Abstract
Plasmodium falciparum apical membrane antigen 1 (PfAMA1), a candidate malaria vaccine, is polymorphic. This polymorphism is believed to be generated predominantly under immune selection pressure and, as a result, may compromise attempts at vaccination. Alignment of 355 PfAMA1 sequences shows that around 10% of the 622 amino acid residues can vary between alleles and that linkages between polymorphic residues occur. Using this analysis, we have designed three diversity-covering (DiCo) PfAMA1 sequences that take account of these linkages and, when taken together, on average incorporate 97% of amino acid variability observed. For each of the three DiCo sequences, a synthetic gene was constructed and used to transform the methylotrophic yeast Pichia pastoris, allowing recombinant expression. All three DiCo proteins were reactive with the reduction-sensitive monoclonal antibody 4G2, suggesting the DiCo sequences had conformations similar to those of naturally occurring PfAMA1. Rabbits were immunized with FVO strain PfAMA1 or with the DiCo proteins either individually or as a mixture. Antibody titers and the ability to inhibit parasite growth in vitro were determined. Animals immunized with the DiCo mix performed similarly to animals immunized with FVO AMA1 when measured against FCR3 strain parasites but outperformed animals immunized with FVO AMA1 when assessed against other strains. The levels of growth inhibition (∼70%) induced by the mix of three DiCo proteins were comparable for FVO, 3D7, and HB3, suggesting that a considerable degree of diversity in AMA1 is adequately covered. This suggests that vaccines based upon the DiCo mix approach provide a broader functional immunity than immunization with a single allele.
Malaria is estimated to cause up to 500 million clinical cases and 2 million deaths annually (28). Most of the severe morbidity and mortality occurs through infection with Plasmodium falciparum in young children and pregnant women of sub-Saharan Africa (30). A vaccine that prevents or reduces infection and minimizes morbidity and mortality would be a very useful additional tool for control and prevention programs. Several potential molecular targets for vaccine development have been identified, one of these being P. falciparum apical membrane antigen 1 (PfAMA1). The evidence for PfAMA1 as a vaccine target has recently been reviewed (27). Briefly, PfAMA1 is encoded by an essential single-copy gene (35), evidence from rodent and nonhuman primate malaria models shows that antibody responses to AMA1 can reduce levels of infection (1, 4, 6, 8, 9, 19), and antibodies to PfAMA1 inhibit asexual parasite multiplication in vitro (12, 15, 16). In areas where malaria is endemic, humans make anti-AMA1 antibodies in response to infection (7, 14, 26, 33) and these may correlate with resistance to clinical malaria (26).
PfAMA1 is initially expressed as an 83-kDa protein, comprising a large N-terminal ectodomain, a transmembrane region, and an ∼50-amino-acid C-terminal cytoplasmic tail (20). The ectodomain contains 16 conserved cysteine residues that form eight intramolecular disulfide bonds; these were used to define a potential three-domain structure (13). The recent elucidation of crystal structures for AMA1 (3, 5, 24) confirms the three-domain structure and shows there is considerable interaction between the domains. Antibody to AMA1 blocks merozoite invasion of erythrocytes (32) and merozoite reorientation at the erythrocyte surface (17), and asexual blood-stage parasites devoid of AMA1 appear not to be viable (35), suggesting that AMA1 plays a critical and nonredundant role in erythrocyte invasion. AMA1 is also present in sporozoite and liver stages of development (29), suggesting that vaccination with AMA1 may target more than just asexual erythrocytic development.
AMA1 has long been known to be polymorphic (34), although unlike many other malarial surface proteins, this polymorphism is not due to repetitive sequence but is entirely due to single amino acid substitutions (5). Where they have been mapped, polymorphisms have been found to be restricted to the surface of AMA1, mapping predominantly to one molecular face (3, 5, 24). Studies of the rodent malaria parasite Plasmodium chabaudi have shown how polymorphism in AMA1 may negatively affect vaccine outcomes (8). Furthermore, immunization studies of rabbits have shown that although antibodies to PfAMA1 obtained from one strain of malaria inhibit the growth of the homologous strain well, other strains are inhibited to various lesser degrees (12, 15, 16). This suggests that PfAMA1 polymorphism may diminish the efficacy of PfAMA1-based vaccines and that the most effective AMA1 vaccines will induce responses to conserved determinants as well as to the broadest possible range of variable determinants.
Cost constraints mean that only a limited number of components can be developed for any one vaccine. As an approach to this, we have designed, produced, and immunologically evaluated a limited number of artificial AMA1 sequences that share conserved amino acids while covering a maximal degree of polymorphism. By using only three PfAMA1 sequences, we have been able to include an average of 97% of the naturally occurring amino acid substitutions.
MATERIALS AND METHODS
Cloning of DiCo AMA1 sequences in P. pastoris.
Three synthetic diversity-covering (DiCo) genes, DiCo1, DiCo2, and DiCo3, with optimized codon usage for Pichia pastoris were designed, essentially comprising domains I, II, and III of PfAMA1 (DNA2.0, San Diego, CA). These sequences were PCR amplified with primers X1 (5′ GCGAATTCATTGAAATTGTTGAAAGATC 3′) and Y1 (5′ GGGGTACCAACATCTTATCGTAAGTTGG 3′), X1 and Y2 (5′ GGGGTACCGACATGTTATCGTAAGTTGGC 3′), and X1 and Y1 for DiCo1, -2, and -3, respectively. The PCR products were cloned into the EcoRI-KpnI sites of the pPicZαA vector (Invitrogen, Groningen, The Netherlands) and used to transform Escherichia coli DH5α cells. After transformation of the E. coli DH5α cells, plasmids were isolated, checked for the presence of the expected restriction sites, and then used to transform P. pastoris KM71H according to the manufacturer's protocols. Transfected P. pastoris was tested for protein production by culture in 10 ml glycerol-containing medium in a 50-ml tube for 48 h at 29 to 30°C under conditions of vigorous shaking. Cells were harvested by centrifugation (5 min, 2,500 rpm, tabletop centrifuge), resuspended in 4 ml methanol-containing medium, and then cultured for 24 h at 29 to 30°C under conditions of vigorous shaking. After low-speed centrifugation, the culture supernatant was harvested and 20 μl was tested on a sodium dodecyl sulfate-polyacrylamide gel for proteins with the expected size. The identity was confirmed by Western blotting with the reduction-sensitive monoclonal antibody 4G2.
Protein production.
Fermentation runs were performed in either 3- or 7-liter fermentors (Applikon, Schiedam, The Netherlands), with initial starting volumes of 1 or 2 liters, respectively. The P. pastoris clones were grown in BMGY (1% yeast extract, 2% peptone, 1.34% yeast nitrogen base, 1% glycerol, 0.4 mg of biotin per liter, 0.1 M K phosphate, pH 6.0) at 30°C for 24 h. An amount of 50 ml/liter was used to inoculate the fermentor containing minimal salt fermentation medium ([per liter of medium] MgSO4·7H2O, 14.9 g; K2SO4, 18.2 g; CaCl2, 0.65 g; KOH, 4.13 g; glycerol, 40 g; 26.7 ml 85% H3PO4; and 12 ml PTM1 trace salt solution) (www.invitrogen.com/content/sfs/manuals/pichiaferm_prot.pdf). During the batch phase, air was sparged at 1 liter per liter initial medium volume. The temperature was kept constant throughout the fermentation at 30°C, and the pH was kept at 6.0, using 25% NH4OH or 85% H3PO4. The fed batch was started after dissolved oxygen levels returned to 21% after being down to (almost) zero, generally between 18 and 24 h. Subsequently, the culture was fed 50% glycerol fortified with 12 ml/liter PTM1 trace salts at a rate of 32 ml/h per liter initial medium volume for 20 to 24 h. During this fed-batch phase of the fermentation, the medium was sparged with 100% oxygen to keep the dissolved oxygen concentration at 21%. Last, the culture was induced by the addition of methanol, for the first 3 h at a rate of 1 ml/h per liter initial medium volume, increasing to 3 ml/h per liter initial medium volume at 3 h, and continuing at the latter rate for approximately 18 h. During the induction phase, in addition to sparging 100% oxygen, air was continuously sparged (at 1 liter per liter initial fermentation medium), as we found that use of pure oxygen alone reduced or prevented expression of some proteins. After induction, the pH of the medium was increased to 7.8 and cooled with a cryostat to 15°C or lower. Cells were removed by centrifugation (25 min, 5,000 × g, 4°C), and culture medium was filtered through a 0.22-μm filter by using a Quixstand hollow-fiber cartridge (GE Healthcare, Etten-Leur, The Netherlands) to remove all remaining yeast cells. Protein in the culture supernatant was concentrated to approximately 100 ml by using a Quixstand cartridge with a 10-kDa-cutoff hollow-fiber column and then diluted with an equal volume of demineralized water and concentrated again, a procedure repeated four times. Proteins were subsequently bound to a hydroxyapatite column (Bio-Rad type I; Bio-Rad Laboratories, Veenendaal, The Netherlands); equilibrated with 1 mM sodium phosphate buffer, pH 8.0; and eluted with 50 mM sodium phosphate buffer, pH 8.0. This fraction was concentrated to less than 1 ml and subjected to Superdex 75 preparative size exclusion chromatography (26/60 prep grade; GE Healthcare, Etten-Leur, The Netherlands). Fractions containing DiCo protein were pooled, concentrated, and sterilized by filtration (0.22 μm).
