Abstract
The regulation of caspase-1 activation in macrophages plays a central role in host defense against bacterial pathogens. The activation of caspase-1 by the detection of bacterial products through Nod-like receptors leads to the secretion of mature interleukin-1β (IL-1β) and IL-18 and the induction of rapid host cell death (pyroptosis). Here, we report that pyroptosis induced by Salmonella enterica serovar Typhimurium can be positively regulated by prior gamma interferon (IFN-γ) stimulation of RAW 264.7 cells. This increase in cell death is dependent on both caspase-1 activation and, in part, Salmonella pathogenicity island 1 (SPI-1) expression by Salmonella. Furthermore, the exogenous expression of the IFN-γ-induced protein guanylate binding protein 5 (GBP-5) is sufficient to induce a heightened susceptibility of RAW 264.7 cells to Salmonella-induced pyroptosis, and the endogenous expression of GBP-5 is important for this phenomenon. RAW 264.7 cells with decreased expression of GBP-5 mRNA (inhibited by short hairpin RNA against GBP-5) release twofold less lactate dehydrogenase (a marker of membrane permeability) upon infection by invasive S. enterica serovar Typhimurium than do infected control cells. Importantly, 3× FLAG-tagged GBP-5 is localized to membrane ruffles, which contact invasive Salmonella, and is found on the membranes of spacious phagosomes containing Salmonella (although it is also found in the cytoplasm and on other cellular membranes), placing 3× FLAG GBP-5 at the interface of secreted SPI-1 effectors and host protein machinery. The regulation of pyroptosis by the IFN-γ-induced protein GBP-5 may play an important role in the host defense against Salmonella enterica serovar Typhimurium and perhaps other invasive bacterial pathogens.
Macrophages surveil the tissues using a large repertoire of pattern recognition receptors to detect tissue damage and invading microorganisms. These receptors consist of cell surface receptors, such as Toll-like receptors (TLRs), which interrogate the external environment (2), and internal receptors, such as Nod-like receptors (NLRs), which interrogate the environment of the cytoplasm (20). Upon the activation of TLRs by pathogen-associated molecular patterns (PAMPs) such as lipopolysaccharide (LPS) and flagellin, TLRs activate the transcription of cytokines and other innate immune regulatory genes (23).
Cytokines such as interleukin-1β (IL-1β) and IL-18 require proteolytic processing of the proform to the mature form by caspase-1 before they are secreted (5, 14, 15, 26, 28, 53). Two signals are thought to be required. The first signal, mediated by TLRs, induces the IL-1β transcript and perhaps other proteins necessary for caspase-1 activation (11, 23, 25). The second signal, mediated by NLRs, activates caspase-1 (32-34). This second signal, the activation of caspase-1, can come in the form of macrophages sensing tissue damage through NLRs or in the form of macrophages sensing PAMPs in their cytoplasm through NLRs (32, 33). NLR signaling stimulates the formation of a multiprotein complex called the inflammasome, which activates caspase-1 (1, 35). Finally, not only does the activation of caspase-1 lead to proteolytic processing and secretion of the inflammatory mediators IL-1β and IL-18, it can also result in rapid host cell death, recently termed pyroptosis (8, 12, 38).
A number of different bacterial species and bacterial toxins result in the activation of caspase-1 upon infection of macrophages (36). For the purpose of this paper, we will focus on the mechanism of caspase-1 activation by Salmonella enterica serovar Typhimurium. Recently, it has been demonstrated that interleukin-converting enzyme-protease-activating factor (IPAF), a member of the NLR family, detects Salmonella flagellin subunits in the cytoplasm and activates caspase-1 (13, 37). An intact Salmonella pathogenicity island 1 (SPI-1) type 3 secretion system (T3SS) is required for flagellin to gain access to the cytoplasm, where IPAF is located (13, 37). The detection of flagellin by IPAF results in the secretion of IL-1β, if IL-1β transcript has been induced, and pyroptosis.
Unlike the secretion of mature IL-1β, the induction of host cell death by Salmonella does not require a priming stimulus through a TLR (6, 39). Pyroptosis induced by S. enterica serovar Typhimurium invasion of macrophages is characterized by a requirement for caspase-1 activation, cellular swelling, rapid membrane permeability, and DNA cleavage (4, 6, 18, 39). Although Salmonella can induce other forms of host cell death, only the form mediated by caspase-1 activation occurs within 2 h of macrophage invasion (17, 19, 40, 55). This form of Salmonella-induced rapid host cell death (pyroptosis) has been studied in a variety of macrophage-like cell lines (6, 39).
The macrophage-like cell line RAW 264.7 displays resistance to Salmonella-induced pyroptosis compared to primary mouse macrophages, and interestingly, prior stimulation of RAW 264.7 cells with LPS or gamma interferon (IFN-γ) confers greater susceptibility to Salmonella-induced pyroptosis (39). While many mechanisms that negatively regulate the activation of caspase-1 have been discovered, little is known about mechanisms that might positively regulate caspase-1 activation (34). Both Listeria monocytogenes and Francisella tularensis can induce caspase-1 activation and host cell death but require host cell activation through IFN-β signaling in order to induce caspase-1 activation (16, 50, 51). It is not clear how IFN-β signaling primes the inflammasome for activation, but it probably requires a host protein induced by IFN-β (16).
The most potent host cell response against intracellular bacterial pathogens such as Salmonella is mediated by IFN-γ. IFN-γ is secreted largely by NK cells and T cells in response to stimulation with IL-12 and IL-18, two cytokines secreted by activated macrophages (48). IFN-γ serves as a ligand for receptors found on most host cell types and elicits the transcription of over 600 genes that play an important role in cellular defense against intracellular pathogens (10). Among the genes induced by IFN-γ are a superfamily of dynamin-related GTPases known as the p47 family and the p65 family (3, 43, 57). Members of the p47 family have been shown to play a specific role in host defense against intracellular pathogens such as mycobacteria, Toxoplasma species, and others, with LRG-47 being the most well-studied member (9, 30, 31, 52).
We, and others, have recently discovered that the p65 family member murine guanylate binding protein 5 (GBP-5) is highly enriched on purified phagosomal membranes in IFN-γ-stimulated RAW 264.7 cells (detected by a proteomic method) and thus could play a role in defense against bacterial pathogens (data not shown) (21). In this paper, we will provide evidence that the IFN-γ-inducible protein GBP-5 positively regulates Salmonella-induced pyroptosis through a mechanism that requires the activation of caspase-1 by secretion through the Salmonella T3SS SPI-1. Furthermore, we demonstrate that 3× FLAG GBP-5 can be found on actin-rich membrane ruffles and spacious phagosomes induced by invading Salmonella, placing GBP-5 at the interface between the host and the pathogen.
MATERIALS AND METHODS
Macrophage cell lines.
RAW 264.7 cells were purchased from the ATCC and maintained in RPMI 1640 medium with 10% fetal calf serum (FCS). BAC1.2F5 cells were obtained from E. R. Stanley and maintained as described previously (41).
Description of Salmonella strains.
Wild-type Salmonella enterica serovar Typhimurium strains used in this study were SL1344 (58), CS401 (47), and 14028s (ATCC). All strains were maintained as pure cultures on Luria-Bertani agar plates and grown with shaking in Luria-Bertani broth at 37°C. SPI-1 mutant strain CS401 ΔprgHIJK was obtained from S. I. Miller (24). The flagellin-deficient ΔfliC ΔfljB strain (14028s fliC::Tn10 fljB5001::Mud-Cm) was obtained from E. A. Miao (37).
Cloning of 3× FLAG GBP-5 and short hairpin RNA (shRNA) expression vectors.
The following oligonucleotides encoding the 3× FLAG epitope (Sigma-Aldrich) were synthesized, annealed, and cloned into BamHI and XhoI sites of the retroviral vector pFB-Neo (Stratagene). The 3× FLAG sense oligonucleotide was 5′-GGA TCC ACC ATG GAC TAC AAA GAC CAT GAC GGT GAT TAT AAA GAT CAT GAC ATC GAT TAC AAG GAT GAC GAT GAC AAG-3′, and the antisense oligonucleotide was 5′-GAG CTC CTT GTC ATC GTC ATC CTT GTA ATC GAT GTC ATG ATC TTT ATA ATC ACC GTC ATG GTC TTT GTA GTC CAT GGT-3′. The following primers were used to clone murine GBP-5 by PCR from cDNA derived from IFN-γ-stimulated RAW 264.7 cells: sense primer 5′-CTC GAG GCC CCA GAG ATT CAC ATG CC-3′ and antisense primer 5′-GCG GCC GCG TAT TAG CTT ATA ACA CAG-3′. The resulting PCR product was cloned into 3× FLAG pFB-Neo using XhoI and NotI restriction sites. The expressed protein was 70 kDa.
shRNAs were cloned into pSM2c (Open Biosystems), a retroviral vector, according to the protocol detailed previously by Paddison and coworkers (44). The following target sequences were used: 5′-CTA TTT GAA CGC CAA AGA A-3′ for GBP-5 and 5′-GAT CGA GGA GAT GTT CGT GTA C-3′ for humanized Renilla green fluorescent protein (hrGFP).
