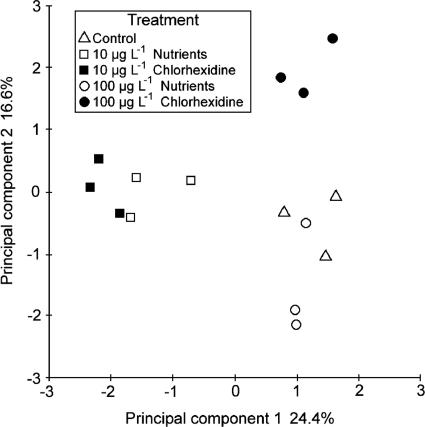
Comparative DGGE analyses of DNA extracted from the control-, nutrient-, and chlorhexidine-treated biofilm communities were performed to assess the effects of the treatments on bacterial community composition (Fig. 5). PCR products targeted by the eubacterial primers were amplified from replicates of all three treatments and separated by DGGE. For the 10-μg liter−1 treatments, visual inspection of the DGGE gels indicated that the greatest change in banding patterns relative to the control was detected for the nutrient treatment (Fig. 5). Principal-component analyses of the DGGE data indicated that at 100 μg liter−1 the control, nutrient equivalent, and chlorhexidine treatments resulted in apparently different communities and that the nutrient treatment grouped more closely with control communities (Fig. 6). The results from the ANOSIM analysis of the PCA results showed that there were significant differences (R = 1.00; P < 0.001) in the structures of microbial communities between the control and chlorhexidine treatment at 10 μg liter−1, the 10-μg liter−1 nutrient equivalent, and when chlorhexidine was applied at 100 μg liter−1. However, there was no significant difference (R = 0.22; P < 0.001) between the microbial community structures of the control and chlorhexidine treatment groups at 100 μg liter−1 nutrient equivalent treatment. Also, there was no significant difference detected between communities developing with chlorhexidine at 10 μg liter−1 and the corresponding 10-μg liter−1 nutrient treatment.
FIG. 5.
Denaturing gradient gel comparison of the effects of treatments with control and 10 or 100 μg liter−1 of nutrients or chlorhexidine (CHX) on the nature of the developing bacterial community. Each bracketed region of the gel contains the results obtained with DNA from three independent replicate reactors.
FIG. 6.
Results of PCA analyses of replicate DGGE data showing the significant differences (P < 0.001; ANOSIM) between chlorhexidine-treated communities and the control and nutrient treatments.
In contrast, our previous studies showed strong and differential impacts of both nutrients and contaminants, such as diclofenac at 10 μg liter−1 (23), on community structure. Despite inherent limitations due to DNA extraction and PCR, DGGE can detect shifts in community composition between treatments and sites (45).
FISH is also an effective tool for these comparisons and provides additional in situ comparative data on community composition. Analyses through application of a suite of FISH probes (Table 1) indicated that there were significant reductions in the numbers of probe-positive cells for the beta- and gammaproteobacterial probes in the nutrient and chlorhexidine treatments, whereas the total EUB338 counts were not significantly affected, in keeping with more general measures. Notably, the number of ARC915 probe-positive cells was reduced by the 10-μg liter−1 chlorhexidine and significantly (P < 0.05) reduced by the molar equivalent nutrient treatments relative to control biofilms. However, in all cases, there was no significant difference between the nutrient and chlorhexidine treatments. This is in contrast to the results detected at 100 μg liter−1 chlorhexidine, where the communities were significantly different (P < 0.05) from each other. In general, the molecular-level analyses indicated changes in community structure as a consequence of the addition of chlorhexidine, including potential effects on the archaeal members.
TABLE 1.
Results of FISH analyses of control-, nutrient-, and chlorhexidine-treated biofilm communities
| Treatment | % Area (mean ± SD)
|
|||
|---|---|---|---|---|
| Betaproteobacteria | Gammaproteobacteria | Arch915 | EUB338 | |
| Control | 2.9 ± 3.2b | 3.2 ± 4.4b | 3.5 ± 3.3b | 6.1 ± 5.9a |
| Nutrient (10 μg liter−1) | 1.9 ± 1.9a | 1.5 ± 1.4a | 1.5 ± 1.2a | 6.5 ± 6.1a |
| Chlorhexidine (10 μg liter−1) | 1.7 ± 1.3a | 1.8 ± 1.4a | 2.8 ± 1.2a,b | 6.1 ± 6.1a |
Not significantly different from values with similar labels (P < 0.05).
