Abstract
PTEN, a tumor suppressor whose function is frequently lost in human cancers, possesses a lipid phosphatase activity that represses phosphatidylinositol 3-kinase (PI3K) signaling, controlling cell growth, proliferation, and survival. The potential for PTEN to regulate the synthesis of RNA polymerase (Pol) III transcription products, including tRNAs and 5S rRNAs, was evaluated. The expression of PTEN in PTEN-deficient cells repressed RNA Pol III transcription, whereas decreased PTEN expression enhanced transcription. Transcription repression by PTEN was uncoupled from PTEN-mediated effects on the cell cycle and was independent of p53. PTEN acts through its lipid phosphatase activity, inhibiting the PI3K/Akt/mTOR/S6K pathway to decrease transcription. PTEN, through the inactivation of mTOR, targets the TFIIIB complex, disrupting the association between TATA-binding protein and Brf1. Kinetic analysis revealed that PTEN initially induces a decrease in the serine phosphorylation of Brf1, leading to a selective reduction in the occupancy of all TFIIIB subunits on tRNALeu genes, whereas prolonged PTEN expression results in the enhanced serine phosphorylation of Bdp1. Together, these results demonstrate a new class of genes regulated by PTEN through its ability to repress the activation of PI3K/Akt/mTOR/S6K signaling.
PTEN (phosphatase and tensin homolog deleted on chromosome 10) is a frequently mutated or deleted gene in human cancer. Somatic inactivating mutations in PTEN are found in multiple sporadic tumor types. Germ line mutations of PTEN result in inherited hamartoma and the cancer predisposition syndrome Cowden disease (5). Key to its tumor suppressor function is the ability of PTEN to negatively regulate the phosphatidylinositol 3-kinase (PI3K) signaling cascade. The lipid phosphatase activity of PTEN dephosphorylates phosphatidylinositol 3,4,5-triphosphate at the plasma membrane, which then inhibits PI3K-mediated signals for growth, proliferation, and survival (10). In addition to this cytoplasmic function, PTEN is also found in the nucleus in many normal and tumor cells, where it regulates the expression of select genes, such as p53, and maintains chromosome stability (23, 24, 33, 37, 43).
RNA polymerase (Pol) III is responsible for the synthesis of a variety of small untranslated RNAs, including tRNAs, 5S rRNAs, 7SL RNA, U6 RNA, and, most recently identified, Alu-associated microRNAs (2). The type 2 class of promoters, typified by tRNA gene promoters, requires the transcription factor complexes TFIIIB and TFIIIC, in addition to RNA Pol III, to specify accurate and efficient transcription (30). This TFIIIB complex, used by both tRNA and 5S rRNA promoters, consists of TATA-binding protein (TBP) and the associated factors Brf1 and Bdp1. In contrast, the U6 RNA gene uses a TFIIIB complex that consists of TBP, Bdp1, and Brf2, a differentially spliced variant of Brf1 (30). Consistent with the idea that a high translational capacity is necessary for the rapid growth and proliferation of tumor cells, RNA Pol III transcription products are elevated in transformed and tumor cells (3, 4, 15, 31, 49). Accordingly, the tumor suppressors p53 (6) and Rb (19, 35) repress, while oncogenic c-myc (15) induces, RNA Pol III-dependent transcription. The ability of these proteins to deregulate RNA Pol III-dependent transcription occurs through their capacity to directly associate with the TFIIIB complex and modify its function.
While a variety of cellular proteins that serve to directly modulate RNA Pol III-dependent transcription have been identified, comparatively little is known regarding the intracellular signaling pathways that serve to regulate this class of genes in mammalian cells. The activation of epidermal growth factor receptor 1 leads to the induction of TBP expression, requiring the activation of Ras and all three classes of mitogen-activated protein kinases (MAPKs) (51). Since TBP is a limiting component for RNA Pol III transcription in certain cell types and contexts, this increase in TBP, alone, can stimulate transcription (51). In addition to MAPK-mediated alterations in cellular TBP amounts, extracellular signal-regulated kinase (ERK) directly phosphorylates Brf1, thereby inducing tRNA gene transcription (12). While PI3K/Akt/mTOR signaling and its effect on RNA Pol III transcription in mammalian cells have not been examined, the TOR inhibitor rapamycin represses RNA Pol III transcription in Saccharomyces cerevisiae, inducing a response similar to that produced by nutrient limitation (39).
Given its importance as a tumor suppressor, we have examined the potential for PTEN to regulate RNA Pol III-dependent transcription. Increased PTEN expression serves to repress the transcription of tRNA and 7SL RNA genes, while decreased PTEN expression induces transcription in a p53-independent manner. Transcriptional repression requires the lipid phosphatase activity of PTEN, suggesting that PTEN acts through its ability to inhibit PI3K signaling. Furthermore, blocking the activation of PI3K, Akt, or mTOR represses RNA Pol III transcription, whereas the activation of PI3K signaling is sufficient for inducing transcription. The expression of constitutively activated S6K, a downstream target of mTOR, is able to alleviate PTEN-mediated transcriptional repression. Although PTEN has been shown to negatively regulate cyclin D1 and induce cell cycle arrest under certain conditions (14, 27, 44), and RNA Pol III transcription is tightly regulated throughout the cell cycle (11, 16, 18), we find that PTEN-mediated repression of RNA Pol III transcription can be uncoupled from PTEN-mediated effects on cyclin D1 and the cell cycle. Analysis of PTEN-mediated effects on the transcription machinery revealed that PTEN negatively regulates the association between TBP and Brf1, thus preventing the formation of functional TFIIIB complexes. In a similar manner, inhibiting the activation of mTOR also reduces the number of TBP-Brf1 complexes. PTEN induction initially induces an increase in the serine phosphorylation of Brf1, which results in the loss of all three TFIIIB subunits from tRNA gene promoters. After prolonged PTEN induction, an increase in the serine phosphorylation of Bdp1 is observed, with no further loss of TFIIIB occupancy from these genes. Consistent with PTEN and the PI3K/Akt/mTOR/S6K pathway targeting Brf1 to repress transcription, the U6 RNA gene, which does not use Brf1 for transcription, is not regulated through this pathway. Together, these results identify a new class of genes negatively regulated by PTEN through its ability to inhibit PI3K/Akt/mTOR/S6K signaling, which targets and prevents the formation of functional TFIIIB complexes.
MATERIALS AND METHODS
Plasmids and reagents.
Expression plasmids, described previously, are as follows: PTEN, PTEN-G129E (40), Flag-cyclin D1, Flag-cyclin D1-T286A (8), PI3K-DN, Akt-DN (50), RasV12, RasV12C40 (45), and S6K-E389 (36). PTEN small interfering RNA (siRNA) and mismatch RNA (mmRNA) were described previously (50). Primary anti-human antibodies used are as follows: mouse polyclonal PTEN (Axel Schönthal, University of Southern California), rabbit polyclonal TBP (Upstate), rabbit polyclonal Bdp1 and TFIIIC102 (Robert Roeder, Rockefeller University), goat polyclonal Brf1, goat polyclonal p27, rabbit polyclonal cyclin D1, rabbit polyclonal Akt, rabbit polyclonal phospho-Akt (Cell Signaling), rabbit polyclonal phosphoserine, mouse monoclonal β-actin (Chemicon), and mouse monoclonal Flag (Sigma-Aldrich). Doxycycline, wortmannin, and rapamycin were obtained from Sigma-Aldrich.
Transcription and promoter assays.
PC-3 and LNCaP prostate cell lines; A172, U87, and LN18 glioblastoma cell lines; and MCF10A and MCF-7 breast epithelial cell lines were obtained from the American Type Culture Collection (ATCC). A172, U87, LN18, and MCF-7 cells were grown in Dulbecco's modified Eagle's medium with 4.5 g/liter glucose and 10% fetal calf serum. PC-3 and LNCaP cells were grown in RPMI 1640 with 10% fetal calf serum. MCF10A cells were grown in medium as specified by the ATCC. PTEN-inducible and PTEN-C124S-inducible U87 cell lines were obtained from M.-M. Georgescu (M. D. Anderson Cancer Center, Houston, TX) (27). To induce PTEN and PTEN-C124S expression, cells were incubated with doxycycline (DOX) at 1 μg/ml for either 6 or 24 h. A172, U87, U87-inducible, and LN18 cells were transfected using Lipofectin (1 μl reagent to 1 μg DNA; Invitrogen). Primary rat hepatocytes were transfected as described previously (21). Lipofectamine 2000 (1 μl reagent to 1 μg DNA/siRNA; Invitrogen) was used for cotransfection of DNA and 100 nM PTEN siRNA or mmRNA into LN18 and MCF-7 cells. For transfections with expression vectors, appropriate empty vectors were used in controls. Cells were incubated with the transfection mix for 4 h and then replaced with fresh medium. Twenty-four hours following transfection, cells were harvested, and total RNA or protein was isolated. RNA Pol III transcription of the reporter gene, a modified tRNAArg, pArg-maxigene, was measured by an RNase protection assay (42). Total RNA from cells was isolated using RNA Stat-60 (TelTest). RNA analyses were normalized to total RNA. For in vitro transcription assays, cell extracts were prepared (9), and the transcription reaction was performed with either 10 to 20 μg of nuclear protein extract as described previously (51). Transcription products were visualized by autoradiography. Resultant autoradiographs were quantified using UN-Scan-It software (Silk Scientific).
