Abstract
The formin family of proteins mediates dynamic changes in actin assembly in eukaryotes, and therefore it is important to understand the function of these proteins in Entamoeba histolytica, where actin forms the major cytoskeletal network. In this study we have identified the formin homologs encoded in the E. histolytica genome based on sequence analysis. Using multiple tools, we have analyzed the primary sequences of the eight E. histolytica formins and discovered three subsets: (i) E. histolytica formin-1 to -3 (Ehformin-1 to -3), (ii) Ehformin-4, and (iii) Ehformin-5 to -8. Two of these subsets (Ehformin-1 to -3 and Ehformin-4) showed significant sequence differences from their closest homologs, while Ehformin-5 to -8 were unique among all known formins. Since Ehformin-1 to -3 showed important sequence differences from Diaphanous-related formins (DRFs), we have studied the functions of Ehformin-1 and -2 in E. histolytica transformants. Like other DRFs, Ehformin-1 and -2 associated with F-actin in response to serum factors, in pseudopodia, in pinocytic and phagocytic vesicles, and at cell division sites. Ehformin-1 and -2 also localized with the microtubular assembly in the nucleus, indicating their involvement in genome segregation. While increased expression of Ehformin-1 and -2 did not affect phagocytosis or motility, it clearly showed an increase in the number of binucleated cells, the number of nuclei in multinucleated cells, and the average DNA content of each nucleus, suggesting that these proteins regulate both mitosis and cytokinesis in E. histolytica.
Polymerization of actin into helical filaments is nucleated by different groups of actin-binding proteins, which in turn are controlled by specific signaling molecules in eukaryotic cells (10, 66). Currently, the three known groups of F-actin nucleating factors are the Arp2/3 complex, formins, and spire (10, 21). Formin homology proteins promote rapid assembly of unbranched actin filaments and local reorganization of higher-order cellular structures to execute specified cytoskeletal functions (reviewed in reference 21). These multidomain proteins have many isoforms that are ubiquitously expressed and perform diverse cellular functions in almost all eukaryotic cells, from vertebrates and plants to unicellular protists (25). The defining feature of formin proteins is the formin homology 2 (FH2) domain, which is required for the nucleation and elongation of nascent actin filaments (24, 43, 46). The FH1 domain, preceding the FH2 domain, is another key region found in most formins and is made up of tandem repeats of proline and other amino acids (24). Polyproline residues in the FH1 domain interact with profilin to recruit assembly-competent actin monomers in the vicinity of the FH2 domain (50, 52, 61). The FH1 domain also interacts with Src homology 3 (SH3) domain- or WW domain-containing proteins (17, 59). Thus, the core FH1-FH2 region nucleates unbranched actin filaments, unlike the Arp2/3 complex, which creates branched filaments (21). Other regulatory domains, such as the FH3 domain and the GTPase binding domain (GBD), are loosely conserved and are not found in all formin homology proteins (24). The FH3 domain harbors information for cellular localization in some formins (28, 45), while the GBD confers the ability to respond to Rho GTPase-mediated extracellular signals (61, 62).
The Diaphanous-related formins (DRFs) are a subfamily of formin homology proteins that include the metazoan Diaphanous, DAAM (the Disheveled-associated activators of morphogenesis), and FRL (the formin-related proteins in leukocytes), as well as the fungal Bni1, Bnr1, and SepA proteins (21). The fungal members are divergent but share similar functional domains with conserved residues (25). The unique feature of DRFs is the presence of a 17- to 20-residue motif at the C-terminal end, known as the Diaphanous autoregulatory domain (DAD) (1). The DAD sequence has been demonstrated to bind intramolecularly to an N-terminal region, the Diaphanous inhibitory domain (DID), allowing the protein to fold back on itself (31, 33, 39, 60) and consequently inhibit Diaphanous function (1, 34). Binding of Rho GTPase to the GBD site, adjacent to the DID, is believed to release DAD-DID binding (31, 42, 51) and induce the formation of stress fibers, actin cables, focal adhesions, and contractile cytokinetic rings (1, 62).
Recent studies have demonstrated the interaction of several DRFs with the spindle microtubules in dividing cells (26, 32, 67) and with filamentous microtubular assemblies in nondividing cells (44, 64). In brief, different formin proteins regulate various dynamic functions such as the formation of stress fibers, actin cables, filopodia, cytokinetic contractile rings, and endosomes, the maintenance of cell polarity, cell growth and movement, embryonic development, cell and tissue morphogenesis, and the microtubular assemblies in different cell types (21).
Entamoeba histolytica is the causative agent for dysentery and the formation of liver abscesses in humans. Its ability to adhere to host cells, penetrate and destroy intestinal tissues, phagocytose epithelial cells, and proliferate after colonization of the gut mucosa is essential for the manifestation of pathogenesis and is dependent on dynamic changes in the actin cytoskeleton (6, 7, 37, 48). Aggressive motility and polymorphic shape changes—characteristic features of this organism—are derived from the signal-induced remodeling of the acto-myosin cytoskeleton (16). The E. histolytica genome encodes several cytoskeleton-associated proteins and signaling molecules that could regulate actin reorganization (35; TIGR Entamoeba histolytica Genome Project [http://www.tigr.org/tdb/e2k1/eha1/]; Pathema Bioinformatics Resource Center, Entamoeba [http://pathema.jcvi.org/cgi-bin/Entamoeba/PathemaHomePage.cgi]). Some of them have been shown to participate in motility through pseudopod and uroid formation, capping, phagocytosis, interaction with extracellular matrix proteins, and signal transduction mechanisms (3, 5, 15, 20, 22, 30, 47, 53, 56, 58, 63, 65). However, unlike those of many metazoan systems, the cytoskeleton components involved in cell division in E. histolytica are not fully understood.
The cell division cycle of E. histolytica trophozoites in axenic culture shows very unusual features (36). Log-phase cultures may accumulate multinucleated cells containing heterogeneous amounts of DNA (12, 18). Our previous observations suggest that genome reduplication without mitosis and irregularity in cytokinesis contribute maximally to the observed heterogeneity (12, 36). We have also observed that sequence homologs of several eukaryotic “checkpoint proteins” are not found in the amoeba genome (36). However, functional analysis of a kinesin-like protein, E. histolytica Klp5 (Klp5Eh), showed that increased expression of this protein, while promoting microtubular spindles, leads to homogenization of the average DNA content in growing cells (13). Thus, in the absence of conserved regulatory proteins, genome content and cell division may be regulated by other proteins in this parasite.
Here we have focused on understanding the properties of formin homology proteins in E. histolytica, both by comparative sequence analysis and by functional studies. Our analyses revealed that three of the eight formins, Ehformin-1 to -3, are related to Diaphanous proteins, with important differences. We have discovered their function in actin reorganization, microtubular assembly, cell division, and nuclear DNA content in E. histolytica. We have also identified a new family of formin homology proteins, Ehformin-5 to -8, that contains the FH2 domain alone.
MATERIALS AND METHODS
Cell culture and maintenance.
E. histolytica HM1:IMSS trophozoites were maintained axenically and subcultured every 72 h in TYIS 33 medium (14) at 37°C. Stable transformants were maintained in the same medium containing G418 at different concentrations, specified below.
Cloning and sequencing of epitope-tagged Ehformin-1 and -2.
Cloning of the hemagglutinin (HA)-tagged Ehformin-1 gene has been reported previously (27). The DNA fragment encoding the six-His and two-HA region (HH) of the multiaffinity epitope tag CHH (13) was cloned in frame at the 3′ end of the Ehformin-2 gene (3.5 kb) in pBlueScript SK(−). Subsequently, the HH-tagged Ehformin-2 gene was subcloned into pJST4. Stable transformants of the HA-tagged Ehformin-1 gene, the HH-tagged Ehformin-2 gene, and an empty vector control (13) were stably maintained in the presence of G418.
Isolation of RNA and quantitative PCR.
