Abstract
The ability of the bacterial pathogen Actinobacillus pleuropneumoniae to grow anaerobically allows the bacterium to persist in the lung. The ArcAB two-component system is crucial for metabolic adaptation in response to anaerobic conditions, and we recently showed that an A. pleuropneumoniae arcA mutant had reduced virulence compared to the wild type (F. F. Buettner, A. Maas, and G.-F. Gerlach, Vet. Microbiol. 127:106-115, 2008). In order to understand the attenuated phenotype, we investigated the ArcA regulon of A. pleuropneumoniae by using a combination of transcriptome (microarray) and proteome (two-dimensional difference gel electrophoresis and subsequent mass spectrometry) analyses. We show that ArcA negatively regulates the expression of many genes, including those encoding enzymes which consume intermediates during fumarate synthesis. Simultaneously, the expression of glycerol-3-phosphate dehydrogenase, a component of the respiratory chain serving as a direct reduction equivalent for fumarate reductase, was upregulated. This result, together with the in silico analysis finding that A. pleuropneumoniae has no oxidative branch of the citric acid cycle, led to the hypothesis that fumarate reductase might be crucial for virulence by providing (i) energy via fumarate respiration and (ii) succinate and other essential metabolic intermediates via the reductive branch of the citric acid cycle. To test this hypothesis, an isogenic A. pleuropneumoniae fumarate reductase deletion mutant was constructed and studied by using a pig aerosol infection model. The mutant was shown to be significantly attenuated, thereby strongly supporting a crucial role for fumarate reductase in the pathogenesis of A. pleuropneumoniae infection.
Actinobacillus pleuropneumoniae is the causative agent of a porcine pleuropneumonia that results in high economic losses worldwide (16). After A. pleuropneumoniae-containing aerosols are inhaled, the pathogen colonizes the respiratory epithelium of its host and persists on tonsils, on healthy lung epithelium, and in sequestered lesions (14). The persistence of bacterial pathogens in the respiratory tracts of clinically healthy carriers is of major importance in the spread of infection in both animals and humans (6, 10, 29).
Previously, we have shown that the ability of A. pleuropneumoniae to adapt to low redox conditions is essential for its long-term persistence on intact and diseased respiratory tract epithelia (4, 26). In particular, the arcA deletion mutant of A. pleuropneumoniae was severely attenuated in this respect (8). A role in virulence for ArcA has also been implicated for intracellular bacterial pathogens such as Mycobacterium tuberculosis (42, 45), invasive pathogens such as Haemophilus influenzae (13, 59), and the enteric pathogens Vibrio cholerae (50) and Shigella flexneri (7). However, the molecular mechanisms responsible for this attenuation are only partially resolved.
In Escherichia coli, the transcriptional regulator ArcA functions primarily as a major metabolic modulator, downregulating many genes involved in respiratory metabolism (24) and upregulating genes necessary for fermentative metabolic activities under anaerobic conditions (48). The importance of basic metabolic pathways for virulence is increasingly being recognized. For example, it was recently shown that deletion of the genes encoding some enzymes of the tricarboxylic acid (TCA) cycle resulted in attenuation in Salmonella enterica serovar Typhimurium (54). The glyoxylate shunt is required for persistence of M. tuberculosis (34) and fungal virulence (32), and genes involved in energy metabolism are differentially expressed in active versus persistent infections with Chlamydia trachomatis (19). Based on these considerations, we set out to investigate whether ArcA-mediated regulation of metabolic functions could be partially responsible for the attenuation and reduced persistence of the A. pleuropneumoniae arcA mutant. Thus, the ArcA regulon of A. pleuropneumoniae was analyzed by whole-genome microarray and two-dimensional difference gel electrophoresis (2D DIGE) analyses. The results suggested that attenuation of the A. pleuropneumoniae arcA mutant was due to its inability to anaerobically adapt its metabolism in order to use fumarate as a terminal electron acceptor and to provide succinate and other essential metabolic intermediates via the reductive branch of the citric acid cycle. This hypothesis was supported by the attenuation of a fumarate reductase (frd) deletion mutant in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, primers, and growth conditions.
The bacterial strains, plasmids, and primers used in this study are listed in Table 1. A. pleuropneumoniae wild-type (wt) and mutant strains were cultured at 37°C and 5% CO2 in PPLO medium or on PPLO agar (Difco GmbH, Augsburg, Germany), both supplemented with NAD (10 μg/ml; Merck AG, Darmstadt, Germany), l-cysteine hydrochloride (260 μg/ml; Sigma Chemical Company, Deisenhofen, Germany), l-cystine dihydrochloride (10 μg/ml; Sigma), and dextrose (1 mg/ml). For cultivation of the complemented mutants, kanamycin (25 μg/ml) was added. For anaerobic growth, supplemented medium (PPLO medium) was preincubated 48 h prior to inoculation in an anaerobic chamber (Don Whitley Scientific, Shipley, England) in an atmosphere containing 5% CO2, 10% H2, and 85% N2 at 37°C. Anaerobicity of the medium was confirmed using a dissolved oxygen sensor (CellOx 325; WTW, Weilheim, Germany) linked to an inoLab instrument (WTW, Weilheim, Germany). For RNA and protein preparations, this medium was inoculated with 1% of an aerobically grown log-phase culture in supplemented PPLO medium with an optical density at 600 nm (OD600) of 0.3, and bacteria were grown anaerobically for 6 h, until they reached late exponential growth phase, and were then harvested by centrifugation. Due to severe autoaggregation under anaerobic conditions, the growth phase was assessed by determination of the total protein content (8).
TABLE 1.