Immunoblot and immunoprecipitation analyses.
Protein lysates were subjected to immunoblot analysis as described previously (6). Immunoprecipitation assays were carried out as previously described (50). Bound primary antibody was visualized using biotinylated secondary antibody complexed with avidin/biotinylated peroxidase (Vector Laboratories) and enhanced chemiluminescence reagents (Pico Western blotting; Pierce). The resultant autoradiographs were quantified using UN-Scan-It software.
Semiquantitative and quantitative RT-PCR analyses.
Total RNA was isolated from cells as described above. RNA was reverse transcribed using the SuperScript III first-strand synthesis system for reverse transcription (RT)-PCR (Invitrogen). Primer sets for pre-tRNATyr, pre-tRNALeu, 7SL RNA, U6 RNA, β-actin, and GAPDH (glyceraldehyde-3-phosphate dehydrogenase) were previously described (17, 22, 49). Semiquantitative PCR was performed on serial dilutions of cDNA, and products were resolved on agarose gels and visualized by ethidium bromide staining. Gels were scanned using an AlphaImager 2000 apparatus (Alpha Innotech) and quantified using UN-Scan-It software. Quantitative PCR was performed as described previously (22).
FACS analysis.
Single-cell suspensions in phosphate-buffered saline (PBS) were fixed with 100% ethanol and then centrifuged and washed with PBS. Cells were centrifuged again, resuspended in PBS containing 40 μg/ml RNase and 20 μg/ml propidium iodide, and then incubated at room temperature for 2 h. Fluorescence-activated cell sorting (FACS) analysis was performed by the University of Southern California Norris Cancer Center Flow Cytometry Core Facility.
ChIP assay.
Chromatin immunoprecipitation (ChIP) was performed as described previously (6, 7, 22). Quantitative PCR was performed using primers for the pre-tRNALeu gene promoter. Threshold cycle values were normalized to the PCR efficiency (between 95 and 105%). Each time point (0, 6, and 24 h) was normalized to its respective input.
RESULTS
Cytoplasmic, but not nuclear, PTEN represses RNA Pol III transcription independently of p53 via its lipid phosphatase activity.
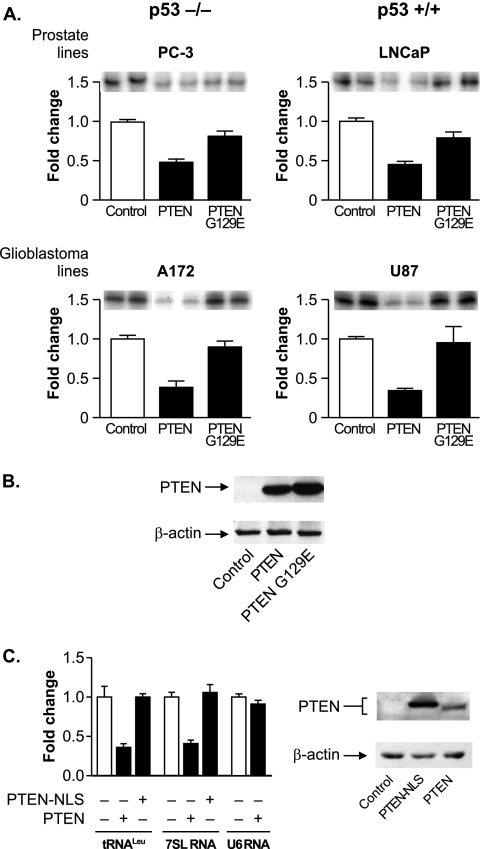
We first determined whether the expression of PTEN in cells lacking endogenous PTEN could regulate RNA Pol III transcription. As PTEN positively regulates p53 (13, 25, 34) and RNA Pol III transcription is negatively regulated by p53 (6), we used cells either lacking or expressing endogenous p53 to determine whether PTEN could repress RNA Pol III transcription in the absence of p53. All of these cell lines are deficient for PTEN. The glioblastoma-derived A172 (p53−/−) and U87 (p53+/+) and prostate-derived PC-3 (p53−/−) and LnCap (p53+/+) cell lines were transiently transfected with a tRNAArg gene reporter plasmid together with expression plasmids for PTEN or a mutant PTEN-G129E lacking lipid phosphatase activity. The expression of PTEN, but not PTEN-G129E, reduced tRNAArg gene transcription in all cell lines (Fig. 1A). Immunoblot analysis confirmed that both PTEN and PTEN G129E were appreciably expressed in these cells (Fig. 1B).
FIG. 1.
PTEN represses RNA Pol III transcription via its lipid phosphatase activity independently of p53. (A) PTEN expression in PTEN null cell lines decreases RNA Pol III transcription. Prostate PC-3 (p53−/−) and LnCap (p53+/+) and glioblastoma A172 (p53−/−) and U87 (p53+/+) cell lines were transiently cotransfected with a tRNA gene reporter and an expression vector for PTEN or PTEN-G129E, a lipid phosphatase-defective mutant. Total RNA was isolated, and RNase protection assays were performed to determine the amounts of transcript. The change in transcription is calculated based on reporter gene activity in cells transfected with an empty vector control. (B) Expression of PTEN and PTEN-G129E in U87 cells. Total protein was isolated from cells transfected as described above (A) and subjected to immunoblot analysis using antibodies against PTEN or β-actin. Representative autoradiographs are shown. (C) Targeting PTEN to the nucleus prevents repression of RNA Pol III transcription. U87 cells were transfected with an expression vector for PTEN or PTEN-NLS. (Left) Total RNA was isolated and reverse transcribed. Quantitative real-time PCR analysis was performed to quantify the level of transcription of endogenous precursor tRNALeu, 7SL RNA, and U6 RNA. For quantification, transcript levels were each normalized to values obtained for GAPDH transcripts. The change in transcription is calculated based on the level of transcripts in cells transfected with an empty vector. Graphs represent determinations from at least three biologically independent samples (means ± standard errors [SE]). (Right) Total protein was isolated from transfected cells and subjected to immunoblot analysis using antibodies against PTEN or β-actin. Representative autoradiographs are shown.
In addition to its well-established role in the cytoplasm, PTEN has been shown to also function in the nucleus (23, 24, 33, 37, 43). Therefore, we examined whether PTEN might directly act to repress RNA Pol III transcription in the nucleus. We first assessed the ability of wild-type PTEN to repress endogenous precursor tRNALeu, 7SL RNA, and U6 RNA in U87 cells. PTEN expression resulted in a decrease in both precursor tRNALeu and 7SL RNA but not U6 RNA (Fig. 1C, left). The ability of wild-type PTEN to repress transcription was compared to that of PTEN containing a nuclear localization signal (PTEN-NLS), which was also effectively expressed in these cells (Fig. 1C, right). As shown in Fig. 1, PTEN-NLS expression failed to repress tRNA and 7SL RNA expression in U87 cells. These results support the idea that only cytoplasmic PTEN functions to repress RNA Pol III transcription. In addition, the inability of PTEN to regulate U6 RNA expression indicates that only specific classes of RNA Pol III promoters may be regulated by PTEN.