The resulting plasmids were sequenced to verify that the inserts had the correct reading frames and sequences.
Generation of replication-defective retrovirus particles and transduction of RAW 264.7 cells.
Retrovirus-containing supernatants were generated by transfecting the Phoenix Ampho cell line (ATCC) (see the Gary Nolan laboratory website [www.stanford.edu/group/nolan] for details of acquisition) at 80 to 90% confluence in a 10-cm tissue culture dish with 10 μg of the appropriate purified retroviral plasmid using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Viral supernatants were then harvested 48 h posttransfection and filtered with a 0.45-μm filter to remove any cells. Polybrene (Sigma) was added to the filtered supernatant to achieve 4 μg/ml. RAW 264.7 cells or BAC1.2F5 cells were plated in six-well tissue culture plates the previous day to achieve 30% confluence on the day of transduction. The medium was removed, and 5 ml of the retrovirus-containing supernatant was overlaid onto the cells. Plates were then centrifuged for 2 h at 1,000 × g at room temperature. Plates were incubated in a 5% CO2 incubator at 37°C overnight. Fresh medium was added the next morning. Selection with 600 μg/ml G418 was started 24 h after fresh medium was added. Forty to eighty percent of cells generally survived selection and were maintained as a stabile pool under selection.
Salmonella-induced pyroptosis assay.
In the case where cell death was measured with a lactate dehydrogenase (LDH) release assay (Cytotox-ONE; Promega), RAW 264.7 cells were plated in a 96-well plate with black walls at 2 × 104 cells per well 48 h before the start of the experiment. Bacterial strains were grown overnight to stationary phase, subcultured with a 1:50 dilution, and grown for 3 h at 37°C with shaking. Bacteria were then washed once with Dulbecco's phosphate-buffered saline (PBS) (DPBS; Cellgro) and resuspended in DPBS, and the optical density at 600 nm was determined. An optical density at 600 nm of 1.0 was equal to 1 × 109 bacterial cells as determined by dilution plating and colony counts.
Plated RAW 264.7 cells were washed one time with Dulbecco's modified Eagle's medium (DMEM) containing 5% FCS and then infected at the indicated multiplicity of infection (MOI) by adding the bacteria in 50 μl of medium and centrifuging the plates for 30 s at 500 × g. When nonmotile bacterial strains were used, plates were centrifuged for 5 min at 500 × g. The plates were then incubated at 37°C for 10 min, washed two times with 37°C DMEM with 5% FCS and 50 μg/ml gentamicin, and 50 μl of 37°C DMEM with 5% FCS and 50 μg/ml gentamicin was then added to each well. Plates were then incubated at 37°C for 60 min. At the end of the incubation, the plates were processed according to the manufacturer's protocol, except that 0.5 volumes of all reagents were used. All experimental wells were tested in quadruplicate, and final values were calculated as follows: (experimental release − spontaneous release)/(Triton X-100 release − spontaneous release) × 100.
LDH release assays were also performed using the apoptosis inducers staurosporine (LC Laboratories) and camptothecin (LC Laboratories). Both drugs were dissolved in dimethyl sulfoxide and added to the cells at the indicated concentrations. Cells were incubated with the drugs 6 h prior to LDH assay. Control wells were treated with the highest added concentration of vehicle.
In the case where cell death was measured by membrane exclusion of propidium iodide, RAW 264.7 cells were plated in 24-well tissue culture plates at 1.5 × 105 cells per well 24 h prior to the experiment. Salmonella strains were cultured as described above and added to the wells in fresh medium. The plate was centrifuged for 30 s at 500 × g and incubated at 37°C for 30 min. Wells were washed three times with PBS and chased in RPMI 1640 medium with 10% FCS and 50 μg/ml gentamicin. After a 2-h chase, cells were lifted with DPBS-1.5 mM EDTA and kept on ice until flow cytometry analysis. Thirty seconds prior to analysis on a Becton Dickinson FACSCalibur flow cytometer, propidium iodide was added to a 1-μg/ml final concentration, and fluorescence was analyzed with channel FL-2 (585/42-nm band-pass filter).
In both of the cases described above, we stimulated cells with IFN-γ (murine; Peprotech) overnight at 50 U/ml (units are defined by the manufacturer [Peprotech]) and kept IFN-γ in the chase medium. We used the caspase-1 inhibitor AC-YVAD-CMK (caspase-1 inhibitor II; Calbiochem) at a 40 μM final concentration and pretreated the cells for 45 min. The inhibitor was used in the chase.
Salmonella uptake assay.
RAW 264.7 cells were plated in 96-well plates as described above, and Salmonella strain 1344 was cultured as described above. Salmonella cells were added to cell culture at an MOI of 12 and briefly centrifuged for 30 s at 500 × g. The plates were incubated for 5 min at 37°C and then washed two times with culture medium, followed by washing two times with PBS. Cells were lysed with 1% Triton X-100 in water, and serial dilutions were then spot plated in triplicate onto LB plates. An LDH assay was performed using part of the cell lysate, and CFU/well was normalized for the cell line with lower average LDH activity by taking the ratio of high to low average LDH activities and multiplying by the colony counts from the low cell line.
Real-time PCR.
RNA was purified using Trizol reagent (Invitrogen) according to the manufacturer's protocol. shRNA GBP-5 and shRNA hrGFP stabile pools were plated at 3.5 × 105 cells per well in six-well tissue culture plates 48 h prior to RNA extraction. IFN-γ was added at 50 U/ml 24 h prior to RNA extraction. Purified RNA was treated with DNase I (RNase-free DNase I; Promega), and first-strand cDNA synthesis was performed with Superscript II (Invitrogen) according to the manufacturer's protocol.
Real-time PCR was performed using Quantitect Sybr green PCR master mix (Qiagen) according to the manufacturer's protocol with sense and antisense primers at 0.3 μM and 200 ng of template in a 25-μl reaction mixture. Reactions were performed in duplicate using a 96-well reaction plate and a Bio-Rad iCycler iQ machine. The primer annealing temperature was 50°C for 30 s, followed by extension at 72°C for 30 s for 40 cycles.
The following primers were used for the detection of GBP-5 cDNA by real-time PCR: sense primer 5′-CAG ACC TAT TTG AAC GCC AAA GA-3′ and antisense primer 5′-TGC CTT GAT TCT ATC AGC CTC T-3′. The following primers were used for the detection of β-actin cDNA by real-time PCR: sense primer 5′-GAA ATC GTG CGT GAC ATC AAA G-3′ and antisense primer 5′-TGT AGT TTC ATG GAT GCC ACA G-3′. Both primer sets gave a single product by PCR of cDNA from control and IFN-γ-treated RAW 264.7 cells at the expected molecular weight (data not shown). Both amplicons spanned introns and did not give detectable signals in reverse transcriptase-negative controls.
The real-time PCR data were analyzed using the 2−ΔΔCT method (29). The primer sets met the assumption that PCR efficiency must be approximately equal for the method to be valid, as a plot of the log template dilution (a 16,000-fold range) versus GBP-5 threshold cycle (CT) minus the β-actin CT had a slope of −0.08.
Microscopy and image acquisition.
BAC1.2F5 cells expressing 3× FLAG GBP-5 were plated onto circular coverslips in 24-well plates to achieve 50% confluence at the time of experimentation. In experiments with SL1344, bacteria were grown as described above for pyroptosis assays and added to cells at an MOI of 20. Plates were centrifuged for 30 s at 500 × g and incubated at 37°C in a CO2 incubator for 15 min. Cells were washed with DPBS and fixed with 4% paraformaldehyde in DPBS (pH 7.4) for 15 min. Cells were washed with DPBS and incubated with blocking buffer (10% FCS, 5% bovine serum albumin, 0.02% [wt/vol] Tween 20, and 0.1% [wt/vol] saponin in DPBS) for 30 min at room temperature. Cells were washed with DPBS and incubated with antibody staining buffer (1:5 dilution of blocking buffer with DPBS and the addition of 0.1% saponin) with the appropriate dilution of antibodies. Antibodies and staining agents used were as follows: M2 anti-FLAG antibody (Sigma), Alexa 546 phalloidin and Oregon Green phalloidin (Molecular Probes), goat anti-Salmonella (Abcam), Cy5 donkey anti-goat, Texas Red donkey anti-mouse, and fluorescein isothiocyanate (FITC) donkey anti-mouse (Jackson Immunoresearch). Decorated coverslips were mounted onto glass slides with ProLong Gold antifade reagent (Invitrogen).