Not significantly different from values with similar labels (P < 0.05).
These changes may have impacts on biogeochemical cycling, specific pathways, or overall community diversity and redundancy. For example, Lawrence et al. (24) reported impacts of nutrients and metals on specific groups, eliminating, for example, the nirS but not the nirK denitrifying bacteria in the community; thus, the denitrification process would continue but redundancy required to adapt to changing conditions might be lost. As hypothesized, in addition to being generally toxic, chlorhexidine may act as a selective agent with differential effects on populations within the microbial biofilm community.
Chlorhexidine as a selective agent.
Chlorhexidine is potentially a highly selective agent that could significantly alter community structure. This phenomenon has been reported for dental and medical studies of oral biofilms (18, 41, 42). Although medical and dental research reports are based on levels of chlorhexidine orders of magnitude higher than those in the current research or concentrations likely to occur in the environment, they support the notion that chlorhexidine has the capacity to act as a highly differentially selective agent on specific members of a microbial community, resulting in alterations in community function and diversity, as indicated by the DGGE and FISH analyses described above.
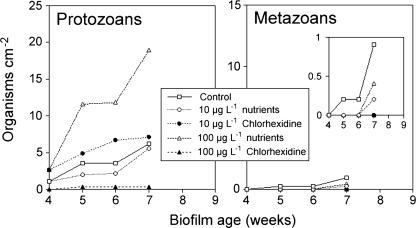
The most obvious significant effect of chlorhexidine may occur as a consequence of its impact on grazer populations. The introduction of chlorhexidine at 100 μg liter−1 resulted in the virtual elimination of protozoans and metazoans in the biofilms (Fig. 7). In contrast, its molar equivalent in nutrients was highly stimulatory for protozoans. At 10 μg liter−1, neither the nutrients nor the chlorhexidine resulted in effects on protozoans that were significantly different from the control communities. The observed effects of 100 μg liter−1 chlorhexidine on the invertebrate populations in the community were in keeping with the limited reports of chlorhexidine being “very highly toxic to aquatic invertebrates” at levels of 32 μg liter−1 for D. magna (2). Interestingly, our toxicological evaluation of chlorhexidine at 10, 50, and 100 μg liter−1 indicated no effects on cultures of protists or rotifera, with the exception of the apparent immobilization of Spirostomum sp. at the 50-μg liter−1 concentration. This is supportive of the claim that community-based evaluations, although more time consuming and complex, may provide greater ecological realism in evaluations of toxicological effects (34). Although the protozoans and micrometazoans are increasingly recognized as important in lotic ecosystems (37), it is unclear which factors influence the abundance and species compositions of these populations.
FIG. 7.
Enumeration of protozoa (ciliates, flagellates, and amoebas) (left) and micrometazoa (rotifers and nematodes) (right) showing the effects of nutrient amendments and chlorhexidine on the cumulative grazer density within river biofilm communities. The inset shows metazoan data with a reduced y axis.
Chlorhexidine degradation.
Microbial degradation of chlorhexidine has not been extensively studied, though its degradation by activated sludge communities has been reported (reviewed in reference 27). In a pure-culture study, a number of intermediates were detected, including chlorhexidine degradation intermediates B and C, p-chlorphenylurea, and p-chloraniline; the last two metabolites have been shown to have no antimicrobial activity (27). Our observations to date using [14C]chlorhexidine indicated that after 120 days of incubation there was no difference between the live biofilm and killed controls in terms of 14CO2 evolved (<1% of added label). The lack of mineralization of chlorhexidine supports our contention that the observed effects on the river biofilm community may be attributed to chlorhexidine or its major metabolites. Reports have indicated that chlorhexidine may be extensively sorbed; however, our observations indicated that only 8% of labeled chlorhexidine was sorbed to the living biofilm. Extensive soft X-ray transmission studies of chlorhexidine distribution in these river biofilms indicated that both bacterial and algal (diatom) biomass was involved in the sorption process and that bacterial and algal lipids were the specific sorption sites (10).