Bacterial strains and primers used in this study
| Strain, plasmid, or primer | Characteristic(s)a | Source and/or reference |
|---|---|---|
| Strains | ||
| E. coli DH5α F′ | F′ endA1hsdR17(rκ− mκ−) supE44thi-1recA1gyrA(Nalr) relA1 Δ(lacZYA-argF)U169deoR [φ80dlacΔ(lacZ)M15] | 44 |
| E. coli β2155 | thrB1004 pro thi strA hsdS lacZΔM15 (F′ lacZΔM15 laqIqtraD36proA+proB+)Δdap::erm(Ermr) recA::RPA-2-tet(Tcr)::Mu-Km (Kmr) λ pir | 11 |
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1deoRaraD139 Δ(araleu)7697 galU galK rpsL(Strr) endA1 nupG | TOPO TA cloning |
| A. pleuropneumoniae wt | A. pleuropneumoniae serotype 7 isolate 76 | 1 |
| A. pleuropneumoniae ΔarcA | Unmarked arcA deletion mutant of A. pleuropneumoniae wt | 8 |
| A. pleuropneumoniae Δfrd | Unmarked frd deletion mutant of A. pleuropneumoniae wt | This work |
| Plasmids | ||
| pCR2.1-TOPO | E. coli cloning vector for fast and efficient cloning of Taq polymerase-amplified PCR products | TOPO TA cloning; Invitrogen, Groningen, The Netherlands (52) |
| pFRD811 | pCR2.1-TOPO-based plasmid containing a 1,300-bp PCR product obtained with primers ofrd_3 and ofrd_4, starting at position 1534 downstream of the frdA start codon and ending 251 bp downstream of the frdC start codon | This work |
| pFRD820 | PCR product obtained with primers ofrd_1 and ofrd_2, digested with BsmBI and PspOMI and ligation of the 1,231-bp fragment into pFRD811 digested with PspOMI and BsmBI, resulting in pFRD820 | This work |
| pEMOC2 | Transconjugation vector based on pBluescript SK with mobRP4, a polycloning site, Cmr, and transcriptional fusion of the omlA promoter with the sacB gene | Accession no. AJ868288 (3) |
| pFRD710 | pEMOC2-based plasmid carrying the truncated frdABC fragment excised from pFRD820 using NotI and PspOMI and ligated into pEMOC2 restricted with NotI and PspOMI | This work |
| pLS88 | Broad host range shuttle vector from Haemophilus ducreyi; Strr Smr Kmr | 58 |
| pFRD1300 | pLS88-based complementation plasmid carrying the PCR product of primers ofrd_5 and ofrd_6 encoding the entire frdABCD operon and 477-bp upstream sequence cloned into the EcoRI restriction site | This work |
| Primers | ||
| ofrd_1 | 5′-ATGCGGGCCCCGGGGTTTCCACTTCTAATA-3′; forward primer containing an internal PspOMI site (underlined) comprising positions 477-458 upstream of the frdA gene start codon | This work |
| ofrd_2 | 5′-ATGCCGTCTCATTCACCGCGACAACCTTCAG-3′; reverse primer containing an internal BsmBI site (underlined) comprising positions 734-753 of the frdA gene | This work |
| ofrd_3 | 5′-ATGCCGTCTCATGAATTCATTTTAGATGTGGCGCA-3′; forward primer containing an internal BsmBI site (underlined) comprising positions 1534-1553 of the frdA gene | This work |
| ofrd_4 | 5′-ATGCGCGGCCGCGCATGGTAAAGTAATGCGGC-3′; reverse primer containing an internal NotI site (underlined) comprising positions 232-251 of the frdC gene | This work |
| ofrd_5 | 5′-ACGTACCAATTGGATTACAGTGCGATTACTGC-3′; forward primer containing an internal MfeI site (underlined) comprising positions 477-458 upstream of the frdA gene start codon | This work |
| ofrd_6 | 5′-ACGTACCAATTGCGGGGTTTCCACTTCTAATA-3′; reverse primer containing an internal MfeI site (underlined) comprising position 18 upstream to position 2 downstream of the frdD stop codon | This work |
| ofrd_7 | 5′-CAATGCGAATGCGGTGGTTA-3′; forward primer comprising positions 522-541 of the frdA gene | This work |
| ofrd_8 | 5′-CGTTTCGCCGGTTGTGATTT-3′; reverse primer comprising positions 1714-1733 of the frdA gene | This work |
Nalr, nalidixic acid resistance; Ermr, erythromycin resistance; Tcr, tetracycline resistance; Kmr, kanamycin resistance; Strr, streptomycin resistance.
Microarray analysis.
RNA isolation was carried out using a FastRNA Pro Blue kit (Qbiogen, Heidelberg, Germany), and further purification was done using an RNeasy Mini-Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. DNA contamination was removed using a Turbo DNA-free kit (Ambion, Austin, TX). RNA concentration and quality were determined spectrophotometrically and by agarose gel electrophoresis, respectively.
For cDNA synthesis, a 10-μl volume containing 2 to 5 μg of total RNA and 2 μg of random hexamers (Gibco-BRL) was heated at 70°C for 5 min and quick-cooled on ice. cDNA synthesis was performed at 42°C for 1.5 h in a 25-μl reaction mixture containing the RNA, the primer mixture, 1× first-strand buffer (Invitrogen), 500 U Superscript II (Invitrogen), 10 mM dithiothreitol (Invitrogen), and 1.7 nmol of either Cy3-dCTP or Cy5-dCTP (Amersham) at 37°C for 90 min. Unincorporated nucleotides were removed using a MinElute kit (Qiagen), according to the manufacturer's protocol. Labeled samples were combined and hybridized to the A. pleuropneumoniae serotype 5b strain L20 microarray (12). This microarray contains PCR products representing each of the 2,025 open reading frames of the genome of A. pleuropneumoniae serotype 5b strain L20. Details on the construction and content of the microarray (AppChip1) are available on the Institute for Biological Sciences website (http://ibs-isb.nrc-cnrc.gc.ca/glycobiology/appchips_e.html). Array hybridization and washing were performed as described previously (12). In total, six hybridizations were performed for six independent biological repeats of the A. pleuropneumoniae wt and the A. pleuropneumoniae ΔarcA mutant. The scanned images were analyzed with GenePix Pro 6.0 to generate a data format compatible for further statistical analysis using Genespring GX7.3 (Agilent). The spot intensities of mutant and wt samples were defined as signal and control channels, respectively. The signal channel was normalized to the control channel using LOWESS (intensity dependent), and the percentile was calculated by using all genes not marked absent by GenePix Pro analysis. The final ratio of expression (mutant/wt) was generated using the means for all biologicals and dye swaps. Differentially expressed genes were selected using “filtering on confidence” (t test P value) plus multiple testing (Benjamini and Hochberg false discovery rate) at a cutoff of ≤0.05 in order to generate a comprehensive list. For biological relevance, a >2-fold change in expression was considered significant; accordingly, discussion was focused on these genes.
2D DIGE.
Anaerobic cultures were cooled on ice, and bacteria were harvested by centrifugation (13,000 × g, 10 min, 4°C) and washed once with a buffer containing 10 mM Tris-HCl (pH 8.0) and 5 mM magnesium acetate. Pellets were resuspended in 50 mM Tris-HCl (pH 7.3) and stored at −70°C overnight. Cells were thawed and then were ruptured using a FastPrep Instrument (Qbiogene, Heidelberg, Germany) three times for 40 s each time at an intensity setting of 5.0, and cell debris was removed by centrifugation (15,000 × g, 30 min). Protein concentration was determined by a MicroBC assay (Uptima Interchim, Montluçon Cedex, France) and confirmed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Proteins were precipitated with trichloroacetic acid, washed with acetone, and solubilized in lysis buffer composed of 30 mM Tris-HCl (pH 8.0), 7 M urea (Roth, Karlsruhe, Germany), 2 M thiourea (Sigma), and 4% (wt/vol) 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS; Roth) to a protein concentration of 5 mg/ml. Labeling with CyDyes and 2D gel electrophoresis was performed according to the manufacturer's instructions (Amersham Biosciences AB, Uppsala, Sweden), and four independent biological repeats of the A. pleuropneumoniae wt and its arcA mutant were used. Gels were scanned with a Typhoon Trio scanner (Amersham), and the intensity of pixel values was adjusted such that the maximum volume of each image was between 50,000 and 90,000. Quantitative evaluation and statistical analyses of the 2D DIGE results was performed using DeCyder 6.5 software (GE Healthcare) according to the manufacturer's instructions. All spots calculated as having an expression difference of statistical significance (unpaired t test, P ≤ 0.05) were highlighted. If, for a certain protein, a horizontal string of spots occurred, these spots were numbered with the lowest number being given to the spot showing the highest calculated difference between the two strains (also see Table S2 in the supplemental material).
Protein identification by Q-TOF MS/MS or MALDI-TOF MS.
