Abstract
Mutant screens and transcriptome studies led us to consider whether the metabolism of glucose polymers, i.e., maltose, maltodextrin, and glycogen, is important for Escherichia coli colonization of the intestine. By using the streptomycin-treated mouse model, we found that catabolism of the disaccharide maltose provides a competitive advantage in vivo to pathogenic E. coli O157:H7 and commensal E. coli K-12, whereas degradation of exogenous forms of the more complex glucose polymer, maltodextrin, does not. The endogenous glucose polymer, glycogen, appears to play an important role in colonization, since mutants that are unable to synthesize or degrade glycogen have significant colonization defects. In support of the hypothesis that E. coli relies on internal carbon stores to maintain colonization during periods of famine, we found that by providing a constant supply of a readily metabolized sugar, i.e., gluconate, in the animal's drinking water, the competitive disadvantage of E. coli glycogen metabolism mutants is rescued. The results suggest that glycogen storage may be widespread in enteric bacteria because it is necessary for maintaining rapid growth in the intestine, where there is intense competition for resources and occasional famine. An important implication of this study is that the sugars used by E. coli are present in limited quantities in the intestine, making endogenous carbon stores valuable. Thus, there may be merit to combating enteric infections by using probiotics or prebiotics to manipulate the intestinal microbiota in such a way as to limit the availability of sugars preferred by E. coli O157:H7 and perhaps other pathogens.
In nature, the nutrients that support bacterial growth are rarely available continuously and are almost always present in limiting amounts, leading to the idea that bacterial existence is one of feast or famine (24). Others have proposed the existence of an in-between state, namely hunger, which is defined by expression of carbon-scavenging systems (18). In the mammalian gastrointestinal tract (9), despite growing in a mucus layer rich in carbohydrates, lipids, and proteins (27), Escherichia coli apparently leads a scavenging lifestyle, using up to seven different sugars to support its colonization (7, 16). It is likely that the availability of one or more of its preferred nutrients in the intestine is limited and variable. Thus, for E. coli to cope with life in a nutrient-limiting environment where it is hungry or occasionally starving, we hypothesize that successful competition and persistence in the intestine requires intracellular energy stores. Glycogen is the primary carbon and energy storage molecule for enteric bacteria, including E. coli (5).
The discovery of glycogen in E. coli is credited to Cedergren and Holme (6). In an early review of bacterial cell composition, it was Neidhardt who first suggested that glycogen degradation might be linked to energy metabolism (34). It is now well established that glycogen is the fundamental carbon and energy storage compound for many bacteria and plays a major role in long-term survival of the cell. Glycogen is synthesized during times when carbon is abundant but other nutrients are limiting (36). GlgA, glycogen synthase, is responsible for the transfer of the glucosyl unit from ADP-glucose to the growing glycogen polymer to form a new α-1,4-glucosidic linkage (37). GlgB is the branching enzyme that catalyzes formation of α-1,6-glucosidic linkages. GlgC, glucose-1-phosphate adenylyltransferase, forms ADP-glucose from glucose-1-phosphate and ATP. GlgS is required for glycogen synthesis, its overproduction stimulates glycogen synthesis, and it is induced upon entry into stationary in an RpoS-dependent manner (21). However, GlgS has no defined role in glycogen synthesis, although its crystal structure suggests involvement in protein-protein interactions (25). Glycogen degradation occurs when carbon sources become limiting (13). GlgP, glycogen phosphorylase, catalyzes glycogen breakdown by removing glucose units from the polysaccharide. GlgX, glycogen debranching enzyme, hydrolyzes α-1,6-glucosidic linkages in limit dextrins generated by GlgP (12).
In salmonellae, the role of glycogen metabolism in colonization and virulence is controversial, with one study suggesting a connection between glycogen storage and virulence (4) and another showing a minor role, at best (30). The contributions of glycogen synthesis and degradation to animal colonization have not been tested for E. coli. In support of the hypothesis that glycogen metabolism is important for colonization, we previously found that one of the genes involved in glycogen metabolism, glgS, is highly induced in E. coli MG1655 when growing on mucus, conditions that are thought to mimic nutrient availability in the intestine (7).
Since glycogen breakdown involves maltose and maltodextrins as intermediates, we also thought to examine whether catabolism of exogenous maltose and maltodextrins supports colonization. This possibility seemed reasonable since maltose metabolism genes are also induced in E. coli MG1655 when growing on mucus (7). Maltose consists of two glucose molecules joined by an α-1,4 linkage; maltodextrins are longer glucose polymers. These sugars are most commonly derived from starch and glycogen. At least nine genes encode proteins that are involved in the utilization of maltose and maltodextrins in E. coli (29). The maltose-inducible porin for maltodextrin transport into the periplasm is encoded by lamB (46). The maltose/maltodextrin transporter is a high-affinity ABC system consisting of a periplasmic maltose-binding protein, encoded by malE, and a transport complex encoded by malF, malG, and malK (5). malQ (amylomaltase) and malP (maltodextrin phosphorylase) encode enzymes for maltose and maltodextrin catabolism, respectively. MalQ uses maltodextrins as maltose acceptors in a reaction that releases glucose, which enters glycolysis after being phosphorylated by glucokinase (5). MalP cleaves glucosyl residues from the nonreducing end of maltodextrins, forming α-glucose-1-phosphate, which enters glycolysis after conversion to glucose 6-phosphate by phosphoglucomutase (5). Mutants lacking MalQ cannot grow on maltose, whereas malP mutants can grow on maltose but not on maltodextrins (14).
We previously identified several monosaccharides utilized during colonization of the mouse intestine by pathogenic E. coli EDL933 (16) and commensal E. coli MG1655 (7). Here, we explore the possibility that complex sugars, i.e., maltose, maltodextrin, and glycogen, support intestinal colonization. The results demonstrate that maltose, but not maltodextrin, is important for colonization of both pathogenic and commensal E. coli strains. In addition, glycogen synthesis and glycogen degradation are important for efficient intestinal colonization, suggesting a hunger or famine lifestyle for E. coli in the mouse intestine. The results increase our understanding of nutrient utilization in vivo and provide new insights into host-pathogen interactions.
MATERIALS AND METHODS
Bacterial strains.
The bacterial strains used in the present study were derived from E. coli MG1655 Strr (streptomycin resistant), a K-12 strain (32), and E. coli EDL933 Strr, a streptomycin-resistant derivative of the prototypical O157:H7 strain (31). The bacterial strains and plasmids used in the present study are listed in Table 1.
TABLE 1.