Total RNA was isolated from control, Ehformin-1, and Ehformin-2 stable transformants using Trizol reagent (Invitrogen) and was purified with an RNeasy kit (Qiagen, Germany). Reverse transcriptase (RT) reactions were performed using a First-Strand cDNA synthesis kit (Roche, Germany), and quantitative PCR analysis was carried out in a 7500 Real-Time system with SDS software and Sybr green PCR master mix (Applied Biosystems) according to the manufacturer's instructions. Primers specific to the unique regions of the Ehformin-1, Ehformin-2, and actinEh genes (Ehformin-1 sense and antisense, 5′-AGGCACAAAGAGAAAAAGAAG-3′ and 5′-AACCCACCAACATTAGCAG-3′, respectively; Ehformin-2 sense and antisense, 5′-AAGCAGACAATGAAATGAAAA-3′ and 5′-CCAATAACAACAGTGGAAGG-3′, respectively; actinEh sense and antisense, 5′-TGGGACGATATGGAAAAGAT-3′ and 5′-ATAGCTGGGGTGTTGAATGT-3′, respectively) were used for the PCR. All reactions were carried out in triplicate, and average threshold cycle values were calculated. Sample values for the Ehformin-1 and Ehformin-2 genes were normalized to that for the actinEh gene (endogenous control), and relative expression was calculated using the relative quantification software (Applied Biosystems).
Cell lysis, antibodies, and Western blot analysis.
Forty-eight-hour-grown cells (3 × 106 cells) of E. histolytica transformants were lysed in a buffer containing 25 mM HEPES-KOH (pH 7.4), 100 mM KCl, 5 mM MgCl2, 5% glycerol supplemented with Complete protease inhibitor (Roche, Germany), 1 mM E-64 (Sigma), 1 mM phenylmethylsulfonyl fluoride (Sigma), and 1% Triton X-100 at 4°C. Fifty micrograms of total protein from each sample was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and Western blots of these proteins were hybridized overnight at 4°C with the monoclonal anti-HA antibody 12CA5 (1:1,000; Roche, Germany) and a polyclonal antibody against E. histolytica β-tubulin (1:500) (13), followed by horseradish peroxidase-conjugated secondary antibodies (1:5,000; Sigma). Signals were detected by chemiluminescence using an ECL kit (Roche, Germany).
Isolation of actin-rich fractions from Ehformin-1 and -2 transformants.
The actin-rich fractions were isolated from Ehformin-1 and -2 transformants as described by Vargas et al. (56). Actin-rich fractions and crude lysates were separated by SDS-PAGE, transferred to Western blots, and hybridized with anti-HA (1:1,000) and anti-β-actin (1:1,000; Sigma) antibodies. Densitometric analysis was performed with ImageQuant, version 5.2, to quantitate band intensities.
Immunofluorescence microscopy.
Ehformin-1 or Ehformin-2 transformants were grown on coverslips in 24-well plates at 37°C, fixed directly with warm 3.7% formaldehyde for 15 min, and permeabilized with 0.1% Triton X-100 for 10 min. Fixed cells were stained with an anti-HA antibody (1:200), followed by an Alexa Fluor 488-conjugated anti-mouse secondary antibody (1:2,500; Molecular Probes) and rhodamine-conjugated phalloidin (1 U per coverslip; Sigma), or a polyclonal anti-E. histolytica β-tubulin antibody (1:200), followed by a tetramethyl rhodamine isocyanate-conjugated anti-rabbit secondary antibody (1:200; Jackson Laboratories). Images were acquired with a 63× Plan-Apochromat 1.4 oil differential interference contrast (DIC) objective (numerical aperture, 1.4) in a Zeiss LSM 510 Meta confocal microscope equipped with a 488-nm argon laser and a 543-nm He/Ne laser and were analyzed with the LSM Meta 510 software package (Zeiss, Germany).
Analysis of DNA content by flow cytometry.
Cells were prepared for flow cytometry according to the work of Dastidar et al. (13), and data were acquired in a FACSCalibur instrument (Becton Dickinson) equipped with a single laser system (6-W Innova 90-6 argon ion laser). For measurement of DNA content, cells were excited with 488-nm light, and emission was measured through a 575-nm-wavelength DF20 filter (for propidium iodide fluorescence; FL2). Data from 10,000 cells were recorded for each experiment and analyzed using CellQuest software (Becton Dickinson).
Scanning cytometry to determine the DNA content of each nucleus.
For measurement of nuclear DNA contents, ethanol-fixed cells were stained with 4′,6′-diamidino-2-phenylindole (DAPI) (0.1 μg/ml) for 10 min and spread on glass slides (13). Each slide was scanned for the DNA contents of individual nuclei under a 40× objective of a Zeiss Axiovert 200M fluorescence microscope fitted with a MetaCyte scanning cytometer and Metafer 4 software (Zeiss, Germany). A minimum of 2,000 nuclei were scanned for each strain. The DAPI fluorescence values were normalized and represented as histograms.
Comparative sequence analysis of E. histolytica formins.
The amino acid sequences of FH2 domains were taken from the NCBI protein database and used as queries for BLAST (2) searches in the E. histolytica genome database (http://www.tigr.org/tdb/e2k1/eha1/) to identify E. histolytica formins. Amino acid sequence alignments of formin homologs were performed using ClustalW with the BLOSUM62 matrix and default settings, followed by manual editing for optimization. An unrooted phylogenetic tree was constructed and visualized using the p-distance matrix of neighbor-joining algorithms through MEGA (version 3.1) (29). To provide confidence levels for the tree topology and the statistical reliability of individual nodes, bootstrap analyses were performed with 1,000 replications. Formin domains were analyzed by using a combination of searches in the Pfam database (http://pfam.janelia.org/), the Conserved Domain Database (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml), and SMART tools (http://smart.embl-heidelberg.de/).
RESULTS
The Entamoeba histolytica genome encodes three classes of formin homology proteins, one subset of which is unique in the eukaryotic kingdom.
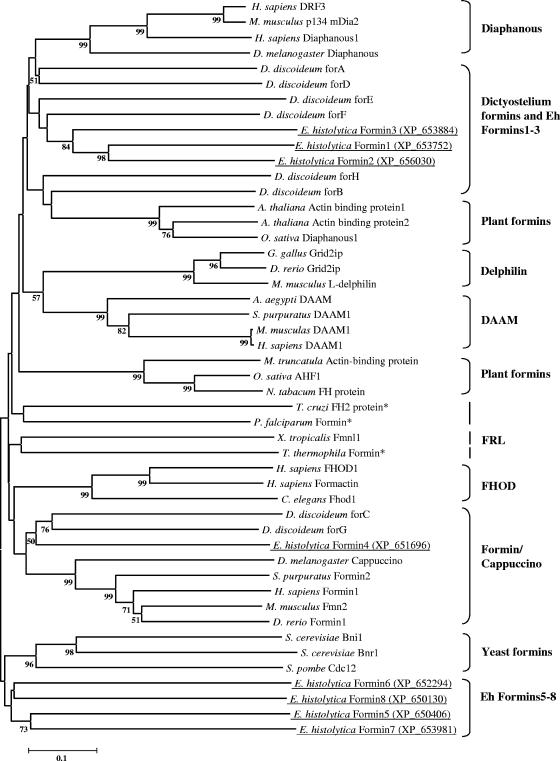
The defining and conserved feature of different isoforms of formins is the FH2 domain, which dimerizes to stabilize intermediates during actin polymerization and elongation (21, 43, 46). Based on the presence of the FH2 domain, we identified nine formin homology proteins in the recently completed E. histolytica genome. Two of these proteins (NCBI accession numbers XP_650406 and XP_652479) were found to be identical in their amino acid and coding nucleotide sequences. One of the eight unique E. histolytica formins (NCBI accession number XP_653752) was described as DiaphanousEh by us earlier (19, 27), but after examining the sequence attributes of all eight E. histolytica formins, we have reannotated DiaphanousEh as Ehformin-1 and the other formin proteins as Ehformin-2 to -8 (Fig. 1). Incidentally, data obtained from microarray hybridization of total RNA showed that all eight formins were expressed in E. histolytica (S. Majumder and A. Lohia, unpublished data).