Rabbit immunizations.
Rabbits were housed and immunized and blood was sampled by Eurogentec SA, Seraing, Belgium, according to national animal welfare regulations. Five groups of five rabbits were immunized on days 0, 28, and 56 with Pichia-expressed DiCo1 (30 μg), DiCo2 (30 μg), or DiCo3 (30 μg); FVO PfAMA1 D123 (30 μg); or a mixture of DiCo1, DiCo2, and DiCo3 (10 μg each, for a total of 30 μg). Montanide ISA 51 (Seppic, Paris, France) was used as the adjuvant (2). Vaccine formulations were prepared according to the manufacturer's instructions (50/50, mass/mass). Antisera obtained 2 weeks after the third immunization (day 70) were tested for reactivity by an enzyme-linked immunosorbent assay (ELISA) and for functional capacity by an in vitro growth inhibition assay (GIA).
ELISA.
ELISA was performed in duplicate on serum samples in 96-well flat-bottomed microtiter plates (Greiner, Alphen a/d Rijn, The Netherlands) coated with 500 ng/ml purified AMA1 antigens according to published methods (16). The secondary antibody was anti-rabbit immunoglobulin G (IgG) conjugated to alkaline phosphatase (Pierce, Rockford, IL). A standard curve was included on each plate, and titers of the unknowns were calculated by a four-parameter fit. Titers are expressed as arbitrary units (AU), where 1 AU yields an optical density (OD) of 1.0. Thus, the amount of AU of a sample is the reciprocal dilution at which an OD at 405 nm of 1 will be achieved.
IgG purification.
Antibodies to be used for GIAs were purified on protein A columns (Sigma, St. Louis, MO) by using standard protocols, exchanged into RPMI 1640 by using Amicon Ultra-15 concentrators (30-kDa cutoff; Millipore, Ireland), filter sterilized, and stored at −20°C until use. IgG concentrations were determined using a Nanodrop ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE).
Parasites.
P. falciparum strains 3D7, FCR3, and HB3 were cultured in vitro using standard P. falciparum culture techniques in an atmosphere of 5% CO2, 5% O2, and 90% N2. FCR3 AMA1 (GenBank accession no. M34553) (34) differs by 1 amino acid (aa) in the prosequence from that of FVO AMA1 (GenBank accession no. AJ277646) (16).
In vitro GIA.
The effect of purified IgG antibodies on parasite invasion was evaluated in triplicate using 96-well flat-bottomed plates (Greiner) with in vitro-matured and -synchronized P. falciparum schizonts at a starting parasitemia of 0.2 to 0.4%, a hematocrit of 2.0%, and a final volume of 100 μl containing 10% normal human serum, 20 μg ml−1 gentamicin in RPMI 1640. After 40 to 42 h, cultures were resuspended, and 50 μl was transferred into 200 μl ice-cold phosphate-buffered saline. The cultures were then centrifuged, the supernatant was removed, and the plates were frozen. Inhibition of parasite growth was estimated using a parasite lactate dehydrogenase assay as previously described (15). Parasite growth inhibition, reported as a percentage, was calculated as follows: 100 − [(ODexperimental − ODbackground)/(ODcontrol − ODbackground) × 100]. Control IgG was isolated from rabbits that had been immunized with adjuvant only. All GIA results reported are the averages of two independent GIA runs. The AMA1 expressed by each of the laboratory strains was verified to be correct by restriction fragment length polymorphism analysis, and all parasite cultures used for GIA tested negative for mycoplasma by PCR.
Protein sequence analysis.
All available AMA1 sequences were aligned; variable amino acid positions and their relative frequencies were identified from the alignment. Linkages, both in the N- and in the C-terminal direction, were determined by separate variable amino acid analyses for each of the potential residues at the variable positions; for example, at position 162 (dimorphic with N or K), the frequencies of N- and C-terminal variable residues were determined for all sequences with an N and all sequences with a K. Positions were considered linked at a frequency of 99% or higher.
Statistical analyses.
All statistical analyses were performed using the R statistics package, version 2.6 (R Foundation for Statistical Computing, Vienna, Austria). IgG titers were compared with analysis of variance (ANOVA), using log-transformed IgG titers as the dependent variable and vaccine antigen as the independent variable. GIA titers were analyzed with nonlinear mixed-effect (NLME) models (23) to allow for the simultaneous comparison of GIA titers at various IgG concentrations while correcting for pseudoreplication (22). A logistic fit was applied for the GIA titer versus the log2-transformed IgG concentration. The upper and lower asymptotes for the logistic curve were set at 0 and 100%, respectively, and slope and midpoint were estimated by restricted maximum likelihood. The parameterization used was 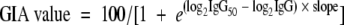 , where IgG50 is the value of the IgG concentration where 50% inhibition is achieved, IgG is the concentration of IgG used in the GIA (0.75, 1.5, 3, and 6 mg/ml, respectively), and slope is a parameter describing the increase in GIA value with increasing IgG concentration, with higher values indicating steeper curves. Various NLME models with different random-effect structures and fixed effects for treatment on slope or IgG50 were fitted for each of the strains tested by GIA, and the best-fitting model was selected based on Akaike's information criterion. In the event the best-fitting models included treatment effects, the statistical significance thereof was evaluated further by ANOVA. To enable easier interpretation of the originally log2-transformed IgG50 values, IgG50 values are presented as
, where IgG50 is the value of the IgG concentration where 50% inhibition is achieved, IgG is the concentration of IgG used in the GIA (0.75, 1.5, 3, and 6 mg/ml, respectively), and slope is a parameter describing the increase in GIA value with increasing IgG concentration, with higher values indicating steeper curves. Various NLME models with different random-effect structures and fixed effects for treatment on slope or IgG50 were fitted for each of the strains tested by GIA, and the best-fitting model was selected based on Akaike's information criterion. In the event the best-fitting models included treatment effects, the statistical significance thereof was evaluated further by ANOVA. To enable easier interpretation of the originally log2-transformed IgG50 values, IgG50 values are presented as 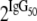 with 95% confidence intervals. Slope values are presented as untransformed estimates with 95% confidence intervals. The statistical significance of group differences in slope and IgG50 is expressed as the P value obtained from the NLME models, without correction for multiple comparisons because the ANOVA P value for treatment effects indicates whether at least two of the treatment groups differ significantly.
with 95% confidence intervals. Slope values are presented as untransformed estimates with 95% confidence intervals. The statistical significance of group differences in slope and IgG50 is expressed as the P value obtained from the NLME models, without correction for multiple comparisons because the ANOVA P value for treatment effects indicates whether at least two of the treatment groups differ significantly.
RESULTS
Analysis of polymorphisms in AMA variants.