Strains and plasmids used in this studya
| Strain or plasmida | Genotypeb | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | Wild-type (CGSC 7740) | CGSCc |
| MG1655 Strr | Spontaneous Strr | 32 |
| MG1655 Strr Nalr | Spontaneous Nalr | 32 |
| EDL933 | Wild-type O157:H7 | Alison O'Brien |
| EDL933 Strr | Spontaneous Strr | 31 |
| EDL933 Strr Nalr | Spontaneous Nalr | 31 |
| Derivatives of strain MG1655 Strr | ||
| glgA | ΔglgA::cat | This study |
| glgP | ΔglgP::cat | This study |
| glgP malP | ΔglgP ΔmalP::cat | This study |
| glgS | ΔglgS::cat | This study |
| lamB | ΔlamB::cat | This study |
| malEFG | Δ(malE-malG)::cat | This study |
| malPQ | Δ(malP-malQ)::cat | This study |
| malX | ΔmalX::cat | This study |
| Derivatives of strain EDL933 Strr | ||
| glgA | ΔglgA::cat | This study |
| glgP | ΔglgP::cat | This study |
| glgP malP | ΔglgP ΔmalP::cat | This study |
| glgS | ΔglgS::cat | This study |
| lamB | ΔlamB::cat | This study |
| malEFG | Δ(malE-malG)::cat | This study |
| malEFG malX | Δ(malE-malG) ΔmalX::cat | This study |
| malP | ΔmalP::cat | This study |
| malPQ | Δ(malP-malQ)::cat | This study |
| malQ | ΔmalQ::cat | This study |
| malX | ΔmalX::cat | This study |
| ppsA pckA | ΔppsA ΔpckA::cat | 31 |
| Plasmids | ||
| pKD3 | bla cat | 11 |
| pKD4 | bla kan | 11 |
| pKD46 | Ts Apr, ParaC-λ-red recombinase | 11 |
| pCP20 | Ts Apr, FLP recombinase | 11 |
| pTag-mini-Tn5Km2 | Apr Knr; mini-Tn5 transposon delivery plasmid | 22 |
For simplicity, strains are named by strain and mutation(s).
Nalr, nalidixic acid resistance; Apr, ampicillin resistance; Strr, streptomycin resistance; Knr, kanamycin resistance; Ts, temperature-sensitive replication.
E. coli Genetic Stock Culture Collection, Yale University.
Mutant constructions.
Null alleles were constructed by using the allelic replacement method of Datsenko and Wanner (11), as described previously (7, 16), such that target genes were deleted and replaced with kanamycin or chloramphenicol resistance cassettes, which served as selectable markers in mouse colonization assays, as described below. The null allele strains are identified in the text by the genes that were deleted; single gene deletions began with the start codon and ended with the stop codon, and consecutive gene deletions began with the start codon of the first gene deleted and ended with the stop codon of the last gene deleted. Strains containing multiple mutations were constructed by sequential allelic replacement; the first inserted cassette was removed with FLP recombinase (11), followed by subsequent allelic replacement(s) and removal of the insertion as necessary, leaving the selectable marker in the last mutation made. Mutant strains were verified by phenotype analysis and DNA sequencing.
Phenotypic analysis.
To test for the ability to use sole carbon and energy sources, mutants were grown overnight in MOPS [3-(N-morpholino) propanesulfonic acid] defined medium (35) containing 0.2% maltose or maltodextrin (maltopentaose to maltoheptaose). Maltose and maltodextrin sensitivity were evaluated by growth on Luria broth containing 0.2% carbohydrate. Cell growth was monitored spectrophotometrically at 600 nm (OD600). To test for the ability to synthesize glycogen, strains were grown overnight on Luria plates containing 2% glucose and stained with iodine vapor (14). In addition, the mutants used in the present study were phenotyped for carbon source oxidation by using Biolog GN2 (carbon nutrition) plates according to the manufacturer's instructions. Briefly, the mutants were grown overnight on tryptic soy agar, washed in Biolog inoculating fluid (gellan gum, 0.2 g/liter; NaCl, 4.0 g/liter; Pluronic F-68, 0.3 g/liter; 2.5 mM thioglycolate), and the plates were inoculated with 150 ml per well of a cell suspension with an OD600 of 0.2 to 0.3. The cells were incubated in Biolog plates for 24 h at 37°C in an Omnilog system, and the amount of reduced tetrazolium violet dye in each well was quantified every 15 min. The amount of dye reduced corresponds to the extent of carbon source oxidized. Dye reduction was plotted versus time, and the area under each resulting kinetic curve was used as an overall measure of the cells’ ability to utilize each carbon source.
Mouse colonization experiments.
The streptomycin-treated mouse model has been used extensively to study colonization of the mouse large intestine by E. coli and Salmonella enterica serovar Typhimurium (8, 9, 27). Briefly, three 6-week-old CD-1 male mice were given drinking water containing streptomycin sulfate (5 g/liter) for 24 h to selectively remove the existing resident facultative microbiota and then starved for food and water for 18 to 24 h. Unless otherwise indicated, the mice were then fed approximately 105 CFU of both wild-type and mutant strains in 1 ml of 20% sucrose. The wild-type strains were streptomycin resistant and nalidixic acid resistant (Strr Nalr) (31); Nalr was used to distinguish the wild-type (reference strain) from the null allele mutants in fecal plate counts. After the bacterial suspension was ingested, food and streptomycin-water (5 g/liter) were restored, and fecal plate counts were determined at 5 h, 24 h, and on every other day thereafter for 15 days. Fecal samples were homogenized and diluted in 1% tryptone broth and plated on MacConkey agar either containing streptomycin (100 μg/ml) and nalidixic acid (50 μg/ml) to count the wild type or containing streptomycin and kanamycin (40 μg/ml) or chloramphenicol (30 μg/ml) to count the null allele mutants. Each colonization experiment was repeated, on separate occasions, and averaged for six (or more) mice, such that the presented values are the log10 mean number of CFU per gram of feces ± the standard error for each strain. In all experiments, a difference between two strains of ≥10-fold CFU/g of feces was statistically significant, i.e., P < 0.05 as determined by the Student t test (two-tailed with unequal variance). The limit of detection in fecal plate counts was 102 CFU/g of feces.
Signature-tagged mutagenesis.
A signature-tagged mutant (STM) library of E. coli EDL933 Strr ppsA pckA was constructed as previously described (43). A total of 1,440 mutants were arrayed in 30 pools each containing 48 uniquely tagged mutants with mini-Tn5Km2 insertions. The mutant library was screened in streptomycin treated mice (as described above) in competition with E. coli MG1655 Strr Nalr. Each pool of mutants was grown in Luria broth supplemented with kanamycin in a microtiter plate at 37°C overnight. The mutants from each microtiter plate were pooled, diluted, and mixed with a 10-fold-concentrated overnight culture of E. coli MG1655 Strr Nalr in 20% sucrose to obtain an inoculum containing approximately 107 CFU of signature-tagged E. coli EDL933 Strr ppsA pckA mutants/ml and 1010 CFU of the E. coli MG1655 Strr Nalr strain/ml. A 100-μl portion of inoculum was fed to two mice. Signature-tagged E. coli EDL933 Strr ppsA pckA mutants unable to persist in the intestine 3 days after inoculation were identified by PCR and hybridization analysis as previously described (43). The competition experiment for colonization was repeated for selected mutants from the screening. The locations of the mini-Tn5Km2 insertions were determined by DNA sequence analysis, as described previously (3).
RESULTS
Colonization assays.
