Abstract
The SerB protein of Porphyromonas gingivalis is a HAD family serine phosphatase that plays a critical role in entry and survival of the organism in gingival epithelial cells. SerB is secreted by P. gingivalis upon contact with epithelial cells. Here it is shown by microarray analysis that SerB impacts the transcriptional profile of gingival epithelial cells, with pathways involving the actin cytoskeleton and cytokine production among those significantly overpopulated with differentially regulated genes. Consistent with the transcriptional profile, a SerB mutant of P. gingivalis exhibited defective remodeling of actin in epithelial cells. Interaction between gingival epithelial cells and isolated SerB protein resulted in actin rearrangement and an increase in the F/G actin ratio. SerB protein was also required for P. gingivalis to antagonize interleukin-8 accumulation following stimulation of epithelial cells with Fusobacterium nucleatum. SerB is thus capable of modulating host cell signal transduction that impacts the actin cytoskeleton and cytokine production.
Porphyromonas gingivalis, a gram-negative anaerobe, is an important pathogen in chronic and severe manifestations of periodontal disease (28). P. gingivalis is also a common inhabitant of the gingival crevice in healthy individuals (44) and thus can be categorized as a host-adapted pathogen. The primary ecological niche of P. gingivalis is in the subgingival crevice, and the organism engages the host epithelial cells that line the gingival crevice in an intricate molecular dialogue. Through manipulation of epithelial cell signaling pathways, P. gingivalis rapidly internalizes within these cells in high numbers (3, 8, 27). Intracellular P. gingivalis cells accumulate in the perinuclear area and remain viable and capable of cell-to-cell spread (3, 54). Despite the burden of large numbers of intracellular P. gingivalis, gingival epithelial cells do not undergo either necrotic or apoptotic cell death, and indeed are rendered resistant to chemically induced apoptosis (31, 34, 53). Adaptation of both P. gingivalis and the gingival epithelial cells to their cohabitation occurs on a genome-wide scale (21, 58). Approximately 40% of the expressed proteome of P. gingivalis is modulated upon internalization within epithelial cells (51). In gingival epithelial cells, several thousand genes are differentially expressed following infection with P. gingivalis, a response that is tailored to P. gingivalis and is markedly different from the transcriptional profiles of epithelial cells infected with Aggregatibacter actinomycetemcomitans, Fusobacterium nucleatum, or Streptococcus gordonii (21, 22). Entry of P. gingivalis into gingival epithelial cells requires remodeling of both the microfilament and microtubule cytoskeleton (27), along with modulation of cytosolic calcium ion concentrations (2), and activation of the mitogen-activated protein kinase (MAPK) family member JNK (49). Infection by P. gingivalis impacts the innate immune functionality of epithelial cells as NF-κB activity is suppressed and secretion of interleukin-8 (IL-8) is inhibited (11, 49). Moreover, invasive P. gingivalis antagonizes the IL-8 secretion stimulated by other oral organisms, such as F. nucleatum, a phenomenon known as local chemokine paralysis (11).
A distinctive feature of P. gingivalis manipulation of host cell physiology is that the process is accomplished in the absence of a conventional type III secretion system, machinery that is utilized by other invasive organisms to deliver effector proteins directly into the cytoplasm of host cells (9). Rather, P. gingivalis secretes effector proteins into the extracellular milieu when stimulated by epithelial cell components (7, 37). One such secreted effector protein is SerB, a phosphoserine phosphatase and a member of the haloacid dehalogenase superfamily of hydrolytic dehalogenases, widespread in both prokaryotes and eukaryotes (46). P. gingivalis mutants unable to express SerB are deficient in internalization and survival in gingival epithelial cells (46). SerB is functional within epithelial cells, interacting with cytoplasmic glyceraldehyde-3-phosphate dehydrogenase and HSP90 and modulating microtubule dynamics. As tightly regulated patterns of protein phosphorylation and dephosphorylation are important for a wide range of eukaryotic cell activities, we hypothesized that SerB may have a more broadly based effect on gingival epithelial cell physiology. To better define the role of SerB in the interaction between P. gingivalis and gingival epithelial cells, we examined its effects on the epithelial cell transcriptome. A microarray analysis revealed that SerB impacts the expression of over 3,000 genes, including those involved in regulating the dynamics of the actin cytoskeleton and in cytokine secretion. These activities of SerB were confirmed at the phenotypic level by confocal microscopy, showing actin rearrangements in response to SerB, and by loss of IL-8 chemokine paralysis in epithelial cells infected with a SerB mutant.
MATERIALS AND METHODS
Bacteria and epithelial cells.
P. gingivalis strains ATCC 33277, SerB mutant, complemented SerB mutant (46), and F. nucleatum ATCC 25586 were cultured in trypticase soy broth, supplemented with yeast extract (1 mg ml−1), hemin (5 μg ml−1), and menadione (1 μg ml−1), anaerobically at 37°C. Escherichia coli strains were grown in LB medium aerobically at 37°C. Human immortalized gingival keratinocytes (HIGKs) (36) were cultured under 5% CO2 in keratinocyte serum-free medium (Gibco/Invitrogen, Carlsbad, CA) supplemented with 0.05 mM calcium chloride and 200 mM l-glutamine (Gibco/Invitrogen). Bacteria-HIGK cell coculture was performed as described elsewhere (30). Briefly, bacteria at mid-log phase were harvested and resuspended in antibiotic-free keratinocyte serum-free medium. Bacteria were added to 70 to 90% confluent HIGK cells to give a multiplicity of infection (MOI) of 100 and incubated at 37°C in 5% CO2.
Microarray hybridization.
After a 2-h challenge with bacteria, quadruplicate cultures of HIGKs were lysed with Trizol (Invitrogen). RNA isolation, cDNA synthesis, labeled cRNA synthesis, and chip hybridization were conducted as previously described (30). Briefly, total RNA was extracted from Trizol-lysed cells, treated with DNase I, purified, and quantified according to standard methods (Qiagen, Valencia, CA, and Affymetrix, Santa Clara, CA). cDNA synthesis was performed according to the Affymetrix protocol (SuperScript double-stranded cDNA synthesis kit; Invitrogen) with 5 to 8 μg of total cellular RNA used as a template to amplify mRNA species for detection. Double-stranded cDNA was purified and used as a template for labeled cRNA synthesis. In vitro transcription was performed using a BioArray high-yield RNA transcript labeling kit (T7; Enzo Life Science, Farmingdale, NY) to incorporate biotinylated nucleotides. cRNA was subsequently fragmented and hybridized onto Genechip human genome U133-A oligonucleotide arrays (Affymetrix) with appropriate controls. Each sample was studied in parallel, and the samples were not pooled. The microarrays were hybridized for 16 h at 45°C, stained with phycoerythrin-conjugated streptavidin, and washed according to the Affymetrix protocol (EukGE-WS2v4) using an Affymetrix fluidics station and scanned with an Affymetrix GeneChip 3000 scanner.
Microarray analysis.