Imaging was performed with a Nikon Eclipse TE300 inverted microscope using epifluorescence and differential interference contrast optics, FITC or Texas Red filter sets, and a 60× Plan Apo differential interference contrast H lens. Confocal imaging was performed using a Nikon Eclipse TE300 inverted microscope and a Bio-Rad Radiance 2000 laser scanning attachment with an argon laser line at 488 nm, a helium neon laser line at 543 nm, and a red diode line at 637 nm.
Postacquisition image processing was performed using ImageJ and Photoshop CS3. Sharpening was performed with Photoshop CS3 by using the unsharp mask feature.
Western blotting.
RAW 264.7 cells expressing the empty pFB-Neo vector or 3× FLAG GBP-5 were lysed in 24-well tissue culture plates with boiling Laemmli buffer. Approximately 1 μg of protein was loaded onto a 10% polyacrylamide gel using the discontinuous buffer system described previously by Laemmli (27). Once proteins had migrated sufficiently on the gel, the proteins were transferred onto a polyvinylidene fluoride transfer membrane, and Western blots were processed using M2 (Sigma-Aldrich) antibody to detect the 3× FLAG tag and anti-tubulin for a loading control. Primary antibodies were detected with anti-mouse IgG horseradish peroxidase (Amersham) and developed with ECL Plus reagent (Amersham). Blots were then exposed to film (Kodak Biomax XAR film) and developed with an automated film processor.
Intracellular antibody staining and flow cytometry.
RAW 264.7 cells expressing the empty pFB-Neo vector or 3× FLAG GBP-5 were lifted from 24-well tissue culture plates with DPBS-1 mM EDTA and processed for staining of 3× FLAG GBP-5 as described above for microscopy, except that an FITC-conjugated M2 antibody (Sigma) was used for the detection of the 3× FLAG tag. Decorated cells were then analyzed using a Becton Dickinson FACSCalibur system in analysis mode. Histograms were generated by gating on intact cells using forward-scatter and side-scatter data and by plotting FL-1 (530/30-nm band-pass filter) intensity.
Statistics.
All stated P values were derived from a Student's t test as calculated in Excel. All values are two-tailed values.
RESULTS
Activation of RAW 264.7 cells with IFN-γ results in increased susceptibility to Salmonella-induced pyroptosis and depends on caspase-1 activation, in part, by the Salmonella T3SS SPI-1.
Previous studies have shown that the activation of caspase-1 may be positively regulated by IFN signaling. The stimulation of RAW 264.7 cells with LPS or IFN-γ results in greater susceptibility to Salmonella-induced pyroptosis, and more recently, it was found that macrophage cell death induced by Francisella and Listeria requires IFN-β signaling in order for caspase-1 to be activated by these organisms (16, 39, 50, 51).
We further characterized the role of IFN-γ in regulating Salmonella-induced pyroptosis in RAW 264.7 cells using an LDH assay. We treated cells overnight with IFN-γ and infected them with S. enterica serovar Typhimurium cells that were conditioned to maximally express SPI-1 genes (see Materials and Methods). The IFN-γ-stimulated RAW 264.7 cells were infected with Salmonella for 10 min, followed by multiple washes and a 60-min chase in the presence of gentamicin. These conditions resulted in rapid membrane permeability as detected by the release of LDH (Fig. 1, 2, and 3).
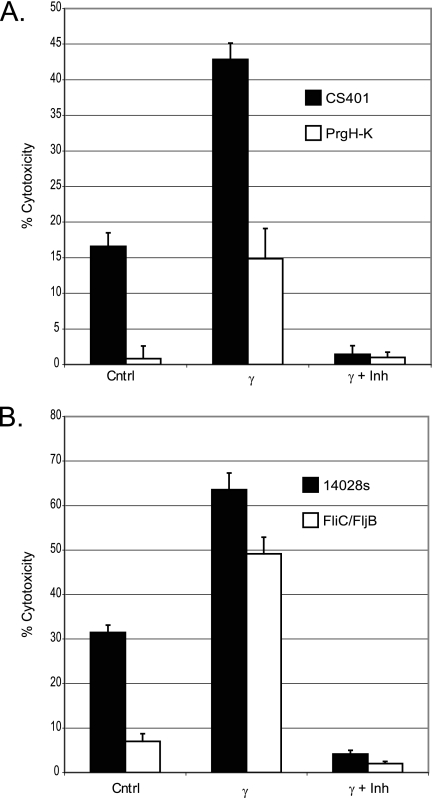
FIG. 1.
Activation of RAW 264.7 cells with IFN-γ results in increased susceptibility to Salmonella-induced pyroptosis and depends on caspase-1 activation, in part, by the Salmonella T3SS SPI-1. An LDH assay was performed as described in Materials and Methods to determine LDH release after infection at an MOI of 12.5 with various S. enterica serovar Typhimurium strains. (A) LDH assay comparing Salmonella WT strain CS401 to the ΔprgHIJK mutant and effect of prior stimulation of RAW 264.7 cells with IFN-γ on Salmonella-induced pyroptosis. (B) LDH assay comparing Salmonella WT strain 14028s to the ΔfliC ΔfljB mutant and effect of prior stimulation of RAW 264.7 cells with IFN-γ on Salmonella-induced pyroptosis. Groups labeled γ received 50 U/ml IFN-γ overnight prior to infection with Salmonella. Groups labeled γ and inh (inhibitor) were stimulated with IFN-γ overnight prior to treatment with 40 μM caspase-1 inhibitor II for 30 min prior to infection with the indicated strains of Salmonella. The y axis indicates the percentage of total LDH release determined by lysing control wells with Triton X-100. The formula (experimental release − spontaneous release)/(Triton X-100 release − spontaneous release) × 100 was used to calculate percent pyroptosis. Bars depict averages of data from four wells ± standard deviations. Cntrl, control.
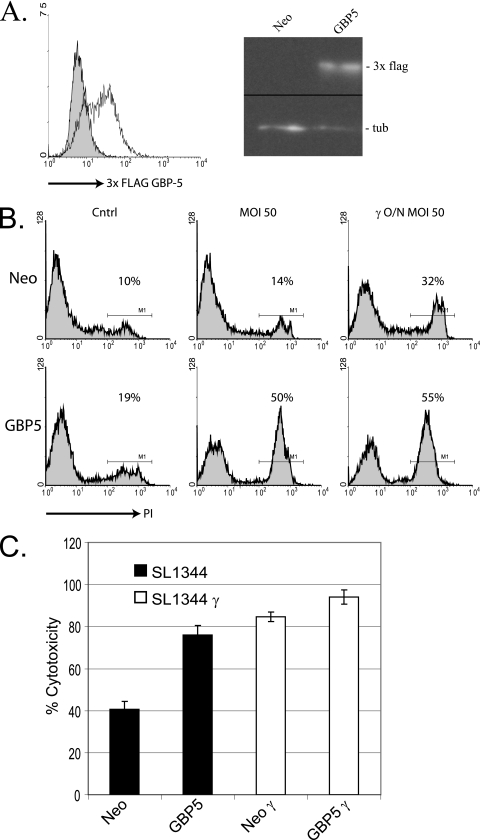
FIG. 2.
Expression of 3× FLAG GBP-5 in RAW 264.7 cells results in increased sensitivity to S. enterica serovar Typhimurium-induced pyroptosis. (A) RAW 264.7 cell pools transduced by retroviral vectors with an internal ribosome entry site neomycin cassette selected with G418 expressed 3× FLAG GBP-5 as detected by flow cytometry (left) and Western blotting (right). In the case of flow cytometric analysis of 3× FLAG GBP-5 expression, cells were permeabilized and decorated with FITC-labeled M2 antibodies. (Left) The shaded histogram depicts FITC staining of control cells expressing an empty vector, while the unshaded histogram depicts FITC staining of 3× FLAG GBP-5-expressing cells. In the case of the Western blot (right), the top row was decorated with an M2 anti-FLAG antibody (3× FLAG), and the bottom row was decorated with an anti-tubulin antibody (tub). Neo equals cell lysate from cells expressing the empty vector, while GBP5 equals cell lysate from 3× FLAG GBP-5-expressing cells. (B) RAW 264.7 control cells and 3× FLAG GBP-5 cells were infected with S. enterica serovar Typhimurium SL1344 as described in Materials and Methods, followed by staining with propidium iodide and analysis of staining by flow cytometry. The percentage of the gated population considered to be positive for membrane permeability is shown under M1 and corresponds to the printed number. Cntrl indicates uninfected cells, and γ indicates cells which were treated overnight with 50 U/ml IFN-γ. Definitions for Neo and GBP5 are the same as those described above (A). (C) An LDH assay was performed as described in Materials and Methods to determine LDH release after infection of control cells and 3× FLAG GBP-5-expressing cells with S. enterica serovar Typhimurium SL1344. The y axis indicates the percentage of total LDH release determined by lysing control wells with Triton X-100. The formula (experimental release − spontaneous release)/(Triton X-100 release − spontaneous release) × 100 was used. Bars depict averages of data from four wells ± standard deviations. γ in x axis identifiers indicates overnight stimulation of those experimental wells with IFN-γ.