All P. falciparum AMA1 sequences, complete sequences and fragments, available on 3 January 2005 (360 in total), were retrieved from a nucleotide search of the PubMed database (www.ncbi.nlm.nih.gov/). Duplicates were removed (PFU84348 [3D7], AU087598 [FVO], PFU33274 [3D7], and AF061332 [KF1916]), and two partial sequences for the NF7 strain were combined (PFU33280 and PFAAMA1B). The ensuing 355 sequences were aligned and polymorphic residues identified. To reduce the risk of incorporating data resultant from sequencing errors, polymorphic positions were defined as those that varied between sequences such that two or more of the 355 sequences carried the same amino acid substitution at that site. Using this definition, 64 of 622 amino acid positions were designated polymorphic: nine in the prosequence (aa 1 to 96; 9.4%), 32 in domain I (aa 97 to 303; 15.5%), 11 in domain II (aa 304 to 440; 8.0%), nine in domain III (aa 441 to 546; 8.5%), none in the transmembrane region (aa 547 to 567; 0%), and three in the cytoplasmic tail (aa 568 to 622; 5.5%). The alignment also showed that the presence of a particular amino acid at one polymorphic position was often linked to the presence of particular amino acids down- as well as upstream (i.e., in the direction of the N terminus or the C terminus, respectively) of the protein. Linkages were determined for all residues that occurred in 10% or more of the sequences. Downstream linkage was observed for 24 residues at 22 polymorphic sites (Table 1), The most-N-terminal member of each downstream linkage group tended not to be the predominant residue at that position (for 21 out of 24 linked residues) but tended to be linked to downstream residues that were the predominant (consensus) residue. At position 197, two of the possible amino acids (G and D) showed downstream linkage. As exceptions, in both position 172 and position 283 the predominant residue linked to downstream residues and in position 200 both the predominant and the minor residues are linked.
TABLE 1.
Downstream-linked residues determined from the AMA1 alignment
| Linking parameter
|
Consensus residue at linked positiona:
|
||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Residue | Frequency | 167 T | 172 E | 173 N | 175 D | 187 N | 190 M | 196 D | 200 D | 204 D | 206 E | 207 Y | 225 N | 230 K | 243 K | 283 S | 285 Q | 296 D | 308 E | 332 N | 407 Q | 448 D | 451 K | 485 K |
| 162 | N | 0.77 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − |
| K | 0.23 | T | E | N | D* | − | − | − | − | − | − | − | − | − | − | − | − | D | − | − | − | − | − | − | |
| 167 | T | 0.74 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| K | 0.26 | G | − | D* | − | − | − | − | − | E | Y* | N* | − | − | − | − | − | − | − | − | − | − | − | ||
| 172 | E | 0.55 | − | D* | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| G | 0.45 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 173 | N | 0.86 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| K | 0.14 | D | N | − | − | − | − | E | − | − | − | − | − | − | − | − | N | − | − | − | − | ||||
| 187 | N | 0.39 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||
| E | 0.37 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||
| K | 0.24 | M* | D* | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||
| 190 | M | 0.67 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||
| I | 0.33 | − | − | − | E | − | N* | − | − | − | − | − | − | − | − | − | − | − | |||||||
| 196 | D | 0.74 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||||
| N | 0.25 | D | − | E | − | N | − | − | − | − | − | − | N | − | − | − | − | ||||||||
| 197 | Q | 0.33 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||||
| G | 0.24 | D | − | E | − | N | − | − | − | − | − | − | N | − | − | − | − | ||||||||
| D | 0.24 | − | − | E | − | − | − | − | − | − | − | − | N | − | − | − | − | ||||||||
| 200 | D | 0.45 | − | E | − | N* | − | − | − | − | − | − | − | − | − | − | − | ||||||||
| H | 0.40 | D | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||||||
| 201 | F | 0.79 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||||
| L | 0.19 | − | E | − | − | − | − | − | − | − | − | N | − | − | − | − | |||||||||
| 204 | D | 0.58 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||||||
| N | 0.42 | E | − | N | − | − | − | − | − | − | − | − | − | − | − | ||||||||||
| 206 | E | 0.78 | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||||||
| K | 0.22 | Y | − | K | − | − | − | D* | − | − | − | − | − | − | |||||||||||
| 225 | N | 0.68 | − | − | − | − | − | − | − | − | − | − | − | ||||||||||||
| I | 0.32 | K | − | − | − | − | − | − | − | − | − | − | |||||||||||||
| 242 | Y | 0.52 | − | − | − | − | − | − | − | − | − | − | |||||||||||||
| D | 0.48 | K | − | − | − | − | − | − | − | − | − | ||||||||||||||
| 243 | K | 0.63 | − | − | − | − | − | − | − | − | − | ||||||||||||||
| N | 0.20 | − | − | − | E | N | − | − | − | − | |||||||||||||||
| E | 0.17 | − | − | − | − | − | − | − | − | − | |||||||||||||||
| 282 | K | 0.67 | − | − | − | − | − | − | − | − | − | ||||||||||||||
| I | 0.33 | S | Q | − | − | − | − | − | − | − | |||||||||||||||
| 283 | S | 0.70 | Q | − | − | − | − | − | − | − | |||||||||||||||
| L | 0.30 | − | − | − | − | − | − | − | − | ||||||||||||||||
| 296 | D | 0.80 | − | − | − | − | − | − | |||||||||||||||||
| H | 0.20 | − | − | − | D | − | − | ||||||||||||||||||
| 332 | N | 0.83 | − | − | − | − | |||||||||||||||||||
| I | 0.17 | Q | − | − | − | ||||||||||||||||||||
| 435 | I | 0.78 | − | − | − | ||||||||||||||||||||
| N | 0.20 | D | K | − | |||||||||||||||||||||
| 448 | D | 0.81 | − | − | |||||||||||||||||||||
| N | 0.19 | M | K | ||||||||||||||||||||||
| 451 | K | 0.59 | − | ||||||||||||||||||||||
| M | 0.41 | K | |||||||||||||||||||||||
An asterisk indicates that the frequency of the designated residue is greater than 99% and less than 100%. A minus sign (−) indicates the absence of linkage for the specified positions.
Upstream linkage was observed for 26 residues at 22 polymorphic sites (Table 2). The most-C-terminal member of the linkage group tended not to be the predominant residue at that position, and the linked upstream residues were generally the predominant (consensus) residues (for 23 out of 26 linked residues). Exceptionally, at position 197, all residues showed upstream linkage with position 196. The E at position 285 showed an unusual upstream linkage in that it is linked to the nonconsensus L283, and R503 is upstream linked to the nonconsensus residue M496.
TABLE 2.
Upstream-linked residues determined from the AMA1 alignment
| Linking parameter
|
Consensus residue at linked positiona:
|
|||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Position | Residue | Frequency | 162 N | 167 T | 172 E | 173 N | 175 D | 187 N | 190 M | 196 D | 200 D | 201 F | 204 D | 206 E | 207 Y | 225 N | 230 K | 242 Y | 282 K | 283 S | 296 D | 407 Q | 435 I | 448 D | 496 I | 503 N |
| 167 | T | 0.74 | − | |||||||||||||||||||||||
| K | 0.26 | N | ||||||||||||||||||||||||
| 172 | E | 0.55 | − | − | ||||||||||||||||||||||
| G | 0.45 | N | − | |||||||||||||||||||||||
| 173 | N | 0.86 | − | − | − | |||||||||||||||||||||
| K | 0.14 | N | − | − | ||||||||||||||||||||||
| 175 | D | 0.90 | − | − | − | − | ||||||||||||||||||||
| Y | 0.10 | − | − | − | N | |||||||||||||||||||||
| 187 | N | 0.39 | − | − | − | − | − | |||||||||||||||||||
| E | 0.37 | − | − | − | N | − | ||||||||||||||||||||
| K | 0.24 | N | T | E | N | D | ||||||||||||||||||||
| 197 | Q | 0.33 | − | − | − | − | D | − | − | D | ||||||||||||||||
| G | 0.24 | − | − | − | − | − | − | − | N* | |||||||||||||||||
| D | 0.24 | − | − | − | − | − | − | − | D* | |||||||||||||||||
| 200 | D | 0.45 | − | − | − | − | − | − | − | − | ||||||||||||||||
| H | 0.41 | − | T* | − | − | − | − | − | D | |||||||||||||||||
| 206 | E | 0.78 | − | − | − | − | − | − | − | − | − | − | − | |||||||||||||
| K | 0.22 | − | T | − | N | − | − | M | D | H | F | D | ||||||||||||||
| 207 | Y | 0.84 | − | − | − | − | − | − | − | − | − | − | − | − | ||||||||||||
| D | 0.16 | − | − | − | − | − | − | − | − | − | − | − | E | |||||||||||||
| 225 | N | 0.68 | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||||||||
| I | 0.32 | − | T* | − | − | − | − | − | D* | − | − | D | − | − | ||||||||||||
| 230 | K | 0.67 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||||||
| E | 0.27 | − | − | − | − | D | − | − | − | − | − | − | E | − | N | |||||||||||
| 243 | K | 0.63 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||||
| N | 0.20 | − | − | − | N | − | − | − | − | − | − | − | E* | − | N | K* | Y | |||||||||
| E | 0.17 | − | − | − | N | D | − | − | − | − | − | − | − | − | − | − | Y | |||||||||
| 282 | K | 0.67 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||||
| I | 0.33 | − | − | − | − | − | − | − | − | − | − | − | − | Y | − | − | − | − | ||||||||
| 283 | S | 0.70 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||||
| L | 0.30 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | K | ||||||||
| 285 | Q | 0.79 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||
| E | 0.21 | − | − | − | N* | − | − | − | − | − | − | − | − | − | − | − | − | K | L | |||||||
| 296 | D | 0.80 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||||
| H | 0.20 | N | − | − | − | − | − | − | − | − | − | − | E* | − | − | − | − | − | − | |||||||
| 332 | N | 0.83 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||||
| I | 0.17 | − | − | − | N | − | − | − | D | − | − | − | − | − | − | − | − | − | − | − | ||||||
| 435 | I | 0.78 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||||
| N | 0.20 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | Q | |||||
| 448 | D | 0.81 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| N | 0.19 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | D | − | I | ||||
| 451 | K | 0.59 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | D | ||
| M | 0.41 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |||
| 503 | N | 0.58 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
| R | 0.41 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | M | ||
| 512 | K | 0.53 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | N* |
| R | 0.47 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | |
An asterisk indicates that the frequency of the designated residue is greater than 99% and less than 100%. A minus sign (−) indicates the absence of linkage for the specified positions.