FIG. 1.
Phylogenetic analysis of E. histolytica formins. The amino acid sequences of the FH2 domains of E. histolytica formins and their closest homologs from the nonredundant database (see the table in the supplemental material) were aligned with ClustalW, and the output file was subsequently edited. A bootstrapped (1,000 times), unrooted phylogenetic tree was constructed using the neighbor-joining method of MEGA (version 3.1). For simplicity, only nodes supported by >50% bootstraps have been indicated. The E. histolytica formins are underlined, and corresponding NCBI accession numbers are given in parentheses. Asterisks indicate formins from other protists. Scale bar, percent substitutions.
(i) Phylogenetic analysis identified a novel group of formins in E. histolytica.
We identified 40 representative formin homologs across species (see the table in the supplemental material) from BLAST searches in the nonredundant protein database using the eight E. histolytica FH2 domain sequences, followed by the construction of an unrooted phylogenetic tree. Two recent reports have grouped metazoan formins into seven major classes, while Dictyostelium, fungal, and plant formins were distinct from metazoan formins (25, 49). Our analysis also showed a similar distribution of different eukaryotic formins into separate classes on the phylogenetic tree (Fig. 1). Our analysis showed that E. histolytica formins could be classified into three distinct subsets: (i) Ehformin-1 to -3, (ii) Ehformin-4, and (iii) Ehformin-5 to -8. In addition, we found that formins from protists such as Trypanosoma cruzi, Plasmodium falciparum, and Tetrahymena thermophila were randomly distributed with metazoan, plant, and fungal formins on the tree (Fig. 1).
(ii) Ehformin-1 to -3.
Ehformin-1 to -3 clustered together with high bootstrap support. Ehformin-1 was 63% similar (47% identical) to Ehformin-2 and 48% similar (28% identical) to Ehformin-3, while Ehformin-2 was 53% similar (33% identical) to Ehformin-3 in their FH2 domains. The closest homologs of Ehformin-1 to-3 were a group of Dictyostelium discoideum formins, although these showed low bootstrap support (Fig. 1). These Dictyostelium formins were characterized as DRFs in different experimental studies (49, 54).
(iii) Ehformin-4.
Ehformin-4 belonged to the group of formins containing D. discoideum ForC and ForG (Fig. 1). Although D. discoideum ForC showed overall homology to the Cappuccino group of formins, it was distinct from all formin proteins in that it lacked a FH1 domain (28).
(iv) Ehformin-5 to -8.
Ehformin-5 to -8, clustered together on the phylogenetic tree and were more divergent than other formin homologs (Fig. 1). Importantly, the amino acid residues crucial for the FH2 domain function of nucleating actin filaments were well conserved in these proteins (see Fig. S1 in the supplemental material). Domain analysis revealed the absence of the polyproline repeat-containing FH1 domain in these amoeba formins except for a single stretch of seven proline residues in Ehformin-5 alone (Fig. 2). The absence of the FH1 domain was first shown in D. discoideum ForC (28). Although D. discoideum ForC did not contain the FH1 domain, it was demonstrated to be functional (28). Thus, the presence of the FH1 domain was not indispensable for formin function. Except for the conserved FH2 domain, other known motifs identified in different formin homology proteins could not be identified in Ehformin-5 to -8 proteins. Therefore, the uncharacterized region of Ehformin-5 to -8 may lead to the identification of novel mechanisms that regulate the functions of these proteins in E. histolytica.
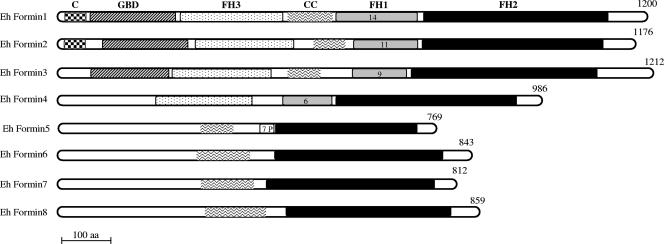
FIG. 2.
Domain architecture of E. histolytica formin-1 to -8. The multiple domains of E. histolytica formins were identified by their matches in the Pfam domain database, the Conserved Domain Database, and SMART tools and are represented schematically. The FH2, FH1, and FH3 domains, the GBD, the predicted coiled-coil region (CC), and the protein kinase C1-like domain (C) are shown. Numbers inside the FH1 boxes are numbers of tandem repeats of polyproline interspersed with glycine and methionine mostly, in Ehformin-1 to -4. “7P” indicates the 7 proline residues preceding the FH2 domain in Ehformin-5. The number of amino acids in each protein is given on the right.
(v) Domains of E. histolytica formins.
Ehformin-1 to -3 contained the conventional GBD and FH3 domain in their N termini, like other DRFs (Fig. 2). Additionally, Ehformin-1 and Ehformin-2 contained the cysteine-rich protein kinase C1 domain at their extreme N-terminal ends (Fig. 2), like some Dictyostelium DRFs (49). Another conserved feature of DRFs is the DAD at the C-terminal end, which is known to interact with the N-terminal DID to regulate DRF function (1, 33, 60). The DID is a part of the GBD and the FH3 domain. Compared to those in the DAD, the amino acids in the DID were loosely conserved in different DRFs (39, 42). The DAD and the DID were identified in Ehformin-1 to -3 but showed important alterations in some conserved amino acids (see Fig. S2 and S3 in the supplemental material). Since the subset of Ehformin-1 to -3 was related to Dictyostelium DRFs but showed important differences in regulatory sequences, we chose to characterize the roles of Ehformin-1 and -2 in remodeling the actin cytoskeleton in E. histolytica.
Ehformin-1 and -2 were colocalized with F-actin structures involved in motility, phagocytosis, and cell division.
In order to study the function of Ehformin-1 in E. histolytica, we generated polyclonal antibodies against the FH1 domain to localize Ehformin-1 (27). Using one of these antibodies, we previously showed the expression of Ehformin-1 in wild-type cells and HA-tagged Ehformin-1 transformants. However, this antibody cross-reacted with other formins (27) and therefore lacked the specificity required for analyzing the functions of Ehformin-1 and -2. Subsequently, epitope-tagged Ehformin-1 and -2 were expressed in stable transformants in order to study their specific localization and function. The DNA fragments carrying the HA-tagged Ehformin-1 and His-HA-tagged Ehformin-2 genes were cloned into the pJST4 expression vector in order to express the recombinant proteins (see Fig. S4 in the supplemental material) in E. histolytica transformants. Stable transformants of HA-tagged Ehformin-1, His-HA-tagged Ehformin-2, and an empty vector control were selected at increasing concentrations of G418 (10 μg/ml to 40 μg/ml) (23). Expression of the recombinant proteins was detected by a monoclonal anti-HA antibody in the transformants maintained at 40 μg/ml of G418 (Fig. 3A). The mRNA levels for both Ehformin-1 and Ehformin-2 were increased 1.6- to 1.8-fold over endogenous expression (Fig. 3B).
FIG. 3.
Expression of Ehformin-1 and -2 in stable transformants of E. histolytica. (A) Western blot analysis of the expression of recombinant Ehformin-1 and -2 in stable transformants of E. histolytica. Cell lysates from control (lane 1), HA-tagged Ehformin-1 (lane 2), and His-HA-tagged Ehformin-2 (lane 3) transformants were separated by 7% SDS-PAGE and transferred by Western blotting (WB). Western blots were hybridized with a monoclonal anti-HA antibody to detect the epitope-tagged Ehformin-1 and -2. A similar blot was hybridized with a polyclonal anti- E. histolytica β-tubulin antibody as a loading control. (B) Quantitative RT-PCR analysis of E. histolytica formins in the respective transformants. Real-time PCR was performed on cDNA prepared from control and forminEh transformants (x axis) using specific forminEh primer sets. ActinEh primers were used as an endogenous control. The relative expression of Ehformin-1 and Ehformin-2 in control, Ehformin-1, and Ehformin-2 transformants was plotted. Error bars, standard deviations (n = 3). (C) Association of Ehformin-1 and -2 with the actin-rich subcellular fraction. Crude lysates (lanes 1 and 2) and the actin-rich fraction (lanes 3 and 4) from Ehformin-1 (lanes 1 and 3) and Ehformin-2 (lanes 2 and 4) transformants were separated by SDS-PAGE, transferred to Western blots, and hybridized with monoclonal anti-HA and anti-β-actin antibodies. M, molecular sizes. Arrow indicates bands for HA-tagged Ehformin-1 and HA-tagged Ehformin-2.