Following initial assessment of the host cell response to each condition, supervised analysis was performed to investigate differences in gene regulation among experimental conditions. For this analysis, the raw signal intensities were log transformed for all probe sets that passed the initial expression filters and were correlated using BRB Array Tools 5.0 (Simon and Peng-Lam, National Cancer Institute, Rockville, MD). In each supervised analysis, biological replicates were grouped into classes according to their infection state during coculture experiments, and probe sets significant at the P < 0.005 level between classes were identified. To test the ability of these significant probe sets to truly distinguish between the classes, leave-one-out cross-validation (LOOCV) studies were preformed. In these LOOCV studies each array was left out in turn and a classifier was derived between the groups by selecting probe sets significant at a P level of <0.005. The significant probe sets were then used with several prediction models (covariate predictor, diagonal linear discriminant analysis, 1-nearest neighbor, 3-nearest neighbors, nearest centroid, support vector machines, and Bayesian compound covariate predictor) to predict the class identity of the array that was left out and not included when the classification model was built. The ability of the classifier to correctly predict the class identity of the left-out array was estimated using Monte Carlo simulations with 2,000 permutations of the data set and compared to the likelihood of a correct prediction by chance alone (P < 0.5).
Functional categorization by gene ontology and bioinformatics analyses.
Kegg pathways were populated using Pathway Express (25), available at http://vortex.cs.wayne.edu/projects.htm.
Quantitative RT-PCR.
Total RNA isolated from control and P. gingivalis-infected HIGK cells was reverse transcribed to cDNA using SuperScript III. Specific DNA standards were synthesized from chromosomal DNA in a standard PCR. Real-time reverse transcription-PCR (RT-PCR) was performed on a Bio-Rad iCycler using Sybr green supermix (Bio-Rad). Results were analyzed with the iCycler iQ optical system software version 3.0a. The melt curve profiles were examined to verify a single peak for each sample, indicating primer specificity, and the transcript copy number was calculated (57). RNA extracts were prepared in duplicate from independent experiments, and cDNA samples were loaded in triplicate.
Expression of recombinant protein.
rSerB 0653 and rPG1107 were expressed in Escherichia coli strain TunerDE3 (Novagen, San Diego, CA) using the pET30b expression system. His tag proteins were purified with a nickel resin (Bio-Rad, Hercules, CA). Purity was >97%, as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Coomassie staining.
Confocal microscopy.
P. gingivalis strains (MOI, 100) or rSerB enzymes (100 μg) were added to HIGK cells and incubated for the appropriate time. Cells were washed three times with phosphate-buffered saline (PBS, pH 7.4) and fixed with 4% paraformaldehyde in PBS pH 7.4 for 30 min at room temperature. Immunostaining was performed after permeabilization with 0.2% saponin in PBS containing 10% goat serum for 20 min. Bacteria were detected with polyclonal rabbit anti-P. gingivalis antibody (1:500), and rSerB was detected with anti-SerB antibody (1:500), followed by fluorescein isothiocyanate-conjugated anti-rabbit secondary antibody (1:500). F actin was stained by phalloidin-Texas Red isothiocyanate (1:200). Images were taken at 40× or 60× on a Bio-Rad MRC1024 CLSM confocal microscope. A series of fluorescent optical x-y sections were collected to create digitally reconstructed images (x-z section and z-projection of x-y sections) with ImageJ 1.35c and Adobe Photoshop 6.0 software. Total actin fluorescent accumulations in the stack of z-projections representing whole cells were quantified with ImageJ 1.35c using the area calculator plug-in.
Quantitative F actin analysis.
HIGK cells were treated with rSerB for 2 h, and the ratio of F actin to G actin was analyzed using an F actin/G actin in vivo assay kit (Cytoskeleton Inc., Denver, CO) based on the manufacturer's protocol. Briefly, cells were lysed, F actin was stabilized in actin stabilization buffer, and the cell lysates were centrifuged at 100,000 × g for 60 min. The supernatants (G actin) were separated from the pellets (F actin). The pellets were resuspended to the same volume as the supernatants using ice-cold distilled water containing 2 μM cytochalasin D and were incubated on ice for 1 h. Equal volumes of the samples were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and analyzed by Western blotting with an antiactin antibody. Actin bands in each fraction were detected using chemiluminescence, and the ratio of F actin to G actin was quantified using the Kodak 1D Image Analysis software v.3.6.1.
IL-8 assay.
HIGK cells were reacted sequentially with F. nucleatum and P. gingivalis at an MOI of 100 at 37°C in 5% CO2 in the presence of a proteinase inhibitor cocktail (Sigma, St. Louis, MO). Supernatant samples were removed, filtered, and analyzed with an enzyme-linked immunosorbent assay kit for IL-8 (R&D Systems, Minneapolis, MN) according to the manufacturer's instructions.
Microarray accession number.
The array results have been deposited in the GEO repository (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE10526.
RESULTS
Transcriptional profiling of gingival epithelial cells infected with P. gingivalis wild-type and SerB mutant strains.
To reveal the range of effects of SerB on host cells, HIGKs were transcriptionally profiled following infection with parental or SerB mutant strains of P. gingivalis. HIGK cells behave similarly to primary low-passage cultures of gingival epithelial cells in terms of responses to P. gingivalis challenge (21). In addition, the contribution of SerB to the invasive process is equivalent in HIGKs and in primary gingival epithelial cells (46). Furthermore, HIGKs can be grown to high cell number without the patient-to-patient transcriptome variability that can occur with primary cells (26); thus, HIGKs represent a useful model for the study of gingival epithelial cell global transcriptional responses to oral organisms (21, 22). Samples of uninfected HIGKs and P. gingivalis 33277-infected and P. gingivalis SerB mutant-infected cells were used to determine the overall similarity of the transcriptional responses. After elimination of probe sets with signals that were not greater than background levels on all arrays, signal intensity data for the 12,699 probe sets that passed the initial expression filters were included in an unsupervised cluster analysis and supervised class prediction analysis. The unsupervised hierarchical cluster analysis (not shown) revealed transcriptional profiles that were characteristic of each infection state, as biological replicates clustered together. A supervised analysis was performed using individual comparisons of the parent and mutant infection states with a common baseline of the uninfected state. At a significance level of P < 0.005, 3,365 probe sets were differentially regulated in HIGKs infected with the SerB mutant of P. gingivalis compared to the parental strain. Assuming normality of the data set, the 3,365 significant genes was 50-fold higher than the 64 probe sets that would be expected by chance alone at P < 0.005, given that 12,699 probe sets passed the expression filter. A TreeView visualization of the 3,365 probe sets differentially expressed between the infection classes is shown in Fig. 1. The ability of the regulated probe sets to correctly differentiate treatment groups was confirmed by a LOOCV analysis. The classifiers performed flawlessly and correctly predicted the treatment group with 100% accuracy using seven prediction models: covariate predictor, diagonal linear discriminant analysis, 1-nearest neighbor, 3-nearest neighbors, nearest centroid, support vector machines, and Bayesian compound covariate predictor.
FIG. 1.