All variable residues had the same codon usage, with the exception of position 201 F (TTT, 79%), which is the consensus residue, and L, the subdominant residue, with three different codons used (CTT, TTG, and TTA). Position 228 N is the consensus residue (AAT, 97%) and K the subdominant residue with two different codons used (AAA and AAG).
Design of artificial genes to cover diversity and incorporate linkages.
To reduce the complexity of downstream vaccine development, it was considered reasonable that a maximum of three DiCo sequences could be accommodated. These DiCo sequences (DiCo1, DiCo2, and DiCo3), each of which comprised domains I, II, and III (aa 97 to 545), were designed such that, when taken together, the maximal number of naturally occurring residues was incorporated, with the proviso that residues with the lowest frequencies were linked, thereby restricting the variability in these sequences. Thus, at least 80% of amino acids present at any given position in naturally occurring sequences and most of the linkages that we have identified are included in the DiCo1, -2, and -3 combination. To restrict it to three DiCo sequences, 12 positions at which less than 10% of the sequences showed variation (residues 121 [99% E], 189 [92% L], 199 [99% R], 224 [99% M], 228 [97% N], 244 [92% D], 245 [96% K], 269 [96% K], 325 [98% H], 330 [91% S], 395 [92% K], and 505 [96% F]) and four positions at which between 10 and 16% of sequences showed variation (residues 173 [85.7% N], 175 [89.6% D], 207 [83.7% Y], and 407 [84.4% Q]) were excluded and consensus amino acids were included at these positions. Thus, of the 52 polymorphic positions in domains I, II, and III, the 36 most variable were included in the design of the DiCo proteins (21/32 positions, 7/11 positions, and 8/9 positions in domains I, II, and III, respectively).
Three backbones were designed, taking linkages into account. DiCo1 was fully compliant to the down- and upstream linkages. DiCo2 broke downstream linkage in that H296 is linked to N448 rather than D448 as in natural AMA1 sequences and upstream linkage in that D296 is restricted by N448 while H296 is included. DiCo3 incorporated the least prevalent amino acids, and two restricted downstream positions were not compliant. Normally, E206 is restricted by D197/D200/L201, while DiCo3 incorporates K206. In addition, N332 is normally restricted by L201; in DiCo3, I332 is incorporated. Two upstream linkages were not compliant: H200 and F201 are restricted by K206, but D200 and L201 are included.
Nonlinked polymorphisms were subsequently incorporated, respectively, into DiCo1, DiCo2, and DiCo3 backbones according to their frequency of occurrence. Because AMA1 is not believed to be N glycosylated in malaria parasites, NxS/NxT motifs were also changed to remove potential N-glycosylation sites during expression in Pichia; nonpolymorphic residues were changed as previously described (16) (T288 → V, S373 → D, N422 → D, S423 → K, and N499 → Q). Because N162 of DiCo1 and DiCo2 is a linked polymorphic residue as well as a potential N-glycosylation site, it was changed to Q162 to avoid introducing restrictions.
Figure 1 shows all resultant DiCo sequences in alignment with HB3, 3D7, and FVO strain AMA1 and with the consensus sequence derived from alignment of all 355 input sequences. The differences between sequences are summarized in Table 3, which includes a profile of differences according to AMA1 domain. DiCo1 is close to the consensus sequence (differing at only three positions [all of which are in domain I] when not considering mutated N-glycosylation sites and differing at nine positions [five, three, and one position in domains I, II, and III, respectively] when also considering mutated N-glycosylation sites); as expected, DiCo2 and DiCo3 sequences differ markedly from the consensus sequence, and overall the DiCo sequences differ quite considerably from one another (Table 3). The overall coverage of naturally occurring polymorphisms calculated as the average of the coverage at 52 variable sites was 97% (a Q at position 162 was considered not different from an N, because it is a potential N-glycosylation site). The average coverage of the whole protein was 95.5% if a Q at position 162 was considered different and coverage at position 162 was then 22.8%. Otherwise, all amino acid positions had coverage above 80% and only five were less than 90%: positions 173 (85.7%), 175 (89.6%), 197 (80.1%), 207 (83.7%), and 407 (84.4%).
FIG. 1.
Alignment of aa 100 to 545 of DiCo1, DiCo2, DiCo3, and the yeast-expressed HB3 AMA1, 3D7 AMA1, and FVO AMA1, as well as the consensus sequence (Cons) that was derived from the alignment of the 355 PfAMA1 sequences obtained from the GenBank database. Residues that were changed in order to prevent N glycosylation in P. pastoris (residues 162, 288, 373, 422, 423, and 499) are underlined.
TABLE 3.
Differences between Pichia-expressed sequences in domains I to III
| Sequence | No. of differences between Pichia-expressed sequencesa
|
||||||
|---|---|---|---|---|---|---|---|
| Consensus | FVO | 3D7 | HB3 | DiCo1 | DiCo2 | DiCo3 | |
| Consensus | 22 | 25 | 16 | 9 | 26 | 21 | |
| FVO | 13, 4, 5 | 24 | 18 | 20 | 9 | 25 | |
| 3D7 | 11, 9, 5 | 17, 5, 2 | 24 | 18 | 27 | 17 | |
| HB3 | 10, 5, 1 | 11, 2, 5 | 14, 5, 5 | 16 | 21 | 18 | |
| DiCo1 | 5, 3, 1 | 15, 1, 4 | 8, 6, 4 | 10, 5, 1 | 23 | 18 | |
| DiCo2 | 14, 5, 7 | 4, 3, 2 | 19, 6, 2 | 13, 3, 5 | 15, 2, 6 | 34 | |
| DiCo3 | 10, 8, 3 | 15, 4, 6 | 8, 3, 6 | 11, 4, 3 | 11, 5, 2 | 19, 7, 8 | |
Entries with a single value show the total differences between alleles; entries with three values show differences according to domains I, II, and III, respectively. Consensus, consensus sequence with N-glycosylation sites derived from the alignment. All Pichia-produced AMA1 proteins have mutated N-glycosylation sites (FVO N, six; 3D7 N, six; HB3 N, five; DiCo1 N, six; DiCo2 N, six; and DiCo3 N, five), namely, positions 162, 288, 373, 422, 423, and 499. HB3 and DiCo3 have a lysine (K) at position 162 and therefore require only five mutations to remove potential N-glycosylation sites.
DiCo expression in P. pastoris.
Expression in small-scale fermentors resulted in protein levels of 40 mg/liter or more before purification. DiCo1 was purified using Ni-immobilized metal affinity chromatography. Although a six-His tag was also incorporated into DiCo2 and DiCo3, these did not bind to Ni-immobilized metal affinity chromatography and were instead purified by sequential hydroxyapatite and size exclusion chromatography.