Of the animal models available for measuring the relative fitness of two bacterial strains for intestinal colonization, the streptomycin-treated mouse is preferred for the following reasons (for a review of intestinal colonization models, see reference 3). Bioreactors (i.e., intestinal simulators) have proven useful for analyzing specific strain interactions but, in addition to their failure to mimic intestinal anatomy, it remains unclear what growth medium will duplicate the complex nutritional makeup of the intestinal contents. Conventional mice are resistant to experimental colonization. The gnotobiotic mouse model is a powerful tool for comparing a finite number of strains in direct competition, e.g., how two strains adapt their substrate utilization in response to one another but, in addition to being expensive and time-consuming, this model lacks the diversity of background functions provided by the “normal microbiota,” and gnotobiotic mice exhibit physiological and anatomical differences from conventional animals. Streptomycin treatment selectively removes facultative anaerobes while leaving the anaerobic microbiota essentially intact, i.e., at pretreatment population size; this opens a previously unavailable niche, which can then be colonized by experimentally introduced microorganisms such as E. coli (23). In this model, streptomycin-treated mice are fed streptomycin resistant strains of bacteria. Usually, these are two isogenic strains that differ only in a single characteristic, i.e., a mutant and its wild-type parent. If one of these strains possesses even a slight advantage over the other, the conditions of the mouse large intestine select for that strain and within a few days, it becomes dominant. If neither strain possesses an advantage, the two strains cocolonize and are maintained at approximately equal levels. Since the population of a strain in mouse feces is a reflection of its number in the mouse large intestine, fecal counts are used to judge the relative colonizing abilities of E. coli strains. The colonization data shown in Table 2 are given for day 1 (initiation) and day 15 (maintenance) for mutations constructed in E. coli MG1655 and E. coli EDL933. Growth from low to high numbers in the initiation stage likely depends on utilization of nonlimiting nutrients made available by removing the streptomycin-sensitive facultative anaerobes, whereas persistence during the maintenance stage most likely depends on competition for limiting nutrient(s) (7). Please note that E. coli EDL933 does not cause disease in streptomycin-treated CD-1 mice (45).
TABLE 2.
Competitive colonization between maltose/glycogen mutants and wild-type E. coli strainsa
| Respiratory enzyme | Mutation | Avg (log10 CFU/g of feces) ± SEM for:
|
|||
|---|---|---|---|---|---|
|
E. coli EDL933 at:
|
E. coli MG1655 at:
|
||||
| Day 1 | Day 15 | Day 1 | Day 15 | ||
| Maltose transport | malEFG | 0.4 ± 0.1 | 3.3 ± 0.2 | 0.0 ± 0.1 | 0.4 ± 0.3 |
| Maltose PTS system | malX | 0.4 ± 0.1 | 0.2 ± 0.2 | 0.6 ± 0.2 | 0.6 ± 0.1 |
| Maltose transport and PTS | malEFG malX | 0.2 ± 0.1 | 2.3 ± 0.2 | 0.6 ± 0.2 | 2.2 ± 0.2 |
| Maltodextrin phosphorylase and amylomaltase | malPQ | 0.8 ± 0.1 | 3.7 ± 0.1 | 0.4 ± 0.2 | 3.5 ± 0.4 |
| Maltodextrin phosphorylase | malP | 0.3 ± 0.1 | 1.4 ± 0.5b | NDc | ND |
| Amylomaltase | malQ | 0.3 ± 0.1 | 3.3 ± 0.4 | ND | ND |
| Maltoporin | lamB | 0.0 ± 0.2 | 0.0 ± 0.2 | 0.0 ± 0.2 | 0.1 ± 0.2 |
| Glycogen phosphorylase | glgP | 0.3 ± 0.1 | 2.4 ± 0.2 | 0.5 ± 0.2 | 3.2 ± 0.1 |
| Glycogen phosphorylase and maltodextrin phosphorylase | glgP malP | 0.3 ± 0.2 | 2.5 ± 0.2 | 0.4 ± 0.1 | 2.5 ± 0.2 |
| Glycogen synthase | glgA | 0.2 ± 0.1 | 2.3 ± 0.2 | 0.2 ± 0.1 | 2.9 ± 0.2 |
| Glycogen synthesis | glgS | 0.2 ± 0.1 | 1.9 ± 0.1 | 0.4 ± 0.2 | 1.6 ± 0.2 |
Mice were fed 105 CFU each of a mutant and its wild-type parent. Mice were transferred to fresh cages every day, and feces samples no older than 24 h were assayed every other day for 15 days. At each time point, for each mouse the log10 CFU/g of feces for the mutant was subtracted from the log10 CFU/g of feces for the wild type. The average ± the standard error of the mean of day 1 and day 15 data from six mice is shown. All values shown in boldface are statistically significant (P < 0.005 [Student t test]).
P < 0.08.
ND, not determined.
Evidence for in vivo glycogen and maltose metabolism by E. coli EDL933.
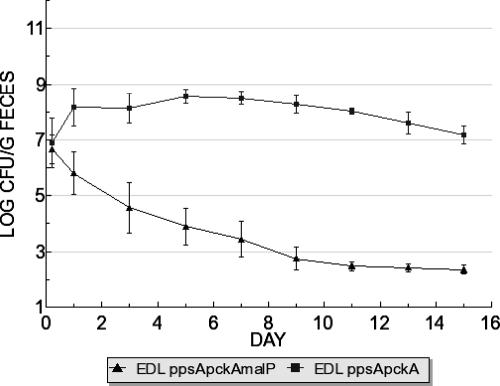
In colonizing the intestine, E. coli EDL933 grows from low to high numbers and can do so in the presence of E. coli MG1655 (31). To do this, it seems likely that E. coli EDL933 uses nutrients that are not used by E. coli MG1655, and we recently demonstrated significant differences in monosaccharide utilization between the two strains during colonization of the mouse intestine (16). Previously, we showed that genes involved in glycogen (glgS), maltose (malE), and maltodextrin (lamB) catabolism were significantly induced in E. coli MG1655 during growth on mucus, conditions that mimic nutrient availability in the intestine (7). More recently, we found that E. coli EDL933 induced malE and glgS during growth on mucus (16). Lastly, an STM screen identified 14 mutants in an E. coli EDL933 ppsA pckA background that were unable to colonize streptomycin treated mice in competition with wild-type E. coli MG1655. The purpose for using the ppsA pckA background was to avoid selection for genes involved in gluconeogenesis (31). One of these insertions was in the malP gene. The E. coli EDL933 ppsA pckA malP::miniTn5-Km2 STM mutant did not grow on maltose (data not shown), suggesting a polar effect on the downstream malQ gene (5). Furthermore, E. coli EDL933 ppsA pckA malP::miniTn5-Km2 had a major colonization defect when competed in mice against its E. coli EDL933 ppsA pckA parent (Fig. 1). Together, these findings led us to consider the importance of maltose and maltodextrin utilization for colonization of the intestine.
FIG. 1.
E. coli EDL933 ppsA pckA malP::mini-Tn5-Km2 STM is outcompeted by E. coli EDL933 ppsA pckA during colonization. Strains were fed to mice at 105 CFU each.
Maltose transport is important for intestinal colonization.
The importance of maltose utilization for colonization was determined by testing maltose/maltodextrin ATP binding cassette (ABC) transport system mutants for their ability to compete with the wild-type in streptomycin-treated mice. Null mutations were constructed that deleted the malEFG operon encoding the maltose-binding protein and the two membrane-spanning subunits of the maltose ABC transport system, without impacting the adjacent malK-lamB-malM operon. malE, malF, and malG mutants are reportedly unable to grow on maltose or maltodextrin (46). Since mutants lacking the maltose ABC transporter can grow on maltose when malX is expressed (39), we also made a malX-null allele strain, which lacks a maltose phosphotransferase system, as well as a malEFG malX double mutant. We verified the phenotypes of these strains before proceeding with the colonization experiments and found that mutation of malX had no effect, whereas mutation of malEFG prevented growth on maltose and maltose oxidation (Fig. 2). Mutation of malEFG significantly reduced but did not completely eliminate growth on maltodextrin (Fig. 2). We note that growth of a malEFG mutant on maltodextrin has not been reported previously (5). Perhaps our E. coli EDL933 malEFG strain grew poorly on maltodextrin by partially degrading it to glucose in the periplasm by using the MalS periplasmic amylase (19). We did not investigate the maltodextrin phenotype of our malEFG mutant further but did confirm the results of the transport mutant colonizations by making independent mutations in the maltose and maltodextrin catabolic pathways, as described below. None of the mutant strains used in the present study affected oxidation of any other carbon sources by comparison to the wild-type on Biolog GN2 plates (data not shown).
