Abstract
Group C rotaviruses have been recognized as a cause of acute gastroenteritis in humans, cattle, and swine, although the true epidemiologic and clinical importance of this virus in these hosts has not yet been fully established. A real-time PCR assay based on a broadly reactive primer pair was developed and used to quantitatively determine the viral load of group C rotaviruses in environmental samples. A total of 35 raw and 35 treated sewage samples collected at the same sampling time in four Hungarian sewage treatment plants during a survey in 2005 were tested for the presence of group C rotaviruses. The overall detection rates were 91% (32 of 35) for the influent and 57% (20 of 35) for the effluent samples. Molecular characterization of the amplified partial VP6 gene revealed the cocirculation of human and animal (i.e., bovine and porcine) strains that were easily distinguishable by melting curve analysis. Human strains yielded relatively high viral loads (mean, 1.2 × 107; median, 6.9 × 105 genome equivalents per liter influent sewage) and appeared to display seasonal activity over the study period, whereas animal strains appeared to circulate throughout the year at much lower average titers (bovine strains mean, 9.9 × 104; median, 3.0 × 104; porcine strains mean, 3.9 × 104; median, 3.1 × 104 genome equivalents per liter influent sewage). Our findings suggest that monitoring of communal sewage may provide a good surrogate for investigating the epidemiology and ecology of group C rotaviruses in humans and animals.
Rotaviruses (family Reoviridae, genus Rotavirus) are common enteric pathogens of humans and animals (7). Rotaviruses are classified into seven (sero) groups, designated A to G (7). A candidate novel rotavirus group has been recently identified (42). Rotaviruses of all serogroups share the capsid morphology and the unique genome structure that is composed of 11 segments of double-stranded RNA. However, rotaviruses among serogroups share no genetic and antigenic relatedness (34).
The three most important human rotavirus serogroups (A, B, and C) have remarkably different epidemiologic features and levels of public health importance. Group A rotaviruses are the main cause of childhood diarrhea worldwide and are responsible for 400,000 to 700,000 deaths per year in children under 5 years of age (30). Group B rotaviruses were first identified in China in the 1980s, where they caused large epidemics of gastroenteritis affecting all age groups, with the preponderance of cases in young healthy adults (12). Recently, group B rotaviruses have been identified among children and adults in Southern Asia (1, 16). Group C rotaviruses were first detected from an Australian infant hospitalized with diarrhea in 1973 (33). Since then, group C rotaviruses have been recognized as a cause of gastroenteritis in all age groups, accounting for either sporadic cases or large outbreaks of gastroenteritis in closed and semiclosed communities (8, 15, 20, 22, 31, 32, 34-36, 38). Cohort and seroepidemiological studies have indicated that virtually all children are infected with group A rotaviruses by 5 years of age (28, 39), while the rate of infection with group C rotaviruses is much lower, and antibody prevalence usually peaks at an age of 40 to 50 years or later (14, 21, 37).
The differences observed for the epidemiological features of different groups of rotaviruses may be accounted for by different mechanisms of transmission or by their different stability levels in the environment and their overall ecology. Rotavirus infections are thought to be acquired primarily via the fecooral route, including person-to-person contact (28), consumption of contaminated water or food (2, 10, 12, 28, 29), and zoonotic transmission (9). Airborne transmission has been hypothesized but has not yet been formally demonstrated (28).
The incidence of group C rotavirus diarrhea in children is generally considered low (3, 6, 27, 35, 38, 41). The low number of identified cases does not permit us to understand the ecology and epidemiology, including the source and mode of transmission of group C rotavirus, in a variety of settings and areas in the world. These shortcomings prompted us to investigate this issue from the viewpoint of environmental virology. Because this group of viruses is difficult to detect in clinical specimens, we sought to determine whether communal sewage serves as a good indicator for group C rotavirus infection. In this study, we provide novel insight into the ecology of this group of rotaviruses by using a broadly reactive primer pair in a sensitive real-time PCR assay.
MATERIALS AND METHODS
Sample collection and treatment plant description.
A total of 35 influent sewage samples were collected by the point sampling approach at four plants (coded as A to D) in Baranya County, Hungary, during sewage monitoring activity in 2005. In addition, 35 effluent samples were collected on the same day of field work. However, because the influent and effluent specimens could not be considered paired samples (Table 1), data presented here apply primarily to the influent samples, unless otherwise indicated. All sewage samples were stored at 4°C for no more than 2 days before the concentration procedure was undertaken.
TABLE 1.
Description of sewage plants involved in the monitoring of group C rotavirusa
| Plant | Population | Sewage characteristics | Treatment procedure
|
Transit time (h) | Daily sewage yield (m3) | ||
|---|---|---|---|---|---|---|---|
| P.t. | S.t. | Chlor. | |||||
| A | 160,000 | Communal | A | A | + | 30 | 27,000 |
| B | 3,100 | Communal | B | B | +/− | 72 | Unknown |
| C | 2,600 | Communal | B | A | + | 8 | 750 |
| D | 3,100 | Communal | B | A | − | 13 | 200 |
P.t., primary treatment (physical process); S.t., secondary treatment (biological process); Chlor., chlorination; P.t. A includes screens, grit chambers, and sedimentation tanks (in that order); P.t. B includes screens only; S.t. A includes aeration tanks and sedimentation tanks; S.t. B includes an aeration tank only; +, applied; −, not applied; +/−, applied occasionally. The aeration tank and the sedimentation tank in the indicated plants are parts of the biological process of the applied sewage treatment technology.