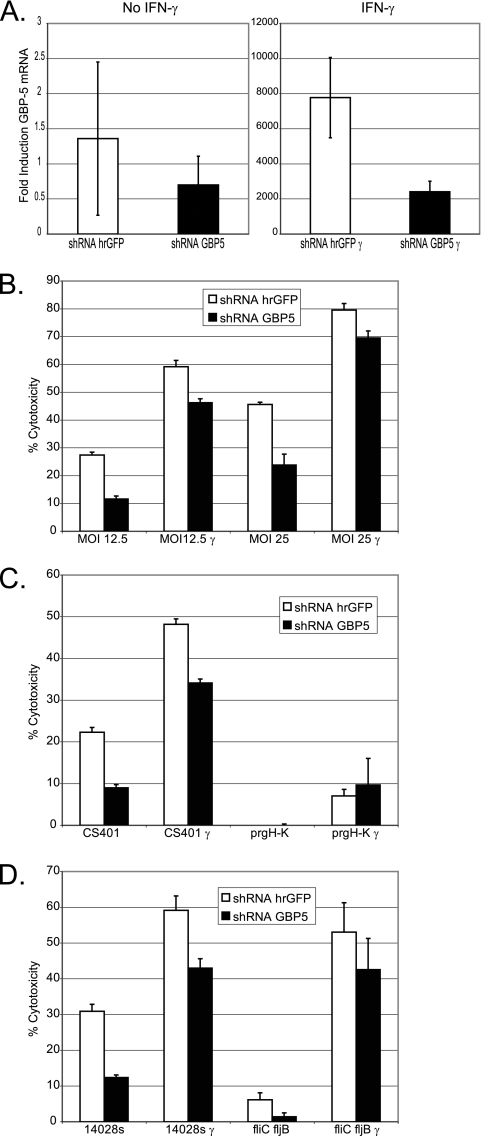
FIG. 3.
Targeting of GBP-5 mRNA expression for downregulation by shRNA against GBP-5 results in a significant decrease in Salmonella-induced pyroptosis. (A) Real-time PCR was performed as described in Materials and Methods using cDNA from RAW 264.7 cells expressing shRNA targeting GBP-5 or hrGFP. γ indicates experimental wells that were treated with 50 U/ml IFN-γ overnight (left is untreated, and right is cells treated with IFN-γ overnight). Bars indicate the average induction of GBP-5 mRNA from three experimental wells compared to unstimulated cells expressing shRNA hrGFP and normalized to β-actin mRNA levels using the 2−ΔΔCT method. (B to D) Salmonella-induced pyroptosis was measured by LDH release as described in Materials and Methods. (B) shRNA hrGFP and shRNA GBP-5 pools were infected with SL1344 at the indicated MOIs. (C and D) shRNA hrGFP and shRNA GBP-5 pools were infected with Salmonella strains as indicated at an MOI of 25. Bars show data from averages of four experimental wells ± standard deviations. The percentage of total LDH release was determined as follows: (experimental release − spontaneous release)/(Triton X-100 release − spontaneous release) × 100. See Materials and Methods for a description of strains and cross-reference to abbreviations used. In the legends, shRNA hrGFP indicates the control pool of RAW 264.7 cells expressing shRNA targeting hrGFP, and shRNA GBP-5 indicates the pool of RAW 264.7 cells expressing shRNA targeting GBP-5. γ indicates wells treated with IFN-γ overnight.
Overnight stimulation of RAW 264.7 cells with IFN-γ followed by infection with Salmonella wild-type strains CS401 and 14028s resulted in a significant increase in Salmonella-induced pyroptosis compared to that in unstimulated cells (Fig. 1A and B) (P < 0.01) (see Materials and Methods). To determine if the SPI-1 T3SS was necessary for increased Salmonella-induced pyroptosis in IFN-γ-treated cells, we infected IFN-γ-treated cells with prgHIJK-deficient S. enterica serovar Typhimurium. Expression of the prgHIJK operon has been shown to be necessary for the formation of the SPI-1 needle complex and secretion of effectors into the host cell cytoplasm (24). We found a significant decrease in Salmonella-induced pyroptosis when IFN-γ-treated cells were infected with prgHIJK-deficient Salmonella compared to isogenic control strain CS401 (Fig. 1A) (P < 0.01). This result suggested that the increase in Salmonella-induced pyroptosis was dependent largely on secretion through the SPI-1 T3SS, although some stimulation of the IFN-γ-activated macrophages through some other SPI-1-independent mechanism appeared to play a smaller role. Pretreatment of the cells with caspase-1 inhibitor II abolished the induction of Salmonella-induced pyroptosis by wild-type control strains CS401 and 14028s (Fig. 1A and B). Thus, the increase in Salmonella-induced pyroptosis that resulted from IFN-γ stimulation of RAW 264.7 cells was partly dependent on SPI-1 secretion by invading Salmonella and completely dependent on caspase-1 activation in the host cell.
Two recent studies demonstrated that flagellar structural genes fliC and fljB were necessary for the induction of Salmonella-induced pyroptosis and caspase-1 activation in macrophages through IPAF (13, 37). In our hands, a ΔfliC ΔfljB mutant was able to induce significant amounts of pyroptosis in RAW 264.7 cells treated with IFN-γ, while this strain induced significantly lower levels of pyroptosis in unstimulated macrophages than did its wild-type parental strain 14028s (Fig. 1B) (P < 0.01 in both cases). This observation suggested to us that an alternative stimulus derived from Salmonella and dependent on SPI-1 secretion (perhaps SipB, another effector protein or cellular component) can also induce pyroptosis in RAW 264.7 cells but requires an IFN-γ-induced protein or signaling pathway to carry out this function.
The stimulation of macrophages by IFN-γ induces multiple proteins. Below, we investigate the role of an IFN-γ-induced protein, GBP-5, in the activation of Salmonella-induced pyroptosis.
Expression of 3× FLAG GBP-5 in RAW 264.7 cells results in increased sensitivity to S. enterica serovar Typhimurium-induced pyroptosis.
Through a separate line of investigation, we discovered that GBP-5, a protein induced by LPS and IFN-γ (42), was highly associated with latex bead-purified phagosomal membranes in IFN-γ-stimulated RAW 264.7 cells (data not shown). To investigate the role of GBP-5 in phagosome biology, we expressed an N-terminal 3× FLAG-tagged form of GBP-5 from a retroviral vector in RAW 264.7 cells. Selection of transductants with G418 resulted in stabile pools of cells with moderate expression levels of 3× FLAG GBP-5. We saw a 1-log increase in the staining intensity of the 3× FLAG tag in 3× FLAG GBP-5-expressing cells compared to a stabile pool of empty vector-expressing cells by flow cytometry (Fig. 2A, left). Western blot analysis revealed an anti-3× FLAG staining band at approximately 70 kDa, as expected (Fig. 2A, right).
When we attempted to determine if 3× FLAG GBP-5 was localized to Salmonella-containing phagosomes, we observed by phase-contrast microscopy that RAW 264.7 cells expressing 3× FLAG GBP-5 seemed to be more sensitive to pyroptosis induced by wild-type Salmonella strain SL1344 than control cells harboring an empty vector (data not shown). This initial observation was confirmed by propidium iodide exclusion experiments analyzed by flow cytometry (Fig. 2B). Infection of 3× FLAG GBP-5-expressing cells with SL1344 resulted in a 7.7-fold increase in propidium iodide-stained cells compared to infected control cells in the particular experiment shown. The propidium iodide staining experiments were performed under different conditions than those used for the LDH release experiments described above and below (see Materials and Methods), and thus, the results should not be directly compared. The trends were the same.
The expression of 3× FLAG GBP-5 in RAW 264.7 cells consistently resulted in increased susceptibility to Salmonella-induced pyroptosis. As seen in Fig. 2C, infection of 3× FLAG GBP-5-expressing cells by SL1344 resulted in a 30% increase in total LDH release compared to that of infected control cells. This result was comparable to the 38% increase in LDH release observed from IFN-γ-stimulated control cells infected by SL1344. Both differences were highly significant (P < 0.01) compared to infected control cells, suggesting that the expression of 3× FLAG GBP-5 was sufficient to cause an increase in susceptibility to Salmonella-induced pyroptosis similar to the increase caused by overnight stimulation with IFN-γ.
We hypothesized that 3× FLAG GBP-5 expression might increase the ability of macrophages to internalize invading Salmonella cells. This did not appear to be the case, as an assay designed to determine the number of internalized SL1344 CFU during a 5-min infection did not reveal any difference between 3× FLAG GBP-5-expressing cells and control cells (control cells internalized 2.41 × 104 ± 2.52 × 103 CFU/well, and 3× FLAG GBP-5 cells internalized 2.60 × 104 ± 5.30 × 103 CFU/well [average ± standard deviation]; n = 8). This result suggested that GBP-5 regulated some aspect of pyroptosis and not the ability of Salmonella to invade macrophages.