In general, our findings suggest that microbial community composition is sensitive to the presence of low levels of chlorhexidine. As noted by Porsbring et al. (34), the approach of community level testing in ecotoxicology is useful because of its realism and reliability in assessing ecological consequences of exposure. In particular, the use of longer time frames allows the evaluation of effects on succession and biological interactions that are important in assessing ecosystem level effects. There is, however, a need to expand and refine techniques that can reveal significant relevant change in microbial communities. One issue in the use of in situ or derived microbial communities is that of seasonal effects on the outcome, as we have reported previously (7), and in the case of chemicals such as chlorhexidine, the influence of temperature differences, turbidity, pH changes, etc., on the direct effects of the compound (42). Community level study of chlorhexidine supports the general conclusion that the chemical poses a risk in aquatic habitats (2). However, additional data on environmental concentrations are required for more conclusive risk assessment.
Acknowledgments
This research was supported by grants from Health Canada and Environment Canada.
Lindsey D. Mooney is acknowledged for excellent technical support.
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Amann, R., L. Krumholz, and D. A. Stahl. 1990. Fluorescent-oligonucleotide probing of whole cells for determinative, phylogenetic, and environmental studies in microbiology. J. Bacteriol. 172:762-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anonymous. 1996. Chlorhexidine diacetate: R.E.D. Facts. U.S. Environmental Protection Agency, Prevention Pesticides and Toxic Substances (7508W). EPA-738-F-96-025. U.S. Environmental Protection Agency, Washington, DC.
- 3.Besemer, K., G. Singer, R. Limberger, A.-K. Chlup, G. Hochedlinger, I. Hödl, C. Baranyi, and T. J. Battin. 2007. Biophysical controls on community succession in stream biofilms. Appl. Environ. Microbiol. 73:4966-4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boon, N., W. D. Windt, W. Verstraete, and E. M. Top. 2002. Evaluation of nested PCR DGGE (denaturing gradient gel electrophoresis) with group-specific 16S rRNA primers for the analysis of bacterial communities from different wastewater treatment plants. FEMS Microbiol. Ecol. 39:101-112. [DOI] [PubMed] [Google Scholar]
- 5.Castillo, J. A., A. Pinazo, J. Carilla, M. R. Infante, M. A. Alsina, I. Haro, and P. Clapés. 2004. Interaction of antimicrobial arginine-based cationic surfactants with liposomes and lipid monolayers. Langmuir 20:3379-3387. [DOI] [PubMed] [Google Scholar]
- 6.Chénier, M. R., D. Beaumier, N. Fortin, R. Roy, B. T. Driscoll, J. R. Lawrence, and C. W. Greer. 2006. Influence of nutrient inputs, hexadecane and temporal variations on denitrification and community composition of river biofilms. Appl. Environ. Microbiol. 72:575-584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chénier, M. R., D. Beaumier, R. Roy, B. T. Driscoll, J. R. Lawrence, and C. W. Greer. 2003. Impact of seasonal variations and nutrient inputs on the cycling of nitrogen and the degradation of hexadecane by replicated river biofilms. Appl. Environ. Microbiol. 69:5170-5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clarke, K. R. 1993. Non-parametric multivariate analyses of changes in community structure. Aust. J. Ecol. 18:117-143. [Google Scholar]
- 9.Davies, G. E., J. Francis, A. R. Martin, F. L. Rose, and G. Swain. 1954. 1:6-Di 4-chlorophenyldiguanidohexane (“Hibitane”). Laboratory investigation of a new antibacterial agent of high potency. Br. J. Pharmacol. 9:192-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dynes, J. J., J. R. Lawrence, D. R. Korber, G. D. W. Swerhone, G. G. Leppard, and A. P. Hitchcock. 2006. Quantitative mapping of chlorhexidine in natural river biofilms. Sci. Total Environ. 369:369-383. [DOI] [PubMed] [Google Scholar]