Since formins are known to bind to the actin assembly, we isolated the subcellular fraction containing actin and actin-bound proteins from cell lysates of Ehformin-1 and -2 transformants. We showed that both Ehformin-1 and Ehformin-2 were associated with the actin-rich fractions (Fig. 3C). The retarded mobility of Ehformin-1 and Ehformin-2 in the actin-rich fraction could be due to the high salt concentrations used during the isolation of this fraction. Although actin was enriched threefold in the actin-rich fraction, Ehformin-1 and -2 did not show significant enrichment. This indicated that Ehformin-1 and -2 were distributed both in the actin-rich fraction and independently in the cytoplasm.
In order to identify the involvement of Ehformin-1 and -2 with various F-actin-based cytoskeletal structures in vivo, we used indirect immunofluorescence confocal microscopy to localize Ehformin-1 and -2 with F-actin (Fig. 4A). Ehformin-1 and -2 colocalized with phalloidin-labeled F-actin at cortical regions, pseudopodial protrusions, cytoplasmic invaginations, and crown-like structures of various sizes (Fig. 4Aa) (Ehformin-1 data not shown), which have been identified for phagocytosis and macropinocytosis in E. histolytica (6, 7, 37, 38). Importantly, Ehformin-1 was localized with actin cables (Fig. 4Ab) running through the constricted cleavage site in predividing cells (9). Ehformin-2 was seen to be colocalized with similar actin assemblies in dividing cells (data not shown). Thus, the association of Ehformin-1 and -2 with polymerized actin in pseudopodia, crown like structures, and cell division sites suggests that Ehformin-1 and -2 may be involved with actin remodeling, which is required for these dynamic processes in E. histolytica. Indeed, mutations or down-regulation in Diaphanous and DRF genes have been shown to abrogate F-actin polymerization and consequent alterations in cell division (8) as well as phagocytosis (11, 55), the formation of filopodial protrusions (54), and other cytoskeletal functions in different organisms (reviewed in reference 21).
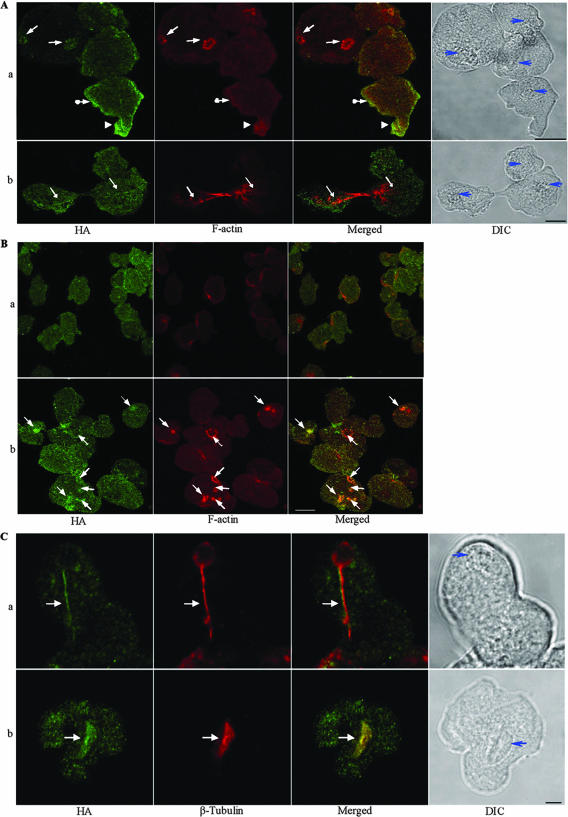
FIG. 4.
Ehformin-1 and Ehformin-2 colocalize with F-actin and microtubular assemblies. (A) Association with F-actin. Trophozoites from Ehformin-2 (a) and Ehformin-1 (b) transformants were stained with a monoclonal anti-HA antibody and rhodamine-conjugated phalloidin. Colocalization of Ehformin-1 and -2 (HA) with F-actin was seen at pseudopodial protrusions (arrowheads), phagocytotic or pinocytotic invaginations (solid-tipped arrows), cortical regions (solid-tipped arrows with circles), and actin cables in dividing cells (arrows). Nuclei are indicated (blue arrows) in the corresponding DIC images. Bar, 10 μm. (B) Association of Ehformin-2 with F-actin is promoted by serum factors. Trophozoites from Ehformin-2 transformants were incubated in serum-free medium (a) for 12 h at 37°C, followed by addition of serum for 30 min (b). Both sets of cells were fixed and hybridized with an anti-HA antibody and rhodamine-conjugated phalloidin. Colocalization of HA-tagged Ehformin-2 with F-actin at phagocytotic or pinocytotic invaginations (solid-tipped arrows) is shown. Bar, 10 μm. (C) Ehformin-1 and -2 colocalize on microtubular assemblies during mitosis. Trophozoites from Ehformin-1 (a) and Ehformin-2 (b) transformants were fixed and stained with an anti-HA antibody and a polyclonal anti-E. histolytica β-tubulin antibody. Colocalization of HA-tagged Ehformin-1 and -2 with spindle microtubular assemblies is shown (solid-tipped arrows). Nuclei are indicated (blue arrows) in the corresponding DIC images. Bar, 5 μm.
(i) Association of Ehformin-1 and -2 with F-actin is promoted by serum factors.
DRFs have been shown to mediate extracellular signals by binding to active Rho GTPases and inducing actin polymerization at specific sites (21, 54, 62). Extensive reorganization of F-actin was observed during the addition of serum or lysophosphatidic acid to serum-starved E. histolytica cells (16, 18a). We next examined whether Ehformin-1 and -2 were involved in the remodeling of actin structures in response to extracellular signals such as serum factors. Stable transformants of Ehformin-1 and -2 were serum starved and fixed at different times after the addition of serum. Characteristically, serum-starved cells are quiescent and therefore show little or no movement. Prominent F-actin structures were absent from serum-starved cells except for a few discrete actin patches. Cytoplasmic Ehformin-2 could be visualized separately from F-actin in serum-starved cells (Fig. 4B). After serum addition, Ehformin-2 was localized at polymerized actin assemblies in multiple cytoskeletal structures including phagocytotic and macropinocytotic rings and crowns, patches, and cell-to-cell adhesion sites (Fig. 4B). Similar changes were seen with Ehformin-1 after the addition of serum to serum-starved cells (data not shown). Thus, association of Ehformin-1 and -2 with F-actin in distinct cellular structures was induced by the addition of serum factors, showing that, like other DRFs, Ehformin-1 and -2 mediated extracellular signals for the reorganization of F-actin.
(ii) Ehformin-1 and -2 were found on microtubular assemblies during mitosis.
In addition to their association with actin assemblies, mammalian Diaphanous proteins mDia1 and mDia3 associate with spindle microtubular assemblies to regulate spindle positioning and proper chromosome alignment to the kinetochore (26, 67). In E. histolytica, microtubular assemblies have been found to be both radial (13, 41, 57) and bipolar spindles (13). Practically nothing is known about the mode of chromosome alignment or segregation on the radial or bipolar assembly in E. histolytica. We have recently shown the association of a kinesin protein, Klp5Eh, with the microtubular assembly (13), but other proteins associated with the microtubular assembly are not yet known. We have now discovered that Ehformin-1 and -2 are associated with both radial and bipolar microtubular assemblies. In the representative images (Fig. 4C), Ehformin-1 is localized at the “tail” region of a telophasic microtubule while Ehformin-2 is localized on a spindle. In order to explore whether the observed association of Ehformin-1 and -2 with the microtubular assemblies affects genome segregation, we examined the effects of changes in the expression of Ehformin-1 and -2 in stable transformants.