Controls.
A human stool sample positive for group C rotavirus was employed as a control in our assay (K. Bányai and G. Szűcs, unpublished data). In addition, a TOPO vector (pBlueBac4.5; Invitrogen, Grand Island, NY) containing the nearly full-length VP6 gene insert (nucleotide [nt] 11 to 1353; B. Jiang, unpublished data) was used to optimize the assay.
Sample concentration.
Concentration of the viruses present in the sewage was performed as described by Minor et al. (24) and Nadan et al. (25), with modifications published recently. Briefly, each sewage sample was clarified by centrifugation for 30 min at 3,000 × g, and the pellet was resuspended in 10 ml of the supernatant. The remaining portion of supernatant was saved. Chloroform was added to the resuspended sample to a concentration of 10% (vol/vol; Sigma, St. Louis, MO) and mixed, and the mixture was centrifuged again for 10 min at 3,000 × g. The combined supernatants were then supplemented with 2.2% (wt/vol) NaCl (Sigma, St. Louis, MO) and 7% (wt/vol) polyethylene glycol 6000 (Fluka, Buchs, Germany). The mixture was stirred at 4°C overnight and then centrifuged for 2 h at 2,000 × g at 4°C. The supernatant was discarded, and the pellet was resuspended in lysis buffer (1/100 of the starting volume). The lysis buffer contained 10 M guanidinium thiocyanate (GuSCN) (Sigma, St. Louis, MO) in 0.1 M Tris-HCl buffer (pH 6.4) as previously described (4).
Viral RNA extraction.
Viral RNA was extracted by using a previously described procedure, with modifications (4). Briefly, the resuspended pellet in lysis buffer was incubated at 56°C for 25 min before silica particles were added, to a final concentration of 0.75% (vol/vol) (silicon dioxide; Sigma, St. Louis, MO). The mixture was stirred to facilitate the binding of nucleic acids to the silica particles for a maximum of 30 min at 25°C, and then it was centrifuged at 11,000 × g for 5 min at 25°C; the supernatant was decanted. The silica pellet was subsequently washed twice with 2 ml of 4 M GuSCN in Tris (0.1 M [pH 6.4]) buffer, then twice with 2 ml of 70% (vol/vol) ethanol (Spektrum 3D, Debrecen, Hungary), and once with 2 ml of acetone (Spektrum 3D, Debrecen, Hungary). After acetone was removed, the pellet was dried at 56°C. Total nucleic acid was eluted from the silica beads in 300 μl of diethylpyrocarbonate-treated water (Q-Bio Gene, Carlsbad, CA) by incubating the suspension for 15 min at 65°C in a heating block. Finally, the eluate was centrifuged at 18,000 × g for 10 min at 25°C, and the supernatant was pipetted off and aliquoted in sterile nuclease-free tubes. This nucleic acid sample was stored at −80°C before use in the molecular detection of group C rotaviruses.
Oligonucleotide primers.
Although broadly reactive primers used to detect group C rotaviruses were described previously (35), utilization of these primer sets in our preliminary quantitative PCR (qPCR) assay revealed considerable nonspecific cross-reactions with nucleic acids present in the sewage samples. Therefore, we designed new primer pairs with the aim of increasing the specificity and sensitivity of the assay for the detection of group C rotavirus in sewage. The new oligonucleotide primers are complementary to the 3′ end region of the group C rotavirus VP6 gene. The expected size of the amplicon was 98 bp. The primer sequences are GRVC-LC-1 (5′-AGCCACATAGTTCACATTTCATCCTYCTG-3′) at nt 1325 to 1351 and GRVC-LC-3 (5′-TAGCATGAATCACGACTGGGTTTAGTC-3′) at nt 1282 to 1256.
RT.
Prior to undergoing reverse transcription (RT), 10 μl nucleic acid extract was heat denatured in 0.5-ml PCR tubes in the presence of 5 μM BMJ44 primer (35) for 5 min at 97°C. To prevent the reannealing of complementary strands, the tubes were immediately chilled in a precooled tube rack, followed by the addition of 15 μl RT mixture that contained a final concentration of 10 mM Tris (pH, 8.3), 50 mM KCl, 3 mM MgCl2, 0.1% bovine serum albumin, 0.4 mM deoxynucleoside triphosphate mixture, 2 U avian myeloblastosis virus reverse transcriptase, 4 U RNase inhibitor, and 50 μM dithiothreitol (all from Promega, Madison, WI). The tubes were then incubated for 1 h at 42°C.
qPCR.