Protein spots identified as differentially expressed by 2D DIGE were excised, trypsinized, and removed from the gel according to the method of Wilm and Mann (51). For electrospray injection quadrupole time-of-flight tandem mass spectrometry (ESI Q-TOF MS/MS), 3 μl of the peptide solution was loaded into a sample tip (Nanoflow Probe Tip, long; Waters, Milford, MA). Peptide sequences were determined from MS/MS fragmentation data recorded with an ESI Q-TOF MS (Q-TOF Ultima, Waters) in positive reflection mode. Proteins were identified using the program ProteinLynx Global Server (version 2.1; Waters) by searching against the A. pleuropneumoniae serotype 5b strain L20 genome database (ftp://ftp.ncbi.nlm.nih.gov./genomes/Bacteria/Actinobacillus_pleuropneumoniae_L20/NC_009053.faa), as downloaded on 10 April 2007. Alternatively, matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS was carried out by using a VoyagerDE Pro (Applied Biosystems, Foster City, CA); 1 μl of the peptide solution was mixed with 1 μl of matrix (α-cyano-4-hydroxy-cinnamic acid [Bruker Daltonics, Billerica, MA], 5 mg/ml, in 50% acetonitrile with 0.1% trifluoroacetic acid) and then spotted on the target plate. Peptide spectra were acquired in positive reflection mode, averaging about 1,000 laser shots per MALDI-TOF spectrum. Mass spectra were calibrated using calibration mixtures CalMix1 and CalMix2 (Applied Biosystems). The peptide mass spectra were analyzed with the peptide mass fingerprint algorithm on the Mascot web site (http://www.matrixscience.com) by searching against the NCBI nonredundant database. The search algorithm was set to allow carbamidomethylation on cysteine residues, oxidation on methionine residues, and a maximum of one missed trypsin cleavage. The peptide mass tolerance was 0.2 Da. Proteins were considered to be positively identified when the probability-based score was above the significance threshold (P ≤ 0.05). Identification was confirmed by comparing the calculated molecular masses and isoelectric point values from the identified proteins with the observed values for the 2D gel.
Construction and in vitro testing of an isogenic A. pleuropneumoniae Δfrd in-frame deletion mutant.
DNA fragments of approximately 1,300 bp located upstream and downstream of the intended deletion were amplified from genomic A. pleuropneumoniae AP76 DNA using primers ofrd_1 and ofrd_2 and primers ofrd_3 and ofrd_4, respectively (Table 1). The downstream fragment was cloned into the vector pCR2.1-TOPO, resulting in pFRD811. Both pFRD811 and the upstream fragment were restricted with BsmBI and PspOMI and ligated, resulting in pFRD820 harboring the truncated frdA gene. Plasmid pFRD820 was restricted with NotI and PspOMI, and the fragment containing the truncated frdA gene was ligated into pEMOC2, resulting in pFRD710 (Table 1). The A. pleuropneumoniae Δfrd mutant was constructed by conjugation with E. coli β2155 harboring the plasmid pFRD710 (Table 1) as described previously (41). Colonies with the correct PCR profile were confirmed by Southern blotting (47), and the recombination site was confirmed by nucleotide sequencing analysis (Seqlab, Göttingen, Germany). The absence of genomic rearrangements was confirmed by pulsed-field gel electrophoresis (41). The specificity of the mutation with respect to the fumarate reductase-negative phenotype was confirmed by complementation in trans with plasmid pFRD1300 (Table 1) and subsequent assaying of fumarate reductase activity (25). Briefly, absorbance at 340 nm was measured for a solution containing 8.4 mM sodium fumarate (Sigma), 116.3 μM NADH (Roth), and 1.25 mM Tris (pH 7.3). The reaction was started by adding an equal volume of bacterial whole-cell lysates. Bacterial cultures and preparation of whole-cell lysates were performed as described for 2D DIGE.
Virulence studies.
Comparisons of the virulence of the A. pleuropneumoniae wt and the frd deletion mutant were assessed by using an aerosol infection model after determining clinical, lung, and reisolation scores as previously described (8). Briefly, A. pleuropneumoniae-free pigs (German Landrace, male, 8 to 9 weeks of age) were randomly assigned to two groups (wt group, 7 pigs; Δfrd group, 8 pigs) and cared for in accordance with the principles outlined by the European Convention for the Protection of Vertebrate Animals used for Experimental and Other Scientific Purposes (European Treaty Series, nos. 123 [http://conventions.coe.int/Treaty/EN/Treaties/Html/123.htm] and 170 [http://conventions.coe.int/Treaty/EN/Treaties/Html/170.htm]).
Based on the specifications for the validation of A. pleuropneumoniae vaccines given in the European Pharmacopoeia (http://www.pheur.org), the occurrence of coughing, dyspnea, and vomiting was assessed daily within the first 7 days postinfection. In order to quantify the results, a score of 1 was given for each of the symptoms, resulting in a maximum score of 3 per pig and day.
The lung lesion score was determined according to the scoring system adopted by the European Pharmacopoeia. Briefly, upon necropsy, each of the seven porcine lung lobes was scored separately with a score of 0 (healthy) to 5 (completely altered), resulting in a maximum score of 35 per pig.
In addition, a histopathological analysis of lung lesions was performed. Briefly, pigs were necropsied on day 7 or day 21 postinfection, and samples were fixed in 10% formaldehyde, embedded in paraffin wax, sectioned at 5 μm, stained with hematoxylin and eosin, and analyzed by light microscopy.
The reisolation score was determined as described previously (33). Briefly, defined locations of each of the seven lung lobes were plated on selective meat blood agar (27). A score for reisolation of 0 was given if no growth of A. pleuropneumoniae occurred or if growth occurred only in the area of the plate inoculated by direct contact with the tissue sample; by using a sterile loop, each sample was fractionated twice, and a score of 1 was given if colonies were also present in the fractionated streaks. To confirm the identity of the challenge strain, one colony for each location or at least three individual colonies were analyzed by PCR using the primers ofrd_7 and ofrd_8. The reisolation score was determined by adding the individual scores for each pig, and the arithmetic means and standard deviations were determined.
Immune response.
The humoral immune response of pigs was determined by two different enzyme-linked immunosorbent assays (ELISAs). Antibody levels directed against the A. pleuropneumoniae ApxII toxin were analyzed using recombinant ApxII protein as the coating antigen (30), and the response is quantified in ELISA units (EU) based on an external standard. Activities of ≤10 EU in the sera are considered negative, those of 11 to 25 EU are intermediate, and those of >25 EU are positive. Antibodies directed against outer membrane-associated proteins were determined by an ELISA using a detergent extract of A. pleuropneumoniae grown under iron restriction as the coating antigen (20). Quantification of the immune response was done by determining the antibody titer in comparison to that of an internal negative control consisting of an equal mixture of all serum samples taken at the arrival of the pigs. The titer was defined as the highest serum dilution resulting in an OD twice as high as that of the negative control serum at a dilution of 1:100.
RESULTS
Identification and functional classification of the A. pleuropneumoniae ArcA regulon.
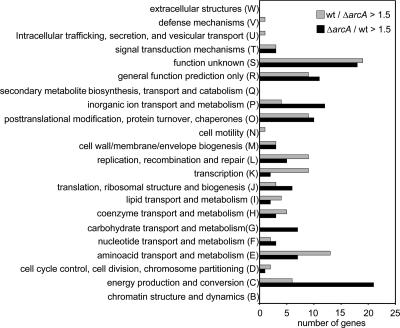
In the presence of ArcA (as in A. pleuropneumoniae wt), among the 2,025 predicted protein-encoding genes on the microarray chip, the expression levels of 16 and 77 genes were upregulated >2-fold and between 1.5- and 2-fold, respectively. The expression levels of 42 and 64 genes were downregulated >2-fold and between 1.5- and 2-fold, respectively (see Table S1 in the supplemental material). An analysis of the ArcA regulon based on the database of Clusters of Orthologous Groups of proteins (COGs) (53) revealed that ArcA controls the expression of genes in almost all prokaryotic COGs, with large differences in the numbers of up- and downregulated genes in some functional categories. Thus, in the category “carbohydrate transport and metabolism,” seven genes were downregulated in the presence of ArcA and none was upregulated. In the “energy production and conversion” category, 21 genes were downregulated and 6 genes were upregulated in the presence of ArcA. In the category “inorganic ion transport and metabolism,” 12 genes were downregulated in the presence of ArcA compared to 4 that were found to be upregulated. In contrast, in the categories “replication, recombination and repair,” “transcription,” and “cell cycle control, cell division, and chromosome partitioning,” considerably more genes were upregulated than downregulated in the presence of ArcA (Fig. 1).
FIG. 1.