FIG. 2.
Phenotypes of E. coli EDL933 mutants used in the present study. Growth on maltose or maltodextrin as the sole carbon source was determined in MOPS medium containing 0.2% carbohydrate. Maltose and maltodextrin sensitivity was determined in LB containing 0.2% carbohydrate. The final OD600 values of cultures at 24 h are shown. Oxidation of maltose was determined on Biolog GN2 phenotype plates, as shown in graphlets of relative amounts of substrate oxidized (y axis) versus time. Glycogen accumulation on LB agar containing 2% glucose was visualized by staining with iodine vapor.
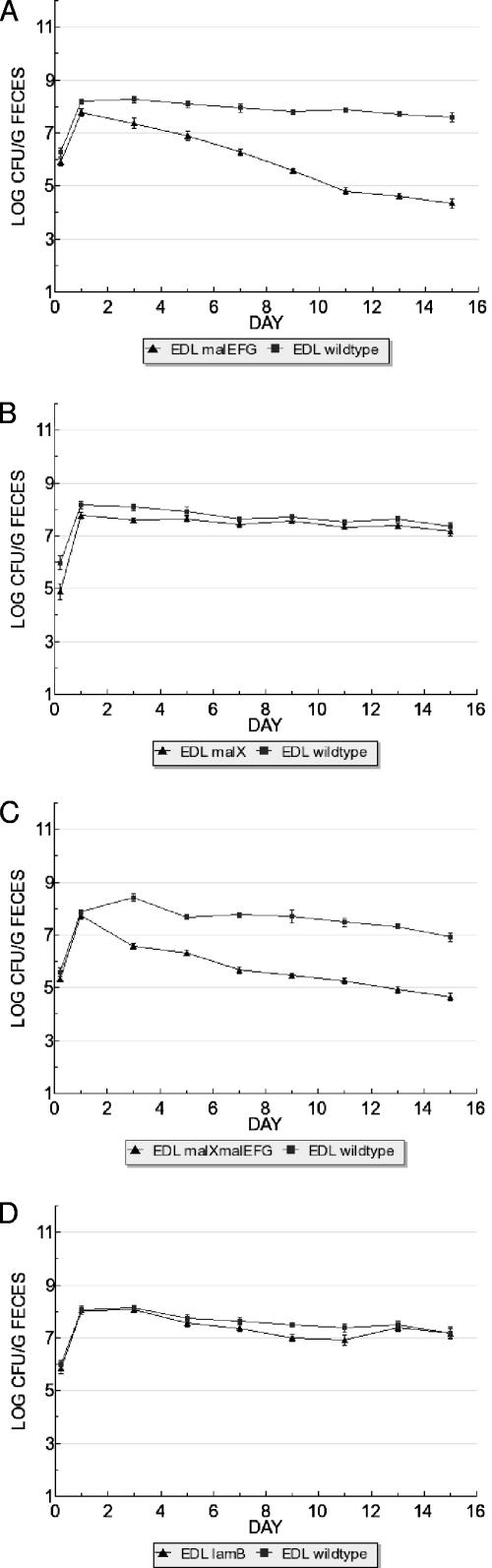
E. coli EDL933 malEFG was fed together with its wild-type parent, each at 105 CFU/mouse. The mutant was able to initiate colonization and reached the same population size as the wild-type on day 1 of the experiment but subsequently declined steadily in numbers, reaching a 3.3-log difference by day 15 (Table 2 and Fig. 3A). E. coli EDL933 malX cocolonized with the wild-type, indicating that the mutation did not cause a colonization defect (Table 2 and Fig. 3B). The E. coli EDL933 malEFG malX strain behaved similarly to E. coli EDL933 malEFG in initiating colonization but subsequently declined in numbers relative to the wild type and reached a 2.3-log difference by the conclusion of the experiment (Table 2 and Fig. 3C). E. coli MG1655 differed from E. coli EDL933 in having no significant impact of the malEFG mutation on colonization (Table 2). Likewise, E. coli MG1655 malX cocolonized with wild-type E. coli MG1655. However, the E. coli MG1655 malEFG malX double mutant had a colonization defect similar to that of E. coli EDL933 malEFG malX (Table 2). We conclude from these results that the transport of maltose and/or maltodextrin is important for maintenance of intestinal colonization. To distinguish the involvement of maltodextrin from maltose transport, we knocked out LamB, encoded by lamB, a maltodextrin-specific outer membrane porin; lamB mutants grow on maltose but cannot grow on maltodextrins (44). This phenotype was verified (Fig. 2). E. coli lamB mutants cocolonized with their respective parent strains when fed together to mice (Table 2 and Fig. 3D). From these results, we conclude that the transport of maltose, but not maltodextrins, is important for E. coli to colonize the murine intestinal tract.
FIG. 3.
Maltose/maltodextrin transport mutants display colonization defects. (A) E. coli EDL933 Δ(malE-malG) has a colonization defect in competition with E. coli EDL933 wild-type. (B) E. coli EDL933 ΔmalX does not exhibit a colonization defect. (C) E. coli EDL933 ΔmalX Δ(malE-malG) has a colonization defect in competition with E. coli EDL933 wild-type. (D) E. coli EDL933 ΔlamB does not exhibit a colonization defect. Strains were fed to mice at 105 CFU each.
Maltose catabolism is important for intestinal colonization.