The quantitative amplification of cDNA was carried out in a LightCycler 2.0 instrument using a LightCycler FastStart DNA MasterPLUS SYBR Green I kit (Roche Diagnostics GmbH, Penzberg, Germany). Briefly, the reaction mixture contained 0.25 μM of each sense- and antisense primer, 1× final concentration of the reaction mixture containing Taq DNA polymerase at the concentration recommended by the manufacturer, 2 μl cDNA and water to give a final reaction mixture volume of 20 μl. The amplification program included a preactivation step for the DNA polymerase at 95°C for 10 min, followed by 40 cycles of denaturation (95°C for 10 s), annealing (64°C for 10 s), and primer extension (72°C for 20 s), with a 20°C/s temperature transition profile between each step. Additional cycles were added when the reaction was anticipated not to cross the inflection point. Fluorescent signals were received at the end of each amplification step.
To quantify the copy numbers contained in the samples, we used an external calibration curve generated by 10-fold serial dilutions (concentration range, 108 to 100 copies) of plasmid DNA containing the target gene. To check the assay specificity, we performed melting point analyses, and subsequently the PCR products were run in agarose gels. The melting point analysis was initiated at 95°C for 1 s and a subsequent cooling step at 40°C for 1 s, followed by continuous sampling of the fluorescence signal until the temperature reached 95°C, with a temperature transition profile of 0.05°C/s. After the qPCR was completed, the amplicons of each sample were run in 2% agarose gels (Cambrex Bio Science Rockland Inc., Rockland, ME) containing ethidium bromide (0.5 μg/ml) and visualized on a UV transilluminator.
Because of the extreme sensitivity of the qPCR, we paid particular attention to preventing cross-contamination. Therefore, the preparation of nucleic acids and reagents, the addition of nucleic acids to aliquoted reagents, the performance of qPCR, and the analysis of the amplicons by electrophoresis were each done in separate rooms. Filtered pipette tips were utilized when the samples were added to the aliquoted reagent mixtures. Disposable gowns and powder-free latex gloves were used and changed for each consecutive step during the assay setup.
Sequencing and phylogenetic analysis.
Amplicons of RT-qPCRs were subjected to sequence analysis. The short amplified sequences used for this analysis were selected for their ability to distinguish strains of various species origin.
The amplicons were purified using minicolumns for the purification of PCR products according to the manufacturer's instructions (Promega, Madison, WI) and then subjected to direct sequencing. Cycle sequencing was performed in both directions with the two qPCR primers using a BigDye Terminator cycle sequencing Ready Reaction kit version 2.0 (Applied Biosystems). Briefly, 2 μl BigDye reagent and 1 μl primer (10 μM) were added to 7 μl purified template. The amplification profile included template denaturation at 96°C for 1 min, followed by 25 cycles of amplification (96°C for 20 s, 50°C for 5 s, and 60°C for 4 min). The dye-labeled product was purified as described previously and analyzed on an ABI Prism 310 DNA Sequencer (Applied Biosystems, Foster City, CA).
Nucleotide sequences were aligned with corresponding fragments of the VP6 gene of human, porcine, and bovine group C rotaviruses using GeneDoc software (26). Phylogenetic relationships were analyzed with MEGA software (version 2.1), using the neighbor-joining algorithm (19).
RESULTS
Assay sensitivity, specificity, and reproducibility.
To determine the viral load in a sample, a standard curve was obtained by plotting the cycle number needed for the amplification of DNA above the threshold (i.e., the crossing point [Cp] value) against the starting concentration of plasmid DNA, ranging from 100 to 108 copies per reaction. A linear relationship was obtained in the range of 101 to 108 copies (PCR efficiency, 1.926; error rate, 0.022). The detection limit, representing the lowest plasmid concentration that could be amplified in independent reactions, was 2 copies of plasmid DNA.
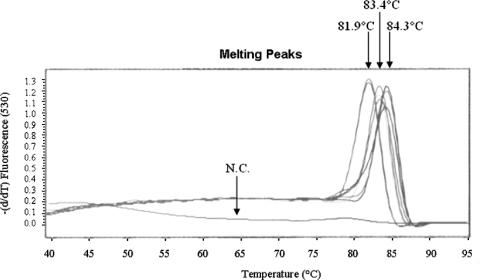
Melting point analysis was carried out to confirm the assay specificity. When testing sewage samples with this assay, three types of melting curves were obtained: one peaked at 81.9°C (with a standard deviation [SD] of 0.06°C), the other two curves peaked at 83.4°C (SD, 0.19°C) and 84.3°C (SD, 0.12°C) (Fig. 1). The agarose gel electrophoresis confirmed that DNA of the expected size (∼100 bp in length) was amplified (data not shown). Nonspecific bands of unknown origin were amplified only in a few of all positive samples (2 of 52 [3.8%]). Almost all samples were positive for at least one additional enteric virus (such as enteric adenoviruses, group A rotaviruses, astroviruses, noroviruses, hepatitis A viruses [E. Meleg and G. Szűcs, unpublished data]). Together, these findings suggest that although sewage samples contain a variety of heterogeneous nucleic acids, no significant cross-amplification of nonhomologous nucleic acids with these newly designed primers occurred.
FIG. 1.
Melting curve analysis of qPCR amplicons. Three amplicon populations of the PCR products were obtained; one peaked at 81.9°C (SD, 0.06°C), and the other two peaked at 83.4°C (SD, 0.19°C) and 84.3°C (SD, 0.12°C). Fluorescence was measured at 530 nm. N.C., negative control.