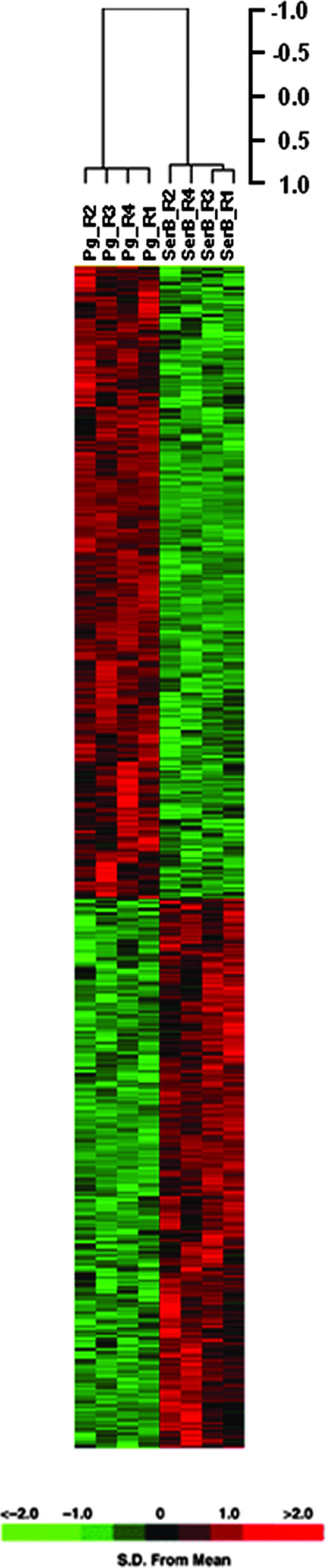
Comparison of transcriptional profiles of HIGK cells following coculture with wild-type P. gingivalis or SerB mutant strain. The expression pattern of the cRNAs analyzed by microarray is represented as a supervised analysis of the variance-normalized data set of differentially expressed genes with the algorithm Cluster and displayed with TreeView. Each row represents an individual DNA element spotted on the array, and each column represents the expression states of cRNAs for the challenge condition indicated. Each expression data point represents the ratio of the fluorescence intensity of the cRNA from P. gingivalis 33277-infected cells (Pg columns, replicates [R] 1 to 4) to the fluorescence intensity of the cRNA from SerB mutant-infected cells (SerB columns, R 1 to 4). The distance matrix used to show the relatedness of samples through gene expression space was the Pearson's correlation coefficient. The cluster is subdivided into three groups consisting of genes that were repressed (green), genes that were induced (red), and genes whose expression did not change (black). The variation in expression for a given gene is expressed as the distance from the mean observation for that gene according to the color scale presented below the heat map.
Ontology analysis of differentially regulated genes.
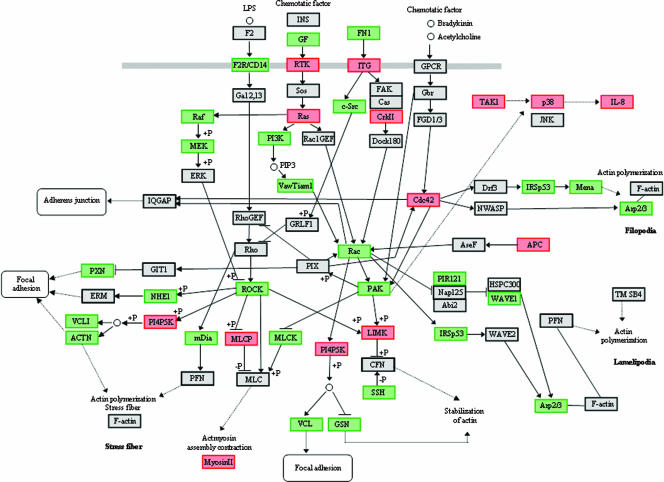
The predictive power of individual differentially regulated genes is limited in the absence of information regarding the expression levels of other genes that also impact related biological processes. Hence, in order to derive biologically relevant information from the array data, an ontology analysis of known metabolic pathways was performed with probe sets at the P < 0.005 significance level using the Pathway Express algorithm (25). The three host cell pathways most significantly impacted by the SerB mutant in comparison with the parental strain were regulation of actin cytoskeleton (Kegg 04810; 49 regulated, 206 pathway genes), focal adhesion (Kegg 04510; 59 regulated, 194 pathway), and MAPK signaling (Kegg 04010; 60 regulated, 273 pathway). In addition to being highly impacted, these processes are interconnected and are relevant to the overall biology of pathogen-host interactions. Differentially regulated genes representing these pathways are depicted in Fig. 2.
FIG. 2.
Summary of epithelial cell actin and MAP kinase-related genes differentially regulated (P < 0.005) following infection with the P. gingivalis SerB mutant compared to the parental strain. The pathway was adapted from Pathway Express and utilizes the Kegg nomenclature. Red represents upregulation, green represent downregulation, and gray represents no change in expression. +P indicates phosphorylation; −P indicates dephosphorylation. Pointed arrows indicate molecular interactions resulting in activation; flat arrows indicate molecular interactions resulting in inhibition. Solid lines show a direct interaction, and dotted lines are interactions involving intermediates.
SerB affects expression of genes that regulate actin dynamics.
In pathways related to actin stability, several downstream genes that are close to the level of actin binding and focal adhesion assembly were differentially regulated. In comparison to the parental strain, the SerB mutant downregulated genes for α-actinin, vinculin, paxillin, Arp2/3, Mena, and mDia. α-Actinin is an F-actin cross-linking protein that can anchor actin to a variety of intracellular structures (52). Vinculin is a prominent component of focal adhesions that can bind actin and mediate barbed end capping (39). Paxillin is a multidomain adaptor that recruits both structural and signaling molecules to focal adhesions (6). Actin-related protein (Arp) 2/3 complex is an actin nucleator that controls polymerization, organization, and recycling of actin filament networks (19). Mena (ENAH/VASP) is an actin-associated protein involved in a range of processes related to cytoskeleton remodeling, and mDia can directly nucleate, elongate, and bundle actin filaments (17, 43). In contrast, LIMK was upregulated in response to the SerB mutant. LIMK is a protein kinase that phosphorylates and inactivates the actin binding/depolymerizing factor cofilin, thereby stabilizing the actin cytoskeleton and preventing actin remodeling. Consistent with upregulation of LIMK was SerB-induced downregulation of SSH (Slingshot homolog), which dephosphorylates and activates cofilin (4, 18). Major nodes in the actin pathways that were downregulated by the SerB mutant included Rho-associated protein kinase (ROCK), Rac, and p21-activated kinase (PAK). ROCK phosphorylates a large number of client proteins that are involved in actin assembly (32). Rac is a Rho family small GTPase that is a central hub in the regulation of actin and adhesion organization (40). PAK is involved in the dissolution of stress fibers and reorganization of focal complexes (59). To confirm differential gene expression of key components of the actin stability pathway and to verify the role of SerB in the process, quantitative real-time RT-PCR was performed following challenge of HIGKs with parental, SerB-deficient, and SerB-complemented (cSerB) strains. As shown in Table 1, the results were consistent with the microarray expression profile. Furthermore, the complemented SerB mutant elicited a pattern of gene expression similar to the parental strain. Collectively, these results suggest that SerB has an overwhelming effect on actin cytoskeletal architecture through differential and consistent regulation of a number of actin binding proteins and through coordinated regulation of central nodes of the pathways that control actin fiber stability and assembly of focal adhesions.