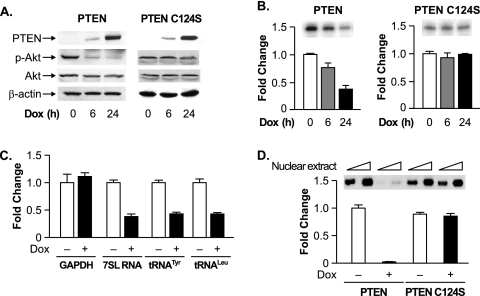
To further examine PTEN-mediated effects on endogenous RNA Pol III-dependent transcription, we used U87 cell lines engineered to express PTEN, or phosphatase-defective PTEN-C124S, in the presence of DOX. Immunoblot analysis revealed that these cells display no detectable expression of PTEN or PTEN-C124S in the absence of an inducer. However, in the presence of DOX, a robust induction of both PTEN and PTEN-C124S was observed (Fig. 2A). The induction of PTEN, but not PTEN-C124S, resulted in a decrease in the activated phosphorylated form of Akt. Furthermore, the induction of PTEN, but not PTEN-C124S, repressed the transient expression of the tRNA reporter gene (Fig. 2B). To measure endogenous RNA Pol III transcription, RT-PCR analysis was performed using total RNA isolated from noninduced and DOX-induced U87 cells. The induction of PTEN reduced endogenous 7SL RNA, precursor tRNATyr, and precursor tRNALeu levels, compared with no observable change in GAPDH mRNA (Fig. 2C). This PTEN-mediated decrease in RNA Pol III-dependent transcription was reproduced in vitro (Fig. 2D). Extracts derived from PTEN-induced cells displayed a substantial decrease in their capacity to transcribe a tRNA gene template compared to extracts derived from PTEN-C124S-induced cells.
FIG. 2.
PTEN selectively represses endogenous RNA Pol III transcription. (A) PTEN expression was induced in stable U87 cell lines. U87-PTEN or U87-PTEN-C124S cells, engineered to induce the expression of PTEN or PTEN-C124S in the presence of DOX, were treated with DOX for 0, 6, or 24 h. Total protein was isolated and subjected to immunoblot analysis using antibodies against PTEN, phospho-Akt (p-Akt), Akt, or β-actin. (B) Induction of PTEN, but not PTEN-C124S, decreases RNA Pol III transcription. Inducible U87-PTEN cells were transfected with the tRNA gene reporter and treated with DOX. Total RNA was isolated, and RNase protection assays were performed. The change in transcription is calculated based on reporter gene activity in untreated cells (0 h). (C) PTEN represses endogenous RNA Pol III transcription. Inducible U87-PTEN cell lines were treated with DOX (24 h), and total RNA was isolated and reverse transcribed. PCR analysis was performed on serial dilutions of cDNAs to quantify the level of transcription of GAPDH mRNA and endogenous precursor transcripts for tRNALeu, tRNATyr, and 7SL RNA. The change in transcription is calculated based on the level of transcripts in untreated cells (0 h). (D) Induction of PTEN, but not PTEN-C124S, decreases RNA Pol III transcription in vitro. Nuclear extracts were prepared from inducible U87-PTEN cell lines treated with DOX (24 h). Transcription assays were carried out using the tRNA gene reporter as a template and 10 or 20 μg of nuclear extract. The change in transcription is calculated based on the level of transcription in untreated cells (0 h). Representative autoradiographs are shown. Graphs represent determinations from at least three biologically independent samples (means ± SE).
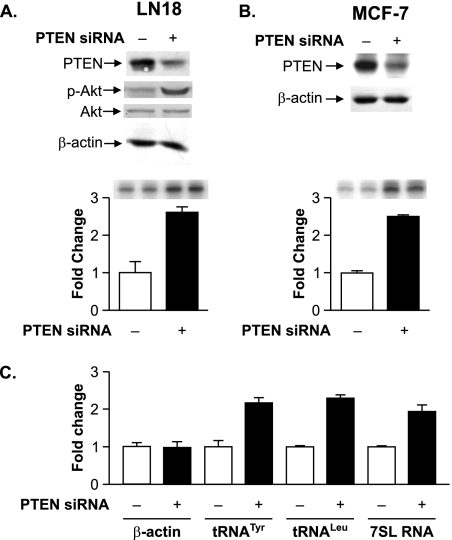
We next determined whether the reduction of PTEN expression in cell lines that contain endogenous PTEN could enhance RNA Pol III transcription. The glioblastoma-derived LN18 cell line and the breast epithelial cell-derived MCF-7 cell line that express wild-type endogenous PTEN were transiently cotransfected with the tRNAArg reporter gene and either an siRNA directed against PTEN or a control mmRNA. Immunoblot analysis illustrates that PTEN expression was decreased approximately two- to fivefold in cells transfected with the PTEN-specific siRNA (Fig. 3A and B). The reduction in PTEN in LN18 cells was accompanied by an increase in the activation state of Akt (Fig. 3A). A reduction in PTEN expression resulted in enhanced transcription of the transfected tRNA gene in both LN18 and MCF-7 cells (Fig. 3A and B). Other PTEN-targeted siRNAs that resulted in a reduction in PTEN expression also served to enhance RNA Pol III transcription (data not shown). Comparison of the amounts of endogenous 7SL RNA, tRNALeu, and tRNATyr precursor transcripts showed a similar increase upon reduced PTEN expression compared with the amount of β-actin mRNA (Fig. 3C). Collectively, these results indicate that cytoplasmic, but not nuclear, PTEN represses RNA Pol III-dependent transcription independently of cell type or p53 status. Furthermore, these results support the idea that PTEN acts to repress transcription through its ability to inhibit PI3K activation through its lipid phosphatase activity.
FIG. 3.
Decreasing PTEN expression selectively enhances RNA Pol III transcription. (A) Reducing expression of PTEN enhances RNA Pol III transcription in LN18 cells. LN18 cells were transfected with PTEN siRNA or control mmRNA. (Top) Total protein was isolated, and immunoblot analysis was performed using antibodies against PTEN, phospho-Akt (p-Akt), Akt, or β-actin. (Bottom) Cells were cotransfected with the tRNA reporter gene and PTEN siRNA or mmRNA. Total RNA was isolated, and RNase protection assays were performed. Representative autoradiographs are shown. The change in transcription is calculated based on reporter gene activity in cells transfected with mmRNA. (B) Reducing expression of PTEN enhances RNA Pol III transcription in MCF-7 cells. MCF-7 cells were transfected as described above (A). (Top) Total protein was isolated, and immunoblot analysis was performed using antibodies against PTEN and β-actin. (Bottom) Total RNA was isolated, and RNase protection assays were performed as described above (A). (C) Decreasing PTEN expression selectively enhances endogenous RNA Pol III transcription. LN18 cells were transfected with PTEN siRNA or mmRNA. Total RNA was isolated and reverse transcribed. PCR analysis was performed on serial dilutions of cDNAs to quantify the expression of β-actin mRNA, precursor tRNATyr, precursor tRNALeu, and 7SL RNA. The change in transcription is calculated based on transcript levels in cells transfected with mmRNA. Graphs represent determinations from at least three biologically independent samples (means ± SE).
RNA Pol III-dependent transcription is regulated through the PI3K/Akt/mTOR/S6K signaling pathway.
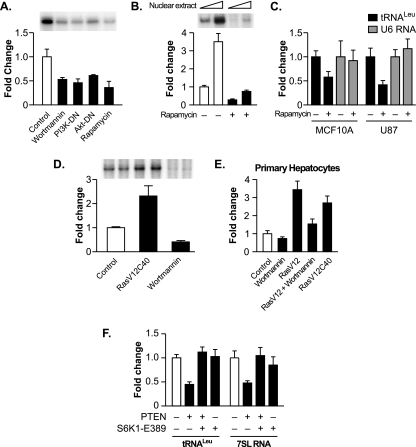
The lipid phosphatase activity of PTEN, which is required for its function as a tumor suppressor, directly antagonizes the PI3K signal transduction pathway by dephosphorylating the membrane lipid that is phosphorylated by PI3K (26). Since the PTEN-mediated repression of RNA Pol III transcription required its lipid phosphatase activity, this suggests that PTEN functions via its antagonism of the PI3K signal transduction pathway. While PI3K signaling has been shown to regulate RNA Pol III-dependent transcription in yeast, the ability of PI3K and its downstream signaling molecules to regulate the transcription of this class of genes in mammalian cells has not been examined. We therefore determined whether RNA Pol III transcription is regulated through this pathway by inhibiting the activation of PI3K and the downstream components Akt and mTOR. LN18 cells transiently transfected with the tRNA gene reporter were treated with either the PI3K inhibitor wortmannin or the mTOR inhibitor rapamycin or were transiently cotransfected with expression plasmids encoding dominant negative mutants of either PI3K or Akt. In each case, a marked decrease in RNA Pol III transcription was observed (Fig. 4A). This can be compared to transcription from the TBP promoter, which was not affected by inhibiting PI3K-mediated signaling (50; data not shown). The regulation of tRNA gene transcription by mTOR was further observed in vitro, as extracts derived from U87 cells treated with rapamycin displayed a substantial decrease in transcriptional capacity compared to those of extracts derived from nontreated cells (Fig. 4B). Furthermore, U87 and MCF10A cells treated with rapamycin displayed a decrease in endogenous precursor tRNALeu but not in U6 RNA (Fig. 4C). Thus, inhibiting the PI3K/Akt/mTOR pathway decreases the transcription of tRNA but not U6 RNA genes. To determine if the activation of PI3K signaling can induce transcription, LnCap cells were transfected with an expression vector containing the Ras effector mutant RasV12C40, which selectively activates PI3K (45). The expression of RasV12C40 was sufficient for inducing tRNA gene transcription, whereas wortmannin treatment reduced transcription (Fig. 4D).