Targeting of GBP-5 mRNA expression for downregulation by shRNA against GBP-5 results in a significant decrease in Salmonella-induced pyroptosis.
We further tested our hypothesis that GBP-5 played a role in regulating Salmonella-induced pyroptosis by knocking down the expression of the endogenous GBP-5 mRNA. We chose to target GBP-5 using a retroviral vector that expresses shRNA with a human miR30 topology containing a sequence to target the gene of interest's mRNA for degradation. This shRNA topology is thought to greatly increase the efficiency of mRNA targeting by binding to Drosha and increasing the efficiency of mRNA delivery to Dicer and RISC (49). We verified the effectiveness of our targeting construct using real-time PCR for the GBP-5 transcript and compared the experimental construct with a construct that targets hrGFP.
We performed real-time PCR for GBP-5 mRNA using β-actin mRNA as a housekeeping control and demonstrated that the GBP-5 targeting vector (shRNA GBP-5) reduced IFN-γ-induced GBP-5 mRNA levels in cells expressing shRNA GBP-5 by an average of 69% in a statistically significant manner compared to IFN-γ-stimulated control cells expressing shRNA hrGFP (Fig. 3A, right) (P < 0.01). Although the real-time signal from the basal expression of GBP-5 mRNA was low, it was clearly detectable as a single band by agarose gel electrophoresis, whereas signal from a no-reverse-transcriptase control was not (data not shown). The GBP-5 targeting vector reduced basal GBP-5 mRNA expression by an average of 49% (Fig. 3A, left). Although this reduction was not statistically significant, subsequent functional analysis suggested that the basal expression of GBP-5 mRNA was important for Salmonella-induced pyroptosis and that the GBP-5 targeting vector reduced the basal expression of GBP-5 mRNA in a functionally significant manner (Fig. 3B).
Subsequent functional analysis using Salmonella-induced pyroptosis as a measure of GBP-5 function revealed that unstimulated cells expressing shRNA GBP-5 released significantly less LDH on average (average decrease of all wild-type strains of 56%) during infection, at an MOI of 25, by wild-type strains SL1344 (48% less), 14028s (60% less), and CS401 (60% less) than infected cells expressing shRNA hrGFP (Fig. 3B, C, and D). Importantly, the decreased susceptibility of shRNA GBP-5-expressing cells to Salmonella-induced pyroptosis was specific, as the treatment of these cells with the apoptosis inducers camptothecin and staurosporine induced apoptosis at levels similar to those of control cells expressing shRNA hrGFP. Total LDH release levels were 5.16% ± 0.52% for shRNA GBP-5-expressing cells and 4.43% ± 0.44% for control cells expressing shRNA hrGFP after treatment with 20 μM camptothecin, and levels were 23.8% ± 2.39% for shRNA GBP-5-expressing cells and 17.6% ± 2.20% for control cells expressing shRNA hrGFP after treatment with 2.5 μM staurosporine (values are means ± standard deviations) (n = 8).
When shRNA GBP-5 cells were treated with IFN-γ, they released on average 23% less LDH than control cells after infection by the above-mentioned strains at an MOI of 25 (Fig. 3B, C, and D). Although a 23% decrease might not seem very robust (the highest was 29% for CS401), one must consider that in the IFN-γ-induced state, a 69% reduction in GBP-5 mRNA still equates to a 2,400-fold increase in GBP-5 mRNA over the basal state. The fact that we saw a significant and highly reproducible decrease in pyroptosis in response to Salmonella under these conditions suggested that the amount of GBP-5 protein being expressed was important for the regulation of Salmonella-induced pyroptosis whether GBP-5 was expressed at high levels or at low levels. All the differences in LDH release between shRNA GBP-5-expressing cells and shRNA hrGFP-expressing cells infected by the above-described wild-type strains were highly significant by a Student's t test (P < 0.001), whether measured in the basal or IFN-γ-stimulated state.
As expected, the SPI-1 secretion complex was necessary to induce Salmonella pyroptosis in these RAW 264.7 pools (Fig. 3C). Importantly, under basal conditions, Salmonella cells deficient in the expression of fliC and fljB induced significantly lower levels of pyroptosis (P < 0.01) in shRNA GBP-5-expressing cells (Fig. 3D). We interpreted this result, a 4.5-fold lower induction of pyroptosis in shRNA GBP-5-expressing cells, as evidence that the flagellin-independent signaling pathway discussed above (Fig. 1B) might also be regulated by GBP-5.
Expressed 3× FLAG GBP-5 is localized to actin-rich membrane ruffles, and M-CSF induced macropinosomes.
To further investigate the intracellular location of GBP-5, we transduced BAC1.2F5 (BAC) cells with retroviral particles encoding 3× FLAG GBP-5. BAC cells are a simian virus 40-transformed murine macrophage cell line clone that depends on the growth factors macrophage colony-stimulating factor (M-CSF) and/or granulocyte-M-CSF for proliferation (41). Transduced cells were selected and maintained as pools with G418 selection. Macropinocytosis can be induced by starving the cells for M-CSF overnight, followed by restimulating the cells with M-CSF (46).
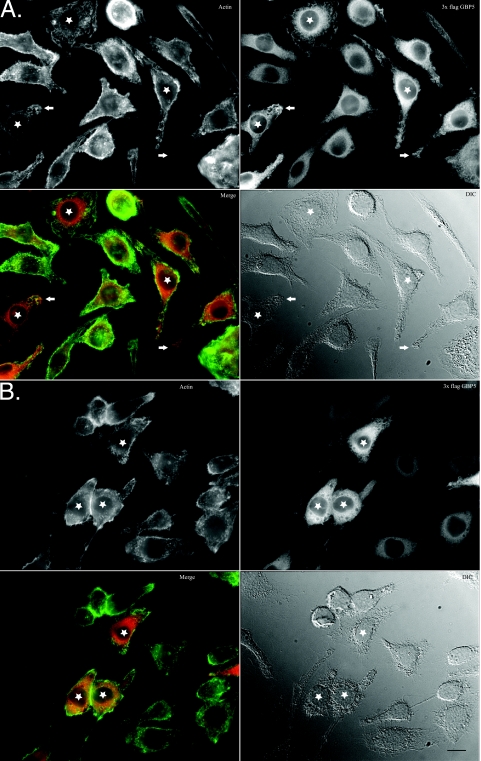
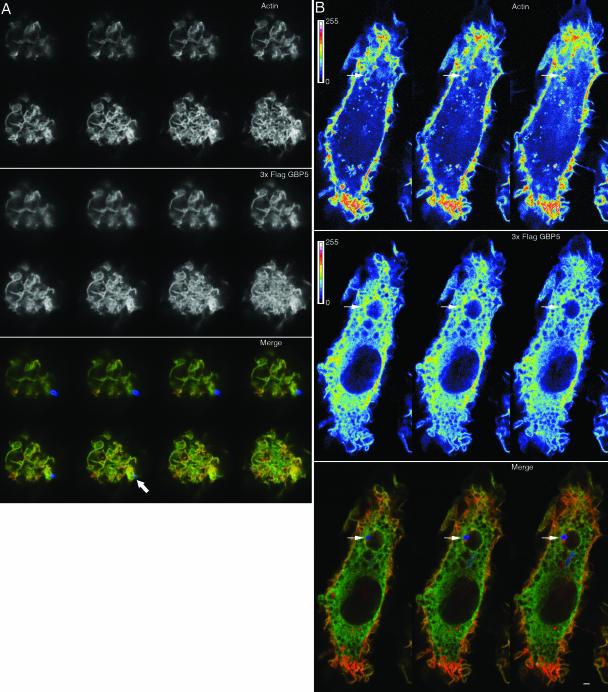
Decoration of 3× FLAG GBP-5-expressing cells with M2 antibody revealed perinuclear staining, which appeared to be vesicular in nature (Fig. 4A). 3× FLAG GBP-5 was colocalized with actin to membrane ruffles (Fig. 4A, stars) and lamellipodia (Fig. 4A, arrows) at the leading edge of cells under culture conditions in the absence of M-CSF. The association of 3× FLAG GBP-5 with filamentous actin was not absolute, as cortical actin did not colocalize with 3× FLAG GBP-5. Not all membrane ruffles appeared to contain 3× FLAG GBP-5, and not all ruffles that stained strongly for 3× FLAG GBP-5 showed strong actin staining. As macrophages often demonstrated heterogenous morphology, this suggested that certain cellular states resulted in a strong colocalization of 3× FLAG GBP-5 with membrane ruffles and that some did not. 3× FLAG GBP-5 might not require filamentous actin for membrane localization, but some connection between 3× FLAG GBP-5 and actin dynamics might exist.
FIG. 4.