We validated our real-time PCR assay by assessing the intra- and interassay variability and demonstrated an overall high reproducibility (Table 2). The coefficient of variation (CV) was determined by measuring the Cp values of various dilutions of plasmid DNA. Independent assays were performed for each dilution in triplicate on three different days at 1- to 4-day intervals. The intra-assay CV ranged from 0.1% to 5.5% in the dilution range of 101 to 108 copies per reaction. The interassay variability showed similar ranges of CV values, from 0.2% to 2.2%. Together, these findings indicate that the assay is highly reproducible.
TABLE 2.
Reproducibility profile of group C rotavirus qPCR assay
| Plasmid concn (copy no./reaction) | Intra-assay variability of mean CPa values ± SD (CV)
|
Interassay variability of mean CP values ± SD (CV) | ||
|---|---|---|---|---|
| Day 1 | Day 2 | Day 3 | ||
| 108 | 9.62 ± 0.53 (5.5) | 9.88 ± 0.01 (0.1) | 10.07 ± 0.07 (0.7) | 9.85 ± 0.22 (2.2) |
| 107 | 13.49 ± 0.07 (0.5) | 13.36 ± 0.08 (0.6) | 13.54 ± 0.09 (0.7) | 13.46 ± 0.09 (0.7) |
| 106 | 16.99 ± 0.08 (0.5) | 16.86 ± 0.02 (0.1) | 17.13 ± 0.11 (0.6) | 16.99 ± 0.13 (0.8) |
| 105 | 20.54 ± 0.04 (0.2) | 20.48 ± 0.02 (0.1) | 20.57 ± 0.06 (0.3) | 20.53 ± 0.04 (0.2) |
| 104 | 23.93 ± 0.09 (0.4) | 23.84 ± 0.03 (0.1) | 24.03 ± 0.05 (0.2) | 23.93 ± 0.09 (0.4) |
| 103 | 27.63 ± 0.18 (0.7) | 27.56 ± 0.11 (0.4) | 27.70 ± 0.20 (0.7) | 27.63 ± 0.07 (0.3) |
| 102 | 31.23 ± 0.57 (1.8) | 31.11 ± 0.51 (1.6) | 31.55 ± 0.24 (0.8) | 31.29 ± 0.22 (0.7) |
| 101 | 34.4 ± 0.98 (2.8) | 34.38 ± 1.21 (3.5) | 35.07 ± 1.03 (2.9) | 34.61 ± 0.39 (1.1) |
CP, crossing point.
Detection and quantitation of group C rotaviruses in sewage samples.
Group C rotavirus was detected in 32 of 35 (91.4%) influent samples and 20 of 35 (57.1%) effluent samples. The detection rates of group C rotavirus varied among the treatment plants; 17 of 17 raw samples and 9 of 17 treated samples were positive in plant A, while the detection rate was 5 of 6 for both the influent and effluent samples from plant B and 4 of 6 for both the raw and treated sewage samples from plant C. In plant D, 6 of 6 raw samples and 3 of 6 treated samples were positive.
The copy number of cDNAs measured by the qPCR assay was used to determine the viral loads in 1 liter of sewage sample collected during the field work. In those cases where the copy number was below the detection limit (corresponding to ∼1.5 to 3.0 × 103 genome equivalents per 1 liter sample), we marked the values as <103. To confirm the reproducibility of the assay, selected samples (n = 7) were reamplified with the qPCR assay; congruent results were obtained with those of the first runs (data not shown).
Molecular characterization of group C rotavirus strains.
The melting curve analysis suggested the existence of at least three distinct amplicon populations. To see if these differences in the melting points were associated with the presence of different strains, 13 qPCR amplicons that included samples from all four plants in each of the three amplicon populations were selected for sequence analysis. Direct inspection of the nucleotide substitution patterns of these short sequences revealed that strains identified in the sewage samples clustered into four sequence groups: one human-, one porcine-, and two bovine-like group C rotaviruses (Fig. 2). Nonetheless, the nucleotide mismatches observed (e.g., two C-to-T transitions between pattern III and pattern IV) did not affect the melting temperatures, perhaps due to the compensatory effect of these inverse substitutions. In general, a good agreement was observed between the sequence and the melting point analysis data. Amplicons with an average melting point 81.9°C were predicted to be of human origin (n = 6), while amplicons of the 83.4°C and 84.3°C melting point profiles corresponded to strains of bovine (n = 3) and porcine origin (n = 4), respectively, suggesting that the origin of strains might be predicted directly from the results of melting point analyses.
FIG. 2.
Alignment of partial nt sequences of the VP6 gene of human and animal group C rotaviruses. A fragment spanning nt 1282 to 1323 of the VP6 gene was amplified and analyzed as described in the text. The four sequence patterns (pat-I to pat-IV) obtained from sewage samples were aligned with strains of the indicated origin of host species (Hu, human; Po, porcine; Bo, bovine). In this alignment, pat-I and pat-II are indistinguishable from the reference human and porcine strains, respectively, while pat-III and pat-IV are more closely related to bovine strains on the basis of some conserved nt bases. Identities to and differences from the consensus sequence (Hu/Jajeri) are indicated with dots and letters, respectively.