FIG. 4.
The PI3K/Akt/mTOR signal transduction pathway regulates RNA Pol III transcription. (A) Inhibiting the PI3K/Akt/mTOR pathway represses RNA Pol III transcription. LN18 cells were transfected with the tRNA gene reporter and expression vectors for dominant negative mutants of PI3K (PI3K-DN) or Akt (Akt-DN) or were treated with 0.5 μM wortmannin (6 h) or 100 nM rapamycin for 3 h prior to harvesting of the cells. Total RNA was isolated, and RNase protection assays were performed. The change in transcription is calculated based on reporter gene activity in untreated cells transfected with an empty vector control. (B) Inhibiting mTOR decreases RNA Pol III transcription in vitro. Nuclear extracts were prepared from LN18 cells following treatment with 100 nM rapamycin for 3 h. Transcription assays were carried out using the tRNA reporter gene as a template and 10 or 20 μg of nuclear extract. (C) Inhibiting mTOR decreases endogenous tRNA but not U6 RNA transcription. MCF-10A and U87 cells were treated with 100 nM rapamycin for 3 h prior to harvesting of cells. Total RNA was isolated and reverse transcribed. Quantitative real-time PCR analysis was performed to quantify the level of transcription of endogenous precursor tRNALeu and U6 RNA. For quantification, transcript levels were each normalized to values obtained for GAPDH transcripts. The change in transcription is calculated based on the level of transcripts in cells treated with dimethyl sulfoxide. (D) Activating the PI3K pathway induces RNA Pol III transcription in LnCap cells. LnCap cells were transfected with the tRNA gene reporter, treated with wortmannin, or cotransfected with RasV12C40, a constitutively activated Ras specific for PI3K activation, as designated. Total RNA was isolated, and RNase protection assays were performed. The change in transcription is calculated based on reporter gene activity in untreated cells transfected with an empty vector control. (E) RNA Pol III transcription is induced by the activation of PI3K in primary rat hepatocytes. Primary rat hepatocytes were cotransfected the tRNA gene reporter and RasV12 or RasV12C40 expression vectors and/or were treated with 0.5 μM wortmannin for 6 h prior to harvesting of the cells. Total RNA was isolated, and RNase protection assays were performed. The change in transcription is calculated based on reporter gene activity in untreated cells transfected with an empty vector control. (F) Expression of constitutively activated S6K1 alleviates PTEN-mediated repression of RNA Pol III-dependent transcription. U87 cells were transfected with an expression vector for PTEN and with either constitutively activated S6K1 (S6K1-E389) or an empty vector. Total RNA was isolated and reverse transcribed. Quantitative real-time PCR analysis was performed to quantify the level of transcription of endogenous precursor tRNALeu and 7SL RNA. For quantification, transcript levels were each normalized to values obtained for GAPDH transcripts. The change in transcription is calculated based on the level of transcripts in cells transfected with an empty vector. All graphs represent determinations from at least three biologically independent samples (means ± SE).
We further examined the regulation of RNA Pol III transcription through the PI3K signaling pathway in primary rat hepatocytes. Consistent with the quiescent nature of primary cells, the treatment of the cells with wortmannin had little effect on the activity of the transfected tRNA gene reporter compared to that of nontreated cells (Fig. 4E). However, the cotransfection of a constitutively activated form of Ras, RasV12, produced an appreciable increase in the level of transcription, which was inhibited upon treatment with wortmannin. Furthermore, the expression of the Ras effector mutant RasV12C40 stimulated tRNA gene transcription. Collectively, these results support the idea that the PI3K/Akt/mTOR signaling pathway regulates RNA Pol III-dependent transcription in mammalian cells.
We further determined whether the ability of PTEN to inhibit the activation of mTOR and its downstream target, S6K, was sufficient for the PTEN-mediated repression of RNA Pol III transcription. U87 cells were cotransfected with expression plasmids for PTEN together with a constitutively activated S6K (S6K1-E389) expression vector or an empty vector. In the absence of PTEN, the expression of S6K1-E389 had no effect on precursor tRNA or 7SL RNA synthesis (Fig. 4F). However, S6K1-E389 expression was able to alleviate PTEN-mediated repression of transcription. Taken together, these results support the idea that PTEN acts to repress RNA Pol III transcription primarily through its ability to repress PI3K/Akt/mTOR/S6K signaling.
PTEN-mediated repression of RNA Pol III transcription is independent of PTEN-mediated effects on the cell cycle.
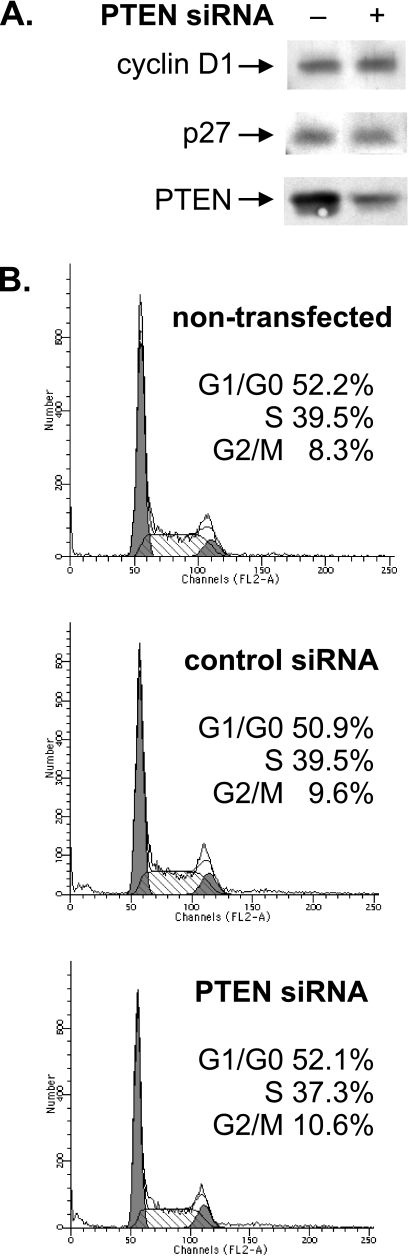
RNA Pol III transcription is regulated throughout the cell cycle and is at a minimum in the M phase (11, 16, 18, 20, 47, 48). PTEN has been found to mediate cell cycle arrest in G1 phase in U87 cells by increasing the protein stability of the cyclin-dependent kinase inhibitor p27 and, conversely, decreasing the protein levels of cyclin D1 and its nuclear localization (27). Therefore, we examined whether the changes observed in RNA Pol III transcription, with the manipulation of PTEN levels, were due to PTEN-mediated changes in the cell cycle. LN18 cells were transiently transfected with an siRNA directed against PTEN or a control mmRNA. Under these conditions, the transfection efficiency was approximately 70 to 80% (data not shown). Immunoblot analysis revealed that reducing PTEN protein levels did not affect the amounts of the cell cycle regulatory proteins cyclin D1 and p27 (Fig. 5A). Furthermore, FACS analysis revealed no discernible difference in the cell cycle profiles between nontransfected cells, cells transfected with mmRNA, and cells transfected with PTEN siRNA (Fig. 5B). These results indicate that under these experimental conditions, PTEN is not affecting cell cycle changes. Thus, PTEN-mediated changes in RNA Pol III transcription in LN18 cells can be uncoupled from PTEN-mediated effects on the cell cycle.
FIG. 5.