Functional classification of ArcA-regulated genes according to the COGs database. The classification was adopted from the genome annotation for A. pleuropneumoniae serotype 5b L20 from the NCBI database (GenBank accession no. CP000569).
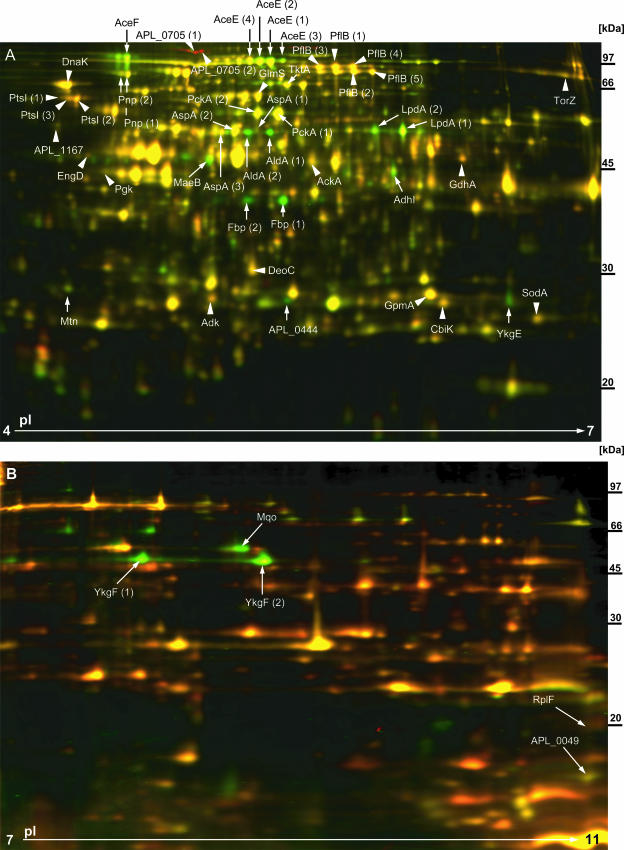
Transcription data were supplemented by 2D DIGE ( Fig. 2), which revealed a total of 156 protein spots that were regulated statistically significantly (P ≤ 0.05) in the presence of ArcA in A. pleuropneumoniae. Definitive protein identification was possible for 52 of these spots, representing 34 different proteins with 16 proteins that were upregulated and 18 that were downregulated (see Table S2 in the supplemental material).
FIG. 2.
2D DIGE analysis. The A. pleuropneumoniae wt (shown here labeled with Cy5 [red]) and A. pleuropneumoniae ΔarcA (shown here labeled with Cy3 [green]) proteins were precipitated directly from whole-cell lysates and separated by using Immobiline DryStrips with pI gradients of 4 to 7 (A) or 7 to 11 (B). The A. pleuropneumoniae wt and A. pleuropneumoniae ΔarcA mutant were grown anaerobically in four independent cultures (each). By using three fluorescent dyes, Cy2 (blue [internal standard]), Cy3 (green [either wt or ΔarcA]) and Cy5 (red [either wt or ΔarcA]), on each gel, an internal standard and a protein preparation of the A. pleuropneumoniae wt and A. pleuropneumoniae ΔarcA mutant were separated simultaneously. Fluorescence was detected using a Typhoon Trio fluorescence scanner. Quantitative and statistical analyses of differences in protein expression levels were performed with DeCyder 2D 6.5 software. Each gel shown here represents one out of four gels that were run and analyzed. Arrowheads highlight protein spots that were significantly (P ≤ 0.05) upregulated in the presence of ArcA. Arrows indicate protein spots that were significantly (P ≤ 0.05) downregulated in the presence of ArcA. Protein spots of interest were excised from preparative gels, trypsinized and, after recovery of peptides from the gel, analyzed by MS.
The microarray data were in accordance with the 2D DIGE data for the proteins downregulated significantly in the presence of ArcA. Thus, for 16 of the 18 proteins identified, the encoding genes were also identified by microarray analysis as being downregulated significantly. For the proteins upregulated significantly in the presence of ArcA, the data were partly confirmed by microarray data. Thus, of the 16 proteins identified, 7 were found by microarray analysis, and 6 of these were significantly upregulated in the presence of ArcA. For the remaining nine proteins, the encoding genes were not identified as being significantly regulated due to variations in signal ratios for the biological repeats. For the gene-protein pairs identified by both microarray and 2D DIGE, the results of both methods correlated with the line of best fit, which showed a slope of 1.9 and an R2 value of 0.65 (see Fig. S1 and Table S3 in the supplemental material).
Predicted impact of ArcA on A. pleuropneumoniae metabolism.
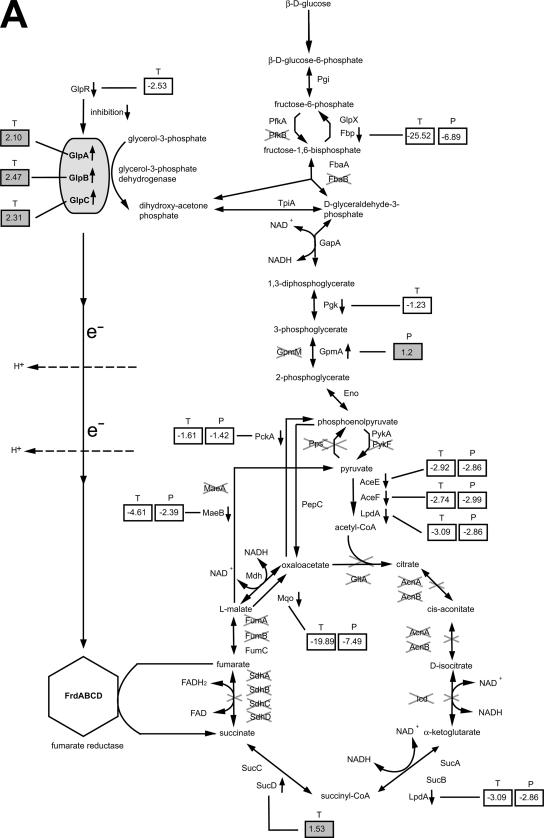
In silico analysis of the ArcA regulon led to the hypothesis that (i) ArcA of A. pleuropneumoniae modulates its metabolism toward the synthesis of fumarate as the terminal electron acceptor and (ii) glycerol-3-phosphate is used as the reduction equivalent for the reduction of fumarate and that, after oxidation to dihydroxyacetone phosphate, it serves as a precursor for fumarate synthesis (Fig. 3A). Transcripts and proteins of the three subunits of the pyruvate dehydrogenase complex, AceE, AceF and LpdA, were downregulated approximately threefold in the presence of ArcA in the A. pleuropneumoniae wt. This result suggests that consumption of pyruvate, by oxidative decarboxylation into acetyl-coenzyme A (acetyl-CoA) for the introduction of reduced carbon atoms into the citric acid cycle, is downregulated. In addition, a BLASTP homology search against E. coli citric acid cycle enzymes revealed that the citric acid cycle of A. pleuropneumoniae is incomplete; it lacks a homologue for citrate synthase (GltA). Therefore, potentially, acetyl-CoA cannot react with oxaloacetate to form citrate. Furthermore, homologues of aconitase and isocitrate dehydrogenase (AcnA, AcnB, Icd), catalyzing the transformation of citrate into α-ketoglutarate, are also missing from the A. pleuropneumoniae genome. These results led us to hypothesize that phosphoenolpyruvate, not being fed into the oxidative branch of the citric acid cycle, is carboxylated to oxaloacetate by phosphoenolpyruvate carboxylase, as can occur with E. coli (2). The absence of a citrate synthase in A. pleuropneumoniae may prevent oxaloacetate consumption by the generation of citrate. Additionally, oxaloacetate is unlikely to be used for gluconeogenesis, as the latter is strongly repressed in the presence of ArcA. Two key enzymes of gluconeogenesis, phosphoenolpyruvate carboxykinase (PckA) and fructose-1,6-bisphosphatase (Fbp), were downregulated in the A. pleuropneumoniae wt compared to the arcA deletion mutant. Therefore, oxaloacetate is most likely converted to l-malate by malate dehydrogenase (Mdh), an enzyme favoring the NADH-consuming reduction of oxaloacetate to malate (39). l-Malate could then be converted into fumarate by FumC, the only fumarase found in A. pleuropneumoniae. An alternative possibility, the irreversible oxidation of malate to oxalacetate by the membrane-bound malate quinone oxidoreductase (Mqo; 28), is unlikely since transcription of mqo is downregulated 20-fold in the presence of ArcA. Also, malic enzyme (MaeB), which catalyzes the oxidative decarboxylation of malate into pyruvate, is downregulated 4.6-fold in the presence of ArcA. The second decarboxylating malate dehydrogenase, E. coli homologue (MaeA), is missing in A. pleuropneumoniae. However, fumarate could serve as the terminal electron acceptor in an electron transfer reaction of the respiratory chain catalyzed by the fumarate reductase enzyme complex (FrdABCD; 5). Fumarate reductase is typically linked with glycerol-3-phosphate dehydrogenase through a simple electron transport chain (49), transferring reduction equivalents directly from glycerol-3-phosphate to fumarate and thereby generating a proton gradient (23). Transcription of the three genes encoding the anaerobic glycerol-3-phosphate dehydrogenase (glpABC) was upregulated >2-fold in the presence of ArcA. This enzyme complex catalyzes the oxidation of glycerol-3-phosphate to dihydroxyacetone phosphate which, in turn, is a glycolysis intermediate and can be converted into fumarate (Fig. 3A).