Seasonal fluctuations in the viral loads of human and animal group C rotaviruses.
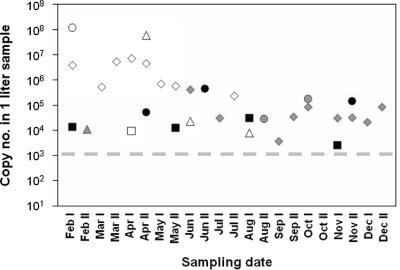
When the viral loads in raw sewage samples were plotted against the time of sample collection, a possible seasonality for the distribution of human group C rotaviruses was observed (Fig. 3). We detected human strains at all four plants during the first 7 months of the investigation period, with titers usually exceeding the 105 genome equivalent in the 1-liter sample (mean, 1.2 × 107; median, 6.9 × 105; range, 8.0 × 103 to 1.2 × 108). In plant A, where most data were collected, the average viral load of human strains between February and May was significantly higher than that in and after May (4.2 × 106 versus 4.9 × 105; P = 0.0006, z-test). In contrast, the seasonal activity of porcine and bovine strains could not be unequivocally determined. In the case of plant A, porcine strains seemed to circulate only when the human strains cleared from the population; however, their quantity was considerably lower than that of human strains during the previous period (mean, 3.9 × 104; median, 3.1 × 104), and thus, it cannot be ruled out that the presence of these animal strains was merely masked during the period when human strains dominated. Bovine strains were identified at low titers (mean, 9.9 × 104; median, 3.0 × 104) only in plants B and D.
FIG. 3.
Seasonal distribution of human, porcine, and bovine group C rotaviruses in sewage samples. Filled symbols: rhombus, plant A; square, plant B; triangle, plant C; circle, plant D; open symbols, human strains; gray symbols, porcine strains; black symbols, bovine strains. Numbers I and II after the months indicate first and second half of the month, respectively. Three influent samples that were negative for group C rotavirus in the qPCR assay were not included in this figure. The dashed line indicates the approximate detection limit of the assay (i.e., ∼1.5 to 3.0 × 103 genome equivalents per 1-liter sample).
DISCUSSION
In this study, new information about the epidemiologic features and ecology of group C rotavirus infection among humans and animals is presented from the examination of sewage samples using a simple, sensitive, specific and reproducible real-time PCR assay. We detected group C rotaviruses from raw sewage samples throughout 2005, suggesting that the virus is in circulation year round. The qPCR assay allowed both the quantification of the viral load and the prediction of the origin of strains by host species. This latter finding might be very helpful for differentiating among animal and human sources of contamination and for obtaining a more in-depth understanding of the ecology and epidemiology of group C rotaviruses. When the viral loads were plotted against the month of detection and the host species origin, a seasonal pattern was observed for human group C rotavirus strains. However, the insufficient amount of data that were collected over a single year from a single plant (plant A) does not allow us to draw firm conclusion about the seasonality of these pathogens. Nonetheless, fluctuation in the viral loads was demonstrated, a finding that raised the question whether the increased virus load might have been related to concomitant local outbreaks in closed and semiclosed communities or to temporal clusters of sporadic episodes of gastroenteritis. However, none of the eight documented outbreaks in the study area over this time frame was associated with human group C rotavirus infections (five were associated with noroviruses, three were associated with salmonellae). Likewise, no significant increase in the number of sporadic cases was observed; the single case we identified by RNA pattern analysis represented only 0.1% of all acute gastroenteritis cases (1 out of 803; K. Bányai, unpublished data).
The interpretation of negative test results is an issue even in the area of environmental microbiology, as identification of the source and the etiology responsible for local outbreaks is crucial to implementing the appropriate public health measures. Given the high detection rate of group C rotavirus in raw sewage samples throughout the study year together with the frequently measured low viral load, we cannot rule out the possibility that the samples that tested negative contained viruses, but their concentration fell below the detection limit. Thus, we need to consider negative test results with caution. Collecting larger amounts of sewage sample (e.g., ≥10 liters per sampling) probably would have provided an opportunity to obtain more precise data about the viral load simply by circumventing the problem associated with the low virus titer.
No specific data are available regarding the minimal infectious dose of group C rotaviruses, information that apparently would be required to more adequately interpret the viral load data from the public health perspective. In addition, group C rotavirus in clinical specimens often appears atypical, broken, and empty (15, 43). Thus, it is difficult to establish a relationship between the rotaviral genome copy number and the particle-to-infectious unit value, an obstacle to assessing the possible health risk represented by the group C rotavirus genome copies in environmental samples. In general, however, in the case of other enteric viruses (e.g., in group A rotaviruses and noroviruses), as few as 1 to 10 infectious virus particles have been shown to be able to induce disease in a susceptible host (17, 23). Thus, if the observation for group A rotaviruses and noroviruses also applies to group C rotaviruses, ingesting a few milliliters of the contaminated water containing ∼103 infectious particles per liter may be a potential health risk. Apparently, communal sewage needs to be considered to contain a high concentration of group C rotaviruses, and it is unknown whether this titer of these viruses is ever reached in various types of surface water resources.