TABLE 1.
Differential expression of HIGK genes by quantitative RT-PCR
| Gene | Expression change (fold)a
|
||
|---|---|---|---|
| 33277 | SerB- | cSerB | |
| ROCK1 | 3.4 | ND | 3.9 |
| Cdc42 | −2.5 | 2.6 | −2.1 |
| LIMK1 | −3.0 | 9.8 | −2.9 |
| ACTN1 | 6.1 | ND | 5.3 |
| VCL | 7.7 | ND | 7.0 |
| Arp2/3 | 5.2 | ND | 4.9 |
| p38 | −12.3 | ND | −10.8 |
Change in expression compared to uninfected HIGK cells after interaction with P. gingivalis 33277, SerB mutant (SerB-), or SerB-complemented mutant (cSerB). All numerical values shown were significant at P < 0.01 (t test). ND, no significant difference.
SerB modulates IL-8 expression.
The actin and MAP kinase signaling pathways interface through Rho family GTPases such as Cdc42 and linking kinases such as PAK. The MAP kinase pathway can ultimately converge on production of cytokines. The array analysis indicated that, in comparison to the parent strain, the SerB mutant elevated IL-8 mRNA levels (Fig. 2). Upregulation of IL-8 by the SerB mutant may result from elevated expression of transforming growth factor β-activated kinase 1 (MAP3K7), which can activate p38 though MKK3/6. Downregulation of p38 by the parental and complemented SerB mutant strains, but not by the SerB mutant, was confirmed by quantitative real-time RT-PCR (Table 1). Both transforming growth factor β-activated kinase 1 and p38 can activate transcription factors, such as NF-κB, which can control IL-8 gene expression (1, 12).
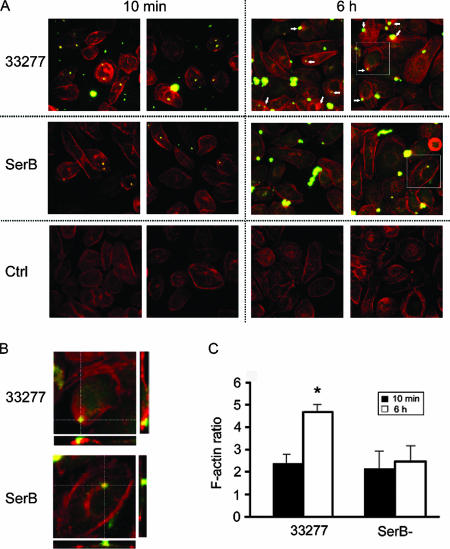
Reorganization of the actin cytoskeleton is deficient in the SerB mutant.
While transcriptional profiling of infected cells can provide insights into their phenotypic status, it does not account for posttranslational modifications, which control much of the information flow through eukaryotic signal transduction pathways. Thus, to verify the array results and to evaluate the extent of actin remodeling that occurs in gingival epithelial cells upon infection with parental or SerB-deficient strains, the actin cytoskeleton was visualized and quantified by confocal microscopy. Actin rearrangements have been shown to be required for P. gingivalis entry into gingival epithelial cells, a rapid process that is complete in approximately 20 min (3). In addition, prolonged cohabitation with intracellular P. gingivalis results in a cortical redistribution and condensation of actin microfilaments (56). As our transcriptional snapshot was taken at 2 h of P. gingivalis infection, the differential gene expression could be a legacy of events that have already occurred at the phenotypic level (e.g., bacterial invasion). Alternatively, as protein levels can lag behind differential mRNA expression by several hours (22), observed transcriptional changes could be an early indication of events that are about to occur at the phenotypic level (e.g., redistribution and condensation of actin microfilaments). To distinguish between these possibilities, actin microfilaments were observed after 10 min and 6 h of infection with parental and SerB mutant strains. No differences in actin microfilament structure or levels were observed after 10 min of interaction of host cells with either P. gingivalis 33277 or SerB mutant strains (Fig. 3A and C). However, after 6 h of association, the parental strain induced an increase in actin microfilaments that had relocated to the cell surface and were associated with external, membrane-bound P. gingivalis (Fig. 3B and C). The SerB mutant, in contrast, failed to induce either an increase or a rearrangement of actin. Hence, SerB appears to ultimately exert its actin-related functionality on the membrane of host epithelial cells and participate in the process that causes actin remodeling over longer time periods. The snapshot of gene expression data depicted in Fig. 2, therefore, can be predicted to make a significant contribution to the mechanism by which SerB modulates the actin cytoskeleton.
FIG. 3.
SerB modulates the actin cytoskeleton in gingival epithelial cells. (A) HIGK cells were infected with P. gingivalis 33277 (WT) or SerB mutant (SerB) for 10 min or 6 h. Control (Ctrl) cells were uninfected. The actin cytoskeleton was visualized with rhodamine-phalloidin (red), and bacteria were stained with P. gingivalis antibody and fluorescein-labeled secondary antibody (green). The image is a confocal 0.2-μm optical x-y projection magnified 40×. Arrows indicate areas of actin redistribution around P. gingivalis. (B) Boxed areas from panel A are shown, along with x-z and y-z projections. (C) Quantitative image analysis of actin fluorescence in the stack of z-projections representing whole cells shown in panel A. Data are expressed as the ratio of actin in infected versus control HIGK cells. *, P < 0.01 (t test) at 6 h compared to 10 min.
SerB protein can induce actin reorganization.
As the SerB mutant is deficient in internalization and intracellular survival (46), it was important to ensure that the actin-related phenotype was not a general consequence of failure to establish an intracellular infection. P. gingivalis secretes SerB when in direct contact with epithelial cells and can also secrete SerB into the culture medium when stimulated by soluble signals from epithelial cells (7). Therefore, extracellular SerB is likely to be functional, and we sought to determine whether cell-free SerB can play a role in actin remodeling. To accomplish this, we first expressed recombinant SerB (rSerB) in E. coli. As a control, we also expressed and purified the rPG1170 protein. PG1170 is another serine phosphatase of P. gingivalis, although it is not involved in the invasion process and has a higher affinity for phosphothreonine than phosphoserine (46). rSerB and rPG1170 were incubated with gingival epithelial cells, probed with rhodamine-phalloidin and antibodies to SerB, and examined by confocal microscopy. As shown in Fig. 4A, SerB attached to the epithelial cell membrane and was not observed within the cells. External SerB protein was capable of inducing actin reorganization, whereas PG1170 interaction with the epithelial cells did not result in visible actin rearrangements. In order to quantitate the degree of actin remodeling induced by SerB, we measured the ratio of F/G actin following exposure to rSerB, through purification of the cellular actin populations and quantitation by densitometric analysis of Western blots. Figure 4B shows that rSerB activity results in approximately double the amount of filamentous actin inside epithelial cells compared to control cells. Hence, SerB can exhibit at least two distinct functional properties with regard to interactions with host cells. When produced by intracellular organisms, SerB interacts with cytosolic glyceraldehyde-3-phosphate dehydrogenase and HSP90, ultimately impacting the integrity of the microtubule cytoskeleton (46). Second, as shown here, over longer periods of time, SerB modulates transcription of actin cytoskeleton-associated genes and induces rearrangements of the actin cytoskeleton. Further, SerB can accomplish actin remodeling when present on the surface of epithelial cells.