Increased expression of Ehformin-1 and -2 leads to an increase in the DNA content of E. histolytica cells.
We selected and maintained stable transformants of Ehformin-1 and -2 at different concentrations of G418 in order to monitor the effects of increasing expression levels (4, 23) of these proteins in E. histolytica. Expression of HA-tagged recombinant Ehformin-1 and Ehformin-2 in the respective transformants showed a gradual increase with increasing concentrations of G418 (Fig. 5A). While the motility of the cells was not affected at different drug concentrations, we observed a remarkable increase in the distribution of the average DNA contents of Ehformin-1 and -2 stable transformants compared to that for control transformants at 40 μg/ml of G418 (Fig. 5B). The average DNA distributions were almost similar for control transformants maintained at 10, 20, and 40 μg of G418/ml (Fig. 5B). The histogram of DNA content obtained by flow cytometry was demarcated into electronic gates to identify 1×, 2×, 4×, and >4× genome contents as described previously (13) (1× represents one genome copy). Data from an average of three independent experiments are shown in a bar graph (Fig. 5C). It can be clearly seen that the numbers of cells with ≥4× DNA contents were significantly increased in both Ehformin-1 and Ehformin-2 transformants over those in control transformants. A concomitant decrease in the 1×-to-2× cell population was seen in Ehformin-1 and Ehformin-2 transformants compared to that in control cells. This effect must be specific for the formin proteins and not due to the HA and His epitopes, since in an earlier study we showed that increased expression of HA- and His-tagged Klp5Eh reduced the heterogeneity and average genome content of E. histolytica (13).
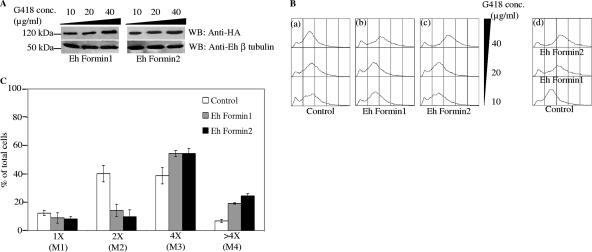
FIG. 5.
Increased expression of Ehformin-1 and -2 leads to increases in the DNA contents of E. histolytica cells. (A) Lysates from 48-h-grown cells of Ehformin-1 and -2 transformants maintained at 10-, 20-, and 40-μg/ml concentrations of G418 were separated by SDS-PAGE, and Western blots (WB) of these lysates were hybridized with an anti-HA antibody to detect the expression of HA-tagged Ehformin-1 and -2 in the respective transformants maintained at different G418 concentrations. Similar blots were hybridized with a polyclonal anti-E. histolytica β-tubulin antibody as a loading control. (B) Forty-eight-hour-grown cells of Ehformin-1, Ehformin-2, and control transformants maintained at 10-, 20-, and 40-μg/ml concentrations of G418 were stained with propidium iodide to analyze the average DNA content. (a to c) Flow cytometric profiles of DNA contents of control (a), Ehformin-1 (b), and Ehformin-2 (c) transformants at increasing concentrations of G418 are shown as overlay diagrams. (d) Histogram overlay shows DNA contents of control, Ehformin-1, and Ehformin-2 transformants maintained at 40 μg/ml of G418. (C) The histogram plots obtained from the flow cytometric analyses of propidium iodide-stained Ehformin-1, Ehformin-2, and control transformants maintained at 40 μg/ml G418 were demarcated into electronic gates M1, M2, M3, and M4, corresponding to cells with 1×, 2×, 4×, and >4× DNA contents, respectively. Bars show percentages of cells in each electronic gate. Error bars, standard deviations (n = 3).
Our previous studies have shown that an increased population of cells with 4× or more genome contents identified by flow cytometry could be due to an increase in the number of multinucleated cells or in the number of genome copies in uninucleated cells (12, 13, 18). An increase in the number of multinucleated cells signifies a delay in cytokinesis, while an increase in the DNA content of individual nuclei suggests inhibition of mitosis. In order to determine if the observed increase in the average genome contents of Ehformin-1 and -2 transformants was due to a delay in cell division or mitosis or both, we analyzed the number of nuclei in individual cells and the average genome contents of individual nuclei in the respective transformants.
Increased expression of Ehformin-1 and -2 delays both cell division and mitosis.
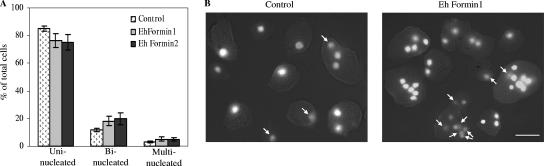
Both Ehformin-1 and -2 stable transformants accumulated 17 to 20% binucleated cells when maintained at 40 μg/ml G418, compared to 10 to 12% binucleated cells in control transformants at 40 μg/ml G418 (Fig. 6A). The proportion of multinucleated cells also increased to 4 to 5% in Ehformin-1 and -2 transformants, compared to 2 to 3% in control transformants (Fig. 6A). Notably, in control transformants, multinucleated cells contained 3 to 4 nuclei, while transformants of Ehformin-1 and -2 contained 3 to 8 nuclei on average (Fig. 6B). This result clearly shows that both Ehformin-1 and Ehformin-2 inhibit cell division at higher expression levels, leading to an accumulation of cells with multiple nuclei.
FIG. 6.
Increased expression of Ehformin-1 and -2 delays cell division. (A) Forty-eight-hour-grown cells of Ehformin-1, Ehformin-2, and control transformants maintained at 40 μg/ml G418 were ethanol fixed, DAPI stained, and counted for the number of nuclei in each cell. Percentages of uninucleated, binucleated, and multinucleated (more than 2 nuclei per cell) cells were plotted on a bar graph. Error bars, standard deviations (n = 3). (B) Representative images of DAPI-stained cells from control and Ehformin-1 transformants maintained at 40 μg/ml G418. In addition to uninucleated and binucleated cells, cells containing multiple (3 to 8) nuclei are often seen in both Ehformin-1 and Ehformin-2 transformants, while multinucleated cells in control transformants contain only 2 to 3 nuclei. Arrows indicate DAPI-stained nuclei that are out of focus in the Z-section shown here. Bar, 20 μm.
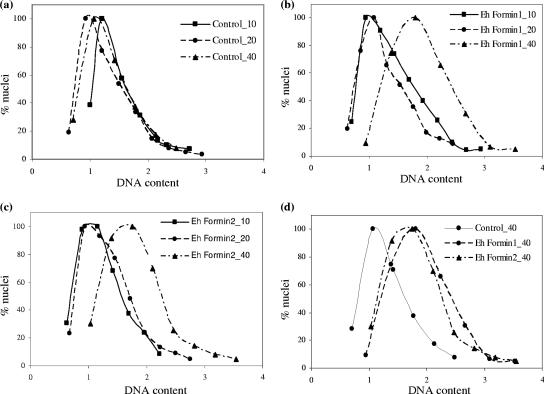
In order to determine if Ehformin-1 and -2 also affected mitosis, we analyzed the genome contents of individual nuclei in both types of transformants and compared them with those for control transformants. We performed scanning cytometry to quantitate the DAPI fluorescence of individual nuclei from each sample and represented the data as histograms as described previously (13). The average DNA contents of individual nuclei in Ehformin-1 and -2 stable transformants were significantly higher than those for control transformants at 40 μg/ml G418 and higher than those for Ehformin-1 and -2 transformants maintained at 10 and 20 μg/ml G418 (Fig. 7). Thus, at higher expression levels of Ehformin-1 or Ehformin-2, mitosis was delayed. We cannot define the exact stage of mitosis where this delay occurs, but the net effect of increased expression of Ehformin-1 and -2 was an increase in the DNA contents of nuclei over those for control transformants grown at the same drug concentrations.