The detection of group C rotavirus RNA of presumed animal origin in human sewage is an intriguing finding and raises further questions about their possible source. The anthropogenic origin of these animal-like rotavirus strains is suspected when there are no recognized animal herds in the service area of these sewage plants (40). No swine or cattle herds are known to be present in the service area of the investigated sewage plants, although porcine and bovine group C rotaviruses were detected throughout the year sporadically in plants B, C, and D and at least during a 4-month period in plant A. When the employees of each plant were interviewed about the possible source of animal virus contamination, we uncovered two surprising facts. In the service area of plant A, 300 m3 of the daily average 27,000 m3 of communal sewage is contributed by slaughterhouse activities. Anecdotal reports obtained from plant B suggest that families who live in the service area and keep swine or cattle for their own use may have illegally released animal manure into the communal sewage. Whether these practices have contributed to the continuous occurrence of animal strains in the sewage remains to be determined. An alternative hypothesis to explain the presence of animal-like strains in the communal sewage is that they might infect humans. The usually low detection rate of group C rotaviruses in childhood gastroenteritis (around 1% for children less than 5 years of age [3, 5, 41]) contrasts with reports of the relative abundance of antibodies against the virus in the same age group, as nearly 20% to 30% of children develop an antibody response to group C rotavirus by 5 years of age (14, 21, 37). An apparent increase in the serological prevalence of group C rotavirus has been observed among people of rural communities, an increase which has been linked to enhanced exposure to animals (13). Our detection of animal strains in urban sewage samples is of interest. More intense studies are required to determine if zoonotic transmission of group C rotaviruses occurs.
Increasing numbers of reports of outbreaks of gastroenteritis associated with the consumption of virus-contaminated food and water (11, 17) suggest that it is important to look for group C rotavirus in food, water, and sewage samples to determine if this virus plays a role in food- and waterborne causes of diarrhea. The outcome of these studies could help implement effective intervention measures and halt the spread of infection in the community. A previous study described a water-borne outbreak associated with a mixture of enteric viruses, based on a traditional epidemiological investigation and microbiological examination of clinical cases (18); group C rotavirus was detected in patients but was not identified in the implicated water sample, perhaps due to the unavailability of an adequate and sensitive detection method. With the advent of molecular viral diagnostic techniques, it is now possible to perform rapid detection and characterization tests of group C rotavirus in specimens suspected to be contaminated. Our simple, sensitive, specific, and reproducible method to quantitatively detect group C rotavirus in sewage samples might serve as a good starting point when polluted water is implicated in gastroenteritis outbreaks of unknown etiology. The application of this method in screening other environmental samples and clinical specimens including both human and animal origins needs to be explored further.
Acknowledgments
Financial support was received from the OTKA (grant no. T049020) and the EVENT program (SP22-CT-2004-502571). K.B. is a recipient of the Bolyai János fellowship of the Hungarian Academy of Sciences.
We thank the epidemiologists of the Baranya County Institute of State Public Health Service for sharing the information on the occurrence of local gastroenteritis outbreaks. We also thank István Gűth for assistance with sample collection, Jon Gentsch for critical reading of the manuscript, and the anonymous reviewers for their valuable comments.
Footnotes
Published ahead of print on 4 April 2008.
REFERENCES
- 1.Ahmed, M. U., N. Kobayashi, M. Wakuda, T. Sanekata, K. Taniguchi, A. Kader, T. N. Naik, M. Ishino, M. M. Alam, K. Kojima, K. Mise, and A. Sumi. 2004. Genetic analysis of group B human rotaviruses detected in Bangladesh in 2000 and 2001. J. Med. Virol. 72:149-155. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, E. J., and S. G. Weber. 2004. Rotavirus infections in adults. Lancet Infect. Dis. 4:91-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bányai, K., B. Jiang, Á. Bogdán, B. Horváth, F. Jakab, E. Meleg, V. Martella, L. Magyari, B. Melegh, and G. Szücs. 2006. Prevalence and molecular characterization of human group C rotaviruses in Hungary. J. Clin. Virol. 37:317-322. [DOI] [PubMed] [Google Scholar]
- 4.Boom, R., C. J. A. Sol, M. M. M. Salimans, C. L. Jansen, P. M. E. Wertheim-Van Dillen, and J. van der Noordaa. 1990. Rapid and simple method for purification of nucleic acids. J. Clin. Microbiol. 28:495-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Castello, A. A., M. H. Arguelles, G. A. Villegas, A. Olthoff, and G. Glikmann. 2002. Incidence and prevalence of human group C rotavirus infections in Argentina. J. Med. Virol. 67:106-112. [DOI] [PubMed] [Google Scholar]
- 6.Cunliffe, N. A., W. Dove, B. Jiang, B. D. Thinwda Cert, R. L. Broadhead, M. E. Molyneux, and C. A. Hart. 2001. Detection of group C rotavirus in children with acute gastroenteritis in Blantyre, Malawi. Pediatr. Infect. Dis. J. 20:1088-1090. [DOI] [PubMed] [Google Scholar]
- 7.Estes, M. K. 2001. Rotaviruses and their replication. p. 1747-1785. In D. M. Knipe and P. M. Howley (ed.), Fields virology, 4th ed. Lippincott-Raven Publishers, Philadelphia, PA.