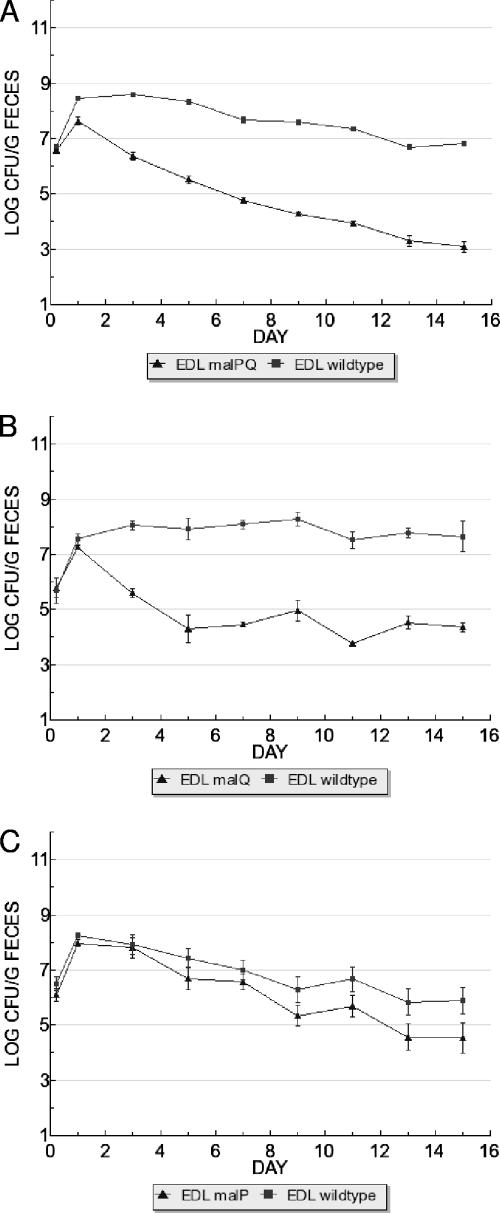
Mutants lacking malP, which encodes maltodextrin phosphorylase are unable to catabolize maltodextrin, and malQ mutants are unable to catabolize maltose (5). To verify the result of the mini-Tn5 insertion in malP (Fig. 1), we constructed malPQ-null mutations in the E. coli EDL933 and E. coli MG1655 backgrounds; both of these malPQ mutants were unable to maintain colonization during competition with their respective wild types and had major colonization defects of 3.7 and 3.5 logs, respectively (Table 2 and Fig. 4A). The inability of E. coli EDL933 malPQ to metabolize either maltose or maltodextrin was confirmed (Fig. 2). It was reported that the growth of malQ mutants is inhibited by maltose (5), which could negatively impact competitive fitness in the intestine if maltose was present. For this reason, we tested the mutant strains used here for maltose and maltodextrin sensitivity; none of the strains tested were sensitive to maltose or maltodextrin (Fig. 2). Since the E. coli EDL933 malPQ population in the competitive colonization assay was only 1 log above the limit of detection (103 CFU/g of feces), we wanted to test whether the observed colonization defect resulted from an inability to grow in the intestine or an inability to compete with the wild type; the latter was confirmed, since both malPQ mutant strains colonized at wild-type levels when fed alone to mice (data not shown). Thus, we conclude that the inability of the E. coli malPQ mutants to catabolize maltose and/or maltodextrin strongly and negatively impacted competitive fitness in the mouse intestine, which explains why the STM experiment revealed a malP insertion mutant.
FIG. 4.
Catabolism of maltose but not maltodextrins is important for competitive colonization of the mouse intestine. (A) E. coli EDL933 Δ(malP-malQ) has colonization defect in competition with E. coli EDL933 wild-type. (B) E. coli EDL933 ΔmalQ has colonization defect in competition with E. coli EDL933 wild-type. (C) E. coli EDL933 ΔmalP does not exhibit a significant colonization defect. Strains were fed to mice at 105 CFU each.
To distinguish the catabolism of maltose from that of maltodextrin, individual null mutations were constructed in malP and malQ, which eliminated growth on maltodextrin and maltose, respectively (5). Before proceeding with the colonizations, we verified these phenotypes (Fig. 2). E. coli EDL933 malQ was fed together with wild-type E. coli EDL933 and observed to have a 3.3-log colonization defect (Table 2 and Fig. 4B), confirming that maltose catabolism is important for colonization. The malP mutation had a modest, but statistically insignificant, impact on colonization of E. coli EDL933 (Table 2 and Fig. 4C). This result, together with the lack of a colonization defect for the lamB mutant (Fig. 3C), suggests that maltodextrins are not important substrates in the intestine. These results confirm the results obtained with the transporter mutants, i.e., that the ability to use maltose, but not maltodextrins, is important for intestinal colonization.
Maltose is necessary for E. coli EDL933 to initiate colonization during competition with E. coli MG1655.
To test whether maltose metabolism is important for the pathogen E. coli EDL933 to compete with the commensal E. coli MG1655 in the mouse intestine, 105 CFU of the E. coli EDL933 malQ mutant was fed to mice together with 1010 CFU of wild-type E. coli MG1655. The results show that E. coli EDL933 malQ was unable to initiate in competition with wild-type E. coli MG1655 during colonization, whereas the wild-type E. coli EDL933 initially outcompetes E. coli MG1655 (compare Fig. 5A and Fig. 5B). This result indicates that maltose is used by pathogenic E. coli EDL933 to grow from low to high numbers while attempting to gain residence in the mouse intestine when higher numbers of commensal E. coli MG1655 are present.
FIG. 5.
Catabolism of maltose is necessary for E. coli EDL933 to compete efficiently with wild-type E. coli MG1655 in the mouse intestine. (A) E. coli EDL933 wild-type (fed to mice at 105 CFU) initially out competes higher numbers of E. coli MG1655 wild-type (fed at 1010 CFU). (B) E. coli EDL933 ΔmalQ (fed at 105 CFU) cannot compete with higher numbers of E. coli MG1655 wild-type (fed at 1010 CFU).
Glycogen synthesis is important for colonization of the intestine.
To test whether the ability to make glycogen (i.e., for carbon storage) provides an advantage in the intestine, null mutations were constructed in two of the genes that are necessary for glycogen synthesis (glgA and glgS), the phenotypes of which were previously described (21, 26). Mutation of glgA, which encodes glycogen synthetase (26), resulted in a glycogen-negative phenotype; mutation of glgS, which encodes a factor that plays and important and yet unknown role in glycogen synthesis (21), resulted in a reduced amount of glycogen; neither mutation affected metabolism of maltose or maltodextrin (Fig. 2). E. coli EDL933 and E. coli MG1655 glgA mutants had colonization defects of 2.3 and 2.9 logs, respectively, in competition with their wild-type parents (Table 2 and Fig. 6). Likewise, E. coli EDL933 and E. coli MG1655 glgS mutants had colonization defects of 1.9 and 1.6 logs, respectively, in competition with their wild-type parents (Table 2 and Fig. 6). Thus, we conclude that that ability to synthesize carbon stores in the form of glycogen is important for E. coli to colonize the mouse intestine.
FIG. 6.
Glycogen synthesis mutants do not compete effectively in the mouse intestine. (A) E. coli EDL933 ΔglgA exhibits colonization defect in competition with E. coli EDL933 wild-type. (B) E. coli EDL933 ΔglgS exhibits colonization defect in competition with E. coli EDL933 wild-type. Strains were fed to mice at 105 CFU each.
Glycogen degradation is important for colonization of the intestine.
We wanted to test whether the ability to degrade glycogen provides a competitive advantage in the mouse intestine. The primary pathway for glycogen degradation involves glycogen phosphorylase, which is encoded by glgP. Although the pathways for metabolism of glycogen and maltodextrin are similar, they are not inextricable, i.e., although they have similar α-glucan phosphorylase activities, GlgP cannot replace MalP in E. coli (5). Mutants were constructed in glgP, which are unable to breakdown glycogen but could still catabolize maltose and maltodextrin, as described previously (1). Before proceeding with the colonization, we tested the phenotype and found the E. coli EDL933 glgP strain was glycogen positive, showed modestly reduced growth on maltodextrin compared to the wild type, and metabolized maltose normally (Fig. 2). E. coli EDL933 and E. coli MG1655 glgP mutants could initiate but not maintain colonization, as indicated by 2.4- and 3.2-log colonization defects, respectively, by day 15 postfeeding (Table 2 and Fig. 7). Apparently, MalP did not contribute to glycogen degradation in vivo, since the malP mutation alone did not significantly affect colonization and the glgP malP double mutants had colonization defects of 2.5 logs at day 15, which were similar to the colonization defects of the glgP mutants (Table 2). These results provide additional evidence that glycogen is an important carbon store that provides a colonization advantage in the intestine.