FIG. 4.
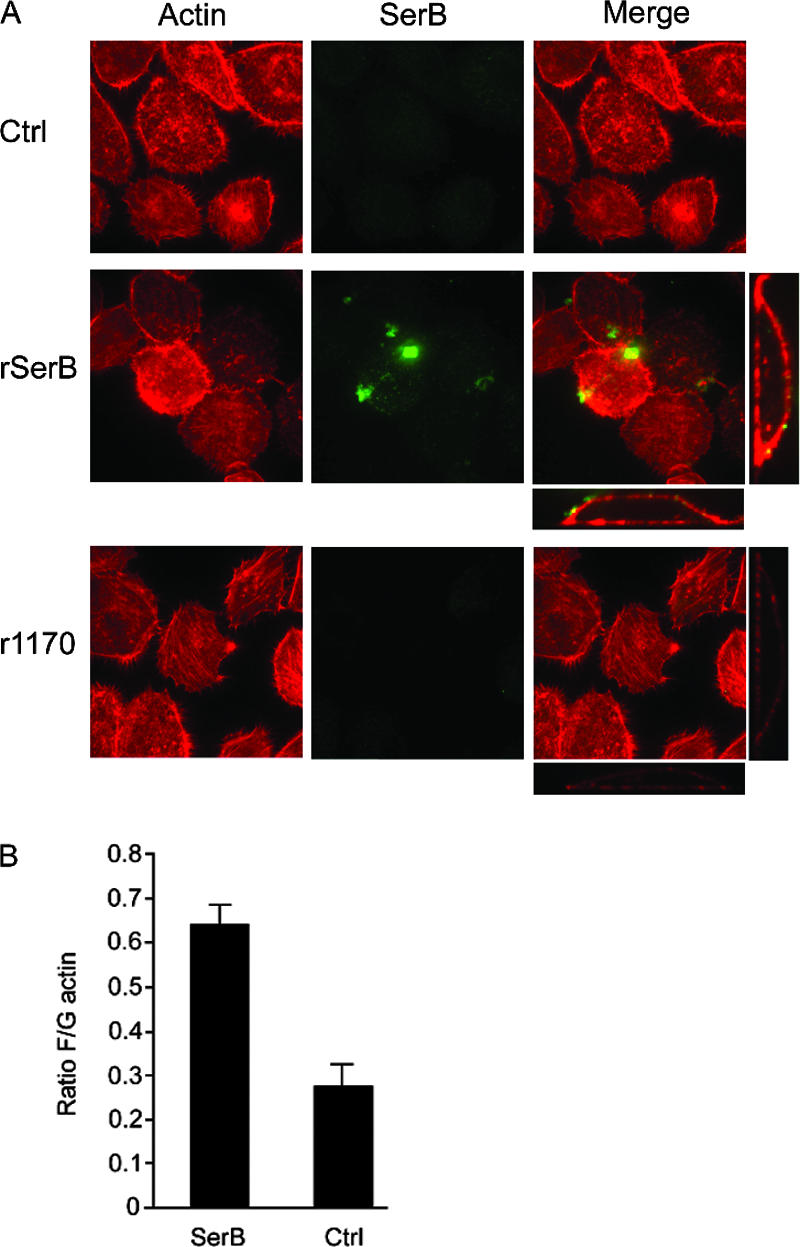
Extracellular SerB regulates F actin in gingival epithelial cells. HIGK cells were reacted with rSerB or rPG1170. The control (Ctrl) was prepared in the absence of phosphatase. (A) Actin was visualized with rhodamine-phalloidin (red), and bacterial phosphatases were visualized with SerB antibody and fluorescein-labeled secondary antibody (green). Confocal x-y projections of a z-stack (×40) are shown, along with x-z and y-z projections for the merged images (×60) with enzyme-reacted cells. (B) F/G actin ratio in HIGKs reacted with SerB. Error bars represent standard deviations. Values are statistically different at P < 0.01 (t test).
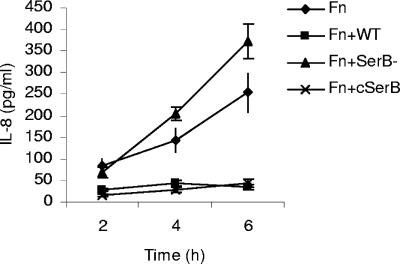
Suppression of IL-8 secretion is diminished with the SerB mutant.
The array analysis indicated that SerB may be involved in local chemokine paralysis (11), the process by which P. gingivalis suppresses IL-8 production by epithelial cells, even after stimulation with other bacteria such as F. nucleatum. To explore this possibility, IL-8 secreted by HIGK cells stimulated with F. nucleatum alone and with P. gingivalis parental and mutant strains was measured. As shown in Fig. 5, wild-type P. gingivalis, but not the SerB mutant strain, antagonized IL-8 secretion induced by F. nucleatum. The complemented SerB mutant strain displayed a phenotype indistinguishable from the parental strain. Thus, the transcriptional response to SerB can converge on IL-8 and inhibit production of this chemokine.
FIG. 5.
Enzyme-linked immunosorbent assay of IL-8 accumulation in HIGK supernatants following stimulation with F. nucleatum (Fn) alone and with P. gingivalis 33277 (WT), SerB mutant (SerB-), or the complemented SerB mutant (cSerB) for the times indicated. Error bars represent standard deviations (n = 3).
DISCUSSION
Bacteria that initiate infections at mucosal membranes are often capable of seizing control of host epithelial cell signal transduction pathways and directing their uptake into the host cell. P. gingivalis is an intensely invasive gram-negative organism that invades gingival epithelial cells rapidly and in high numbers and accumulates in the perinuclear area (3). Moreover, P. gingivalis can suppress host cell apoptosis, modulate MAPK activity, and block maturation of autophagosomes (16, 31, 49). While all of these activities are consistent with type III secretion system functionality (9), the genome of P. gingivalis does not contain obvious homologs of the structural components of the type III secretion system organelle (35). Hence, the nature of the molecules that mediate P. gingivalis cross talk with host cells and the means of their presentation to the host cell have become subjects of interest.