FIG. 3.
Impact of ArcA-regulated genes on metabolic pathways. (A) Glycolysis and citric acid cycle. (B) Dehydrogenases, terminal reductases, and membrane transporter. (C) Fermentation. (A to C) T, transcript; P, protein. The degree of ArcA-dependent regulation is given within the boxes when results obtained by microarray analysis (transcript) or by 2D DIGE (protein) were statistically significant (P ≤ 0.05; gray-highlighted box, upregulated in the presence of ArcA; unhighlighted box, downregulated in the presence of ArcA). Dehydrogenases are indicated by a rounded rectangle, terminal reductases are indicated by hexagons, and membrane transporters are indicated by rounded rectangle pairs. Biochemical pathways were adopted from those for E. coli K-12 (BioCyc, http://www.biocyc.org). The sequence and annotation of the A. pleuropneumoniae serotype 5b strain L20 genome have recently been published (GenBank accession no. CP000569). If A. pleuropneumoniae carries no homologue to the respective E. coli protein, its name is crossed out.
The hypothesized use of glycerol-3-phosphate as a reduction equivalent and fumarate as an alternative electron acceptor was supported by the simultaneous downregulation of other enzymes commonly involved in anaerobic metabolism, including (i) enzymes involved in fermentation, such as an alcohol dehydrogenase (AdhI) and an aldehyde dehydrogenase (AldA), which were repressed 5- and 16-fold, respectively (Fig. 3C); (ii) dehydrogenases of the respiratory chain, which require l-lactate (LldD), d-lactate (Dld), proline (PutA), or formate (FdhD, FdxG [APL_0892], FdxG [APL_0893], FdxH, FdnI, FdnE) as reduction equivalents; and (iii) terminal reductases using nitrite (NrfA, NrfB, NrfC, NrfD, NrfE), nitrate (NapA, NapF), or oxygen (CydA, CydB) as oxidation equivalents in the respiratory chain (Fig. 3B).
Impact of the frdABCD genes, encoding fumarate reductase, on virulence.
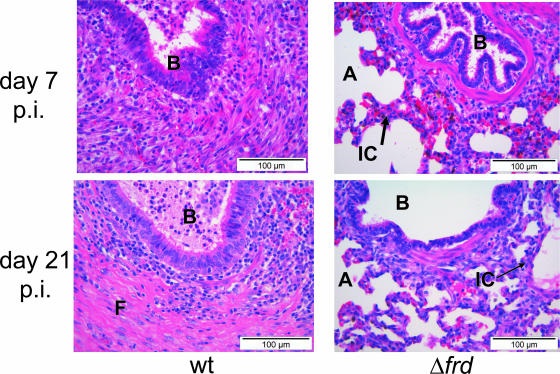
To test whether fumarate reductase may be partly responsible for the attenuation of the A. pleuropneumoniae ΔarcA mutant, fumarate reductase was deleted by genomic exchange against an frdABCD operon harboring an in-frame-deleted version of the frdA sequence. The deletion mutant was confirmed by PCR, Southern blot analysis, sequencing, and pulsed-field gel electrophoresis (data not shown). By use of an enzymatic assay, it was shown that NADH consumption in the presence of fumarate was considerably lowered in the mutant and could be restored by complementation in trans (data not shown). Pigs infected with the A. pleuropneumoniae Δfrd mutant had clearly but not significantly reduced clinical and reisolation scores. The lung lesion score was reduced by an amount that was statistically significant (P ≤ 0.05) (Table 2), and the quality of lung lesions was completely different than those seen with the wt. Thus, pigs infected with the A. pleuropneumoniae Δfrd mutant had a mild bronchiolo-interstitial pneumonia, whereas animals infected with the A. pleuropneumoniae wt showed a severe fibrinous pleuropneumonia and a moderate focal purulent pneumonia with abscess formation. Similar observations were made on day 7 postinfection and day 21 postinfection (Fig. 4). Histopathology of lung tissue obtained from an uninfected control pig revealed a diffuse interstitial pneumonia comparable to that in animals infected with the A. pleuropneumoniae Δfrd mutant (data not shown).
TABLE 2.
Virulence of A. pleuropneumoniae parent and frd mutant strain following aerosol challenge
| Challenge strain | No. of animals | Challenge dose (OD600 [aerosolized] per 4 pigs)a | Day of necropsy | Serological response withb:
|
Arithmetic mean (± SD) forc:
|
|||
|---|---|---|---|---|---|---|---|---|
| Detergent wash | ApxII | Clinical score | Lung lesion score | Reisolation score | ||||
| AP76 wt | 1 | 0.5 | 2d | ND | ND | 21 | 15.4 | 7 |
| 3 | 0.5 | 7 | ND | ND | 6 ± 3 | 21.21 ± 11.07 | 5 ± 2.65 | |
| 3 | 0.5 | 21 | 21,333 ± 6,034 | 64 ± 9 | 3.33 ± 2.31 | 14.92 ± 14.18 | 1.67 ± 2.89 | |
| AP76 Δfrd | 4 | 0.45 | 7 | ND | ND | 1.5 ± 1.29 | 6 ± 4.77 | 1.5 ± 1.73 |
| 4 | 0.45 | 21 | 22,400 ± 5,543 | 66 ± 19 | 2.75 ± 0.03 | 0.73 ± 0.57 | 0.25 ± 0.5 | |
| P valuee | 0.093 | 0.046 | 0.106 | |||||
Bacteria were grown aerobically in the presence of Tween 80 (0.1%) to the respective OD600 and diluted 1:30,000 with sterile saline; 13 ml of this dilution was aerosolized in the aerosol chamber.
Detergent wash, arithmetic mean of the highest serum dilution resulting in an OD twice as high as that of the negative control serum at a dilution of 1:100 ± standard deviation. ApxII, arithmetic mean (in EU) of the serum activity ± standard deviation. ND, not determined.