FIG. 7.
Colonization defects of glycogen degradation mutants are rescued by adding 2% gluconate to drinking water of mice. (A) E. coli EDL933 ΔglgP and (C) E. coli MG1655 ΔglgP exhibit colonization defects in competition with their respective wild-type parents. (B and D) These colonization defects are rescued by addition of gluconate to the drinking water . Strains were fed to mice at 105 CFU each.
Evidence for a hunger or famine existence in the intestine.
Since glycogen is known to promote the survival of E. coli (42), our finding that glycogen synthesis and degradation is necessary for efficient colonization suggests that E. coli encounters times of carbon limitation in the intestine that can be offset by glycogen carbon stores. To confirm this possibility, we used a strategy designed to provide the microbiota with a steady, high level of a rapidly metabolized carbon source. We previously showed that gluconate, which supports the growth of E. coli but is not absorbed by the host, is able to affect the outcome of experiments that depend on competition for gluconate (28). We reasoned that the constant availability of an excess supply of gluconate, provided by adding 2% gluconate to the drinking water of mice throughout the experiment, would rescue the dependence of colonized E. coli on stored glycogen. Indeed, the E. coli EDL933 and E. coli MG1655 glgP mutants were equally fit for colonization in competition with their wild-type parents when gluconate was supplied in the drinking water (Fig. 7). Likewise, the addition of 2% gluconate to the drinking water rescued the colonization defect of E. coli EDL933 glgA (data not shown). These results support the conclusion that glycogen is important for maintaining the highest possible energy levels during times of carbon limitation in the intestine.
DISCUSSION
In this study, we examined the metabolism of maltose, maltodextrin, and glycogen by pathogenic and commensal E. coli in the mouse intestine. The results show that catabolism of the disaccharide maltose provides a competitive advantage in vivo, whereas degradation of exogenous forms of the more complex glucose polymer, maltodextrin, does not. Furthermore, we found that the utilization of maltose in vivo gives the pathogen E. coli EDL933 a competitive advantage during the initiation stage of colonization when competing with commensal E. coli MG1655. The endogenous glucose polymer, glycogen, appears to play a very important role in colonization. In support of the hypothesis that E. coli relies on internal carbon stores to maintain colonization during periods of famine, we found that by providing a constant supply of a readily metabolized sugar, i.e., gluconate, in the drinking water, the competitive disadvantage of E. coli glycogen metabolism mutants (e.g., glgP) was rescued.
The hypothesis that glycogen provides a source of internal carbon and energy storage and thus is advantageous for animal colonization and virulence was considered previously. However, the results from the two published studies are in conflict and neither provided a direct test of the hypothesis. Bonafonte et al. (4) showed a correlation between the amount of glycogen stores in Salmonella enterica serovar Enteritidis and the 50% lethal dose in 1-day-old chicks. In that study, there was no way to know whether the stored glycogen was used by the bacteria during the course of infection. In another study, McMeechan et al. (30) focused on the role of ADP-glucose pyrophosphorylase, encoded by glgC, in Salmonella enterica serovar Typhimurium. These authors found no significant differences in intestinal colonization or virulence (in 1-day-old chicks) between strains carrying glgC mutations or wild-type alleles in three different serovars (Gallinarum, Pullorum, or Typhimurium) (30). The glgC strains were thought not to accumulate glycogen because they cannot make the glycogen precursor, ADP-glucose. However, that conclusion is brought into question by the recent finding that glgC mutants of E. coli and S. enterica serovar Typhimurium possess an alternative pathway for ADP glucose formation and can accumulate glycogen under certain conditions (33).
The experiments described here directly tested the hypothesis that synthesis and utilization of glycogen is necessary for efficient colonization of the intestine by E. coli. Mutants lacking GlgA, which cannot synthesize glycogen, had significant colonization defects in competition with the wild-type strain that accumulates glycogen normally. Likewise, mutants lacking GlgP, which cannot degrade glycogen, had similarly significant colonization defects. Loss of GlgS, which is known to impair glycogen metabolism, resulted in a colonization defect similar to those of mutants lacking GlgA or GlgP. These data provide strong evidence in support of the hypothesis that glycogen metabolism is important in colonized E. coli. The need for glycogen stores may reflect a physiological status of nutrient limitation in the intestine. Accordingly, glycogen stores might be consumed by colonized bacteria as they cometabolize mixtures of severely limiting carbon sources or switch from one substrate to another. Alternatively, carbon availability in the intestine might be intermittent, requiring the cell to obtain maintenance energy from glycogen during brief periods of famine. If it is true that glycogen provides a source of carbon and energy during times of nutrient limitation, we reasoned that by providing a constant source of a readily metabolized carbon source the colonization defect of a glgP mutant would be rescued. We previously established that gluconate, which is not absorbed in the mammalian gastrointestinal tract, is available to strains that can use it and can impact colonization, strongly suggesting that gluconate provided in the drinking water of mice is metabolized by E. coli in the intestine (28). Indeed, we found that addition of gluconate to the drinking water made glgP mutants better colonizers, equal to wild-type E. coli (Fig. 7). These results indicate that glycogen stores not only support the long-term survival of E. coli outside of the host (42) but also enhance persistence in the intestine.
The biochemistry of glycogen metabolism is ideally suited to provide energy during brief respites from exogenous carbon and energy sources. The genes for glycogen synthesis and degradation are arranged together in an operon and are coregulated, such that the enzymes of both glycogen synthesis and degradation are coexpressed (37). At first, this may seem paradoxical but, in fact, since both sets of enzymes are present, the cell is poised at any given moment to either make glycogen or degrade it, as conditions allow. Recall that glycogen is formed when carbon is in excess but some other nutrient is limiting and glycogen is degraded when carbon is limiting (13, 36). Moreover, glycogen is synthesized from glucose-1-phosphate, which is also the breakdown product of glycogen. Since the enzymes of glycogen synthesis and degradation are coexpressed, their activities (i.e., accumulation or catabolism of glycogen) are reciprocally controlled by the physiological constraints of carbon source excess and depletion, both of which are reflected in the intracellular pool of glucose-1-phosphate, which in turn reflects the energy status of the cell (5). If nutrient limitation prevails in the intestine and if this limitation includes, periodically, nutrients other than carbon sources (i.e., nitrogen), then the biochemical regulation of glycogen metabolism is ideal for responding to the instantaneous excesses and limitations of carbon sources and other nutrients. Perhaps the reason that glycogen storage is widespread in enteric bacteria is because this is a necessary strategy for rapid growth in the intestine, where there is intense competition for resources.
While E. coli apparently depends on endogenous stores of the polysaccharide, glycogen, it cannot metabolize exogenous polysaccharides, with the exception of maltodextrin (29). The data presented here indicate that maltodextrin does not support colonization. Thus, the in vivo catabolic capacity of E. coli must be limited primarily to mono- and disaccharides. Previous in vivo studies of E. coli catabolism focused on monosaccharides (7). We have, however, now studied a few disaccharides and found that lactose does not support colonization by E. coli EDL933 and E. coli MG1655 (16), nor do β-glucuronides support colonization by E. coli MG1655 (7). The in vivo catabolism of α-glycosides, i.e., trehalose and sucrose, and α-galactosides, i.e., melibiose, has not been studied and should be, but at least one disaccharide, maltose, supports colonization by E. coli EDL933 and E. coli MG1655.