Expressed 3× FLAG GBP-5 is localized to actin-rich membrane ruffles and M-CSF-induced macropinosomes. Bac1.2F5 cells expressing 3× FLAG GBP-5 were starved for M-CSF overnight (A) or pulsed with M-CSF after starvation for 10 min (B) prior to fixation and decoration with fluorescent phalloidin and antibodies as indicated. Stars highlight some cells with membrane ruffling or macropinosomes. Arrows highlight 3× FLAG GBP-5 enrichment at sites of cellular protrusions. In merged panels, filamentous actin staining is shown in green, while 3× FLAG GBP-5 is shown in red. Bar, 5 μm. DIC, differential interference contrast.
The addition of M-CSF for 10 min resulted in the formation of numerous macropinosomes. 3× FLAG GBP-5 localized to actin-rich membranes of forming and formed macropinosomes (Fig. 4B, stars). Since Salmonella SPI-1 effectors regulate the actin cytoskeleton to induce membrane ruffling and macropinocytosis, the above-described results suggested that 3× FLAG GBP-5 would be associated with Salmonella-induced cellular structures (7, 45).
3× FLAG GBP-5 is localized to actin-rich pedestals induced by S. enterica serovar Typhimurium and to the membranes of spacious phagosomes induced by S. enterica serovar Typhimurium.
We discovered that 3× FLAG GBP-5 did indeed localize to cellular structures formed by invasive S. enterica serovar Typhimurium (Fig. 5A). 3× FLAG GBP-5 colocalized with actin at the base of membrane-attached Salmonella, suggesting that 3× FLAG GBP-5 could be involved in the initial signal transduction events during the process of Salmonella invasion (Fig. 5A). Analysis of macrophages in which S. enterica serovar Typhimurium had formed spacious vacuoles demonstrated that 3× FLAG GBP-5 was found on the vacuolar membrane and colocalized with actin-rich pedestals that associated with invasive bacteria inside the vacuoles (Fig. 5B). These observations placed 3× FLAG GBP-5 at the interface between the invading Salmonella cells and the host and suggested that 3× FLAG GBP-5 could be involved in regulating some pathway that was stimulated by invading Salmonella and which activated caspase-1, perhaps the IPAF inflammasome. Table 1 summarizes the results of our confocal analysis of the association of 3× FLAG GBP-5 and Salmonella in cells. Although a majority of bacteria (within cells or touching cells) appeared to be strongly associated with 3× FLAG GBP-5 staining, it must be noted that 3× FLAG GBP-5 was clearly associated with other cellular membranes and found in the cytoplasm, facts which made the assignment of 3× GBP-5 localization with Salmonella-associated structures difficult.
FIG. 5.
3× FLAG GBP-5 is localized to actin-rich pedestals induced by S. enterica serovar Typhimurium and to the membranes of spacious phagosomes induced by S. enterica serovar Typhimurium. Bac1.2F5 cells that expressed 3× FLAG GBP-5 were allowed to interact with S. enterica serovar Typhimurium (cultured to maximize the expression of SPI-1) for 5 min, followed by fixation and decoration with fluorescent phalloidin, antibodies to 3× FLAG GBP-5, and Salmonella. (A) Confocal series starting with the cellular crown and ending at an actin-rich pedestal (indicated with a white arrow in the merge pane) with attached Salmonella bacterium. (B) Three consecutive confocal sections through a spacious vacuole containing a Salmonella bacterium (indicated with white arrows). Both the top and middle panels were reproduced with a false color lookup table with 16 colors, as indicated in the panels, to emphasize the enrichment of actin around the bacterium and 3× FLAG GBP-5 on the vacuolar membrane. In merged panels, filamentous actin staining is shown in red, while 3× FLAG GBP-5 is shown in green. Salmonella cells are shown in blue. Bar, 1 μm (A and B).
TABLE 1.
Sixty-seven percent of invasive Salmonella cells were strongly associated with 3× FLAG GBP-5
| Parameter | Total no. of cells | % of total cells |
|---|---|---|
| Total analyzed cells | 22 | |
| Total associated bacteria | 46 | |
| Association with 3× FLAG GBP-5 | ||
| Strong | 31 | 67.40 |
| Moderate | 12 | 26.10 |
| Weak | 3 | 6.50 |
DISCUSSION
Recent studies strongly suggest that IFN signaling positively regulates the detection of PAMPs in the cytoplasm and activation of the inflammasome, leading to cell death of innate immune cells infected by F. tularensis and L. monocytogenes (16, 50, 51). In addition to those observations, we have extended an observation made previously by Monack and colleagues that prior activation of RAW 264.7 cells by LPS or IFN-γ results in an increase in susceptibility to S. enterica serovar Typhimurium-induced pyroptosis (39). We have demonstrated that the increase in susceptibility to Salmonella-induced pyroptosis due to activation by IFN-γ is dependent on caspase-1 activation and is partly dependent on the Salmonella T3SS SPI-1 (Fig. 1).
Furthermore, although Salmonella flagellin subunits have been shown to activate the IPAF inflammasome, another Salmonella PAMP is detected during infection by the ΔfliC ΔfljB strain and results in Salmonella-induced pyroptosis in IFN-γ-stimulated RAW 264.7 cells (Fig. 1B and 3D) (13, 37). Miao and colleagues also found that the ΔfliC ΔfljB strain was capable of inducing IL-1β production at high MOIs and also hypothesized that another Salmonella PAMP may be detected by bone marrow-derived macrophages, but they did not perform experiments with IFN-γ-activated cells (37). Although Franchi and coworkers did not report a flagellin-independent stimulation of caspase-1 activity, they also did not report experiments with high MOIs or activation of macrophages with IFN-γ (13). A number of PAMPs have been shown to activate caspase-1 if access to the cytoplasm is given (22). In the case of infection by the ΔfliC ΔfljB strain, the SPI-1 T3SS provides a route of access for other PAMPs, and in the case of RAW 264.7 cells, gene induction by IFN-γ may provide protein machinery to detect or amplify the detection of those PAMPs. Importantly, we have discovered that the augmentation of Salmonella-induced pyroptosis by prior stimulation of RAW 264.7 cells with IFN-γ is dependent, in part, on the expression of the IFN-inducible gene GBP-5 (Fig. 2).
In summary, three major forms of evidence suggest that GBP-5 interacts with signal transduction events induced by S. enterica serovar Typhimurium and that GBP-5 regulates those signals to modulate the process of Salmonella-induced pyroptosis. First, the expression of 3× FLAG GBP-5 in RAW 264.7 cells causes increased susceptibility to S. enterica serovar Typhimurium-induced pyroptosis (Fig. 2). This observation suggests that the expression of 3× FLAG GBP-5 is sufficient to confer the phenotype of increased sensitivity to Salmonella-induced pyroptosis conferred by the stimulation of RAW 264.7 cells with IFN-γ. We hypothesize that GBP-5 expression must regulate some aspect of pyroptosis, either upstream or downstream of caspase-1 activation. This role does not appear to be increased internalization of invading Salmonella.
Second, the downregulation of endogenous GBP-5 mRNA by specific shRNA targeting of its transcript causes a significant decrease in the susceptibility of RAW 264.7 cells to Salmonella-induced pyroptosis under both basal expression of GBP-5 and IFN-γ-induced conditions (Fig. 3). The fact that shRNA GBP-5-expressing cells are not less susceptible to apoptosis induced by camptothecin or staurosporine suggests that GBP-5 may specifically regulate pyroptosis as opposed to cell death in general. Furthermore, these data suggest that the basal state of GBP-5 expression is important for the regulation of the inflammasome. In our hands, primary bone marrow-derived macrophages do not become more susceptible to Salmonella-induced pyroptosis upon stimulation with IFN-γ, but others have shown that caspase-1 activity is very high in bone marrow-derived macrophages, especially compared to RAW 264.7 cells (56). We hypothesize that basal expression levels of GBP-5 in primary macrophages may be sufficient to perform its function. Further research will be needed to confirm or reject this hypothesis. Thus, we have shown, through both the exogenous expression of 3× FLAG GBP-5 and the downregulation of endogenous GBP-5 mRNA, that GBP-5 is both sufficient and partially necessary for the regulation of Salmonella-induced pyroptosis in RAW 264.7 cells.
Third, 3× FLAG GBP-5 localizes to actin-rich membrane ruffles, macropinosomes induced by M-CSF, and spacious phagosomes induced by invasive S. enterica serovar Typhimurium (Fig. 4 and 5). 3× FLAG GBP-5 colocalizes with actin-rich pedestals at nascent contact sites with Salmonella, strategically placing it where Salmonella effectors are regulating the cellular actin remodeling machinery. 3× FLAG GBP-5 remains associated with S. enterica serovar Typhimurium-containing spacious vacuoles, where it may continue to regulate signals from the vacuole membrane.