Animal died on day 2; these results are not included in the statistical comparison of scores between wt- and Δfrd-infected pigs.
The statistical analysis comparing A. pleuropneumoniae wt- and A. pleuropneumoniae Δfrd mutant-infected animals was performed using the Wilcoxon signed-rank test, and results were considered significant when the P value was ≤0.05. Animals infected with the A. pleuropneumoniae wt and the A. pleuropneumoniae Δfrd mutant, respectively, were considered one group, independent of the day of necropsy, as half of the animals was sacrificed on day 7 and on day 21 in either group. The A. pleuropneumoniae wt-infected animal, which died on day 2 postinfection, was not included in order to not overassess the scores for the A. pleuropneumoniae wt group.
FIG. 4.
Histopathological examination of lung lesions. Sections were obtained from pigs necropsied on day 7 postinfection or on day 21 postinfection. Animals were infected with either the A. pleuropneumoniae wt or the A. pleuropneumoniae Δfrd mutant. A, alveole; B, bronchus; F, fibrin; IC, interstitial consolidation.
DISCUSSION
The role of central metabolic enzymes in bacterial virulence and persistence is an area of burgeoning research (19, 32, 34, 54). In particular, in view of gene conservation in many bacterial species, it may be a fruitful research field for the identification of potential targets for new therapeutic drugs. A suitable approach to defining metabolic pathways involved in virulence is the analysis of isogenic mutants in global regulators that are attenuated for virulence, such as ArcA. ArcA is an established global regulator that affects metabolic genes in Enterobacteriaceae (31, 46). The inactivation of arcA has been shown to cause attenuation in a number of pathogens, such as M. tuberculosis (45), V. cholerae (50), S. flexneri (7), H. influenzae (13, 59), and A. pleuropneumoniae (8). However, the consequences of arcA-induced changes of metabolic enzymes for virulence have not been investigated to date.
The ArcA regulon of A. pleuropneumoniae is considerably smaller than that of E. coli (31, 46). Possible reasons include the twofold-smaller genome size of A. pleuropneumoniae and the more-restricted environmental niche it inhabits, the porcine respiratory tract. In accordance with the ArcA regulon of its close relative H. influenzae (59), ArcA of A. pleuropneumoniae appears to act primarily as a repressor of transcription. In A. pleuropneumoniae, as well as in H. influenzae (59), the gene most strongly downregulated by the presence of ArcA was the l-lactate permease gene. This downregulation under anaerobic conditions is energetically efficient, since the transport is at the expense of the proton gradient and l-lactate seems not to be a favored substrate. Genes like the l-lactate dehydrogenase gene lldD or the formate dehydrogenase genes fdnI, fdxH, and fdhE were also downregulated in A. pleuropneumoniae as well as in H. influenzae by the presence of ArcA. However, overall, the ArcA regulon of A. pleuropneumoniae is larger than that described for H. influenzae (59), and there was no correlation between the genes upregulated by the presence of ArcA in both species. The three genes most strongly upregulated in A. pleuropneumoniae exhibit homology to a methylation subunit of a type III restriction modification system, a restriction enzyme, and a hypothetical DNA/RNA helicase. These genes are located next to each other in the genome of A. pleuropneumoniae and, to our knowledge, the homologues in other species have not previously been described as being regulated by ArcA. As in Shewanella oneidensis (21), genes encoding dimethyl sulfoxide reductase (dmsABC) and cytochrome d oxidase (cydAB) were up- and downregulated in the presence of ArcA, respectively. This contrasts with E. coli, where ArcA-dependent dms gene regulation is unknown and cydAB is upregulated by ArcA.
We analyzed the ArcA regulon of A. pleuropneumoniae with respect to its possible effect on virulence and persistence in the porcine respiratory tract. The results suggested an ArcA-dependent coordinated regulation of genes involved in the pathway leading to fumarate synthesis. This implied that fumarate is used as the terminal electron acceptor, with glycerol-3-phosphate being the most likely reduction equivalent (49). We assume that fumarate is synthesized from dihydroxyacetone phosphate, the oxidation product of glycerol-3-phosphate, which is processed during glycolysis, and that phosphoenolpyruvate is then introduced into the reductive branch of the citric acid cycle to generate fumarate. In support of this hypothesis, the transcription of glycerol-3-phosphate dehydrogenase was upregulated, whereas several competing steps (the pyruvate dehydrogenase, malate:quinone reductase, and malic enzyme steps) were strongly downregulated. These findings emphasize the necessity of a thorough pathway analysis, as genes encoding enzymes with crucial functions (phosphoenolpyruvate carboxylase, fumarate reductase) may be constitutively expressed.
The midpoint potentials for the glycerol-3-phosphate/dihydroxyacetone phosphate and the fumarate/succinate redox pairs are −0.19 V and 0 V, respectively. The coupling of these two systems has been calculated for E. coli to yield a −ΔG° of about 8.8 kcal (37). This energy facilitates the export of approximately two protons from the cytosol into the periplasm, thereby generating a proton gradient (38). If a stoichiometry of three to four protons is transported per ATP synthesized (56), the oxidation of two molecules of glycerol-3-phosphate coupled to the reduction of two molecules of fumarate leads to synthesis of at least one ATP by oxidative phosphorylation plus two ATPs by substrate chain phosphorylation. Importantly, the NAD+ balance is neutral, facilitating ongoing glycolysis.
A. pleuropneumoniae, H. pylori (43), and H. influenzae (NC_000907) lack citric acid cycle enzymes. In these organisms, the citric acid cycle functions as a noncyclic branched pathway, as has been reported for several anaerobic organisms (57). Homology analyses revealed that A. pleuropneumoniae lacks the initial steps of the oxidative branch of the TCA cycle but can synthesize TCA cycle intermediates through to α-ketoglutarate via the reductive branch. Therefore, in A. pleuropneumoniae, the TCA cycle appears to be directed toward (i) synthesis of the metabolic intermediates succinate, succinyl-CoA, and α-ketoglutarate, which cannot be produced by the TCA cycle upon deletion of frd; (ii) energy production by reduction of fumarate with glycerol-3-phosphate as the reduction equivalent (49), as proposed for H. pylori (35); and (iii) fermentation to achieve a neutral NAD+ balance (9).
In order to support a predicted role for fumarate reductase in virulence, A. pleuropneumoniae fumarate reductase was inactivated by deletion. The deletion mutant was attenuated for infection so that it caused less-severe lung lesions than the parent strain. An essential role for fumarate reductase in bacterial virulence has previously been described for H. pylori, where it was hypothesized that inactivation would abolish a crucial energy-producing source required to penetrate the mucus layer of the gastric epithelia (18). For A. pleuropneumoniae, the reduction of fumarate might likewise be an efficient energy source in its environment on the intact respiratory epithelium. In both environments, the gastric mucus and the epithelial lining fluid, phospholipids are abundant and phospholipases might provide a rich source of glycerol-3-phosphate (15), which is predicted to serve directly as the reduction equivalent.
The fumarate reductase of H. pylori is a potential therapeutic target (17, 36), but none of the three known inhibitory compounds available were suitable for therapeutic use (36). However, more recently, nafuredin, a novel noncommercial anthelminthic substance inhibiting fumarate reductase (55), has been used successfully to treat experimental Haemonchus contortus infections in sheep (40). Since higher eukaryotes do not carry homologues to the bacterial or helminthal fumarate reductase enzyme complex, nafuredin could have potential as a novel therapeutic drug to treat not only helminthal but also at least some bacterial infections.
Taken together, these data lead us to propose that fumarate reductase activity may be crucial not only for virulence of A. pleuropneumoniae but also for a number of other bacterial pathogens that effectively colonize mucous membranes in the respiratory and gastrointestinal tracts, by providing either energy by anaerobic respiration or important metabolic intermediates or both of the above. Therefore, it might be worthwhile to investigate the use of nafuredin or other fumarate reductase inhibitors as novel therapeutics for the treatment of bacterial infections of mucosal surfaces.