Clearly, growth on intestinal polysaccharides is the domain of the anaerobes that secrete polysaccharide hydrolyzing enzymes (10, 15, 41). Some, if not all, dominant intestinal anaerobes preferentially utilize oligosaccharides over mono- and disaccharides (2). Moreover, at least some minor members of the intestinal microbiota grow by cross-feeding on sugars released by the polysaccharide degraders (17, 40). For example, there is recent evidence that vancomycin-resistant Enterococcus colonizes the intestine by growing on monosaccharides released by breakdown of complex polysaccharides by the indigenous microbiota (38). As we suggested previously (7) and in accordance with the findings summarized here, E. coli almost certainly colonizes the intestine by cross-feeding on simple sugars provided by anaerobic polysaccharide degradation. These sugars must be of limited availability, since E. coli relies on endogenous carbon stores to efficiently colonize the intestine. Therefore, changes in the microbiota and/or polysaccharide availability that further limit the availability of the nutrients preferred by E. coli would limit its colonization. Importantly, we take this to mean that there is merit to combating enteric infections by using probiotics or prebiotics (20) to manipulate the intestinal microbiota in such a way as to limit the availability of sugars preferred by E. coli O157:H7 (16) and perhaps other pathogens. Accordingly, prebiotics would have to stimulate populations that consume the sugars favored by invading pathogens, while probiotics could accomplish this directly, since organisms could be engineered to specifically consume preferred nutrients.
Studies of the physiology of carbohydrate metabolism and its contribution to pathogenesis are yielding novel, broadly applicable insights into host-pathogen interactions, which are specifically related to the underlying mechanisms that enable pathogens to successfully colonize their hosts. Many of the nutrient acquisition systems used by E. coli are similar to those used by other mucosal pathogens. Therefore, such studies will aid in finding new and effective treatments. In conclusion, we wish to emphasize that therapies designed to specifically reduce intestinal nutrient availability may be effective because organisms such as E. coli O157:H7 live a hunger or famine existence, as indicated by their requirement for glycogen stores, which places the pathogens in peril of starvation before infection can begin, if administration of prebiotics or probiotics depletes their preferred nutrients.
Acknowledgments
This research was supported by Public Health Service grant AI48945 to T.C. and P.S.C. Partial support for C.S. was provided by Danish Research Agency Grant 2052-03-0013.
We thank April B. Anderson for the construction of some strains used in this study and Susanne Schjøring for assisting with colonization experiments.
Editor: A. J. Bäumler
Footnotes
Published ahead of print on 17 March 2008.
REFERENCES
- 1.Alonso-Casajus, N., D. Dauvillee, A. M. Viale, F. J. Munoz, E. Baroja-Fernandez, M. T. Moran-Zorzano, G. Eydallin, S. Ball, and J. Pozueta-Romero. 2006. Glycogen phosphorylase, the product of the glgP Gene, catalyzes glycogen breakdown by removing glucose units from the nonreducing ends in Escherichia coli. J. Bacteriol. 1885266-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amaretti, A., E. Tamburini, T. Bernardi, A. Pompei, S. Zanoni, G. Vaccari, D. Matteuzzi, and M. Rossi. 2006. Substrate preference of Bifidobacterium adolescentis MB 239: compared growth on single and mixed carbohydrates. Appl. Microbiol. Biotechnol. 73654-662. [DOI] [PubMed] [Google Scholar]
- 3.Autieri, S. M., J. J. Lins, M. P. Leatham, D. C. Laux, T. Conway, and P. S. Cohen. 2007. l-Fucose stimulates utilization of d-ribose by Escherichia coli MG1655 ΔfucAO and E. coli Nissle 1917 ΔfucAO mutants in the mouse intestine and in M9 minimal medium. Infect. Immun. 755465-5475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonafonte, M. A., C. Solano, B. Sesma, M. Alvarez, L. Montuenga, D. Garcia-Ros, and C. Gamazo. 2000. The relationship between glycogen synthesis, biofilm formation and virulence in Salmonella enteritidis. FEMS Microbiol. Lett. 19131-36. [DOI] [PubMed] [Google Scholar]
- 5.Boos, W., and H. Shuman. 1998. Maltose/maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62204-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cedergren, B., and T. Holme. 1959. On the glycogen in Escherichia coli B; electron microscopy of ultrathin sections of cells. J. Ultrastruct. Res. 370-73. [DOI] [PubMed] [Google Scholar]
- 7.Chang, D. E., D. J. Smalley, D. L. Tucker, M. P. Leatham, W. E. Norris, S. J. Stevenson, A. B. Anderson, J. E. Grissom, D. C. Laux, P. S. Cohen, and T. Conway. 2004. Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. USA 1017427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conway, T., K. A. Krogfelt, and P. S. Cohen. 2007. Escherichia coli at the intestinal mucosal surface, p. 175-196. In K. A. Brogden, F. C. Minion, N. Cornick, T. B. Stanton, Q. Zhang, L. K. Nolan, and M. J. Wannemuehler (ed.), Virulence mechanisms of bacterial pathogens, 4th ed. ASM Press, Washington, DC.