FIG. 7.
Increased expression of Ehformin-1 and -2 leads to increases in the DNA contents of E. histolytica nuclei. Forty-eight-hour-grown cells of Ehformin-1, Ehformin-2, and control transformants maintained at 10, 20, and 40 μg of G418/ml were stained with DAPI to analyze the DNA contents of individual nuclei in a Metacyte scanning cytometer using Metafer4. (a to c) Normalized DAPI fluorescence values for nuclei from cells of control (a), Ehformin-1 (b), and Ehformin-2 (c) transformants maintained at different G418 concentrations (10, 20, and 40 μg/ml) were plotted as individual histograms. (d) Histograms corresponding to the set of nuclei from control, Ehformin-1, and Ehformin-2 transformants maintained at 40 μg of G418/ml.
DISCUSSION
Phagocytosis and the formation of pseudopodia are important cellular activities of the E. histolytica trophozoite. Dynamic remodeling of the actin cytoskeleton occurs during the formation of the phagocytotic cup and pseudopodia. Earlier studies have shown the association of proteins such as Myosin-IBEh, several actin binding proteins, and signaling molecules in phagosomes in E. histolytica (38, 40, 58). Recent studies of different organisms have shown the association of formins in both phagocytic and macropinocytic actin structures (11, 28, 55). Proteomic analyses showed the presence of Ehformin-1 and -2 (38) and Ehformin-3 (40) in phagosomes of E. histolytica. In this study, we have shown that in response to serum factors, Ehformin-1 and -2 associate with the actin cytoskeleton at phagocytic cups, macropinocytotic vesicles, pseudopodial protrusions, cell-cell adhesion sites, actin cables, and cell division sites. While phagocytosis and motility were not affected, distinct changes were observed in the cell division processes of transformants when the expression of Ehformin-1 and -2 was increased.
The appearance of multinucleated cells in untransformed E. histolytica HM1:IMSS (12) and control transformants indicates that cell division is uncoupled from mitosis in this organism. Since DRFs are known to recruit monomeric actin to the site of cell division in other organisms, we had expected that increased expression of Ehformin-1 or -2 might increase the frequency of cell division in amoeba trophozoites, thereby reducing the numbers of bi- and multinucleated cells. Contrary to our expectations, we found that increased expression of these proteins led to an increase in the average number of binucleated cells and, more significantly, in the number of nuclei in multinucleated cells compared to that for the control. Clearly, increased expression of Ehformin-1 or -2 delays cell division. The observed delay in cell division could happen if increased formin concentrations aberrantly induced the formation of multiple actin fibers in an unregulated manner that interfered with cell division. Alternatively, increased formin concentrations may sequester other factors that facilitate cell division, leading to the observed “dominant-negative” phenotype. In any case, the effect points to a tight regulatory role of these proteins in cell division.
Previous studies have shown that the expression of a constitutively active mutant of RacGEh (RacGGly12ValEh), a dominant-negative mutant of RabA, and the kinase domain of PAK2Eh led to cytokinetic defects and consequently to the accumulation of multinucleated cells (5, 22, 63). All these observations indicate that successful completion of cell division in E. histolytica is controlled by the integrated activity of several signaling molecules and proteins such as Ehformin-1 and -2 that mediate extracellular signals for F-actin remodeling.
The localization of mDia1 and mDia3 (26, 67) to the spindle microtubular assembly and kinetochores suggests a regulatory role for formin proteins in genome segregation and mitosis. For E. histolytica, we observed that Ehformin-1 and -2 localized on the microtubular assembly and that one of the effects of overexpressing these proteins was an increase in the DNA content of each nucleus. From these data we inferred that overexpression of Ehformin-1 and -2 delayed either genome segregation or some stage of mitosis in E. histolytica. Since both cell division and mitosis were delayed in response to an increase in the expression of Ehformin-1 and -2, we wondered if F-actin-mediated cytokinesis and genome segregation on microtubular assemblies were coregulated. In metazoan systems, spire proteins are known to mediate cross talk between F-actin and microtubules (10). However, sequence analysis did not identify spire homologs in E. histolytica (S. Majumder and A. Lohia, unpublished data), and therefore the two processes may be regulated by different mechanisms in this organism.
The discovery of a novel group of formins, Ehformin-5 to -8, in E. histolytica opens the possibility of identifying novel interactors and functions of these formins, which will be addressed in future studies. Although Ehformin-1 to -3 and Ehformin-4 have been classified into two groups, they are still divergent enough from their related homologs to warrant characterization of each of their functions. Ehformin-1 and -2 contain different domains identified in DRFs and act in response to extracellular signals associated with F-actin at multiple subcellular structures like other DRFs, but their primary effect was identified in mitosis and cell division in E. histolytica.
Supplementary Material
Acknowledgments
This study was supported by FIRCA subgrants from the University of Virginia (GC11289-123722; primary award 1RO3TW007314) and Stanford University (16846170-33918-A; primary award 1RO3TW007421-01). Partial support was received from the Department of Biotechnology, Government of India (BT/IN/FRG/AL/2003-2004).
Confocal microscopy was carried out at the Facility for Genomics and Proteomics at Bose Institute, sponsored by IRHPA, DST.
Editor: J. F. Urban, Jr.
Footnotes
Published ahead of print on 17 March 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Alberts, A. S. 2001. Identification of a carboxyl-terminal diaphanous-related formin homology protein autoregulatory domain. J. Biol. Chem. 2762824-2830. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215403-410. [DOI] [PubMed] [Google Scholar]
- 3.Arhets, P., P. Gounon, P. Sansonetti, and N. Guillen. 1995. Myosin II is involved in capping and uroid formation in the human pathogen Entamoeba histolytica. Infect. Immun. 634358-4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arhets, P., J. C. Olivo, P. Gounon, P. Sansonetti, and N. Guillen. 1998. Virulence and functions of myosin II are inhibited by overexpression of light meromyosin in Entamoeba histolytica. Mol. Biol. Cell 91537-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Arias-Romero, L. E., M. de Jesus Almaraz-Barrera, J. D. Diaz-Valencia, A. Rojo-Dominguez, R. Hernandez-Rivas, and M. Vargas. 2006. EhPAK2, a novel p21-activated kinase, is required for collagen invasion and capping in Entamoeba histolytica. Mol. Biochem. Parasitol. 14917-26. [DOI] [PubMed] [Google Scholar]