Although murine GBPs have been shown to be coordinately regulated by IFN-γ (42), only GBP-5 is highly enriched on purified phagosomal membranes from RAW 264.7 cells, suggesting that the spatial location of GBP-5 may be important for its function (data not shown) (21). Another study of the subcellular localization of human GBPs also suggested that although GBPs share high amino acid homology, their different subcellular locations may confer diverse cellular functions (54). While we did not attempt to determine the nature of the other membranes that interact with 3× FLAG GBP-5, Tripal and associates argued previously that human GBP-5 is associated constitutively with the Golgi apparatus in endothelial cells (54). Clearly, 3× FLAG GBP-5 associates with other cellular membranes, perhaps the Golgi apparatus, but it is also localized to cellular ruffles and spacious phagosomes induced by Salmonella.
Finally, the pathway regulated by GBP-5 is caspase-1 dependent and partly SPI-1 dependent. These data strongly implicate GBP-5 as being a regulator of the rapid Salmonella-induced pyroptosis that others have described previously (4, 6, 18, 39). Many other pathways that lead to cell death in murine macrophages exist, but none of these pathways promote cell death within the first 2 h of infection. Pathways induced by LPS and SPI-2 secretion of SpvB cytotoxin require 3 to 4 h to induce signs of apoptosis (17, 19, 40).
Although recent publications suggested that an IFN-β-induced protein may positively regulate pathogen-induced pyroptosis, our research is the first demonstration of an IFN-regulated protein, GBP-5, that regulates Salmonella-induced pyroptosis. This novel function of GBP-5 suggests that GBP-5 might play an important role in host defense against bacterial pathogens such as S. enterica serovar Typhimurium and others that activate caspase-1 in macrophages.
Acknowledgments
This work was supported by grant R21AI 069111 to J.A.C. from the National Institute of Allergy and Infectious Diseases.
We thank Ed Miao for helpful suggestions with the manuscript and the kind gift of Salmonella strains CS401 and 14028s and the ΔprgHIJK and ΔfliC ΔfljB strains. Strains CS401 and CS401 ΔprgHIJK were sent with permission of S. I. Miller. We also thank E. R. Stanley for his gift of the BAC1.2F5 cell line.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 24 March 2008.
REFERENCES
- 1.Agostini, L., F. Martinon, K. Burns, M. F. McDermott, P. N. Hawkins, and J. Tschopp. 2004. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity 20319-325. [DOI] [PubMed] [Google Scholar]
- 2.Akira, S., S. Uematsu, and O. Takeuchi. 2006. Pathogen recognition and innate immunity. Cell 124783-801. [DOI] [PubMed] [Google Scholar]
- 3.Boehm, U., L. Guethlein, T. Klamp, K. Ozbek, A. Schaub, A. Futterer, K. Pfeffer, and J. C. Howard. 1998. Two families of GTPases dominate the complex cellular response to IFN-gamma. J. Immunol. 1616715-6723. [PubMed] [Google Scholar]
- 4.Brennan, M. A., and B. T. Cookson. 2000. Salmonella induces macrophage death by caspase-1-dependent necrosis. Mol. Microbiol. 3831-40. [DOI] [PubMed] [Google Scholar]
- 5.Cerretti, D. P., C. J. Kozlosky, B. Mosley, N. Nelson, K. Van Ness, T. A. Greenstreet, C. J. March, S. R. Kronheim, T. Druck, L. A. Cannizzaro, et al. 1992. Molecular cloning of the interleukin-1 beta converting enzyme. Science (New York) 25697-100. [DOI] [PubMed] [Google Scholar]
- 6.Chen, L. M., K. Kaniga, and J. E. Galan. 1996. Salmonella spp. are cytotoxic for cultured macrophages. Mol. Microbiol. 211101-1115. [DOI] [PubMed] [Google Scholar]
- 7.Chen, L. M., S. Hobbie, and J. E. Galan. 1996. Requirement of CDC42 for Salmonella-induced cytoskeletal and nuclear responses. Science (New York) 2742115-2118. [DOI] [PubMed] [Google Scholar]
- 8.Chen, Y., M. R. Smith, K. Thirumalai, and A. Zychlinsky. 1996. A bacterial invasin induces macrophage apoptosis by binding directly to ICE. EMBO J. 153853-3860. [PMC free article] [PubMed] [Google Scholar]
- 9.Collazo, C. M., G. S. Yap, G. D. Sempowski, K. C. Lusby, L. Tessarollo, G. F. Woude, A. Sher, and G. A. Taylor. 2001. Inactivation of LRG-47 and IRG-47 reveals a family of interferon gamma-inducible genes with essential, pathogen-specific roles in resistance to infection. J. Exp. Med. 194181-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrt, S., D. Schnappinger, S. Bekiranov, J. Drenkow, S. Shi, T. R. Gingeras, T. Gaasterland, G. Schoolnik, and C. Nathan. 2001. Reprogramming of the macrophage transcriptome in response to interferon-gamma and Mycobacterium tuberculosis: signaling roles of nitric oxide synthase-2 and phagocyte oxidase. J. Exp. Med. 1941123-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fenton, M. J., B. D. Clark, K. L. Collins, A. C. Webb, A. Rich, and P. E. Auron. 1987. Transcriptional regulation of the human prointerleukin 1 beta gene. J. Immunol. 1383972-3979. [PubMed] [Google Scholar]
- 12.Fink, S. L., and B. T. Cookson. 2007. Pyroptosis and host cell death responses during Salmonella infection. Cell. Microbiol. 92562-2570. [DOI] [PubMed] [Google Scholar]
- 13.Franchi, L., A. Amer, M. Body-Malapel, T. D. Kanneganti, N. Ozoren, R. Jagirdar, N. Inohara, P. Vandenabeele, J. Bertin, A. Coyle, E. P. Grant, and G. Nunez. 2006. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in Salmonella-infected macrophages. Nat. Immunol. 7576-582. [DOI] [PubMed] [Google Scholar]
- 14.Ghayur, T., S. Banerjee, M. Hugunin, D. Butler, L. Herzog, A. Carter, L. Quintal, L. Sekut, R. Talanian, M. Paskind, W. Wong, R. Kamen, D. Tracey, and H. Allen. 1997. Caspase-1 processes IFN-gamma-inducing factor and regulates LPS-induced IFN-gamma production. Nature 386619-623. [DOI] [PubMed] [Google Scholar]
- 15.Gu, Y., K. Kuida, H. Tsutsui, G. Ku, K. Hsiao, M. A. Fleming, N. Hayashi, K. Higashino, H. Okamura, K. Nakanishi, M. Kurimoto, T. Tanimoto, R. A. Flavell, V. Sato, M. W. Harding, D. J. Livingston, and M. S. Su. 1997. Activation of interferon-gamma inducing factor mediated by interleukin-1beta converting enzyme. Science (New York) 275206-209. [DOI] [PubMed] [Google Scholar]
- 16.Henry, T., A. Brotcke, D. S. Weiss, L. J. Thompson, and D. M. Monack. 2007. Type I interferon signaling is required for activation of the inflammasome during Francisella infection. J. Exp. Med. 204987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez, L. D., M. Pypaert, R. A. Flavell, and J. E. Galan. 2003. A Salmonella protein causes macrophage cell death by inducing autophagy. J. Cell Biol. 1631123-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hersh, D., D. M. Monack, M. R. Smith, N. Ghori, S. Falkow, and A. Zychlinsky. 1999. The Salmonella invasin SipB induces macrophage apoptosis by binding to caspase-1. Proc. Natl. Acad. Sci. USA 962396-2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hueffer, K., and J. E. Galan. 2004. Salmonella-induced macrophage death: multiple mechanisms, different outcomes. Cell. Microbiol. 61019-1025. [DOI] [PubMed] [Google Scholar]
- 20.Inohara, N., M. Chamaillard, C. McDonald, and G. Nuñez. 2005. NOD-LRR proteins: role in host-microbial interactions and inflammatory disease. Annu. Rev. Biochem. 74355-383. [DOI] [PubMed] [Google Scholar]
- 21.Jutras, I., M. Houde, N. Currier, J. Boulais, S. Duclos, S. Laboissiere, E. Bonneil, P. Kearney, P. Thibault, E. Paramithiotis, P. Hugo, and M. Desjardins. 2007. Modulation of the phagosome proteome by interferon-gamma. Mol. Cell. Proteomics 7697-715. [DOI] [PubMed] [Google Scholar]
- 22.Kanneganti, T. D., M. Lamkanfi, Y. G. Kim, G. Chen, J. H. Park, L. Franchi, P. Vandenabeele, and G. Nunez. 2007. Pannexin-1-mediated recognition of bacterial molecules activates the cryopyrin inflammasome independent of Toll-like receptor signaling. Immunity 26433-443. [DOI] [PubMed] [Google Scholar]
- 23.Kawai, T., and S. Akira. 2006. TLR signaling. Cell Death Differ. 13816-825. [DOI] [PubMed] [Google Scholar]