- 11.Haak, S. K., and G. A. McFeters. 1982. Microbial dynamics of an epilithic mat community in a high alpine stream. Appl. Environ. Microbiol. 43:702-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haak, S. K., and G. A. McFeters. 1982. Nutritional relationships among microorganisms in an epilithic biofilm community. Microb. Ecol. 8:115-126. [DOI] [PubMed] [Google Scholar]
- 13.Herbert, V. R., J. R. Middleton, E. Tomaszewska, and L. K. Fox. 2003. Methodology for quantifying residues of chlorhexidine in raw dairy milk. J. Agric. Food Chem. 51:567-570. [DOI] [PubMed] [Google Scholar]
- 14.Hespinall, J. A., and R. L. Fuller. 1994. Periphyton reactions to different light and nutrient levels and the response of bacteria to these manipulations. Arch. Hydrobiol. 131:161-173. [Google Scholar]
- 15.Hope, C. K., and M. Wilson. 2004. Analysis of the effects of chlorhexidine on oral biofilm vitality and structure based on viability profiling and an indicator of membrane integrity. Antimicrob. Agents Chemother. 48:1461-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones, C. G., J. H. Lawton, and M. Shackak. 1994. Organisms as ecosystem engineers. Oikos 69:373-386. [Google Scholar]
- 17.Kodama, H., T. Hashimoto, M. Tsuruoka, Y. Kido, M. Uyeda, and M. Shibata. 1988. Microbial degradation of disinfectants. IV. Treatment by activated sludge of chlorhexidine. Eisei Kagaku 34:408-413. [Google Scholar]
- 18.Koljalg, S., P. Naaber, and M. Mikelsaar. 2002. Antibiotic resistance as an indicator of bacterial chlorhexidine susceptibility. J. Hosp. Infect. 51:106-113. [DOI] [PubMed] [Google Scholar]
- 19.Konopka, A., A. L. Oliver, and R. F. Turco. 1998. The use of carbon substrate utilization patterns in environmental and ecological microbiology. Microb. Ecol. 35:103-115. [DOI] [PubMed] [Google Scholar]
- 20.Larson, E. L., and the APIC Guideline Committee. 1995. APIC guidelines for handwashing and hand antisepsis in healthcare settings. Am. J. Infect. Control 23:251-269. [DOI] [PubMed] [Google Scholar]
- 21.Lawrence, J. R., G. D. W. Swerhone, and T. R. Neu. 2000. Design and evaluation of a simple rotating annular reactor for replicated biofilm studies. J. Microbiol. Methods 42:215-224. [DOI] [PubMed] [Google Scholar]
- 22.Lawrence, J. R., G. D. W. Swerhone, L. I. Wassenaar, and T. R. Neu. 2005. Effects of selected pharmaceuticals on riverine biofilm communities. Can. J. Microbiol. 51:655-669. [DOI] [PubMed] [Google Scholar]
- 23.Lawrence, J. R., G. D. W. Swerhone, E. Topp, D. R. Korber, T. R. Neu, and L. I. Wassenaar. 2007. Structural and functional responses of river biofilm communities to the non-steroidal anti-inflammatory diclofenac. Environ. Toxicol. Chem. 26:236-245. [DOI] [PubMed] [Google Scholar]
- 24.Lawrence, J. R., M. Chénier, R. Roy, D. Beaumier, N. Fortin, G. D. W. Swerhone, T. R. Neu, and C. W. Greer. 2004. Microscale and molecular assessment of the impacts of nickel, nutrients and oxygen level on river biofilm communities. Appl. Environ. Microbiol. 70:4326-4339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manz, W., R. I. Amann, W. Ludwig, M. Wagner, and K.-H. Schleifer. 1992. Phylogenetic oligodeoxynucleotide probes for the major subclasses of proteobacteria: problems and solutions. Syst. Appl. Microbiol. 15:593-600. [Google Scholar]
- 26.Manz, W., K. Wendt-Potthoff, T. R. Neu, U. Szewzyk, and J. R. Lawrence. 1999. Phylogenetic composition, spatial structure and dynamics of lotic bacterial biofilms investigated by fluorescent in situ hybridization and confocal scanning laser microscopy. Microb. Ecol. 37:225-237. [DOI] [PubMed] [Google Scholar]
- 27.Murayama, T., S. N. Tuda, M. Nishiyama, K. Nakagawa, Y. Matsuo, Y. Isohama, and Y. Kido. 2005. Microbial degradation of disinfectants. A new chlorhexidine degradation intermediate (CHDI), CHID-C, produced by Pseudomonas SP. strain No. A-3. J. Health Sci. 51:357-361. [Google Scholar]