- 6.Bailey, G. B., D. B. Day, and J. W. Gasque. 1985. Rapid polymerization of Entamoeba histolytica actin induced by interaction with target cells. J. Exp. Med. 162546-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bailey, G. B., D. B. Day, C. Nokkaew, and C. C. Harper. 1987. Stimulation by target cell membrane lipid of actin polymerization and phagocytosis by Entamoeba histolytica. Infect. Immun. 551848-1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Castrillon, D. H., and S. A. Wasserman. 1994. Diaphanous is required for cytokinesis in Drosophila and shares domains of similarity with the products of the limb deformity gene. Development 1203367-3377. [DOI] [PubMed] [Google Scholar]
- 9.Chavez-Munguia, B., P. Talamas-Rohana, A. Rios, M. Gonzalez-Lazaro, and A. Martinez-Palomo. 2008. Entamoeba histolytica: fibrillar aggregates in dividing trophozoites. Exp. Parasitol. 118280-284. [DOI] [PubMed] [Google Scholar]
- 10.Chhabra, E. S., and H. N. Higgs. 2007. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 91110-1121. [DOI] [PubMed] [Google Scholar]
- 11.Colucci-Guyon, E., F. Niedergang, B. J. Wallar, J. Peng, A. S. Alberts, and P. Chavrier. 2005. A role for mammalian diaphanous-related formins in complement receptor (CR3)-mediated phagocytosis in macrophages. Curr. Biol. 152007-2012. [DOI] [PubMed] [Google Scholar]
- 12.Das, S., and A. Lohia. 2002. Delinking of S phase and cytokinesis in the protozoan parasite Entamoeba histolytica. Cell. Microbiol. 455-60. [DOI] [PubMed] [Google Scholar]
- 13.Dastidar, P. G., S. Majumder, and A. Lohia. 2007. Eh Klp5 is a divergent member of the kinesin 5 family that regulates genome content and microtubular assembly in Entamoeba histolytica. Cell. Microbiol. 9316-328. [DOI] [PubMed] [Google Scholar]
- 14.Diamond, L. S., D. R. Harlow, and C. C. Cunnick. 1978. A new medium for the axenic cultivation of Entamoeba histolytica and other Entamoeba. Trans. R. Soc. Trop. Med. Hyg. 72431-432. [DOI] [PubMed] [Google Scholar]
- 15.Diaz-Valencia, J. D., M. D. Almaraz-Barrera, D. Jay, N. A. Hernandez-Cuevas, E. Garcia, C. H. Gonzalez-De la Rosa, L. E. Arias-Romero, R. Hernandez-Rivas, A. Rojo-Dominguez, N. Guillen, and M. Vargas. 2007. Novel structural and functional findings of the ehFLN protein from Entamoeba histolytica. Cell Motil. Cytoskelet. 64880-896. [DOI] [PubMed] [Google Scholar]
- 16.Franco-Barraza, J., H. Zamudio-Meza, E. Franco, M. del Carmen Dominguez-Robles, N. Villegas-Sepulveda, and I. Meza. 2006. Rho signaling in Entamoeba histolytica modulates actomyosin-dependent activities stimulated during invasive behavior. Cell Motil. Cytoskelet. 63117-131. [DOI] [PubMed] [Google Scholar]
- 17.Fujiwara, T., A. Mammoto, Y. Kim, and Y. Takai. 2000. Rho small G-protein-dependent binding of mDia to an Src homology 3 domain-containing IRSp53/BAIAP2. Biochem. Biophys. Res. Commun. 271626-629. [DOI] [PubMed] [Google Scholar]
- 18.Gangopadhyay, S. S., S. S. Ray, K. Kennady, G. Pande, and A. Lohia. 1997. Heterogeneity of DNA content and expression of cell cycle genes in axenically growing Entamoeba histolytica HM1:IMSS clone A. Mol. Biochem. Parasitol. 909-20. [DOI] [PubMed] [Google Scholar]
- 18a.Ganguly, A. 2002. Ph.D thesis. Jadavpur University, Kolkata, India.
- 19.Ganguly, A., and A. Lohia. 2000. The diaphanous protein from Entamoeba histolytica controls cell motility and cytokinesis. Arch. Med. Res. 31S137-S139. [DOI] [PubMed] [Google Scholar]
- 20.Ghosh, S. K., and J. Samuelson. 1997. Involvement of p21racA, phosphoinositide 3-kinase, and vacuolar ATPase in phagocytosis of bacteria and erythrocytes by Entamoeba histolytica: suggestive evidence for coincidental evolution of amebic invasiveness. Infect. Immun. 654243-4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goode, B. L., and M. J. Eck. 2007. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76593-627. [DOI] [PubMed] [Google Scholar]
- 22.Guillen, N., P. Boquet, and P. Sansonetti. 1998. The small GTP-binding protein RacG regulates uroid formation in the protozoan parasite Entamoeba histolytica. J. Cell Sci. 1111729-1739. [DOI] [PubMed] [Google Scholar]
- 23.Hamann, L., R. Nickel, and E. Tannich. 1995. Transfection and continuous expression of heterologous genes in the protozoan parasite Entamoeba histolytica. Proc. Natl. Acad. Sci. USA 928975-8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Higgs, H. N. 2005. Formin proteins: a domain-based approach. Trends Biochem. Sci. 30342-353. [DOI] [PubMed] [Google Scholar]
- 25.Higgs, H. N., and K. J. Peterson. 2005. Phylogenetic analysis of the formin homology 2 domain. Mol. Biol. Cell 161-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kato, T., N. Watanabe, Y. Morishima, A. Fujita, T. Ishizaki, and S. Narumiya. 2001. Localization of a mammalian homolog of diaphanous, mDia1, to the mitotic spindle in HeLa cells. J. Cell Sci. 114775-784. [DOI] [PubMed] [Google Scholar]
- 27.Kaur, G., and A. Lohia. 2004. Inhibition of gene expression with double strand RNA interference in Entamoeba histolytica. Biochem. Biophys. Res. Commun. 3201118-1122. [DOI] [PubMed] [Google Scholar]
- 28.Kitayama, C., and T. Q. Uyeda. 2003. ForC, a novel type of formin family protein lacking an FH1 domain, is involved in multicellular development in Dictyostelium discoideum. J. Cell Sci. 116711-723. [DOI] [PubMed] [Google Scholar]
- 29.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 30.Labruyere, E., C. Zimmer, V. Galy, J. C. Olivo-Marin, and N. Guillen. 2003. EhPAK, a member of the p21-activated kinase family, is involved in the control of Entamoeba histolytica migration and phagocytosis. J. Cell Sci. 1661-71. [DOI] [PubMed] [Google Scholar]
- 31.Lammers, M., R. Rose, A. Scrima, and A. Wittinghofer. 2005. The regulation of mDia1 by autoinhibition and its release by Rho*GTP. EMBO J. 244176-4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, L., S. K. Klee, M. Evangelista, C. Boone, and D. Pellman. 1999. Control of mitotic spindle position by the Saccharomyces cerevisiae formin Bni1p. J. Cell Biol. 144947-961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li, F., and H. N. Higgs. 2005. Dissecting requirements for auto-inhibition of actin nucleation by the formin, mDia1. J. Biol. Chem. 2806986-6992. [DOI] [PubMed] [Google Scholar]
- 34.Li, F., and H. N. Higgs. 2003. The mouse formin mDia1 is a potent actin nucleation factor regulated by autoinhibition. Curr. Biol. 131335-1340. [DOI] [PubMed] [Google Scholar]
- 35.Loftus, B., I. Anderson, R. Davies, U. C. Alsmark, J. Samuelson, P. Amedeo, P. Roncaglia, M. Berriman, R. P. Hirt, B. J. Mann, T. Nozaki, B. Suh, M. Pop, M. Duchene, J. Ackers, E. Tannich, M. Leippe, M. Hofer, I. Bruchhaus, U. Willhoeft, A. Bhattacharya, T. Chillingworth, C. Churcher, Z. Hance, B. Harris, D. Harris, K. Jagels, S. Moule, K. Mungall, D. Ormond, R. Squares, S. Whitehead, M. A. Quail, E. Rabbinowitsch, H. Norbertczak, C. Price, Z. Wang, N. Guillen, C. Gilchrist, S. E. Stroup, S. Bhattacharya, A. Lohia, P. G. Foster, T. Sicheritz-Ponten, C. Weber, U. Singh, C. Mukherjee, N. M. El-Sayed, W. A. Petri, Jr., C. G. Clark, T. M. Embley, B. Barrell, C. M. Fraser, and N. Hall. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433865-868. [DOI] [PubMed] [Google Scholar]
- 36.Lohia, A., C. Mukherjee, S. Majumder, and P. G. Dastidar. 2007. Genome re-duplication and irregular segregation occur during the cell cycle of Entamoeba histolytica. Biosci. Rep. 27373-384. [DOI] [PubMed] [Google Scholar]