One of the major invasion effectors of P. gingivalis is the long (or major) fimbrial structure comprised of the FimA protein (50). FimA binds to integrins on the surface of gingival epithelial cells to initiate the entry process (55). However, FimA mutants possess residual invasive ability and also rearrange actin microfilaments (56), indicating the involvement of additional, nonfimbrial invasins. The serine phosphatase SerB was first identified by a proteomic screen of molecules secreted by P. gingivalis when in contact with epithelial cell components (7), and a SerB mutant is deficient in both entry and survival within gingival epithelial cells (46). Although bacteria more frequently utilize phosphorylation/dephosphorylation of histidine for signal transduction, serine/threonine kinases and phosphatases have been described in Yersinia spp., Pseudomonas aeruginosa, Listeria monocytogenes, Streptococcus agalactiae, and Streptococcus mutans, where they contribute to virulence (13, 23, 38). However, while serine/threonine kinases and inositol phosphatase have been shown to play a role in the molecular dialog between bacteria and host cells (20), SerB is the first serine phosphatase with a documented role in bacterial internalization.
The transcriptome of gingival epithelial cells infected with parental and SerB mutant strains demonstrated that SerB contributes to a variety of host cell responses to P. gingivalis infection. In the absence of SerB, over 60 genes are differentially regulated (at a significance level of P < 0.005) in the actin and focal adhesion pathways alone. The activity of SerB thereby overwhelms the host cell transcriptome and impacts actin architecture through multilevel regulation of actin binding proteins, including α-actinin, vinculin, actin-related protein (Arp2/3), paxillin, Mena, and mDia. Many of these proteins are frequent targets of intracellular pathogens. For example, the L. monocytogenes ActA protein recruits and activates Arp2/3 in order to nucleate actin and initiate actin-based motility of intracellular organisms (10). ActA also binds Mena, and the actin tails that develop on the organism are stabilized with α-actinin (10, 15). Shigella spp. recruit vinculin in order to induce actin polymerization, a process that also requires Arp2/3 (10). Signal transduction through paxillin is stimulated by Campylobacter jejuni and group A streptococci (33, 48) and by P. gingivalis (56). The results of the current study show that an intracellular organism such as P. gingivalis is capable of modulating these proteins at the mRNA level. Similarly, transcriptional regulation of genes encoding α-actinin and Arp2/3 was also reported in endothelial cells in response to Neisseria meningitidis (42).
The temporal progression of actin rearrangements induced by SerB, with a significant difference observed at 6 h, would tend to exclude a direct role in entry of the organism which is complete after 20 min (3). The question then arises as to the functional significance of longer-term actin rearrangements induced by SerB. Part of the answer may reside in the ability of actin per se to play a role in signal transduction (29). It has been proposed that actin remodeling can spatially organize enzymes such as ERK and their substrates and thus regulate enzyme kinetics (41). In addition, binding of enzymes to actin can regulate enzyme activity, and this binding is dependent on the polymerization state of actin (24). G-actin can shuttle in and out of the nucleus, where it regulates chromatin structure and transcription (47). Cytoskeletal tension can also regulate the activity of the Rho family of small GTPases (5). Interestingly, the Rho family kinases, along with phosphatidyl inositol 3-kinase and p21-activated kinase were transcriptionally regulated by SerB, an effect that could reverberate throughout the host cell transcriptome.
Secretion of SerB by P. gingivalis upon stimulation with epithelial cell components would appear to be through an as-yet-undefined mechanism. In addition to lacking a type III secretory system, P. gingivalis does not possess functional homologs of type II secretion machinery. While P. gingivalis is capable of transferring DNA through type IV secretion (45), disruption of the genes required for type IV secretion does not prevent SerB secretion (unpublished observations). However, SerB does not appear to be delivered into the host cell and can exert an effect on the cell surface. The interaction of SerB with the gingival epithelial cell membrane may be a property of the two potential membrane-spanning domains of the enzyme (46). Alternatively, SerB may interact with, and potentially dephosphorylate, an as-yet-unidentified receptor(s) on the epithelial cell membrane.
Periodontal diseases, with which P. gingivalis is associated, involve inflammatory destruction of the supporting tissues of the teeth. Neutrophils are a key component of innate immune responses to periodontal bacteria, and individuals with neutrophil defects are at elevated risk of periodontal disease (14). The recruitment of neutrophils into the gingival crevice is antagonized by P. gingivalis through disruption of the IL-8 gradient. The ability of P. gingivalis to suppress production of IL-8 from epithelial cells stimulated with other oral bacteria causes local chemokine paralysis and may contribute to periods where the immune response is incapable of controlling the subgingival bacterial challenge (11). SerB was found to be required for P. gingivalis to inhibit HIGK IL-8 accumulation in response to F. nucleatum. As an effector molecule of local chemokine paralysis, SerB may play an important role in the virulence of P. gingivalis through modulation of local innate immunity.
In summary, SerB is a multifunctional invasin and modulin of P. gingivalis. In addition to a role in intracellular invasion through control of microtubule dynamics (46), secreted SerB acts on epithelial surfaces to impact actin cytoskeletal structure and cytokine production. These properties are associated with broadly based regulation of the epithelial cell transcriptome.
Acknowledgments
This work was supported by DE11111 (R.J.L.) and DE 16715 (M.H.).
Editor: J. B. Bliska
Footnotes
Published ahead of print on 7 April 2008.