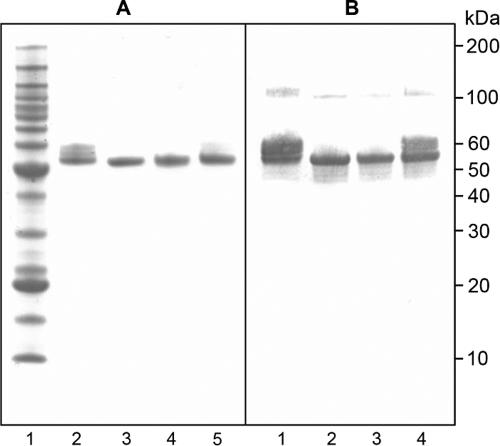
Purified DiCo proteins were assessed for integrity and purity by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and for antigenicity by Western blot analysis using the reduction-sensitive monoclonal antibody 4G2 (Fig. 2). The main band for all three DiCo proteins migrated with the expected size (50 kDa) and reacted with 4G2. A proportion of DiCo1 appears in a more slowly migrating band. Analysis using a stain specific for glycosylation moieties (Emerald Q glyco-stain; Molecular Probes, Leiden, The Netherlands) revealed that part of the protein is glycosylated (data not shown). Because incubation of the protein with PNGase did not result in a band shift, it is likely that DiCo1 is partially O glycosylated, as has previously been observed for other Pichia-produced proteins. Western blot analysis also revealed a small amount of dimerization for all three DiCo proteins; this has previously been observed with the full-length-ectodomain (aa 25 to 545) GMP product, where it did not result in loss of potency (B. W. Faber, E. J. Remarque, C. H. Kocken, P. Cheront, D. Cingolani, F. Xhonneux, M. Jurado, M. Haumont, S. Jespen, O. Leroy, and A. W. Thomas, submitted for publication).
FIG. 2.
SDS-PAGE and Western blot analyses of purified DiCo proteins. (A) SDS-PAGE analysis. Lane 1, benchmark marker; lane 2, DiCo1; lane 3, DiCo2; lane 4, DiCo3; lane 5, DiCo mix. (B) Western blot analysis with 4G2 as the primary antibody. Lane 1, DiCo1; lane 2, DiCo2; lane 3, DiCo3; lane 4, DiCo mix. Samples were not reduced.
Immunological and functional assessment.
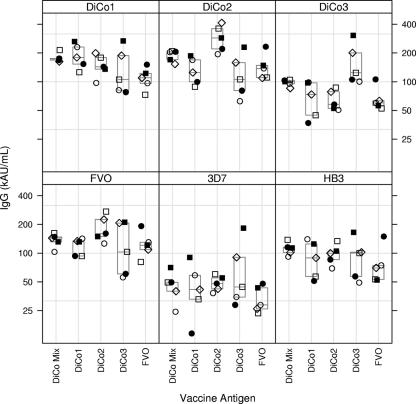
Groups of five rabbits were immunized three times with 30 μg of Pichia-expressed and purified AMA1 from FVO strain, DiCo1, DiCo2, or DiCo3 or with a mix of 10 μg of each of the three DiCo proteins (DiCo mix). Levels of antibodies to the immunizing antigen and to AMA1 from three laboratory strains were determined by ELISA (Fig. 3). Although none of these DiCo sequences have been observed in nature, they were all well-recognized by antibodies from rabbits that had been immunized with FVO PfAMA1; they all elicited antibodies that were strongly reactive with the FVO, 3D7, and HB3 antigens, indicating that they attained appropriate conformation. Moreover, the DiCo proteins showed reactivities indistinguishable from those with naturally occurring alleles when tested with sera from humans naturally exposed to malaria (data not shown). Rabbits immunized with DiCo mix showed responses that overall were comparable to those obtained by immunization with single components. Notably, for all antigens under investigation, the variation in IgG titers was smallest in the group immunized with the DiCo mix. Titers against FVO, 3D7, and HB3 PfAMA1 did not differ significantly between the DiCo1, -2, and -3 treatment groups (P = 0.27, P = 0.48, and P = 0.35, respectively). There was a tendency for the FVO-immunized animals to have lower titers against DiCo1 than the DiCo1 and DiCo mix groups (P = 0.09). Animals immunized with DiCo2 had higher titers overall than those immunized with DiCo mix or DiCo1. DiCo3-immunized animals had lower titers than animals immunized with DiCo2 (P = 0.0035) and had significantly higher DiCo3 titers than animals immunized with DiCo1, DiCo2, or DiCo mix (P = 0.002).
FIG. 3.
IgG antibody levels to various AMA1 variants. The box at the top of each panel indicates the ELISA coating antigen, and the antigen(s) used for immunization is listed on the x axis. The box indicates medians and 25 and 75 percentiles. For each treatment group, open circles (○) represent rabbit 1, filled circles (•) rabbit 2, open squares (□) rabbit 3, filled squares (▪) rabbit 4, and open diamonds (⋄) rabbit 5.
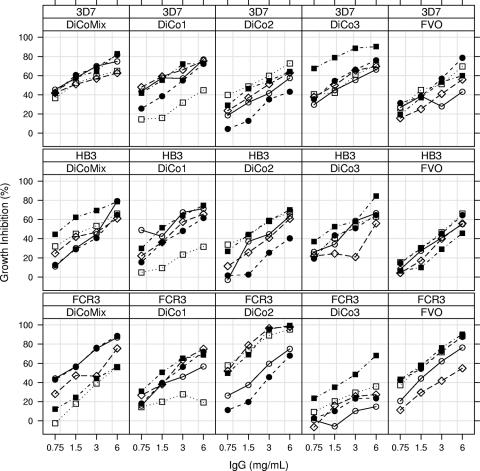
To assess antiparasitic effects in a GIA, IgG purified from rabbit sera obtained 2 weeks after the final immunization was added to synchronized asexual blood-stage cultures of 3D7, FCR3, and HB3 strains (Fig. 4). The DiCo mix was always among the three best-performing antigens, with about 70% inhibition at 6 mg/ml for all three strains tested in the inhibition assay (Fig. 4 and 5). Moreover, the DiCo mix performed nearly as well as the homologous (FVO) antigen in inhibiting growth of FCR3. All separate DiCo antigens elicited antibodies in rabbits that were active in inhibiting parasite growth. The DiCo3 antigen was less effective in inducing antibodies that inhibited FCR3 but induced good functional responses to 3D7 and HB3, despite differing from them by 17 (3D7) or 18 (HB3) aa. These observations further suggest that all antigens attained an appropriate conformation.
FIG. 4.
Growth-inhibiting titers versus IgG concentration in individual rabbits according to vaccine antigen and test strain. The top line of each panel indicates the test strain used for the GIA, and the second line indicates the antigen(s) used for immunization. For each treatment group, open circles (○) represent rabbit 1, filled circles (•) rabbit 2, open squares (□) rabbit 3, filled squares (▪) rabbit 4, and open diamonds (⋄) rabbit 5.
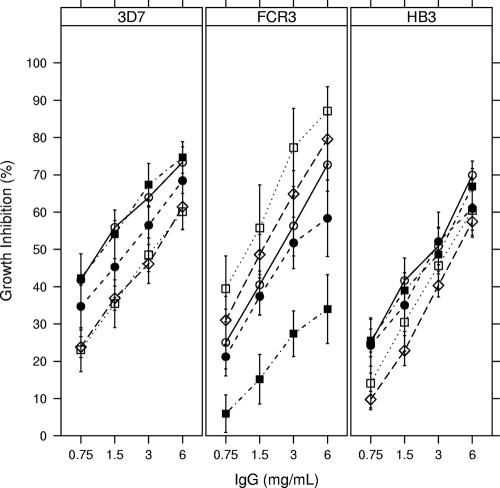
FIG. 5.
Growth-inhibiting titers versus IgG concentration in groups of rabbits according to vaccine antigen and test strain. The top line of each panel indicates the test strain used for the GIA, and the symbols indicate the antigen(s) used for immunization. For each treatment group, open circles (○) represent DiCo mix, filled circles (•) DiCo1, open squares (□) DiCo2, filled squares (▪) DiCo3, and open diamonds (⋄) FVO AMA1.