- 8.Gabbay, Y. B., B. Jiang, C. S. Oliveira, J. D. Mascarenhas, J. P. Leite, R. I. Glass, and A. C. Linhares. 1999. An outbreak of group C rotavirus gastroenteritis among children attending a day-care center in Belem, Brazil. J. Diarrhoeal Dis. Res. 2:69-74. [PubMed] [Google Scholar]
- 9.Gentsch, J. R., A. R. Laird, B. Bielfelt, D. D. Griffin, K. Bányai, M. Ramachandran, V. Jain, N. A. Cunliffe, O. Nakagomi, C. D. Kirkwood, T. K. Fischer, U. D. Parashar, J. S. Bresee, B. Jiang, and R. I. Glass. 2005. Serotype diversity and reassortment between human and animal rotavirus strains: implications for rotavirus vaccine programs. J. Infect. Dis. 192:S146-S159. [DOI] [PubMed] [Google Scholar]
- 10.Hamano, M., M. Kuzuya, R. Fujii, H. Ogura, T. Mori, T. Nakayama, E. Yuen, K. Katayama, Y. Mitsunobu, and K. Inoue. 1999. Outbreak of acute gastroenteritis caused by human group C rotavirus in a primary school. Jpn. J. Infect. Dis. 52:170-171. [PubMed] [Google Scholar]
- 11.Hedberg, C. W., and M. T. Osterholm. 1993. Outbreaks of food-borne and waterborne viral gastroenteritis. Clin. Microbiol. Rev. 6:199-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung, T., G. Chen, C. Wang, H. Yao, Z. Fang, T. Chao, Z. Chou, W. Ye, X. Chang, S. Den, X. Liong, and W. Chang. 1984. Waterborne outbreak of rotavirus diarrhoea in adults in China caused by a novel rotavirus. Lancet i:1139-1142. [PubMed] [Google Scholar]
- 13.Iturriza-Gomara, M., I. Clarke, U. Desselberger, D. Brown, D. Thomas, and J. Gray. 2004. Seroepidemiology of group C rotavirus infection in England and Wales. Eur. J. Epidemiol. 19:589-595. [DOI] [PubMed] [Google Scholar]
- 14.James, V. L. A., P. R. Lambden, E. O. Caul, S. J. Cooke, and I. N. Clarke. 1997. Seroepidemiology of human group C rotavirus in the UK. J. Med. Virol. 52:86-91. [DOI] [PubMed] [Google Scholar]
- 15.Jiang, B., P. H. Dennehy, S. Spangenberger, J. R. Gentsch, and R. I. Glass. 1995. First detection of group C rotavirus in fecal specimens of children with diarrhea in the United States. J. Infect. Dis. 172:45-50. [DOI] [PubMed] [Google Scholar]
- 16.Kelkar, S. D., and J. K. Zade. 2004. Group B rotaviruses similar to strain CAL-1 have been circulating in Western India since 1993. Epidemiol. Infect. 132:745-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Koopmans, M., C. H. von Bonsdorff, J. Vinje, D. de Medici, and S. Monroe. 2002. Foodborne viruses. FEMS Microbiol. Rev. 26:187-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kukkula, M., P. Arstila, M. L. Klossner, L. Maunula, C. H. Bonsdorff, and P. Jaatinen. 1997. Waterborne outbreak of viral gastroenteritis. Scand. J. Infect. Dis. 29:415-418. [DOI] [PubMed] [Google Scholar]
- 19.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 20.Kuzuya, M., R. Fujii, M. Hamano, M. Yamada, K. Shinozaki, A. Sasagawa, S. Hasegawa, H. Kawamoto, K. Matsumoto, A. Kawamoto, A. Itagaki, S. Funatsumaru, and S. Urasawa. 1998. Survey of human group C rotaviruses in Japan during the winter of 1992 to 1993. J. Clin. Microbiol. 36:6-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuzuya, M., R. Fujii, M. Hamano, R. Ohata, H. Ogura, and M. Yamada. 2001. Seroepidemiology of human group C rotavirus in Japan based on blocking enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 8:161-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuzuya, M., M. Hamano, M. Nishijima, R. Fujii, H. Ogura, M. Tanaka, A. Oda, S. Kusaka, and M. Naitou. 2005. An outbreak of acute gastroenteritis caused by human group C rotavirus in a welfare institution in Okayama prefecture. Jpn. J. Infect. Dis. 58:255-257. [PubMed] [Google Scholar]
- 23.Leclerc, H., L. Schwartzbrod, and E. Dei-Cas. 2002. Microbial agents associated with waterborne diseases. Crit. Rev. Microbiol. 28:371-409. [DOI] [PubMed] [Google Scholar]
- 24.Minor, P. D. 1985. Growth, assay and purification of picornaviruses, p. 25-41. In B. W. J. Mahy (ed.), Virology: a practical approach. IRL Press Ltd., Oxford, United Kingdom.