Supplementary Material
Acknowledgments
This work was supported by Sonderforschungsbereich 587 (project A4) of the Deutsche Forschungsgemeinschaft (DFG), Bonn, Germany. F.F.R.B. is a fellow of Graduate College 745 (project A1) of the DFG. I.M.B. was supported by the Ministry of Education, Libya. Funding for J.T.B. was provided by a grant to P.R.L. from the Biotechnology and Biological Sciences Research Council, United Kingdom (grant no. BBS/B/08132).
For histopathology, we thank Dirk Schaudien and Andreas Beineke from the Institute for Pathology, University of Veterinary Medicine Hannover, Hannover, Germany.
Editor: J. L. Flynn
Footnotes
Published ahead of print on 31 March 2008.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Anderson, C., A. A. Potter, and G.-F. Gerlach. 1991. Isolation and molecular characterization of spontaneously occurring cytolysin-negative mutants of Actinobacillus pleuropneumoniae serotype 7. Infect. Immun. 594110-4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashworth, J. M., and H. L. Kornberg. 1966. The anaplerotic fixation of carbon dioxide by Escherichia coli. Proc. R. Soc. Lond. B 165179-188. [DOI] [PubMed] [Google Scholar]
- 3.Baltes, N., I. Hennig-Pauka, I. Jacobsen, A. D. Gruber, and G.-F. Gerlach. 2003. Identification of dimethyl sulfoxide reductase in Actinobacillus pleuropneumoniae and its role in infection. Infect. Immun. 716784-6792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baltes, N., M. N′diaye, I. D. Jacobsen, A. Maas, F. F. Buettner, and G.-F. Gerlach. 2005. Deletion of the anaerobic regulator HlyX causes reduced colonization and persistence of Actinobacillus pleuropneumoniae in the porcine respiratory tract. Infect. Immun. 734614-4619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boonstra, J., J. A. Downie, and W. N. Konings. 1978. Energy supply for active transport in anaerobically grown Escherichia coli. J. Bacteriol. 136844-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosse, J. T., H. Janson, B. J. Sheehan, A. J. Beddek, A. N. Rycroft, K. J. Simon, and P. R. Langford. 2002. Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. 4225-235. [DOI] [PubMed] [Google Scholar]
- 7.Boulette, M. L., and S. M. Payne. 2007. Anaerobic regulation of Shigella flexneri virulence: ArcA regulates fur and iron acquisition genes. J. Bacteriol. 1896957-6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buettner, F. F., A. Maas, and G.-F. Gerlach. 2008. An Actinobacillus pleuropneumoniae arcA deletion mutant is attenuated and deficient in biofilm formation. Vet. Microbiol. 127106-115. [DOI] [PubMed] [Google Scholar]
- 9.Clark, D. P. 1989. The fermentation pathways of Escherichia coli. FEMS Microbiol. Rev. 5223-234. [DOI] [PubMed] [Google Scholar]
- 10.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 2841318-1322. [DOI] [PubMed] [Google Scholar]
- 11.Dehio, C., and M. Meyer. 1997. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179538-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deslandes, V., J. H. Nash, J. Harel, J. W. Coulton, and M. Jacques. 2007. Transcriptional profiling of Actinobacillus pleuropneumoniae under iron-restricted conditions. BMC Genomics 872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Souza-Hart, J. A., W. Blackstock, V. Di Modugno, I. B. Holland, and M. Kok. 2003. Two-component systems in Haemophilus influenzae: a regulatory role for ArcA in serum resistance. Infect. Immun. 71163-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dom, P., F. Haesebrouck, R. Ducatelle, and G. Charlier. 1994. In vivo association of Actinobacillus pleuropneumoniae serotype 2 with the respiratory epithelium of pigs. Infect. Immun. 621262-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dorrell, N., M. C. Martino, R. A. Stabler, S. J. Ward, Z. W. Zhang, A. A. McColm, M. J. Farthing, and B. W. Wren. 1999. Characterization of Helicobacter pylori PldA, a phospholipase with a role in colonization of the gastric mucosa. Gastroenterology 1171098-1104. [DOI] [PubMed] [Google Scholar]
- 16.Fenwick, B., and S. Henry. 1994. Porcine pleuropneumonia. J. Am. Vet. Med. Assoc. 2041334-1340. [PubMed] [Google Scholar]
- 17.Ge, Z. 2002. Potential of fumarate reductase as a novel therapeutic target in Helicobacter pylori infection. Expert Opin. Ther. Targets 6135-146. [DOI] [PubMed] [Google Scholar]
- 18.Ge, Z., Y. Feng, C. A. Dangler, S. Xu, N. S. Taylor, and J. G. Fox. 2000. Fumarate reductase is essential for Helicobacter pylori colonization of the mouse stomach. Microb. Pathog. 29279-287. [DOI] [PubMed] [Google Scholar]
- 19.Gerard, H. C., J. Freise, Z. Wang, G. Roberts, D. Rudy, B. Krauss-Opatz, L. Kohler, H. Zeidler, H. R. Schumacher, J. A. Whittum-Hudson, and A. P. Hudson. 2002. Chlamydia trachomatis genes whose products are related to energy metabolism are expressed differentially in active vs. persistent infection. Microbes Infect. 413-22. [DOI] [PubMed] [Google Scholar]
- 20.Goethe, R., O. F. Gonzales, T. Lindner, and G. F. Gerlach. 2000. A novel strategy for protective Actinobacillus pleuropneumoniae subunit vaccines: detergent extraction of cultures induced by iron restriction. Vaccine 19966-975. [DOI] [PubMed] [Google Scholar]
- 21.Gralnick, J. A., C. T. Brown, and D. K. Newman. 2005. Anaerobic regulation by an atypical Arc system in Shewanella oneidensis. Mol. Microbiol. 561347-1357. [DOI] [PubMed] [Google Scholar]
- 22.Hannan, P. C., B. S. Bhogal, and J. P. Fish. 1982. Tylosin tartrate and tiamutilin effects on experimental piglet pneumonia induced with pneumonic pig lung homogenate containing mycoplasmas, bacteria and viruses. Res. Vet. Sci. 3376-88. [PubMed] [Google Scholar]
- 23.Ingledew, W. J., and R. K. Poole. 1984. The respiratory chains of Escherichia coli. Microbiol. Rev. 48222-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iuchi, S., and E. C. Lin. 1988. arcA (dye), a global regulatory gene in Escherichia coli mediating repression of enzymes in aerobic pathways. Proc. Natl. Acad. Sci. USA 851888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobs, N. J., and P. J. VanDemark. 1960. Comparison of the mechanism of glycerol oxidation in aerobically and anaerobically grown Streptococcus faecalis. J. Bacteriol. 79532-538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacobsen, I., I. Hennig-Pauka, N. Baltes, M. Trost, and G. F. Gerlach. 2005. Enzymes involved in anaerobic respiration appear to play a role in Actinobacillus pleuropneumoniae virulence. Infect. Immun. 73226-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jacobsen, M. J., and J. P. Nielsen. 1995. Development and evaluation of a selective and indicative medium for isolation of Actinobacillus pleuropneumoniae from tonsils. Vet. Microbiol. 47191-197. [DOI] [PubMed] [Google Scholar]