REFERENCES
- 1.Arsura, M., G. R. Panta, J. D. Bilyeu, L. G. Cavin, M. A. Sovak, A. A. Oliver, V. Factor, R. Heuchel, F. Mercurio, S. S. Thorgeirsson, and G. E. Sonenshein. 2003. Transient activation of NF-κB through a TAK1/IKK kinase pathway by TGF-beta1 inhibits AP-1/SMAD signaling and apoptosis: implications in liver tumor formation. Oncogene 22412-425. [DOI] [PubMed] [Google Scholar]
- 2.Belton, C. M., P. C. Goodwin, S. Fatherazi, M. M. Schubert, R. J. Lamont, and K. T. Izutsu. 2004. Calcium oscillations in gingival epithelial cells infected with Porphyromonas gingivalis. Microbes Infect. 6440-447. [DOI] [PubMed] [Google Scholar]
- 3.Belton, C. M., K. T. Izutsu, P. C. Goodwin, Y. Park, and R. J. Lamont. 1999. Fluorescence image analysis of the association between Porphyromonas gingivalis and gingival epithelial cells. Cell Microbiol. 1215-223. [DOI] [PubMed] [Google Scholar]
- 4.Bernard, O. 2007. Lim kinases, regulators of actin dynamics. Int. J. Biochem. Cell Biol. 391071-1076. [DOI] [PubMed] [Google Scholar]
- 5.Bhadriraju, K., M. Yang, S. Alom Ruiz, D. Pirone, J. Tan, and C. S. Chen. 2007. Activation of ROCK by RhoA is regulated by cell adhesion, shape, and cytoskeletal tension. Exp. Cell Res. 3133616-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, M. C., and C. E. Turner. 2004. Paxillin: adapting to change. Physiol. Rev. 841315-1339. [DOI] [PubMed] [Google Scholar]
- 7.Chen, W., K. E. Laidig, Y. Park, K. Park, J. R. Yates III, R. J. Lamont, and M. Hackett. 2001. Searching the Porphyromonas gingivalis genome with peptide fragmentation mass spectra. Analyst 12652-57. [DOI] [PubMed] [Google Scholar]
- 8.Colombo, A. V., C. M. da Silva, A. Haffajee, and A. P. Colombo. 2007. Identification of intracellular oral species within human crevicular epithelial cells from subjects with chronic periodontitis by fluorescence in situ hybridization. J. Periodontal Res. 42236-243. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis, G. R. 2006. The type III secretion injectisome. Nat. Rev. Microbiol. 4811-825. [DOI] [PubMed] [Google Scholar]
- 10.Cossart, P. 2000. Actin-based motility of pathogens: the Arp2/3 complex is a central player. Cell Microbiol. 2195-205. [DOI] [PubMed] [Google Scholar]
- 11.Darveau, R. P., C. M. Belton, R. A. Reife, and R. J. Lamont. 1998. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 661660-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delaney, J. R., and M. Mlodzik. 2006. TGF-beta activated kinase-1: new insights into the diverse roles of TAK1 in development and immunity. Cell Cycle 52852-2855. [DOI] [PubMed] [Google Scholar]
- 13.DeVinney, I., I. Steele-Mortimer, and B. B. Finlay. 2000. Phosphatases and kinases delivered to the host cell by bacterial pathogens. Trends Microbiol. 829-33. [DOI] [PubMed] [Google Scholar]
- 14.Dixon, D. R., B. W. Bainbridge, and R. P. Darveau. 2004. Modulation of the innate immune response within the periodontium. Periodontology 2000 3553-74. [DOI] [PubMed] [Google Scholar]
- 15.Dold, F. G., J. M. Sanger, and J. W. Sanger. 1994. Intact alpha-actinin molecules are needed for both the assembly of actin into the tails and the locomotion of Listeria monocytogenes inside infected cells. Cell Motil. Cytoskeleton 2897-107. [DOI] [PubMed] [Google Scholar]
- 16.Dorn, B. R., W. A. Dunn, Jr., and A. Progulske-Fox. 2002. Bacterial interactions with the autophagic pathway. Cell Microbiol. 41-10. [DOI] [PubMed] [Google Scholar]
- 17.Eisenmann, K. M., E. S. Harris, S. M. Kitchen, H. A. Holman, H. N. Higgs, and A. S. Alberts. 2007. Dia-interacting protein modulates formin-mediated actin assembly at the cell cortex. Curr. Biol. 17579-591. [DOI] [PubMed] [Google Scholar]
- 18.Endo, M., K. Ohashi, and K. Mizuno. 2007. LIM kinase and Slingshot are critical for neurite extension. J. Biol. Chem. 28213692-13702. [DOI] [PubMed] [Google Scholar]
- 19.Goley, E. D., and M. D. Welch. 2006. The ARP2/3 complex: an actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 7713-726. [DOI] [PubMed] [Google Scholar]
- 20.Gruenheid, S., and B. B. Finlay. 2003. Microbial pathogenesis and cytoskeletal function. Nature 422775-781. [DOI] [PubMed] [Google Scholar]
- 21.Handfield, M., J. J. Mans, G. Zheng, M. C. Lopez, S. Mao, A. Progulske-Fox, G. Narasimhan, H. V. Baker, and R. J. Lamont. 2005. Distinct transcriptional profiles characterize oral epithelium-microbiota interactions. Cell Microbiol. 7811-823. [DOI] [PubMed] [Google Scholar]
- 22.Hasegawa, Y., J. J. Mans, S. Mao, M. C. Lopez, H. V. Baker, M. Handfield, and R. J. Lamont. 2007. Gingival epithelial cell transcriptional responses to commensal and opportunistic oral microbial species. Infect. Immun. 752540-2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hussain, H., P. Branny, and E. Allan. 2006. A eukaryotic-type serine/threonine protein kinase is required for biofilm formation, genetic competence, and acid resistance in Streptococcus mutans. J. Bacteriol. 1881628-1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji, Y., G. Ferracci, A. Warley, M. Ward, K. Y. Leung, S. Samsuddin, C. Leveque, L. Queen, V. Reebye, P. Pal, E. Gkaliagkousi, M. Seager, and A. Ferro. 2007. β-Actin regulates platelet nitric oxide synthase 3 activity through interaction with heat shock protein 90. Proc. Natl. Acad. Sci. USA 1048839-8844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khatri, P., S. Sellamuthu, P. Malhotra, K. Amin, A. Done, and S. Draghici. 2005. Recent additions and improvements to the Onto-Tools. Nucleic Acids Res. 33W762-W765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kinane, D. F., H. Shiba, P. G. Stathopoulou, H. Zhao, D. F. Lappin, A. Singh, M. A. Eskan, S. Beckers, S. Waigel, B. Alpert, and T. B. Knudsen. 2006. Gingival epithelial cells heterozygous for Toll-like receptor 4 polymorphisms Asp299Gly and Thr399Ile are hypo-responsive to Porphyromonas gingivalis. Genes Immun. 7190-200. [DOI] [PubMed] [Google Scholar]
- 27.Lamont, R. J., A. Chan, C. M. Belton, K. T. Izutsu, D. Vasel, and A. Weinberg. 1995. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 633878-3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lamont, R. J., and H. F. Jenkinson. 1998. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol. Mol. Biol. Rev. 621244-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maniotis, A. J., C. S. Chen, and D. E. Ingber. 1997. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. USA 94849-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mans, J. J., R. J. Lamont, and M. Handfield. 2006. Microarray analysis of human epithelial cell responses to bacterial interaction. Infect. Disord. Drug Targets 6299-309. [DOI] [PubMed] [Google Scholar]
- 31.Mao, S., Y. Park, Y. Hasegawa, G. D. Tribble, C. E. James, M. Handfield, M. F. Stavropoulos, O. Yilmaz, and R. J. Lamont. 2007. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 91997-2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maruta, H., T. V. Nheu, H. He, and Y. Hirokawa. 2003. Rho family-associated kinases PAK1 and Rock. Prog. Cell Cycle Res. 5203-210. [PubMed] [Google Scholar]