Reduction of PTEN levels in LN18 cells does not induce cell cycle changes. (A) Reduction of PTEN expression levels does not affect the levels of cyclin D1 or p27. LN18 cells were transfected with PTEN siRNA or mmRNA. Total protein was isolated 24 h posttransfection and subjected to immunoblot analysis with antibodies against cyclin D1, p27, or PTEN. (B) Reduction of PTEN levels does not alter the cell cycle profile. A portion of the LN18 cells transfected with PTEN siRNA or mmRNA (A) and LN18 cells that were not transfected were prepared for FACS analysis, and the numbers of cells in G0/G1, S, and G2/M phases were measured.
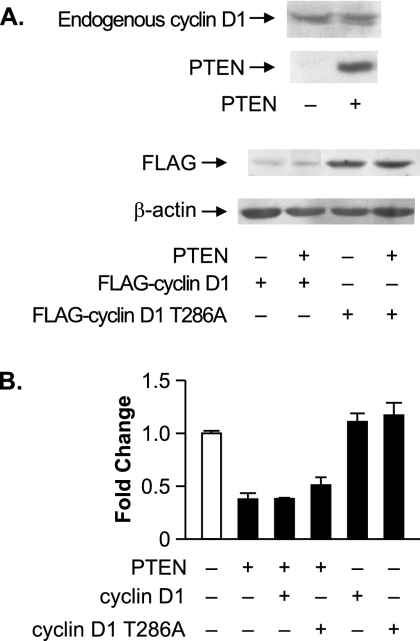
We further assessed whether PTEN-mediated effects on the cell cycle can be uncoupled from its ability to regulate RNA Pol III transcription in the U87 cell line. PTEN expression in these cells has been shown to lead to cell cycle arrest as a result of the decreased expression and decreased nuclear localization of cyclin D1 (27). Therefore, we determined if PTEN-mediated repression of tRNA gene transcription in U87 cells could be abrogated by increased expression of cyclin D1 or a nucleus-persistent form of cyclin D1, cyclin D1-T286A (8). U87 cells were transfected with or without PTEN and either cyclin D1 or cyclin D1-T286A (Fig. 6A). PTEN did not change the level of expression of endogenous or exogenous cyclin D1 in U87 cells. The expression of either cyclin D1 or cyclin D1-T286A alone did not appreciably enhance tRNA gene reporter activity (Fig. 6B). Importantly, PTEN-mediated repression of transcription could not be alleviated by the expression of either cyclin D1 or cyclin D1-T286A. Collectively, these results support the idea that PTEN-mediated RNA Pol III transcription repression can be uncoupled from PTEN-mediated effects on the cell cycle. These results suggest that PTEN represses RNA Pol III transcription through a mechanism that is independent from PTEN-mediated effects on the cell cycle.
FIG. 6.
Increased expression of cyclin D1 or nucleus-persistent cyclin D1-T286A does not alleviate PTEN-mediated repression of RNA Pol III transcription in U87 cells. (A) PTEN expression does not alter cyclin D1 expression. U87 cells were transfected with expression vectors for PTEN and/or Flag-cyclin D1 or Flag-cyclin D1-T286A. Total protein was isolated, and immunoblot analysis was performed using antibodies against PTEN, cyclin D1, a Flag epitope, or β-actin. (B) Ectopic expression of cyclin D1 or cyclin D1-T286A does not affect PTEN-mediated repression of RNA Pol III transcription. Cotransfections were performed as described above (A) with the tRNA gene reporter. Total RNA was isolated, and RNase protection assays were performed. The change in transcription is calculated based on reporter gene activity in cells transfected with an empty vector control. Graphs represent determinations from at least three biologically independent samples (means ± SE).
PTEN targets the TFIIIB complex to repress the formation of TBP-Brf1 complexes.
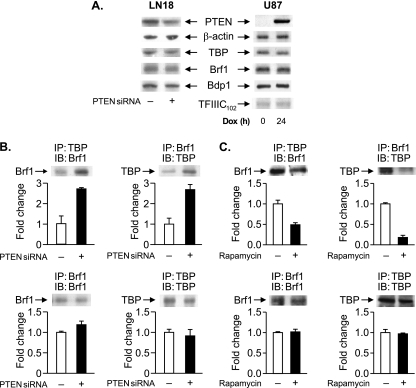
To elucidate the mechanism for PTEN-mediated repression of RNA Pol III transcription, we examined potential changes in TFIIIB, which was previously shown to be targeted by the tumor suppressors Rb (19, 35) and p53 (6). Neither a reduced expression of PTEN in LN18 cells nor the expression of PTEN in U87 cells affected the expression of the TFIIIB components TBP, Brf1, and Bdp1 or the TFIIIC subunit TFIIIC102 (Fig. 7A). As the direct interaction of TBP with Brf1 is critical for the formation of a functional TFIIIB complex, we examined potential changes in TBP-Brf1 interactions when PTEN expression was decreased in LN18 cells. Coimmunoprecipitation analysis demonstrated that decreased PTEN expression resulted in an increase in the association between TBP and Brf1 (Fig. 7B). As our results described above indicated that PTEN represses transcription through its ability to inhibit PI3K/Akt/mTOR signaling, the effect of inhibiting mTOR activation on TBP-Brf1 interactions was determined. The treatment of U87 cells with rapamycin resulted in a decrease in coimmunoprecipitated TBP-Brf1 complexes (Fig. 7C). These results show that PTEN, and the inhibition of mTOR, disrupts the interaction between TBP and Brf1.
FIG. 7.
PTEN targets TFIIIB. (A) PTEN does not alter the protein levels of the TFIIIB components. (Left) LN18 cells were transfected with PTEN siRNA or mmRNA. (Right) PTEN-inducible U87 cells were treated with DOX (24 h). Total protein was isolated, and immunoblot analysis was performed using antibodies against the proteins indicated. (B) PTEN disrupts the association between TBP and Brf1. LN18 cells were transfected with PTEN siRNA or mmRNA, and total protein was isolated. Coimmunoprecipitation assays (IP) and immunoblot analyses (IB) were performed using antibodies against TBP or Brf1. The change in association is calculated based on values from cells transfected with mmRNA. (C) Rapamycin induces dissociation between TBP and Brf1. U87 cells were treated with 100 nM rapamycin (3 h), and total protein was isolated. Coimmunoprecipitation assays and immunoblot analyses were performed as described above (B). The change in association is calculated based on values from cells treated with dimethyl sulfoxide. Representative autoradiographs are shown. Graphs represent determinations from at least three biologically independent samples (means ± SE).
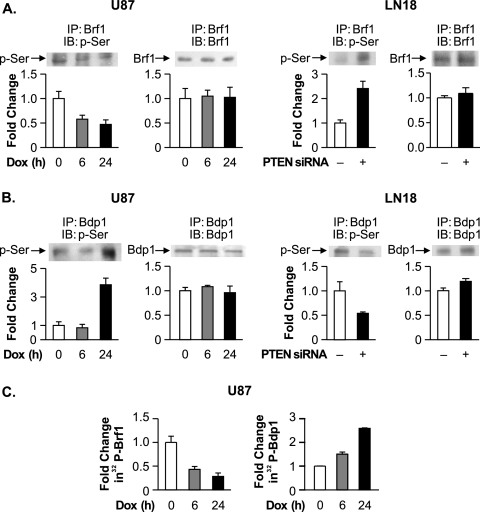
We next considered that PTEN-induced changes in the phosphorylation state of the TFIIIB components might impair their association. Since previous results demonstrated that PTEN does not affect the phosphorylation state of TBP (50), we examined potential changes in Brf1 phosphorylation. Brf1 immunoprecipitation and analysis of the phosphorylation state of the protein showed that the reduction of PTEN expression levels in LN18 cells resulted in enhanced serine phosphorylation, whereas the induced expression of PTEN in U87 cells resulted in diminished serine phosphorylation (Fig. 8A). No detectable tyrosine phosphorylation of Brf1 was observed (data not shown). These results support the idea that the serine phosphorylation state of Brf1 may regulate its association with TBP. In contrast to what was observed with Brf1, reducing PTEN expression in LN18 cells resulted in a decrease in Bdp1 serine phosphorylation (Fig. 8B). When PTEN expression was induced in the U87 cells for 6 h, no change in the serine phosphorylation state of Bdp1 was observed. However, following prolonged PTEN induction, a significant enhancement of Bdp1 phosphorylation was detected. To further confirm the PTEN-mediated changes in the phosphorylation of Brf1 and Bdp1, U87 cells were induced to express PTEN and labeled with 32P. Brf1 and Bdp1 were each immunoprecipitated from the resultant cell lysates. Upon PTEN induction, the phosphorylation state of Brf1 was decreased, while the phosphorylation state of Bdp1 was increased (Fig. 8C). Together, these results support the idea that PTEN alters the phosphorylation states of both Brf1 and Bdp1.