- 37.Manning-Cela, R., and I. Meza. 1997. Up-regulation of actin mRNA and reorganization of the cytoskeleton in Entamoeba histolytica trophozoites. J. Eukaryot. Microbiol. 4418-24. [DOI] [PubMed] [Google Scholar]
- 38.Marion, S., C. Laurent, and N. Guillen. 2005. Signalization and cytoskeleton activity through myosin IB during the early steps of phagocytosis in Entamoeba histolytica: a proteomic approach. Cell. Microbiol. 71504-1518. [DOI] [PubMed] [Google Scholar]
- 39.Nezami, A. G., F. Poy, and M. J. Eck. 2006. Structure of the autoinhibitory switch in formin mDia1. Structure 14257-263. [DOI] [PubMed] [Google Scholar]
- 40.Okada, M., C. D. Huston, M. Oue, B. J. Mann, W. A. Petri, Jr., K. Kita, and T. Nozaki. 2006. Kinetics and strain variation of phagosome proteins of Entamoeba histolytica by proteomic analysis. Mol. Biochem. Parasitol. 145171-183. [DOI] [PubMed] [Google Scholar]
- 41.Orozco, E., F. J. Solis, J. Dominguez, B. Chavez, and F. Hernandez. 1988. Entamoeba histolytica: cell cycle and nuclear division. Exp. Parasitol. 6785-95. [DOI] [PubMed] [Google Scholar]
- 42.Otomo, T., C. Otomo, D. R. Tomchick, M. Machius, and M. K. Rosen. 2005. Structural basis of Rho GTPase-mediated activation of the formin mDia1. Mol. Cell 18273-281. [DOI] [PubMed] [Google Scholar]
- 43.Otomo, T., D. R. Tomchick, C. Otomo, S. C. Panchal, M. Machius, and M. K. Rosen. 2005. Structural basis of actin filament nucleation and processive capping by a formin homology 2 domain. Nature 433488-494. [DOI] [PubMed] [Google Scholar]
- 44.Palazzo, A. F., T. A. Cook, A. S. Alberts, and G. G. Gundersen. 2001. mDia mediates Rho-regulated formation and orientation of stable microtubules. Nat. Cell Biol. 3723-729. [DOI] [PubMed] [Google Scholar]
- 45.Petersen, J., O. Nielsen, R. Egel, and I. M. Hagan. 1998. FH3, a domain found in formins, targets the fission yeast formin Fus1 to the projection tip during conjugation. J. Cell Biol. 1411217-1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruyne, D., M. Evangelista, C. Yang, E. Bi, S. Zigmond, A. Bretscher, and C. Boone. 2002. Role of formins in actin assembly: nucleation and barbed-end association. Science 297612-615. [DOI] [PubMed] [Google Scholar]
- 47.Rahim, Z., A. Raymond-Denise, P. Sansonetti, and N. Guillen. 1993. Localization of myosin heavy chain A in the human pathogen Entamoeba histolytica. Infect. Immun. 611048-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ravdin, J. I., and R. L. Guerrant. 1981. Role of adherence in cytopathogenic mechanisms of Entamoeba histolytica. Study with mammalian tissue culture cells and human erythrocytes. J. Clin. Investig. 681305-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rivero, F., T. Muramoto, A. K. Meyer, H. Urushihara, T. Q. Uyeda, and C. Kitayama. 2005. A comparative sequence analysis reveals a common GBD/FH3-FH1-FH2-DAD architecture in formins from Dictyostelium, fungi and metazoa. BMC Genomics 628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Romero, S., C. Le Clainche, D. Didry, C. Egile, D. Pantaloni, and M. F. Carlier. 2004. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell 119419-429. [DOI] [PubMed] [Google Scholar]
- 51.Rose, R., M. Weyand, M. Lammers, T. Ishizaki, M. R. Ahmadian, and A. Wittinghofer. 2005. Structural and mechanistic insights into the interaction between Rho and mammalian Dia. Nature 435513-518. [DOI] [PubMed] [Google Scholar]
- 52.Sagot, I., A. A. Rodal, J. Moseley, B. L. Goode, and D. Pellman. 2002. An actin nucleation mechanism mediated by Bni1 and profilin. Nat. Cell Biol. 4626-631. [DOI] [PubMed] [Google Scholar]
- 53.Sahoo, N., E. Labruyere, S. Bhattacharya, P. Sen, N. Guillen, and A. Bhattacharya. 2004. Calcium binding protein 1 of the protozoan parasite Entamoeba histolytica interacts with actin and is involved in cytoskeleton dynamics. J. Cell Sci. 1173625-3634. [DOI] [PubMed] [Google Scholar]
- 54.Schirenbeck, A., T. Bretschneider, R. Arasada, M. Schleicher, and J. Faix. 2005. The Diaphanous-related formin dDia2 is required for the formation and maintenance of filopodia. Nat. Cell Biol. 7619-625. [DOI] [PubMed] [Google Scholar]
- 55.Seth, A., C. Otomo, and M. K. Rosen. 2006. Autoinhibition regulates cellular localization and actin assembly activity of the diaphanous-related formins FRLα and mDia1. J. Cell Biol. 174701-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vargas, M., P. Sansonetti, and N. Guillen. 1996. Identification and cellular localization of the actin-binding protein ABP-120 from Entamoeba histolytica. Mol. Microbiol. 22849-857. [DOI] [PubMed] [Google Scholar]
- 57.Vayssie, L., M. Vargas, C. Weber, and N. Guillen. 2004. Double-stranded RNA mediates homology-dependent gene silencing of gamma-tubulin in the human parasite Entamoeba histolytica. Mol. Biochem. Parasitol. 13821-28. [DOI] [PubMed] [Google Scholar]
- 58.Voigt, H., J. C. Olivo, P. Sansonetti, and N. Guillen. 1999. Myosin IB from Entamoeba histolytica is involved in phagocytosis of human erythrocytes. J. Cell Sci. 1121191-1201. [DOI] [PubMed] [Google Scholar]
- 59.Wallar, B. J., and A. S. Alberts. 2003. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 13435-446. [DOI] [PubMed] [Google Scholar]
- 60.Wallar, B. J., B. N. Stropich, J. A. Schoenherr, H. A. Holman, S. M. Kitchen, and A. S. Alberts. 2006. The basic region of the diaphanous-autoregulatory domain (DAD) is required for autoregulatory interactions with the diaphanous-related formin inhibitory domain. J. Biol. Chem. 2814300-4307. [DOI] [PubMed] [Google Scholar]
- 61.Watanabe, N., P. Madaule, T. Reid, T. Ishizaki, G. Watanabe, A. Kakizuka, Y. Saito, K. Nakao, B. M. Jockusch, and S. Narumiya. 1997. p140mDia, a mammalian homolog of Drosophila diaphanous, is a target protein for Rho small GTPase and is a ligand for profilin. EMBO J. 163044-3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Watanabe, N., T. Kato, A. Fujita, T. Ishizaki, and S. Narumiya. 1999. Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1136-143. [DOI] [PubMed] [Google Scholar]
- 63.Welter, B. H., R. R. Powell, M. Leo, C. M. Smith, and L. A. Temesvari. 2005. A unique Rab GTPase, EhRabA, is involved in motility and polarization of Entamoeba histolytica cells. Mol. Biochem. Parasitol. 140161-173. [DOI] [PubMed] [Google Scholar]
- 64.Wen, Y., C. H. Eng, J. Schmoranzer, N. Cabrera-Poch, E. J. Morris, M. Chen, B. J. Wallar, A. S. Alberts, and G. G. Gundersen. 2004. EB1 and APC bind to mDia to stabilize microtubules downstream of Rho and promote cell migration. Nat. Cell Biol. 6820-830. [DOI] [PubMed] [Google Scholar]
- 65.Wender, N., E. Villalobo, and D. Mirelman. 2007. EhLimA, a novel LIM protein localizes to the plasma membrane in Entamoeba histolytica. Eukaryot. Cell 61646-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Winder, S. J., and K. R. Ayscough. 2005. Actin-binding proteins J. Cell Sci. 118651-654. [DOI] [PubMed] [Google Scholar]
- 67.Yasuda, S., F. Oceguera-Yanez, T. Kato, M. Okamoto, S. Yonemura, Y. Terada, T. Ishizaki, and S. Narumiya. 2004. Cdc42 and mDia3 regulate microtubule attachment to kinetochores. Nature 428767-771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.