- 28.Kather, B., K. Stingl, M. E. van der Rest, K. Altendorf, and D. Molenaar. 2000. Another unusual type of citric acid cycle enzyme in Helicobacter pylori: the malate:quinone oxidoreductase. J. Bacteriol. 1823204-3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kramer, A., I. Schwebke, and G. Kampf. 2006. How long do nosocomial pathogens persist on inanimate surfaces? A systematic review. BMC Infect. Dis. 6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leiner, G., B. Franz, K. Strutzberg, and G. F. Gerlach. 1999. A novel enzyme-linked immunosorbent assay using the recombinant Actinobacillus pleuropneumoniae ApxII antigen for diagnosis of pleuropneumonia in pig herds. Clin. Diagn. Lab. Immunol. 6630-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu, X., and P. De Wulf. 2004. Probing the ArcA-P modulon of Escherichia coli by whole genome transcriptional analysis and sequence recognition profiling. J. Biol. Chem. 27912588-12597. [DOI] [PubMed] [Google Scholar]
- 32.Lorenz, M. C., and G. R. Fink. 2001. The glyoxylate cycle is required for fungal virulence. Nature 41283-86. [DOI] [PubMed] [Google Scholar]
- 33.Maas, A., I. D. Jacobsen, J. Meens, and G. F. Gerlach. 2006. Use of an Actinobacillus pleuropneumoniae multiple mutant as a vaccine that allows differentiation of vaccinated and infected animals. Infect. Immun. 744124-4132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKinney, J. D., K. Honer zu Bentrup, E. J. Muñoz-Elías, A. Miczak, B. Chen, W.-T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406735-738. [DOI] [PubMed] [Google Scholar]
- 35.Mendz, G. L., and S. L. Hazell. 1993. Fumarate catabolism in Helicobacter pylori. Biochem. Mol. Biol. Int. 31325-332. [PubMed] [Google Scholar]
- 36.Mendz, G. L., S. L. Hazell, and S. Srinivasan. 1995. Fumarate reductase: a target for therapeutic intervention against Helicobacter pylori. Arch. Biochem. Biophys. 321153-159. [DOI] [PubMed] [Google Scholar]
- 37.Miki, K., and E. C. Lin. 1975. Anaerobic energy-yielding reaction associated with transhydrogenation from glycerol 3-phosphate to fumarate by an Escherichia coli system. J. Bacteriol. 1241282-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miki, K., and T. H. Wilson. 1978. Proton translocation associated with anaerobic transhydrogenation from glycerol 3-phosphate to fumarate in Escherichia coli. Biochem. Biophys. Res. Commun. 831570-1575. [DOI] [PubMed] [Google Scholar]
- 39.Molenaar, D., M. E. van der Rest, A. Drysch, and R. Yucel. 2000. Functions of the membrane-associated and cytoplasmic malate dehydrogenases in the citric acid cycle of Corynebacterium glutamicum. J. Bacteriol. 1826884-6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Omura, S., H. Miyadera, H. Ui, K. Shiomi, Y. Yamaguchi, R. Masuma, T. Nagamitsu, D. Takano, T. Sunazuka, A. Harder, H. Kolbl, M. Namikoshi, H. Miyoshi, K. Sakamoto, and K. Kita. 2001. An anthelmintic compound, nafuredin, shows selective inhibition of complex I in helminth mitochondria. Proc. Natl. Acad. Sci. USA 9860-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oswald, W., W. Tonpitak, G. Ohrt, and G.-F. Gerlach. 1999. A single-step transconjugation system for the introduction of unmarked deletions into Actinobacillus pleuropneumoniae serotype 7 using a sucrose sensitivity marker. FEMS Microbiol. Lett. 179153-160. [DOI] [PubMed] [Google Scholar]
- 42.Parish, T., D. A. Smith, G. Roberts, J. Betts, and N. G. Stoker. 2003. The senX3-regX3 two-component regulatory system of Mycobacterium tuberculosis is required for virulence. Microbiology 1491423-1435. [DOI] [PubMed] [Google Scholar]
- 43.Pitson, S. M., G. L. Mendz, S. Srinivasan, and S. L. Hazell. 1999. The tricarboxylic acid cycle of Helicobacter pylori. Eur. J. Biochem. 260258-267. [DOI] [PubMed] [Google Scholar]
- 44.Raleigh, F. A., K. Lech, and R. Brent. 1989. Selected topics from classical bacterial genetics, p.1.4.1-1.4.14. In F. M. Ausubel, R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, et al. (ed.), Current protocols in molecular biology. Wiley Interscience, New York, NY. [DOI] [PubMed]
- 45.Rickman, L., J. W. Saldanha, D. M. Hunt, D. N. Hoar, M. J. Colston, J. B. Millar, and R. S. Buxton. 2004. A two-component signal transduction system with a PAS domain-containing sensor is required for virulence of Mycobacterium tuberculosis in mice. Biochem. Biophys. Res. Commun. 314259-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Salmon, K. A., S. P. Hung, N. R. Steffen, R. Krupp, P. Baldi, G. W. Hatfield, and R. P. Gunsalus. 2005. Global gene expression profiling in Escherichia coli K12: effects of oxygen availability and ArcA. J. Biol. Chem. 28015084-15096. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Sawers, G., and B. Suppmann. 1992. Anaerobic induction of pyruvate formate-lyase gene expression is mediated by the ArcA and FNR proteins. J. Bacteriol. 1743474-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schryvers, A., and J. H. Weiner. 1981. The anaerobic sn-glycerol-3-phosphate dehydrogenase of Escherichia coli: purification and characterization. J. Biol. Chem. 2569959-9965. [PubMed] [Google Scholar]
- 50.Sengupta, N., K. Paul, and R. Chowdhury. 2003. The global regulator ArcA modulates expression of virulence factors in Vibrio cholerae. Infect. Immun. 715583-5589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shevchenko, A., M. Wilm, O. Vorm, and M. Mann. 1996. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68850-858. [DOI] [PubMed] [Google Scholar]
- 52.Shuman, S. 1994. Novel approach to molecular cloning and polynucleotide synthesis using vaccinia DNA topoisomerase. J. Biol. Chem. 26932678-32684. [PubMed] [Google Scholar]
- 53.Tatusov, R. L., E. V. Koonin, and D. J. Lipman. 1997. A genomic perspective on protein families. Science 278631-637. [DOI] [PubMed] [Google Scholar]
- 54.Tchawa Yimga, M., M. P. Leatham, J. H. Allen, D. C. Laux, T. Conway, and P. S. Cohen. 2006. Role of gluconeogenesis and the tricarboxylic acid cycle in the virulence of Salmonella enterica serovar Typhimurium in BALB/c mice. Infect. Immun. 741130-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ui, H., K. Shiomi, Y. Yamaguchi, R. Masuma, T. Nagamitsu, D. Takano, T. Sunazuka, M. Namikoshi, and S. Omura. 2001. Nafuredin, a novel inhibitor of NADH-fumarate reductase, produced by Aspergillus niger FT-0554. J. Antibiot. (Tokyo) 54234-238. [DOI] [PubMed] [Google Scholar]
- 56.Van Walraven, H. S., H. Strotmann, O. Schwarz, and B. Rumberg. 1996. The H+/ATP coupling ratio of the ATP synthase from thiol-modulated chloroplasts and two cyanobacterial strains is four. FEBS Lett. 379309-313. [DOI] [PubMed] [Google Scholar]
- 57.Weitzman, P. D. 1987. Patterns of diversity of citric acid cycle enzymes. Biochem. Soc. Symp. 5433-43. [PubMed] [Google Scholar]
- 58.Willson, P. J., W. L. Albritton, L. Slaney, and J. K. Setlow. 1989. Characterization of a multiple antibiotic resistance plasmid from Haemophilus ducreyi. Antimicrob. Agents Chemother. 331627-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wong, S. M., K. R. Alugupalli, S. Ram, and B. J. Akerley. 2007. The ArcA regulon and oxidative stress resistance in Haemophilus influenzae. Mol. Microbiol. 641375-1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.