- 24.Kimbrough, T. G., and S. I. Miller. 2000. Contribution of Salmonella typhimurium type III secretion components to needle complex formation. Proc. Natl. Acad. Sci. USA 9711008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koide, S., and R. M. Steinman. 1987. Induction of murine interleukin 1: stimuli and responsive primary cells. Proc. Natl. Acad. Sci. USA 843802-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuida, K., J. A. Lippke, G. Ku, M. W. Harding, D. J. Livingston, M. S. Su, and R. A. Flavell. 1995. Altered cytokine export and apoptosis in mice deficient in interleukin-1 beta converting enzyme. Science (New York) 2672000-2003. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227680-685. [DOI] [PubMed] [Google Scholar]
- 28.Li, P., H. Allen, S. Banerjee, S. Franklin, L. Herzog, C. Johnston, J. McDowell, M. Paskind, L. Rodman, J. Salfeld, et al. 1995. Mice deficient in IL-1 beta-converting enzyme are defective in production of mature IL-1 beta and resistant to endotoxic shock. Cell 80401-411. [DOI] [PubMed] [Google Scholar]
- 29.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta C(T)) method. Methods (San Diego) 25402-408. [DOI] [PubMed] [Google Scholar]
- 30.MacMicking, J. D. 2004. IFN-inducible GTPases and immunity to intracellular pathogens. Trends Immunol. 25601-609. [DOI] [PubMed] [Google Scholar]
- 31.MacMicking, J. D., G. A. Taylor, and J. D. McKinney. 2003. Immune control of tuberculosis by IFN-gamma-inducible LRG-47. Science (New York) 302654-659. [DOI] [PubMed] [Google Scholar]
- 32.Mariathasan, S., K. Newton, D. M. Monack, D. Vucic, D. M. French, W. P. Lee, M. Roose-Girma, S. Erickson, and V. M. Dixit. 2004. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 430213-218. [DOI] [PubMed] [Google Scholar]
- 33.Mariathasan, S., D. S. Weiss, K. Newton, J. McBride, K. O'Rourke, M. Roose-Girma, W. P. Lee, Y. Weinrauch, D. M. Monack, and V. M. Dixit. 2006. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 440228-232. [DOI] [PubMed] [Google Scholar]
- 34.Mariathasan, S., and D. M. Monack. 2007. Inflammasome adaptors and sensors: intracellular regulators of infection and inflammation. Nat. Rev. 731-40. [DOI] [PubMed] [Google Scholar]
- 35.Martinon, F., K. Burns, and J. Tschopp. 2002. The inflammasome: a molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 10417-426. [DOI] [PubMed] [Google Scholar]
- 36.Martinon, F., and J. Tschopp. 2007. Inflammatory caspases and inflammasomes: master switches of inflammation. Cell Death Differ. 1410-22. [DOI] [PubMed] [Google Scholar]
- 37.Miao, E. A., C. M. Alpuche-Aranda, M. Dors, A. E. Clark, M. W. Bader, S. I. Miller, and A. Aderem. 2006. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 7569-575. [DOI] [PubMed] [Google Scholar]
- 38.Miura, M., H. Zhu, R. Rotello, E. A. Hartwieg, and J. Yuan. 1993. Induction of apoptosis in fibroblasts by IL-1 beta-converting enzyme, a mammalian homolog of the C. elegans cell death gene ced-3. Cell 75653-660. [DOI] [PubMed] [Google Scholar]
- 39.Monack, D. M., B. Raupach, A. E. Hromockyj, and S. Falkow. 1996. Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc. Natl. Acad. Sci. USA 939833-9838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monack, D. M., C. S. Detweiler, and S. Falkow. 2001. Salmonella pathogenicity island 2-dependent macrophage death is mediated in part by the host cysteine protease caspase-1. Cell. Microbiol. 3825-837. [DOI] [PubMed] [Google Scholar]
- 41.Morgan, C., J. W. Pollard, and E. R. Stanley. 1987. Isolation and characterization of a cloned growth factor dependent macrophage cell line, BAC1.2F5. J. Cell. Physiol. 130420-427. [DOI] [PubMed] [Google Scholar]
- 42.Nguyen, T. T., Y. Hu, D. P. Widney, R. A. Mar, and J. B. Smith. 2002. Murine GBP-5, a new member of the murine guanylate-binding protein family, is coordinately regulated with other GBPs in vivo and in vitro. J. Interf. Cytok. Res. 22899-909. [DOI] [PubMed] [Google Scholar]
- 43.Olszewski, M. A., J. Gray, and D. J. Vestal. 2006. In silico genomic analysis of the human and murine guanylate-binding protein (GBP) gene clusters. J. Interf. Cytok. Res. 26328-352. [DOI] [PubMed] [Google Scholar]
- 44.Paddison, P. J., M. Cleary, J. M. Silva, K. Chang, N. Sheth, R. Sachidanandam, and G. J. Hannon. 2004. Cloning of short hairpin RNAs for gene knockdown in mammalian cells. Nat. Methods 1163-167. [DOI] [PubMed] [Google Scholar]
- 45.Patel, J. C., and J. E. Galan. 2006. Differential activation and function of Rho GTPases during Salmonella-host cell interactions. J. Cell Biol. 175453-463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Racoosin, E. L., and J. A. Swanson. 1992. M-CSF-induced macropinocytosis increases solute endocytosis but not receptor-mediated endocytosis in mouse macrophages. J. Cell Sci. 102867-880. [DOI] [PubMed] [Google Scholar]
- 47.Rakeman, J. L., H. R. Bonifield, and S. I. Miller. 1999. A HilA-independent pathway to Salmonella typhimurium invasion gene transcription. J. Bacteriol. 1813096-3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schroder, K., P. J. Hertzog, T. Ravasi, and D. A. Hume. 2004. Interferon-gamma: an overview of signals, mechanisms and functions. J. Leukoc. Biol. 75163-189. [DOI] [PubMed] [Google Scholar]
- 49.Silva, J. M., M. Z. Li, K. Chang, W. Ge, M. C. Golding, R. J. Rickles, D. Siolas, G. Hu, P. J. Paddison, M. R. Schlabach, N. Sheth, J. Bradshaw, J. Burchard, A. Kulkarni, G. Cavet, R. Sachidanandam, W. R. McCombie, M. A. Cleary, S. J. Elledge, and G. J. Hannon. 2005. Second-generation shRNA libraries covering the mouse and human genomes. Nat. Genet. 371281-1288. [DOI] [PubMed] [Google Scholar]
- 50.Stockinger, S., T. Materna, D. Stoiber, L. Bayr, R. Steinborn, T. Kolbe, H. Unger, T. Chakraborty, D. E. Levy, M. Muller, and T. Decker. 2002. Production of type I IFN sensitizes macrophages to cell death induced by Listeria monocytogenes. J. Immunol. 1696522-6529. [DOI] [PubMed] [Google Scholar]
- 51.Stockinger, S., B. Reutterer, B. Schaljo, C. Schellack, S. Brunner, T. Materna, M. Yamamoto, S. Akira, T. Taniguchi, P. J. Murray, M. Muller, and T. Decker. 2004. IFN regulatory factor 3-dependent induction of type I IFNs by intracellular bacteria is mediated by a TLR- and Nod2-independent mechanism. J. Immunol. 1737416-7425. [DOI] [PubMed] [Google Scholar]
- 52.Taylor, G. A., C. G. Feng, and A. Sher. 2004. p47 GTPases: regulators of immunity to intracellular pathogens. Nat. Rev. 4100-109. [DOI] [PubMed] [Google Scholar]
- 53.Thornberry, N. A., H. G. Bull, J. R. Calaycay, K. T. Chapman, A. D. Howard, M. J. Kostura, D. K. Miller, S. M. Molineaux, J. R. Weidner, J. Aunins, et al. 1992. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 356768-774. [DOI] [PubMed] [Google Scholar]
- 54.Tripal, P., M. Bauer, E. Naschberger, T. Mortinger, C. Hohenadl, E. Cornali, M. Thurau, and M. Sturzl. 2007. Unique features of different members of the human guanylate-binding protein family. J. Interf. Cytok. Res. 2744-52. [DOI] [PubMed] [Google Scholar]
- 55.van der Velden, A. W., S. W. Lindgren, M. J. Worley, and F. Heffron. 2000. Salmonella pathogenicity island 1-independent induction of apoptosis in infected macrophages by Salmonella enterica serotype Typhimurium. Infect. Immun. 685702-5709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Verhoef, P. A., S. B. Kertesy, K. Lundberg, J. M. Kahlenberg, and G. R. Dubyak. 2005. Inhibitory effects of chloride on the activation of caspase-1, IL-1beta secretion, and cytolysis by the P2X7 receptor. J. Immunol. 1757623-7634. [DOI] [PubMed] [Google Scholar]
- 57.Vestal, D. J. 2005. The guanylate-binding proteins (GBPs): proinflammatory cytokine-induced members of the dynamin superfamily with unique GTPase activity. J. Interf. Cytok. Res. 25435-443. [DOI] [PubMed] [Google Scholar]
- 58.Wray, C., and W. J. Sojka. 1978. Experimental Salmonella typhimurium infection in calves. Res. Vet. Sci. 25139-143. [PubMed] [Google Scholar]