For the FCR3 strain, the highest levels of growth inhibition were observed for the DiCo2-immunized group, followed by the FVO-, DiCo mix-, DiCo1-, and DiCo3-immunized groups, respectively (Fig. 4 and 5). NLME analysis revealed significant treatment effects on slope and IgG50 (P = 0.016 and P = 0.0003, respectively, by ANOVA); the DiCo2-immunized group had the steepest curve compared to all other groups, while the other groups did not differ significantly between one another (Table 4). The IgG concentration required to reach 50% GIA activity was lowest in the animals immunized with the FVO and DiCo2 antigens (1.6 and 1.2 mg/ml, respectively) (Table 4). DiCo mix-immunized animals had an IgG50 of 2.2 mg/ml, which was not statistically different from that of FVO- or DiCo2-immunized animals (P values of 0.52 and 0.19 for FVO and DiCo2, respectively). IgG50 values were significantly higher for animals immunized with DiCo1 (3.4 mg/ml) and DiCo3 (11.5 mg/ml) than for the DiCo2 group and significantly higher for animals immunized with DiCo3 than for the FVO group (Table 4).
TABLE 4.
Slope and IgG50 estimates from NLME models
| Strain | Parameter and group | Estimate (95% CI)a | Slope or IgG50P value |
P value with indicated groupb
|
||||
|---|---|---|---|---|---|---|---|---|
| DiCo mix | DiCo1 | DiCo2 | DiCo3 | FVO | ||||
| FCR3 | Slope | |||||||
| DiCo mix | 0.750 (0.624-0.876) | 0.0001 | 0.1291 | 0.0337 | 0.3773 | 0.9427 | ||
| DiCo1 | 0.619 (0.506-0.733) | 0.0001 | 0.1290 | 0.0008 | 0.7101 | 0.1088 | ||
| DiCo2 | 0.980 (0.810-1.149) | 0.0001 | 0.0336 | 0.0008 | 0.0086 | 0.0377 | ||
| DiCo3 | 0.657 (0.491-0.824) | 0.0001 | 0.3772 | 0.7100 | 0.0086 | 0.3430 | ||
| FVO | 0.756 (0.632-0.880) | 0.0001 | 0.9427 | 0.1088 | 0.0377 | 0.3430 | ||
| IgG50 | ||||||||
| DiCo mix | 2.229 (1.145-4.341) | 0.0191 | 0.3918 | 0.1861 | 0.0017 | 0.5150 | ||
| DiCo1 | 3.356 (1.712-6.579) | 0.0006 | 0.3918 | 0.0319 | 0.0171 | 0.1350 | ||
| DiCo2 | 1.187 (0.610-2.310) | 0.6094 | 0.1861 | 0.0319 | 0.0001 | 0.4981 | ||
| DiCo3 | 11.467 (5.451-24.123) | 0.0001 | 0.0017 | 0.0171 | 0.0001 | 0.0002 | ||
| FVO | 1.637 (0.841-3.187) | 0.1446 | 0.5150 | 0.1350 | 0.4981 | 0.0002 | ||
| HB3 | Slope | |||||||
| All groups | 0.684 (0.625-0.743) | |||||||
| IgG50 | ||||||||
| All groups | 3.233 (2.560-4.083) | |||||||
| 3D7 | Slope | |||||||
| All groups | 0.520 (0.467-0.572) | |||||||
| IgG50 | ||||||||
| DiCo mix | 1.229 (0.713-2.120) | 0.4525 | 0.2719 | 0.0137 | 0.9558 | 0.0069 | ||
| DiCo1 | 1.885 (1.094-3.247) | 0.0230 | 0.2719 | 0.1596 | 0.2493 | 0.0966 | ||
| DiCo2 | 3.256 (1.896-5.591) | 0.0001 | 0.0137 | 0.1596 | 0.0119 | 0.7872 | ||
| DiCo3 | 1.203 (0.697-2.077) | 0.5018 | 0.9558 | 0.2493 | 0.0119 | 0.0059 | ||
| FVO | 3.614 (2.092-6.243) | 0.0001 | 0.0069 | 0.0966 | 0.7872 | 0.0059 | ||
95% CI, 95% confidence interval. IgG50 values are given in milligrams per milliliter.
P values for the slope represent the probability that the slope equals 0, and P values for IgG50 represent the probability that the log2 IgG50 equals 0, as the estimates are presented as the antilog values. The P value for IgG50 thus represents the probability that IgG50 = 2° = 1 (see Materials and Methods). All other columns represent P values for comparisons between the various treatment groups.
For the HB3 strain, the highest levels of growth inhibition were observed for the DiCo mix group, followed by the DiCo3, DiCo1, FVO, and DiCo2 groups, respectively (Fig. 4 and 5). A picture for IgG50 determined by GIA similar to that of FCR3 emerged, albeit not statistically significant, with the IgG50 for the DiCo mix group estimated at 2.5 mg/ml, that for the DiCo1, -2, and -3 groups approximately 3.1 mg/ml, and that for the FVO group at 3.8 mg/ml.
For the 3D7 strain, the highest levels of growth inhibition were observed for the DiCo3 and DiCo mix groups, followed by the DiCo1, FVO, and DiCo2 groups, respectively (Fig. 4 and 5). NLME analysis revealed significant treatment effects on IgG50 (P = 0.0096 by ANOVA); the DiCo3 and DiCo mix groups had the lowest IgG50 value (1.2 mg/ml for both). These differed significantly from those in the FVO and DiCo2 groups (all P values were <0.02 [Table 4]). The IgG50 value for the DiCo1 group (1.9 mg/ml) did not differ significantly from those for all other groups, although there was a trend with the FVO group (P = 0.10).
The DiCo1 sequence, which is closest to the overall consensus, performed well against all three strains tested by GIA, with IgG50 values of 3.4, 3.1, and 1.9, respectively, for FCR3, HB3, and 3D7. The IgG50 values for the DiCo mix were consistently lower, at 2.2, 2.5, and 1.2 mg/ml, respectively, for FCR3, HB3, and 3D7, although this trend was not statistically significant.
DISCUSSION
Despite the promise shown by many animal models, the development of an effective P. falciparum AMA1-based vaccine may be problematic because of the polymorphic nature of the vaccine candidate. The polymorphism is thought to be due in large part to immune selection pressure (25). An effective vaccine will probably need to induce responses to conserved determinants as well as to the broadest range of allele-specific determinants. Failure to cover a sufficient number of the allele-specific determinants found in AMA1 will, most likely, favor the survival of parasites carrying AMA1 alleles that differ from the vaccine, compromising vaccine efficacy. AMA1-based vaccines currently in development are based on one or two naturally occurring allelic forms of AMA1 (15, 16, 21, 31) and may therefore not cover an amount of variation sufficient to prevent the escape of certain allelic forms.
Coverage may be improved by simply extending the number of alleles included, but in practice there is a limit to the number of alleles that can be included, and it is not clear how responses to one allele may affect responses to others. This becomes even more important when several different vaccine target molecules are to be combined, as is generally envisaged for an ultimate vaccine. An alternative to improved coverage is through the use of a limited number of artificial variants optimally designed to cover polymorphism. Here we show that DiCo sequences, including a sequence that is very close to the consensus, can be produced. The DiCo proteins all react with a conformation-sensitive monoclonal antibody recognizing a conserved epitope. Moreover, sera elicited by the DiCo sequences all react with naturally occurring alleles and elicit antibodies that inhibit parasite growth, indicating that each DiCo protein has an authentic conformation.
The DiCo approach could be considered somewhat naïve, as many epitopes within AMA1 are conformational and may also be discontinuous (3, 5, 24). However, by incorporating most of the linked residues, stretches of naturally occurring amino acids are present in each single DiCo and therefore a number of conformational epitopes are likely to be preserved. This is corroborated by the observation that all individual DiCo sequences induce growth-inhibiting antibodies to the three laboratory strains tested. In addition, there is added value in combining the DiCo sequences because, unlike for immunization with any single DiCo, animals immunized with the DiCo mix showed comparable levels of growth inhibition for all three strains tested. Because the ultimate target population of our vaccine is immunologically naïve young children in regions of malaria endemicity, the optimal strategy may well be to evoke as wide a response as possible by immunizing with a broad variant mix. In addition to providing immediate benefits in terms of coverage, this strategy may help to avoid imprinting against one antigenic form of PfAMA1 (original antigenic sin), which may occur when immunizing with a more limited number of naturally occurring alleles (18). This also begs the question as to which naturally occurring alleles are the most suitable to include in a vaccine to adequately cover naturally occurring polymorphisms.