FIG. 8.
PTEN alters the serine phosphorylation state of Brf1 and Bdp1. (A) PTEN decreases Brf1 serine phosphorylation. (Left) PTEN-inducible U87 cells were treated with DOX for 0, 6, or 24 h. (Right) LN18 cells were transfected with PTEN siRNA or mmRNA. Total protein was isolated and subjected to immunoprecipitation (IP) with Brf1 antibody. Immunoblot analysis (IB) was performed using antibodies against Brf1 or phosphoserine (p-Ser). (B) PTEN increases Bdp1 serine phosphorylation. Total protein was isolated from PTEN-inducible U87 cells (left) and LN18 cells (right), each treated as described above (A). Immunoprecipitation was performed using Bdp1 antibody (IP), followed by immunoblot analysis with antibodies against Bdp1 or phosphoserine (IB). Representative autoradiographs are shown. The change in association is calculated based on values from cells left untreated (0 h) or transfected with mmRNA, where indicated. (C) PTEN alters the phosphorylation states of Brf1 and Bdp1. PTEN-inducible U87 cells were treated with DOX for 0, 6, or 24 h. Prior to being harvested, cells were incubated with 32Pi (0.5 mCi/ml) for 3 h. The resultant cell lysates were immunoprecipitated with Brf1 (left) or Bdp1 (right) antibodies and subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Autoradiography was used to examine the amount of 32P-incorporated Brf1 and Bdp1. All graphs represent determinations from at least three biologically independent samples (means ± SE).
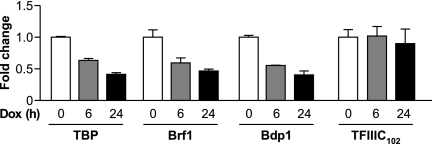
To determine whether these PTEN-mediated changes in TFIIIB might affect the occupancy of these components on RNA Pol III genes, ChIP analysis was used. Chromatin was isolated from U87 cells that had been treated with DOX for 0, 6, or 24 h to induce PTEN expression. Antibodies against TBP, Brf1, Bdp1, or TFIIIC102 were used to immunoprecipitate chromatin, and primers specific for the tRNALeu promoter were used for quantitative PCR to quantify the amount of these proteins present in the absence and in the presence of PTEN expression. The induction of PTEN expression for 6 h reduced the occupancy of TBP, Brf1, and Bdp1 on tRNALeu genes (Fig. 9). A slight additional reduction in promoter occupancy was seen for each TFIIIB subunit when PTEN expression was induced for 24 h. In contrast, the occupancy of TFIIIC102 was unchanged upon the induction of PTEN. Furthermore, PTEN was not found to be associated with the tRNALeu genes (data not shown). Together, these results support the idea that PTEN negatively regulates the assembly and recruitment of the TFIIIB complex to RNA Pol III promoters.
FIG. 9.
PTEN expression selectively decreases the promoter occupancy of TFIIIB. ChIP analysis was performed with PTEN-inducible U87 cells that were treated with DOX for 0, 6, or 24 h using antibodies against TBP, Brf1, Bdp1, or TFIIIC102 and subjected to quantitative PCR with primers specific for the tRNALeu gene promoter. Each time point (0, 6, and 24 h) is normalized to its respective input. The change in occupancy is calculated based on values from untreated cells (0 h). Graphs represent determinations from at least three biologically independent samples (means ± SE).
DISCUSSION
These studies have defined a new class of genes that are targeted by PTEN, one of the most commonly mutated tumor suppressor genes in human cancer. It is now clear that deregulated protein synthesis plays an important role in human cancer (28). RNA Pol III transcription is elevated in a broad range of transformed cell lines and in human tumors (46), and both qualitative and quantitative changes in protein synthesis are seen as a result of the loss of PTEN and the activation of mTOR (1). The lipid phosphatase activity of PTEN antagonizes the activity of PI3K in the cytoplasm and plays a critical role in the tumor suppressor function of PTEN. PI3K itself is a human oncogene, as the amplification or mutation of one of its catalytic subunits has been observed in many human tumors (29, 32). Our studies are the first to demonstrate that the PI3K/Akt/mTOR/S6K signaling pathway in mammalian cells regulates RNA Pol III transcription. Consistent with these results, a loss of the lipid phosphatase activity of PTEN abrogates PTEN-mediated transcriptional repression. While PTEN (37, 43) and components of the PI3K/Akt/mTOR pathway (38) have also been shown to function in the nucleus, our collective results support the idea that PTEN acts in the cytoplasm to regulate RNA Pol III gene activity by blocking the activation of PI3K. As previous studies demonstrated that oncogenic Ras induces RNA Pol III transcription through Raf-1, an immediate downstream target (41), our present studies demonstrate that the activation of PI3K, another key downstream effector of Ras, is an important contributor to the Ras-mediated induction of RNA Pol III transcription. Thus, oncogenic Ras induces at least two distinct pathways that can independently regulate the RNA Pol III transcriptional capacity of cells.
RNA Pol III transcription is highly regulated throughout the cell cycle, being lowest in mitosis (11, 16, 18, 20, 47, 48). The overexpression of PTEN can induce G1-phase cell cycle arrest, which can be reversed in U87 cells by the expression of a nucleus-persistent cyclin D1 mutant (27). Thus, we considered the possibility that PTEN-mediated effects on the cell cycle were responsible for the RNA Pol III transcription repression observed. However, several lines of evidence demonstrate that the ability of PTEN to repress transcription can be uncoupled from its effects on the cell cycle. First, while the reduction of PTEN in LN18 cells resulted in enhanced RNA Pol III transcription, no discernible changes in the cell cycle profile were observed. Second, PTEN-mediated repression of RNA Pol III transcription in U87 cells could not be alleviated by the expression of either cyclin D1 or the nucleus-persistent cyclin D1-T286A. Third, the inactivation or activation of PTEN-regulated PI3K, Akt, or mTOR signaling pathways altered RNA Pol III transcription in nonreplicating primary hepatocytes. Together, these results suggest that PTEN and the PI3K signaling pathway can regulate RNA Pol III transcription independently of their effects on cell proliferation. Alternatively, these results may reflect the different sensitivities of the cell cycle and RNA Pol III transcription to PTEN.
The function of PTEN as a lipid phosphatase that inhibits the PI3K signal transduction pathway distinguishes it from the tumor suppressors p53 and Rb, which function as transcription factors to directly regulate RNA Pol III-dependent transcription through their interaction with the TFIIIB complex. Rb associates with Brf1 (19, 35), while p53 interacts with TBP (6), resulting in a dysfunctional TFIIIB complex that is unable to associate with TFIIIC or RNA Pol III (6, 19, 35). In contrast, PTEN indirectly targets TFIIIB, inducing the dissociation between Brf1 and TBP and preventing the formation of a functional TFIIIB complex. Interestingly, inhibiting the activation of mTOR can reproduce the PTEN-mediated decrease in TBP-Brf1 complexes. These results support the idea that the PTEN-mediated regulation of mTOR is responsible for the disruption of TBP-Brf1 complexes and the decrease in transcription observed. Importantly, these results represent the first example where RNA Pol III transcription is indirectly regulated by the disruption of the TFIIIB complex.
To further examine how PTEN might affect the TFIIIB components, we kinetically examined potential changes in the transcription components following PTEN induction. As no apparent changes in the expression of the TFIIIB components were observed, potential changes in the phosphorylation states of the proteins were examined. Previous studies revealed that PTEN does not alter the phosphorylation state of TBP (50). However, a decrease in the serine phosphorylation of Brf1 was detected at 6 h following PTEN induction, together with a concomitant decrease in the occupancy of all three TFIIIB subunits on endogenous tRNALeu gene promoters. As PTEN was shown to negatively affect TBP-Brf1 interactions, these results are consistent with the idea that PTEN mediates the dephosphorylation of Brf1, facilitates the dissociation between Brf1 and TBP, and decreases the number of TFIIIB complexes that are recruited to tRNA gene promoters. At 24 h following PTEN induction, the repression of RNA Pol III transcription is more pronounced. While no further decrease in Brf1 phosphorylation was observed, an increase in the serine phosphorylation of Bdp1 was now detected, with a small decrease in the occupancy of TFIIIB components on the tRNA genes. These results support the idea that PTEN works to repress transcription through multiple events. PTEN initially induces the dephosphorylation of Brf1, concomitant with the disruption of the TFIIIB complex. The importance of this molecular event as a key mechanism responsible for RNA Pol III transcription repression is further underscored by the fact that the U6 RNA gene promoter does not use Brf1 for transcription, and PTEN and mTOR signaling do not regulate U6 RNA gene transcription. Following Brf1 dephosphorylation, a prolonged expression of PTEN leads to the phosphorylation of Bdp1, which could facilitate a further dissociation of the TFIIIB complex or additionally lead to the disruption of the interaction of Bdp1 with other transcription components.