- 25.Nadan, S., J. E. Walter, W. O. K. Grabow, D. K. Mitchell, and M. B. Taylor. 2003. Molecular characterization of astroviruses by reverse transcriptase PCR and sequence analysis: comparison of clinical and environmental isolates from South Africa. Appl. Environ. Microbiol. 69:747-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nicholas, K. B., H. B. Nicholas, and D. W. I. Deerfield. 1997. GeneDoc: analysis and visualization of genetic variation. EMBNET News 4:1-4. [Google Scholar]
- 27.Nilsson, M., B. Svenungsson, K. O. Hedlund, I. Uhnoo, A. Lagergren, T. Akre, and L. Svensson. 2000. Incidence and genetic diversity of group C rotavirus among adults. J. Infect. Dis. 182:678-684. [DOI] [PubMed] [Google Scholar]
- 28.Offit, P. A., and H. F. Clark. 2000. Rotavirus, p. 1696-1703. In G. L. Mandel, J. E. Bennet, and R. Dolin (ed.), Principles and practice of infectious diseases, 5th ed. Churchill Livingstone, New York, NY.
- 29.Otsu, R. 1998. A mass outbreak of gastroenteritis associated with group C rotaviral infection in schoolchildren. Comp. Immunol. Microbiol. Infect. Dis. 1:75-80. [DOI] [PubMed] [Google Scholar]
- 30.Parashar, U. D., C. J. Gibson, J. S. Bresee, and R. I. Glass. 2006. Rotavirus and severe childhood diarrhea. Emerg. Infect. Dis. 12:304-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Phan, T. G., S. Nishimura, M. Okame, T. A. Nguyen, P. Khamrin, S. Okitsu, N. Maneekarn, and H. Ushijima. 2004. Virus diversity and an outbreak of group C rotavirus among infants and children with diarrhoea in Maizuru city, Japan during 2002-2003. J. Med. Virol. 74:173-179. [DOI] [PubMed] [Google Scholar]
- 32.Rahman, M., S. Banik, A. S. Faruque, K. Taniguchi, D. A. Sack, M. Van Ranst, and T. Azim. 2005. Detection and characterization of human group C rotaviruses in Bangladesh. J. Clin. Microbiol. 43:4460-4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rodger, S. M., R. F. Bishop, and I. H. Holmes. 1982. Detection of a rotavirus-like agent associated with diarrhea in an infant. J. Clin. Microbiol. 16:724-726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saif, L. J., and B. Jiang. 1994. Nongroup A rotaviruses of humans and animals. Curr. Top. Microbiol. Immunol. 185:339-371. [DOI] [PubMed] [Google Scholar]
- 35.Sánchez-Fauquier, A., E. Roman, J. Colomina, I. Wilhelmi, R. I. Glass, and B. Jiang. 2003. First detection of group C rotavirus in children with acute diarrhea in Spain. Arch. Virol. 148:399-404. [DOI] [PubMed] [Google Scholar]
- 36.Schnagl, R. D., K. Boniface, P. Cardwell, D. McCarthy, C. Ondracek, B. Coulson, J. Erlich, and F. Morey. 2004. Incidence of group C human rotavirus in central Australia and sequence variation of the VP7 and VP4 genes. J. Clin. Microbiol. 42:2127-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Steele, A. D., and V. L. A. James. 1999. Seroepidemiology of human group C rotavirus in South Africa. J. Clin. Microbiol. 37:4142-4144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szucs, G., M. Kende, and M. Uj. 1987. Atypical human rotaviruses in Hungary. Ann. Inst. Pasteur Virol. 138:391-395. [Google Scholar]
- 39.Velazquez, F. R., D. O. Matson, J. J. Calva, L. Guerrero, A. L. Morrow, S. Carter-Campbell, R. I. Glass, M. K. Estes, L. K. Pickering, and G. M. Ruiz-Palacios. 1996. Rotavirus infection in infants as a protection against subsequent infections. N. Engl. J. Med. 335:1022-1028. [DOI] [PubMed] [Google Scholar]
- 40.Villena, C., W. M. El-Senousy, F. X. Abad, R. M. Pinto, and A. Bosch. 2003. Group A rotavirus in sewage samples from Barcelona and Cairo: emergence of unusual genotypes. Appl. Environ. Microbiol. 69:3919-3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.von Bosdorff, C. H., and L. Svensson. 1988. Human serogroup C rotavirus in Finland. Scand. J. Infect. Dis. 20:475-478. [DOI] [PubMed] [Google Scholar]
- 42.Yang, H., E. V. Makeyev, Z. Kang, S. Ji, D. H. Bamford, and A. A. van Dijk. 2004. Cloning and sequence analysis of dsRNA segments 5, 6 and 7 of a novel nongroup A, B, C adult rotavirus that caused an outbreak of gastroenteritis in China. Virus Res. 106:15-26. [DOI] [PubMed] [Google Scholar]
- 43.Yee, E. L., B. Jiang, R. S. Kendall, C. Humphrey, and R. I. Glass. 2006. Group C rotavirus in a pediatric kidney transplant patient with diarrhea. J. Clin. Virol. 36:306-308. [DOI] [PubMed] [Google Scholar]