Here we show that immunization with a combination of three artificial alleles induces levels of growth-inhibiting antibodies to three diverse strains (FCR3, 3D7, and HB3) that are comparable to levels induced when immunizing and testing by GIA with homologous antigen/parasite systems (e.g., immunization with PfAMA1 FVO and testing on FCR3), suggesting that a considerable degree of diversity is covered by the combination. Moreover, the DiCo3 antigen yielded high levels (±70%) of growth-inhibiting antibody to the 3D7 and HB3 strains, despite being 17 and 18 aa different from the 3D7 and HB3 antigens, respectively (Table 3). Immunization with the DiCo1 antigen alone, which is close to the consensus sequence, showed very promising broad-acting immune responses. However, there was a notably consistent trend for improved performance by the DiCo mix, as reflected by lower IgG50 values. Addition of DiCo2 and DiCo3 clearly was not deleterious to the performance, and given the breadth of allelic variation in natural populations, the DiCo mix is likely to provide significant extra coverage. Having chosen the DiCo mix as our preferred platform, we have recently evaluated this in rhesus monkeys, employing either a water-in-oil or an oil-in-water adjuvant, and obtained essentially the same breadth of response as in the rabbits (E. J. Remarque, B. W. Faber, and A. Thomas, unpublished data). Based on these combined results, the three DiCo proteins are entering our pipeline for clinical evaluation.
Kennedy et al. have shown that immunization of rabbits with two naturally occurring alleles (FVO and 3D7) yields a functional response pattern that is essentially a combination of the best responses elicited by both antigens (15). However, strains that are inhibited poorly by either component are not improved by combining, which has the potential to result in escape and diminished vaccine efficacy. The data presented in this paper show that although this “best of either” principle applies to the DiCo mix when analyzed with the 3D7 and HB3 strains, it does not hold for the DiCo mix when analyzed with FCR3. In fact, for FCR3, the breadth of the response induced by the DiCo mix is best illustrated by a difference in growth inhibition levels between the highest and the lowest (DiCo2 and -3) of over 50%. It seems quite possible that the DiCo mix could yield antibody levels that inhibit growth of parasites with other PfAMA1 alleles to a degree similar to that observed here, and this is an area that is being addressed.
A recent paper by Dutta et al. (10) suggests that two regions with antigenic escape residues (AER) account for much of the escape advantage of the parasite. The highest-impact cluster for 3D7 (C1-L) comprised 5 aa; 3 are found in DiCo3, the only differences being D at position 197 and K at 206 (with E at 197 occurring only in 8% of natural sequences from the database, and with E at 206 included in DiCo3). In the second most important cluster in domain II, 3 out of 4 aa are identical to those of DiCo3 (with P at 330 occurring in only 9% of natural sequences from the database). The corresponding FVO-type AER in clusters 1 and 2 are identical to those in DiCo2 (10). It is notable that, despite these differences in the 3D7 AER, we find a high inhibitory capacity against 3D7 for antibodies elicited by both DiCo1 and DiCo mix.
When comparing heterologous GIA responses as presented here with those obtained by others (10, 15), it appears that the amount of GIA activity against heterologous alleles (viz., FCR3 versus 3D7 for FVO-immunized animals) is somewhat larger in our system. Despite close similarities in the assay procedures, heterologous GIA responses in rabbits have consistently been somewhat higher in our studies than in others (16). This may well be due to the rabbits used. To address this question, substantial effort is currently being put into harmonization efforts between the laboratories involved, incorporating the same standards into comparative assays.
A further aspect of the DiCo proteins is that they may be combined with one another into recombinant fusion proteins, thereby further reducing the number of components in a candidate malaria vaccine. This fusion protein may also be extended further by the inclusion of other, preferably small, vaccine candidates. One such candidate is MSP119, for which a fusion protein with domains I and II of AMA1 has recently been produced and shown to induce parasite-inhibitory antibodies (11). Given the likely size constraint for Pichia of proteins up to 150 kDa, a fusion protein comprising AMA1 domains I and II of the three DiCo proteins combined with two MSP119 alleles (140 kDa) seems feasible, and preliminary evidence from our lab suggests that such a protein can be expressed.
In summary, a combination of three artificial sequences representing to the greatest extent possible the naturally occurring polymorphism of the PfAMA1 ectodomain has been shown to induce immune responses that are functional against a range of parasites carrying diverse PfAMA1 alleles. This approach may offer a means by which vaccines targeting PfAMA1 can be produced such that a strong and functional protection against the broad range of naturally occurring PfAMA1 alleles can be induced.
Acknowledgments
We thank Anneke Blom, Ella Sick, Wanda Douwenga, and Joost van den Muijsenberg for expert technical assistance.
This work was funded by The European Malaria Vaccine Initiative and The Biomedical Primate Research Centre.
Editor: W. A. Petri, Jr.
Footnotes
Published ahead of print on 31 March 2008.
REFERENCES
- 1.Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16240-247. [DOI] [PubMed] [Google Scholar]
- 2.Aucouturier, J., L. Dupuis, S. Deville, S. Ascarateil, and V. Ganne. 2002. Montanide ISA 720 and 51: a new generation of water in oil emulsions as adjuvants for human vaccines. Exp. Rev. Vaccines 1111-118. [DOI] [PubMed] [Google Scholar]
- 3.Bai, T., M. Becker, A. Gupta, P. Strike, V. J. Murphy, R. F. Anders, and A. H. Batchelor. 2005. Structure of AMA1 from Plasmodium falciparum reveals a clustering of polymorphisms that surround a conserved hydrophobic pocket. Proc. Natl. Acad. Sci. USA 10212736-12741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns, J. M. J., P. R. Flaherty, P. Nanavati, and W. P. Weidanz. 2004. Protection against Plasmodium chabaudi malaria induced by immunization with apical membrane antigen 1 and merozoite surface protein 1 in the absence of gamma interferon or interleukin-4. Infect. Immun. 725605-5612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chesne-Seck, M. L., J. C. Pizarro, B. Vulliez-Le Normand, C. R. Collins, M. J. Blackman, B. W. Faber, E. J. Remarque, C. H. M. Kocken, A. W. Thomas, and G. A. Bentley. 2005. Structural comparison of apical membrane antigen 1 orthologues and paralogues in apicomplexan parasites. Mol. Biochem. Parasitol. 14455-67. [DOI] [PubMed] [Google Scholar]
- 6.Collins, W. E., D. Pye, P. E. Crewther, K. L. Vandenberg, G. G. Galland, A. J. Sulzer, D. J. Kemp, S. J. Edwards, R. L. Coppel, and J. S. Sullivan. 1994. Protective immunity induced in squirrel monkeys with recombinant apical membrane antigen-1 of Plasmodium fragile. Am. J. Trop. Med. Hyg. 51711-719. [DOI] [PubMed] [Google Scholar]
- 7.Cortés, A., M. Mellombo, R. Masciantonio, V. J. Murphy, J. C. Reeder, and R. F. Anders. 2005. Allele specificity of naturally acquired antibody responses against Plasmodium falciparum apical membrane antigen 1. Infect. Immun. 73422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crewther, P. E., M. L. Matthew, R. H. Flegg, and R. F. Anders. 1996. Protective immune responses to apical membrane antigen 1 of Plasmodium chabaudi involve recognition of strain-specific epitopes. Infect. Immun. 643310-3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deans, J. A., A. M. Knight, W. C. Jean, A. P. Waters, S. Cohen, and G. H. Mitchell. 1988. Vaccination trials in rhesus monkeys with a minor, invariant, Plasmodium knowlesi 66 kD merozoite antigen. Parasite Immunol. 10535-552. [DOI] [PubMed] [Google Scholar]