While these and other studies place TFIIIB as a central target for regulation by both oncogenes and tumor suppressors, remarkably little is yet known regarding specific modifications that affect its function. Mitotic repression of RNA Pol III transcription in Xenopus extracts was shown to involve the hyperphosphorylation of TFIIIB through a cdc2-dependent kinase (11, 16). In mitotic HeLa cells, the repression of RNA Pol III transcription is correlated with Brf1 hyperphosphorylation, independent of cdc2 kinase activity (11). In addition, CK2-mediated phosphorylation of Bdp1, at potentially multiple sites, has been shown to mediate the mitotic repression of RNA Pol III transcription (20). Paradoxically, the inhibition of CK2 in mitotic extracts alleviates RNA Pol III transcription repression, while the inhibition of CK2 in transcription-competent S-phase extracts represses transcription. These results suggest that CK2 may phosphorylate multiple targets in the transcription machinery that serve to either positively or negatively regulate gene activity. The MAPK ERK2 directly phosphorylates Brf1 (12). The ERK2-mediated phosphorylation of Brf1 does not affect the association between TBP and Brf1 but does enhance TFIIIB-TFIIIC and TFIIIB-RNA Pol III interactions. Our results suggest that the PTEN-mediated regulation of Brf1 phosphorylation controls the association between TBP and Brf1. The residues within Brf1 that are subject to phosphorylation and the kinases and phosphatases that regulate these phosphorylation events to ultimately dictate the interactions of Brf1 with TBP, and specific TFIIIC and RNA Pol III subunits, are yet to be identified.
RNA Pol I transcription, which is responsible for the synthesis of large rRNAs, is also subject to regulation by tumor suppressors and oncogenic agents. Coordinately controlling the rates of RNA Pol I and Pol III transcription serves to modulate the number of ribosomes and the biosynthetic capacity of cells. Accordingly, previous studies have shown that PTEN represses RNA Pol I transcription by inhibiting PI3K/Akt/mTOR/S6K signaling (50). Interestingly, PTEN targets the TBP-containing SL1 complex, inducing the disruption of the complex. Unlike with the simultaneous dissociation of the TFIIIB subunits from tRNA gene promoters upon PTEN induction, PTEN facilitates a sequential reduction in the occupancy of each of the SL1 subunits from the RNA Pol I promoter that are kinetically separable. Thus, PTEN targets TBP-associated factors, leading to the disruption of the TBP-containing TFIIIB and SL1 complexes, resulting in the coordinate repression of RNA Pol I and Pol III transcription. The ability of PTEN to potentially regulate other TBP-TBP-associated factor interactions that direct RNA Pol II transcription and mRNA synthesis remains to be determined.
RNA Pol III transcription can be regulated by a multitude of different mechanisms that serve to target the transcription machinery either directly or indirectly. Our studies provide new evidence that the formation of the TFIIIB complex is regulated by the phosphorylation of TBP-associated factors and that this is an important regulatory step that contributes to the rate of RNA Pol III transcription. It is likely that the ability of PTEN to regulate RNA Pol III transcription, and the biosynthetic capacity of cells, contributes to its function as a tumor suppressor and its ability to maintain growth control.
Acknowledgments
We gratefully acknowledge Maria-Magdalena Georgescu for her gift of the PTEN- and PTEN-C124S-inducible U87 cell lines, Chuck Sherr for providing the cyclin D1-T286A expression plasmid, and Bill Sellers for the PTEN-NLS expression construct. We thank Bangyan Stiles and Beth Palian for critical reading of the manuscript.
This work was supported by Public Health Service grant CA108614 from the National Cancer Institute to D.L.J. and grants from the Ligue Contre Le Cancer Comité de la Gironde to M.T.
Footnotes
Published ahead of print on 7 April 2008.
REFERENCES
- 1.Bilanges, B., and D. Stokoe. 2007. Mechanisms of translational deregulation in human tumors and therapeutic intervention strategies. Oncogene 265973-5990. [DOI] [PubMed] [Google Scholar]
- 2.Borchert, G. M., W. Lanier, and B. L. Davidson. 2006. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 131097-1101. [DOI] [PubMed] [Google Scholar]
- 3.Chen, W., W. Bocker, J. Brosius, and H. Tiedge. 1997. Expression of neural BC200 RNA in human tumours. J. Pathol. 183345-351. [DOI] [PubMed] [Google Scholar]
- 4.Chen, W., J. Heierhorst, J. Brosius, and H. Tiedge. 1997. Expression of neural BC1 RNA: induction in murine tumours. Eur. J. Cancer 33288-292. [DOI] [PubMed] [Google Scholar]
- 5.Chow, L. M. L., and S. J. Baker. 2006. PTEN function in normal and neoplastic growth. Cancer Lett. 241184-196. [DOI] [PubMed] [Google Scholar]
- 6.Crighton, D., A. Woiwode, C. Zhang, N. Mandavia, J. P. Morton, L. J. Warnock, J. Milner, R. J. White, and D. L. Johnson. 2003. p53 represses RNA polymerase III transcription by targeting TBP and inhibiting promoter occupancy by TFIIIB. EMBO J. 222810-2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Di Cristofano, A., B. Pesce, C. Cordon-Cardo, and P. P. Pandolfi. 1998. Pten is essential for embryonic development and tumour suppression. Nat. Genet. 19348-355. [DOI] [PubMed] [Google Scholar]
- 8.Diehl, J. A., F. Zindy, and C. J. Sherr. 1997. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 11957-972. [DOI] [PubMed] [Google Scholar]
- 9.Dingermann, T., S. Sharp, B. Appel, D. DeFranco, S. Mount, R. Heiermann, O. Pongs, and D. Soll. 1981. Transcription of cloned tRNA and 5S RNA genes in a Drosophila cell free extract. Nucleic Acids Res. 93907-3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Engelman, J. A., J. Luo, and L. C. Cantley. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7606-619. [DOI] [PubMed] [Google Scholar]
- 11.Fairley, J. A., P. H. Scott, and R. J. White. 2003. TFIIIB is phosphorylated, disrupted and selectively released from tRNA promoters during mitosis in vivo. EMBO J. 225841-5850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felton-Edkins, Z. A., J. A. Fairley, E. L. Graham, I. M. Johnston, R. J. White, and P. H. Scott. 2003. The mitogen-activated protein (MAP) kinase ERK induces tRNA synthesis by phosphorylating TFIIIB. EMBO J. 222422-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freeman, D. J., A. G. Li, G. Wei, H. H. Li, N. Kertesz, R. Lesche, A. D. Whale, H. Martinez-Diaz, N. Rozengurt, R. D. Cardiff, X. Liu, and H. Wu. 2003. PTEN tumor suppressor regulates p53 protein levels and activity through phosphatase-dependent and -independent mechanisms. Cancer Cell 3117-130. [DOI] [PubMed] [Google Scholar]
- 14.Furnari, F. B., H. J. Huang, and W. K. Cavenee. 1998. The phosphoinositol phosphatase activity of PTEN mediates a serum-sensitive G1 growth arrest in glioma cells. Cancer Res. 585002-5008. [PubMed] [Google Scholar]
- 15.Gomez-Roman, N., C. Grandori, R. N. Eisenman, and R. J. White. 2003. Direct activation of RNA polymerase III transcription by c-Myc. Nature 421290-294. [DOI] [PubMed] [Google Scholar]
- 16.Gottesfeld, J. M., V. J. Wolf, T. Dang, D. J. Forbes, and P. Hartl. 1994. Mitotic repression of RNA polymerase III transcription in vitro mediated by phosphorylation of a TFIIIB component. Science 26381-84. [DOI] [PubMed] [Google Scholar]