- 9.Conway, T., K. A. Krogfelt, and P. S. Cohen. 29 December 2004, posting date. Chapter 8.3.1.2, The life of commensal Escherichia coli in the mammalian intestine. In R. Curtiss III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://ecosal.org. [DOI] [PubMed]
- 10.Cummings, J. H., and H. N. Englyst. 1987. Fermentation in the human large intestine and the available substrates. Am. J. Clin. Nutr. 451243-1255. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 976640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dauvillee, D., I. S. Kinderf, Z. Li, B. Kosar-Hashemi, M. S. Samuel, L. Rampling, S. Ball, and M. K. Morell. 2005. Role of the Escherichia coli glgX gene in glycogen metabolism. J. Bacteriol. 1871465-1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dawes, E. A., and D. W. Ribbons. 1965. Studies on the endogenous metabolism of Escherichia coli. Biochem. J. 95332-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dippel, R., T. Bergmiller, A. Bohm, and W. Boos. 2005. The maltodextrin system of Escherichia coli: glycogen-derived endogenous induction and osmoregulation. J. Bacteriol. 1878332-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Englyst, K. N., and H. N. Englyst. 2005. Carbohydrate bioavailability. Br. J. Nutr. 941-11. [DOI] [PubMed] [Google Scholar]
- 16.Fabich, A. J., S. A. Jones, F. Z. Chowdhury, A. Cernosek, A. Anderson, D. Smalley, J. W. McHargue, G. A. Hightower, J. T. Smith, S. M. Autieri, M. P. Leatham, J. J. Lins, R. L. Allen, D. C. Laux, P. S. Cohen, and T. Conway. 2008. Comparison of carbon nutrition for pathogenic and commensal Escherichia coli strains in the mouse intestine. Infect. Immun. 761143-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Falony, G., A. Vlachou, K. Verbrugghe, and L. De Vuyst. 2006. Cross-feeding between Bifidobacterium longum BB536 and acetate-converting, butyrate-producing colon bacteria during growth on oligofructose. Appl. Environ. Microbiol. 727835-7841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ferenci, T. 2001. Hungry bacteria: definition and properties of a nutritional state. Environ. Microbiol. 3605-611. [DOI] [PubMed] [Google Scholar]
- 19.Freundlieb, S., and W. Boos. 1986. Alpha-amylase of Escherichia coli, mapping and cloning of the structural gene, malS, and identification of its product as a periplasmic protein. J. Biol. Chem. 2612946-2953. [PubMed] [Google Scholar]
- 20.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota: introducing the concept of prebiotics. J. Nutr. 1251401-1412. [DOI] [PubMed] [Google Scholar]
- 21.Hengge-Aronis, R., and D. Fischer. 1992. Identification and molecular analysis of glgS, a novel growth-phase-regulated and rpoS-dependent gene involved in glycogen synthesis in Escherichia coli. Mol. Microbiol. 61877-1886. [DOI] [PubMed] [Google Scholar]
- 22.Hensel, M., J. E. Shea, C. Gleeson, M. D. Jones, E. Dalton, and D. W. Holden. 1995. Simultaneous identification of bacterial virulence genes by negative selection. Science 269400-403. [DOI] [PubMed] [Google Scholar]
- 23.Hentges, D. J., J. U. Que, S. W. Casey, and A. J. Stein. 1984. The influence of streptomycin on colonization in mice. Microecol. Theor. 1453-62. [Google Scholar]
- 24.Koch, A. L. 1971. The adaptive responses of Escherichia coli to a feast and famine existence. Adv. Microb. Physiol. 6147-217. [DOI] [PubMed] [Google Scholar]
- 25.Kozlov, G., D. Elias, M. Cygler, and K. Gehring. 2004. Structure of GlgS from Escherichia coli suggests a role in protein-protein interactions. BMC Biol. 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kumar, A., C. E. Larsen, and J. Preiss. 1986. Biosynthesis of bacterial glycogen: primary structure of Escherichia coli ADP-glucose:α-1,4-glucan, 4-glucosyltransferase as deduced from the nucleotide sequence of the glgA gene. J. Biol. Chem. 26116256-16259. [PubMed] [Google Scholar]
- 27.Laux, D. C., P. S. Cohen, and T. Conway. 2005. Role of the mucus layer in bacterial colonization of the intestine, p. 199-212. In J. P. Nataro, H. L. T. Mobley, and P. S. Cohen (ed.), Colonization of mucosal surfaces. ASM Press, Washington, DC.
- 28.Leatham, M. P., S. J. Stevenson, E. J. Gauger, K. A. Krogfelt, J. J. Lins, T. L. Haddock, S. M. Autieri, T. Conway, and P. S. Cohen. 2005. Mouse intestine selects nonmotile flhDC mutants of Escherichia coli MG1655 with increased colonizing ability and better utilization of carbon sources. Infect. Immun. 738039-8049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mayer, C., and W. Boos. 29 March 2005, posting date. Chapter 3.4.1, Hexose/pentose and hexitol/pentitol metabolism. In R. Curtis III et al. (ed.), EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. http://www.ecosal.org.
- 30.McMeechan, A., M. A. Lovell, T. A. Cogan, K. L. Marston, T. J. Humphrey, and P. A. Barrow. 2005. Glycogen production by different Salmonella enterica serotypes: contribution of functional glgC to virulence, intestinal colonization and environmental survival. Microbiology 1513969-3977. [DOI] [PubMed] [Google Scholar]
- 31.Miranda, R. L., T. Conway, M. P. Leatham, D. E. Chang, W. E. Norris, J. H. Allen, S. J. Stevenson, D. C. Laux, and P. S. Cohen. 2004. Glycolytic and gluconeogenic growth of Escherichia coli O157:H7 (EDL933) and E. coli K-12 (MG1655) in the mouse intestine. Infect. Immun. 721666-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moller, A. K., M. P. Leatham, T. Conway, P. J. Nuijten, L. A. de Haan, K. A. Krogfelt, and P. S. Cohen. 2003. An Escherichia coli MG1655 lipopolysaccharide deep-rough core mutant grows and survives in mouse cecal mucus but fails to colonize the mouse large intestine. Infect. Immun. 712142-2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moran-Zorzano, M. T., N. Alonso-Casajus, F. J. Munoz, A. M. Viale, E. Baroja-Fernandez, G. Eydallin, and J. Pozueta-Romero. 2007. Occurrence of more than one important source of ADP-glucose linked to glycogen biosynthesis in Escherichia coli and Salmonella. FEBS Lett. 5814423-4429. [DOI] [PubMed] [Google Scholar]
- 34.Neidhardt, F. C. 1963. Effects of environment on the composition of bacterial cells. Annu. Rev. Microbiol. 1761-86. [DOI] [PubMed] [Google Scholar]
- 35.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Preiss, J. 1984. Bacterial glycogen synthesis and its regulation. Annu. Rev. Microbiol. 38419-458. [DOI] [PubMed] [Google Scholar]
- 37.Preiss, J. 1996. Regulation of glycogen synthesis, p. 1015-1024. In F. C. Neidhardt, R. Curtiss III, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. ASM Press, Washington, DC.
- 38.Pultz, N. J., L. C. Hoskins, and C. J. Donskey. 2006. Vancomycin-resistant Enterococci may obtain nutritional support by scavenging carbohydrate fragments generated during mucin degradation by the anaerobic microbiota of the colon. Microb. Drug Resist. 1263-67. [DOI] [PubMed] [Google Scholar]
- 39.Reidl, J., and W. Boos. 1991. The malX malY operon of Escherichia coli encodes a novel enzyme II of the phosphotransferase system recognizing glucose and maltose and an enzyme abolishing the endogenous induction of the maltose system. J. Bacteriol. 1734862-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rossi, M., C. Corradini, A. Amaretti, M. Nicolini, A. Pompei, S. Zanoni, and D. Matteuzzi. 2005. Fermentation of fructooligosaccharides and inulin by bifidobacteria: a comparative study of pure and fecal cultures. Appl. Environ. Microbiol. 716150-6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salyers, A. A., J. K. Palmer, and T. D. Wilkins. 1978. Degradation of polysaccharides by intestinal bacterial enzymes. Am. J. Clin. Nutr. 31S128-S130. [DOI] [PubMed] [Google Scholar]
- 42.Strange, R. E. 1968. Bacterial “glycogen” and survival. Nature 220606-607. [DOI] [PubMed] [Google Scholar]
- 43.Struve, C., C. Forestier, and K. A. Krogfelt. 2003. Application of a novel multi-screening signature-tagged mutagenesis assay for identification of Klebsiella pneumoniae genes essential in colonization and infection. Microbiology 149167-176. [DOI] [PubMed] [Google Scholar]
- 44.Szmelcman, S., M. Schwartz, T. J. Silhavy, and W. Boos. 1976. Maltose transport in Escherichia coli K-12. A comparison of transport kinetics in wild-type and lambda-resistant mutants as measured by fluorescence quenching. Eur. J. Biochem. 6513-19. [DOI] [PubMed] [Google Scholar]
- 45.Wadolkowski, E. A., J. A. Burris, and A. D. O'Brien. 1990. Mouse model for colonization and disease caused by enterohemorrhagic Escherichia coli O157:H7. Infect. Immun. 582438-2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wandersman, C., M. Schwartz, and T. Ferenci. 1979. Escherichia coli mutants impaired in maltodextrin transport. J. Bacteriol. 1401-13. [DOI] [PMC free article] [PubMed] [Google Scholar]