- 28.Muyzer, G., E. C. De Waal, and A. G. Uitterlinden. 1993. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 59:695-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muyzer, G., and N. B. Ramsing. 1995. Molecular methods to study the organization of microbial communities. Water Sci. Technol. 32:1-9. [Google Scholar]
- 30.Neu, T. R., G. D. W. Swerhone, U. Bockelmann, and J. R. Lawrence. 2005. Effect of carbon, nitrogen and phosphorus on the nature and development of lectin-specific glycoconjugates in lotic biofilms. Aquat. Microb. Ecol. 38:283-294. [Google Scholar]
- 31.Neu, T. R., S. Wölfl, and J. R. Lawrence. 2004. 3-Dimensional differentiation of phototrophic biofilm constituents by multi channel confocal and 2-photon laser scanning microscopy. J. Microbiol. Methods 56:161-172. [DOI] [PubMed] [Google Scholar]
- 32.Neu, T. R., G. D. W. Swerhone, and J. R. Lawrence. 2001. Assessment of lectin-binding-analysis for in situ detection of glycoconjugates in biofilm systems. Microbiology 147:299-313. [DOI] [PubMed] [Google Scholar]
- 33.Packroff, G., J. R. Lawrence, and T. R. Neu. 2002. In situ confocal laser scanning microscopy of protozoans in pure cultures and complex communities. Acta Protozool. 41:245-253. [Google Scholar]
- 34.Porsbring, T., A. Arrhenius, T. Backhaus, M. Kuylenenstierna, M. Scholze, and H. Blanck. 2007. The SWIFT periphyton test for high-capacity assessments of toxicant effects on microalgal community development. J. Exp. Mar. Biol. Ecol. 349:299-312. [Google Scholar]
- 35.Pratten, J., P. Barnett, and M. Wilson. 1998. Composition and susceptibility to chlorhexidine of multispecies biofilms of oral bacteria. Appl. Environ. Microbiol. 64:3515-3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ranganathan, N. S. 1996. Chlorhexidine, p. 235-264. In J. M. Ascenzi (ed.), Handbook of disinfection and antiseptics. Marcel Dekker Inc., New York, NY.
- 37.Reiss, J., and J. M. Schmid-Araya. 2007. Existing in plenty: abundance, biomass and diversity of ciliates and meiofauna in small streams. Freshw. Biol. 53:652-668. [Google Scholar]
- 38.Robarts, R. D., and T. Zohary. 1993. Fact or fiction—bacterial growth rates and production as determined by (methyl-3H) thymidine? Adv. Microb. Ecol. 19:371-425. [Google Scholar]
- 39.Robarts, R. D., and R. J. Wicks. 1989. [Methyl-3H]thymidine macromolecular incorporation and lipid labeling: their significance to DNA labelling during measurements of aquatic bacterial growth rate. Limnol. Oceanogr. 34:213-222. [Google Scholar]
- 40.Ros, M., M. Goberna, J. A. Pascual, S. Klammer, and H. Insam. 2008. 16S rDNA analysis reveals low microbial diversity in community level physiological profile assays. J. Microbiol. Methods 72:221-226. [DOI] [PubMed] [Google Scholar]
- 41.Russell, A. D., and M. J. Day. 1993. Antibacterial activity of chlorhexidine. J. Hosp. Infect. 25:229-238. [DOI] [PubMed] [Google Scholar]
- 42.Sreenivasan, P. K., and E. Gittins. 2004. The effects of chlorhexidine mouthrinse on culturable organisms of the tongue and saliva. Microbiol. Res. 159:365-370. [DOI] [PubMed] [Google Scholar]
- 43.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes in bacteria systematics, p. 205-248. In E. Stackebrandt and M. Goodfellow (ed.), Sequencing and hybridization techniques in bacterial systematics. John Wiley, Chichester, United Kingdom.
- 44.Teece, M. A., and M. L. Vogel. 2004. Preparation of ecological and biochemical samples for isotope analysis, p. 177-202. In P. A. de Groot (ed.), Handbook of stable isotope analytical techniques. Elsevier, Amsterdam, The Netherlands.
- 45.Webster, N. S., and A. P. Negri. 2006. Site-specific variation in Antarctic marine biofilms established on artificial surfaces. Environ. Microbiol. 8:1177-1190. [DOI] [PubMed] [Google Scholar]
- 46.Wilson, B. A., V. H. Smith, F. de Noyelles, Jr., and C. K. Larive. 2003. Effects of three pharmaceutical and personal care products on natural freshwater algal assemblages. Environ. Sci. Technol. 37:1713-1719. [DOI] [PubMed] [Google Scholar]