- 10.Dutta, S., S. Y. Lee, A. H. Batchelor, and D. E. Lanar. 2007. Structural basis of antigenic escape of a malaria vaccine candidate. Proc. Natl. Acad. Sci. USA 10412488-12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faber, B. W., E. J. Remarque, W. D. Morgan, C. H. Kocken, A. A. Holder, and A. W. Thomas. 2007. Malaria vaccine-related benefits of a single protein comprising Plasmodium falciparum apical membrane antigen 1 domains I and II fused to a modified form of the 19-kilodalton C-terminal fragment of merozoite surface protein 1. Infect. Immun. 755947-5955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Healer, J., V. Murphy, A. N. Hodder, R. Masciantonio, A. W. Gemmill, R. F. Anders, A. F. Cowman, and A. Batchelor. 2004. Allelic polymorphisms in apical membrane antigen-1 are responsible for evasion of antibody-mediated inhibition in Plasmodium falciparum. Mol. Microbiol. 52159-168. [DOI] [PubMed] [Google Scholar]
- 13.Hodder, A. N., P. E. Crewther, M. L. Matthew, G. E. Reid, R. L. Moritz, R. J. Simpson, and R. F. Anders. 1996. The disulfide bond structure of Plasmodium apical membrane antigen-1. J. Biol. Chem. 27129446-29452. [DOI] [PubMed] [Google Scholar]
- 14.Johnson, A. H., R. G. F. Leke, N. R. Mendell, D. Shon, Y. J. Suh, D. Bomba-Nkolo, V. Tchinda, S. Kouontchou, L. W. Thuita, A. M. van der Wel, A. Thomas, A. Stowers, A. Saul, A. Zhou, D. W. Taylor, and I. A. Quakyi. 2004. Human leukocyte antigen class II alleles influence levels of antibodies to the Plasmodium falciparum asexual-stage apical membrane antigen 1 but not to merozoite surface antigen 2 and merozoite surface protein 1. Infect. Immun. 722762-2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kennedy, M. C., J. Wang, Y. Zhang, A. P. Miles, F. Chitsaz, A. Saul, C. A. Long, L. H. Miller, and A. W. Stowers. 2002. In vitro studies with recombinant Plasmodium falciparum apical membrane antigen 1 (AMA1): production and activity of an AMA1 vaccine and generation of a multiallelic response. Infect. Immun. 706948-6960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kocken, C. H. M., C. Withers-Martinez, M. A. Dubbeld, A. van der Wel, F. Hackett, A. Valderrama, M. J. Blackman, and A. W. Thomas. 2002. High-level expression of the malaria blood-stage vaccine candidate Plasmodium falciparum apical membrane antigen 1 and induction of antibodies that inhibit erythrocyte invasion. Infect. Immun. 704471-4476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitchell, G. H., A. W. Thomas, G. Margos, A. R. Dluzewski, and L. H. Bannister. 2004. Apical membrane antigen 1, a major malaria vaccine candidate, mediates the close attachment of invasive merozoites to host red blood cells. Infect. Immun. 72154-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muller, S. 2004. Avoiding deceptive imprinting of the immune response to HIV-1 infection in vaccine development. Int. Rev. Immunol. 23423-436. [DOI] [PubMed] [Google Scholar]
- 19.Narum, D. L., S. A. Ogun, A. W. Thomas, and A. A. Holder. 2000. Immunization with parasite-derived apical membrane antigen 1 or passive immunization with a specific monoclonal antibody protects BALB/c mice against lethal Plasmodium yoelii yoelii YM blood-stage infection. Infect. Immun. 682899-2906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Narum, D. L., and A. W. Thomas. 1994. Differential localization of full-length and processed forms of PF83/AMA-1 an apical membrane antigen of Plasmodium falciparum merozoites. Mol. Biochem. Parasitol. 6759-68. [DOI] [PubMed] [Google Scholar]
- 21.Pan, W., D. Huang, Q. Zhang, L. Qu, D. Zhang, X. Zhang, X. Xue, and F. Qian. 2004. Fusion of two malaria vaccine candidate antigens enhances product yield, immunogenicity, and antibody-mediated inhibition of parasite growth in vitro. J. Immunol. 1726167-6174. [DOI] [PubMed] [Google Scholar]
- 22.Paterson, S., and J. Lello. 2003. Mixed models: getting the best use of parasitological data. Trends Parasitol. 19370-375. [DOI] [PubMed] [Google Scholar]
- 23.Pinheiro, J. C., and D. M. Bates. 2000. Mixed-effects models in S and S-plus. Springer-Verlag, New York, NY.
- 24.Pizarro, J. C., B. Vulliez-Le Normand, M. L. Chesne-Seck, C. R. Collins, C. Withers-Martinez, F. Hackett, M. J. Blackman, B. W. Faber, E. J. Remarque, C. H. M. Kocken, A. W. Thomas, and G. A. Bentley. 2005. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science 308408-411. [DOI] [PubMed] [Google Scholar]
- 25.Polley, S. D., and D. J. Conway. 2001. Strong diversifying selection on domains of the Plasmodium falciparum apical membrane antigen 1 gene. Genetics 1581505-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Polley, S. D., T. Mwangi, C. H. M. Kocken, A. W. Thomas, S. Dutta, D. E. Lanar, E. Remarque, A. Ross, T. N. Williams, G. Mwambingu, B. Lowe, D. J. Conway, and K. Marsh. 2004. Human antibodies to recombinant protein constructs of Plasmodium falciparum apical membrane antigen 1 (AMA1) and their associations with protection from malaria. Vaccine 23718-728. [DOI] [PubMed] [Google Scholar]
- 27.Remarque, E. J., B. W. Faber, C. H. Kocken, and A. W. Thomas. 2008. Apical membrane antigen 1: a malaria vaccine candidate in review. Trends Parasitol. 2474-84. [DOI] [PubMed] [Google Scholar]
- 28.Sachs, J., and P. Malaney. 2002. The economic and social burden of malaria. Nature 415680-685. [DOI] [PubMed] [Google Scholar]
- 29.Silvie, O., J. F. Franetich, S. Charrin, M. S. Mueller, A. Siau, M. Bodescot, E. Rubinstein, L. Hannoun, Y. Charoenvit, C. H. Kocken, A. W. Thomas, G. J. Van Gemert, R. W. Sauerwein, M. J. Blackman, R. F. Anders, G. Pluschke, and D. Mazier. 2004. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J. Biol. Chem. 2799490-9496. [DOI] [PubMed] [Google Scholar]
- 30.Snow, R. W., C. A. Guerra, A. M. Noor, H. Y. Myint, and S. I. Hay. 2005. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature 434214-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thera, M. A., O. K. Doumbo, D. Coulibaly, D. A. Diallo, A. K. Kone, A. B. Guindo, K. Traore, A. Dicko, I. Sagara, M. S. Sissoko, M. Baby, M. Sissoko, I. Diarra, A. Niangaly, A. Dolo, M. Daou, S. I. Diawara, D. G. Heppner, V. A. Stewart, E. Angov, E. S. Bergmann-Leitner, D. E. Lanar, S. Dutta, L. Soisson, C. L. Diggs, A. Leach, A. Owusu, M. C. Dubois, J. Cohen, J. N. Nixon, A. Gregson, S. L. Takala, K. E. Lyke, and C. V. Plowe. 2008. Safety and immunogenicity of an AMA-1 malaria vaccine in Malian adults: results of a phase 1 randomized controlled trial. PLoS ONE 3e1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomas, A. W., J. A. Deans, G. H. Mitchell, T. Alderson, and S. Cohen. 1984. The Fab fragments of monoclonal IgG to a merozoite surface antigen inhibit Plasmodium knowlesi invasion of erythrocytes. Mol. Biochem. Parasitol. 13187-199. [DOI] [PubMed] [Google Scholar]
- 33.Thomas, A. W., J. F. Trape, C. Rogier, A. Goncalves, V. E. Rosario, and D. L. Narum. 1994. High prevalence of natural antibodies against Plasmodium falciparum 83-kilodalton apical membrane antigen (PF83/AMA-1) as detected by capture-enzyme-linked immunosorbent assay using full-length baculovirus recombinant PF83/AMA-1. Am. J. Trop. Med. Hyg. 51730-740. [DOI] [PubMed] [Google Scholar]
- 34.Thomas, A. W., A. P. Waters, and D. Carr. 1990. Analysis of variation in PF83, an erythrocytic merozoite vaccine candidate antigen of Plasmodium falciparum. Mol. Biochem. Parasitol. 42285-287. [DOI] [PubMed] [Google Scholar]
- 35.Triglia, T., J. Healer, S. R. Caruana, A. N. Hodder, R. F. Anders, B. S. Crabb, and A. F. Cowman. 2000. Apical membrane antigen 1 plays a central role in erythrocyte invasion by Plasmodium species. Mol. Microbiol. 38706-718. [DOI] [PubMed] [Google Scholar]