- 17.Gridasova, A. A., and R. W. Henry. 2005. The p53 tumor suppressor protein represses human snRNA gene transcription by RNA polymerases II and III independently of sequence-specific DNA binding. Mol. Cell. Biol. 253247-3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartl, P., J. Gottesfeld, and D. J. Forbes. 1993. Mitotic repression of transcription in vitro. J. Cell Biol. 120613-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hirsch, H. A., G. W. Jawdekar, K. A. Lee, L. Gu, and R. W. Henry. 2004. Distinct mechanisms for repression of RNA polymerase III transcription by the retinoblastoma tumor suppressor protein. Mol. Cell. Biol. 245989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu, P., K. Samudre, S. Wu, Y. Sun, and N. Hernandez. 2004. CK2 phosphorylation of Bdp1 executes cell cycle-specific RNA polymerase III transcription repression. Mol. Cell 1681-92. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, S. A., N. Mandavia, H. D. Wang, and D. L. Johnson. 2000. Transcriptional regulation of the TATA-binding protein by Ras cellular signaling. Mol. Cell. Biol. 205000-5009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, S. S., C. Zhang, J. Fromm, I. M. Willis, and D. L. Johnson. 2007. Mammalian Maf1 is a negative regulator of transcription by all three nuclear RNA polymerases. Mol. Cell 26367-379. [DOI] [PubMed] [Google Scholar]
- 23.Li, A. G., L. G. Piluso, X. Cai, G. Wei, W. R. Sellers, and X. Liu. 2006. Mechanistic insights into maintenance of high p53 acetylation by PTEN. Mol. Cell 23575-587. [DOI] [PubMed] [Google Scholar]
- 24.Lian, Z., and A. Di Cristofano. 2005. Class reunion: PTEN joins the nuclear crew. Oncogene 247394-7400. [DOI] [PubMed] [Google Scholar]
- 25.Mayo, L. D., J. E. Dixon, D. L. Durden, N. K. Tonks, and D. B. Donner. 2002. PTEN protects p53 from Mdm2 and sensitizes cancer cells to chemotherapy. J. Biol. Chem. 2775484-5489. [DOI] [PubMed] [Google Scholar]
- 26.Myers, M. P., I. Pass, I. H. Batty, J. Van der Kaay, J. P. Stolarov, B. A. Hemmings, M. H. Wigler, C. P. Downes, and N. K. Tonks. 1998. The lipid phosphatase activity of PTEN is critical for its tumor suppressor function. Proc. Natl. Acad. Sci. USA 9513513-13518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Radu, A., V. Neubauer, T. Akagi, H. Hanafusa, and M. M. Georgescu. 2003. PTEN induces cell cycle arrest by decreasing the level and nuclear localization of cyclin D1. Mol. Cell. Biol. 236139-6149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruggero, D., and P. P. Pandolfi. 2003. Does the ribosome translate cancer? Nat. Rev. Cancer 3179-192. [DOI] [PubMed] [Google Scholar]
- 29.Samuels, Y., and V. E. Velculescu. 2004. Oncogenic mutations of PIK3CA in human cancers. Cell Cycle 31221-1224. [DOI] [PubMed] [Google Scholar]
- 30.Schramm, L., and N. Hernandez. 2002. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 162593-2620. [DOI] [PubMed] [Google Scholar]
- 31.Scott, M. R., K. H. Westphal, and P. W. Rigby. 1983. Activation of mouse genes in transformed cells. Cell 34557-567. [DOI] [PubMed] [Google Scholar]
- 32.Shayesteh, L., Y. Lu, W. L. Kuo, R. Baldocchi, T. Godfrey, C. Collins, D. Pinkel, B. Powell, G. B. Mills, and J. W. Gray. 1999. PIK3CA is implicated as an oncogene in ovarian cancer. Nat. Genet. 2199-102. [DOI] [PubMed] [Google Scholar]
- 33.Shen, W. H., A. S. Balajee, J. Wang, H. Wu, C. Eng, P. P. Pandolfi, and Y. Yin. 2007. Essential role for nuclear PTEN in maintaining chromosomal integrity. Cell 128157-170. [DOI] [PubMed] [Google Scholar]
- 34.Stambolic, V., D. MacPherson, D. Sas, Y. Lin, B. Snow, Y. Jang, S. Benchimol, and T. W. Mak. 2001. Regulation of PTEN transcription by p53. Mol. Cell 8317-325. [DOI] [PubMed] [Google Scholar]
- 35.Sutcliffe, J. E., T. R. Brown, S. J. Allison, P. H. Scott, and R. J. White. 2000. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol. Cell. Biol. 209192-9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tee, A. R., D. C. Fingar, B. D. Manning, D. J. Kwiatkowski, L. C. Cantley, and J. Blenis. 2002. Tuberous sclerosis complex-1 and -2 gene products function together to inhibit mammalian target of rapamycin (mTOR)-mediated downstream signaling. Proc. Natl. Acad. Sci. USA 9913571-13576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trotman, L. C., X. Wang, A. Alimonti, Z. Chen, J. Teruya-Feldstein, H. Yang, N. P. Pavletich, B. S. Carver, C. Cordon-Cardo, H. Erdjument-Bromage, P. Tempst, S. G. Chi, H. J. Kim, T. Misteli, X. Jiang, and P. P. Pandolfi. 2007. Ubiquitination regulates PTEN nuclear import and tumor suppression. Cell 128141-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tsang, C. K., and X. F. Zheng. 2007. TOR-in(g) the nucleus. Cell Cycle 625-29. [DOI] [PubMed] [Google Scholar]
- 39.Upadhya, R., J. Lee, and I. M. Willis. 2002. Maf1 is an essential mediator of diverse signals that repress RNA polymerase III transcription. Mol. Cell 101489-1494. [DOI] [PubMed] [Google Scholar]
- 40.Vazquez, F., S. Ramaswamy, N. Nakamura, and W. R. Sellers. 2000. Phosphorylation of the PTEN tail regulates protein stability and function. Mol. Cell. Biol. 205010-5018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, H. D., A. Trivedi, and D. L. Johnson. 1997. Hepatitis B virus X protein induces RNA polymerase III-dependent gene transcription and increases cellular TATA-binding protein by activating the Ras signaling pathway. Mol. Cell. Biol. 176838-6846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang, H. D., A. Trivedi, and D. L. Johnson. 1998. Regulation of RNA polymerase I-dependent promoters by the hepatitis B virus X protein via activated Ras and TATA-binding protein. Mol. Cell. Biol. 187086-7094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, X., L. C. Trotman, T. Koppie, A. Alimonti, Z. Chen, Z. Gao, J. Wang, H. Erdjument-Bromage, P. Tempst, C. Cordon-Cardo, P. P. Pandolfi, and X. Jiang. 2007. NEDD4-1 is a proto-oncogenic ubiquitin ligase for PTEN. Cell 128129-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weng, L. P., W. M. Smith, P. L. Dahia, U. Ziebold, E. Gil, J. A. Lees, and C. Eng. 1999. PTEN suppresses breast cancer cell growth by phosphatase activity-dependent G1 arrest followed by cell death. Cancer Res. 595808-5814. [PubMed] [Google Scholar]
- 45.White, M. A., C. Nicolette, A. Minden, A. Polverino, L. Van Aelst, M. Karin, and M. H. Wigler. 1995. Multiple Ras functions can contribute to mammalian cell transformation. Cell 80533-541. [DOI] [PubMed] [Google Scholar]
- 46.White, R. J. 2005. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 669-78. [DOI] [PubMed] [Google Scholar]
- 47.White, R. J., T. M. Gottlieb, C. S. Downes, and S. P. Jackson. 1995. Cell cycle regulation of RNA polymerase III transcription. Mol. Cell. Biol. 156653-6662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White, R. J., T. M. Gottlieb, C. S. Downes, and S. P. Jackson. 1995. Mitotic regulation of a TATA-binding-protein-containing complex. Mol. Cell. Biol. 151983-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winter, A. G., G. Sourvinos, S. J. Allison, K. Tosh, P. H. Scott, D. A. Spandidos, and R. J. White. 2000. RNA polymerase III transcription factor TFIIIC2 is overexpressed in ovarian tumors. Proc. Natl. Acad. Sci. USA 9712619-12624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang, C., L. Comai, and D. L. Johnson. 2005. PTEN represses RNA polymerase I transcription by disrupting the SL1 complex. Mol. Cell. Biol. 256899-6911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhong, S., C. Zhang, and D. L. Johnson. 2004. Epidermal growth factor enhances cellular TATA binding protein levels and induces RNA polymerase I- and III-dependent gene activity. Mol. Cell. Biol. 245119-5129. [DOI] [PMC free article] [PubMed] [Google Scholar]