- 33.Monteville, M. R., J. E. Yoon, and M. E. Konkel. 2003. Maximal adherence and invasion of INT 407 cells by Campylobacter jejuni requires the CadF outer-membrane protein and microfilament reorganization. Microbiology 149153-165. [DOI] [PubMed] [Google Scholar]
- 34.Nakhjiri, S. F., Y. Park, O. Yilmaz, W. O. Chung, K. Watanabe, A. El-Sabaeny, K. Park, and R. J. Lamont. 2001. Inhibition of epithelial cell apoptosis by Porphyromonas gingivalis. FEMS Microbiol. Lett. 200145-149. [DOI] [PubMed] [Google Scholar]
- 35.Nelson, K. E., R. D. Fleischmann, R. T. DeBoy, I. T. Paulsen, D. E. Fouts, J. A. Eisen, S. C. Daugherty, R. J. Dodson, A. S. Durkin, M. Gwinn, D. H. Haft, J. F. Kolonay, W. C. Nelson, T. Mason, L. Tallon, J. Gray, D. Granger, H. Tettelin, H. Dong, J. L. Galvin, M. J. Duncan, F. E. Dewhirst, and C. M. Fraser. 2003. Complete genome sequence of the oral pathogenic bacterium Porphyromonas gingivalis strain W83. J. Bacteriol. 1855591-5601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oda, D., L. Bigler, P. Lee, and R. Blanton. 1996. HPV immortalization of human oral epithelial cells: a model for carcinogenesis. Exp. Cell Res. 226164-169. [DOI] [PubMed] [Google Scholar]
- 37.Park, Y., and R. J. Lamont. 1998. Contact-dependent protein secretion in Porphyromonas gingivalis. Infect. Immun. 664777-4782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rajagopal, L., A. Clancy, and C. E. Rubens. 2003. A eukaryotic type serine/threonine kinase and phosphatase in Streptococcus agalactiae reversibly phosphorylate an inorganic pyrophosphatase and affect growth, cell segregation, and virulence. J. Biol. Chem. 27814429-14441. [DOI] [PubMed] [Google Scholar]
- 39.Ramarao, N., C. Le Clainche, T. Izard, R. Bourdet-Sicard, E. Ageron, P. J. Sansonetti, M. F. Carlier, and G. Tran Van Nhieu. 2007. Capping of actin filaments by vinculin activated by the Shigella IpaA carboxyl-terminal domain. FEBS Lett. 581853-857. [DOI] [PubMed] [Google Scholar]
- 40.Ridley, A. J. 2001. Rho family proteins: coordinating cell responses. Trends Cell Biol. 11471-477. [DOI] [PubMed] [Google Scholar]
- 41.Rosengart, M. R., S. Arbabi, G. J. Bauer, I. Garcia, S. Jelacic, and R. V. Maier. 2002. The actin cytoskeleton: an essential component for enhanced TNFα production by adherent monocytes. Shock 17109-113. [DOI] [PubMed] [Google Scholar]
- 42.Schubert-Unkmeir, A., O. Sokolova, U. Panzner, M. Eigenthaler, and M. Frosch. 2007. Gene expression pattern in human brain endothelial cells in response to Neisseria meningitidis. Infect. Immun. 75899-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scott, J. A., A. M. Shewan, N. R. den Elzen, J. J. Loureiro, F. B. Gertler, and A. S. Yap. 2006. Ena/VASP proteins can regulate distinct modes of actin organization at cadherin-adhesive contacts. Mol. Biol. Cell 171085-1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Socransky, S. S., A. D. Haffajee, L. A. Ximenez-Fyvie, M. Feres, and D. Mager. 1999. Ecological considerations in the treatment of Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis periodontal infections. Periodontology 2000 20341-362. [DOI] [PubMed] [Google Scholar]
- 45.Tribble, G. D., G. J. Lamont, A. Progulske-Fox, and R. J. Lamont. 2007. Conjugal transfer of chromosomal DNA contributes to genetic variation in the oral pathogen Porphyromonas gingivalis. J. Bacteriol. 1896382-6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tribble, G. D., S. Mao, C. E. James, and R. J. Lamont. 2006. A Porphyromonas gingivalis haloacid dehalogenase family phosphatase interacts with human phosphoproteins and is important for invasion. Proc. Natl. Acad. Sci. USA 10311027-11032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vartiainen, M. K., S. Guettler, B. Larijani, and R. Treisman. 2007. Nuclear actin regulates dynamic subcellular localization and activity of the SRF cofactor MAL. Science 3161749-1752. [DOI] [PubMed] [Google Scholar]
- 48.Wang, B., S. Li, S. Dedhar, and P. P. Cleary. 2007. Paxillin phosphorylation: bifurcation point downstream of integrin-linked kinase (ILK) in streptococcal invasion. Cell Microbiol. 91519-1528. [DOI] [PubMed] [Google Scholar]
- 49.Watanabe, K., O. Yilmaz, S. F. Nakhjiri, C. M. Belton, and R. J. Lamont. 2001. Association of mitogen-activated protein kinase pathways with gingival epithelial cell responses to Porphyromonas gingivalis infection. Infect. Immun. 696731-6737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xia, Q., T. Wang, F. Taub, Y. Park, C. A. Capestany, R. J. Lamont, and M. Hackett. 2007. Quantitative proteomics of intracellular Porphyromonas gingivalis. Proteomics 74323-4337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xu, J., D. Wirtz, and T. D. Pollard. 1998. Dynamic cross-linking by alpha-actinin determines the mechanical properties of actin filament networks. J. Biol. Chem. 2739570-9576. [DOI] [PubMed] [Google Scholar]
- 53.Yilmaz, O., T. Jungas, P. Verbeke, and D. M. Ojcius. 2004. Activation of the phosphatidylinositol 3-kinase/Akt pathway contributes to survival of primary epithelial cells infected with the periodontal pathogen Porphyromonas gingivalis. Infect. Immun. 723743-3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yilmaz, O., P. Verbeke, R. J. Lamont, and D. M. Ojcius. 2006. Intercellular spreading of Porphyromonas gingivalis infection in primary gingival epithelial cells. Infect. Immun. 74703-710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yilmaz, O., K. Watanabe, and R. J. Lamont. 2002. Involvement of integrins in fimbriae-mediated binding and invasion by Porphyromonas gingivalis. Cell Microbiol. 4305-314. [DOI] [PubMed] [Google Scholar]
- 56.Yilmaz, O., P. A. Young, R. J. Lamont, and G. E. Kenny. 2003. Gingival epithelial cell signalling and cytoskeletal responses to Porphyromonas gingivalis invasion. Microbiology 1492417-2426. [DOI] [PubMed] [Google Scholar]
- 57.Yin, J. L., N. A. Shackel, A. Zekry, P. H. McGuinness, C. Richards, K. V. Putten, G. W. McCaughan, J. M. Eris, and G. A. Bishop. 2001. Real-time reverse transcriptase-polymerase chain reaction (RT-PCR) for measurement of cytokine and growth factor mRNA expression with fluorogenic probes or SYBR Green I. Immunol. Cell Biol. 79213-221. [DOI] [PubMed] [Google Scholar]
- 58.Zhang, Y., T. Wang, W. Chen, O. Yilmaz, Y. Park, I. Y. Jung, M. Hackett, and R. J. Lamont. 2005. Differential protein expression by Porphyromonas gingivalis in response to secreted epithelial cell components. Proteomics 5198-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhao, Z. S., E. Manser, X. Q. Chen, C. Chong, T. Leung, and L. Lim. 1998. A conserved negative regulatory region in αPAK: inhibition of PAK kinases reveals their morphological roles downstream of Cdc42 and Rac1. Mol. Cell. Biol. 182153-2163. [DOI] [PMC free article] [PubMed] [Google Scholar]