Abstract
Boundary elements have been found in the regulatory region of the Drosophila melanogaster Abdominal-B (Abd-B) gene, which is subdivided into a series of iab domains. The best-studied Fab-7 and Fab-8 boundaries flank the iab-7 enhancer and isolate it from the four promoters regulating Abd-B expression. Recently binding sites for the Drosophila homolog of the vertebrate insulator protein CTCF (dCTCF) were identified in the Fab-8 boundary and upstream of Abd-B promoter A, with no binding of CTCF to the Fab-7 boundary being detected either in vivo or in vitro. Taking into account the inability of the yeast GAL4 activator to stimulate the white promoter when its binding sites are separated by a 5-kb yellow gene, we have tested the functional interactions between the Fab-7 and Fab-8 boundaries and between these boundaries and the upstream promoter A region containing a dCTCF binding site. It has been found that dCTCF binding sites are essential for pairing between two Fab-8 insulators. However, a strong functional interaction between the Fab-7 and Fab-8 boundaries suggests that additional, as yet unidentified proteins are involved in long-distance interactions between them. We have also shown that Fab-7 and Fab-8 boundaries effectively interact with the upstream region of the Abd-B promoter.
Eukaryotic genomes are highly organized into functional units containing individual genes or gene groups together with the corresponding regulatory elements. Regulatory elements, enhancers/silencers, may be separated from the promoters by dozens of thousands of base pairs (10, 15, 16, 35, 63). Most recent data (14, 18, 42, 52, 67) support the looping model (53), which postulates that enhancers and distant promoters are in physical contact with each other while the intervening sequences loop out. Accordingly, one of the key questions is how distant enhancers communicate with their target promoters. The complexity of higher eukaryotic regulatory systems, which contain many distantly located enhancers that nevertheless properly activate the target promoters, has prompted the hypothesis that the action of enhancers should be restricted by elements called insulators (6, 32, 64, 66, 68). Generally, insulators are defined by two properties: the enhancer-blocking activity, preventing communication between an enhancer and a promoter separated by the insulator, and the boundary function (barrier activity), preventing repressive chromatin spreading. In recent years, however, experimental evidences have been accumulated that insulator proteins may be involved in supporting long-distance interactions between regulatory elements located either within the same complex locus or in distantly located loci (12, 13, 31, 40, 42, 43, 57).
One of the best model systems for studying the role of insulators in long-distance enhancer-promoter communication is the regulatory region of the homeotic Abdominal-B (Abd-B) gene of the bithorax complex (41, 47, 63). The three homeotic genes of the bithorax complex—Ultrabithorax (Ubx), abdominal-A (abd-A), and Abd-B—are responsible for specifying the identity of parasegments 5 to 14 (PS5 to PS14), which form the posterior half of the thorax and all abdominal segments of an adult fly (36, 41, 47, 55). The PS-specific expression patterns of Ubx, abd-A, and Abd-B are determined by a complex cis-regulatory region that spans a 300-kb DNA segment (41, 44, 63). Genetic analysis has indicated that this large regulatory region can be divided into nine discrete segment-specific domains, which are aligned on the chromosome in the same order as are the body segments in which they operate (1, 36, 45). For example, Abd-B expression in PS10, PS11, PS12, and PS13 is controlled by the iab-5, iab-6, iab-7, and iab-8 cis-regulatory domains, respectively (5, 9, 17, 28, 36, 48, 56). Each iab domain appears to contain at least one enhancer that initiates Abd-B expression in the early embryo, as well as a Polycomb response element (PRE) silencer element that maintains the expression pattern throughout development (4, 7, 8, 24, 25, 45, 46, 48, 70, 71). It has been proposed that boundaries flank each iab region and organize the Abd-B regulatory DNA into a series of separate chromatin domains (4, 19, 23, 47, 48). To date, three boundary elements have been defined by deletion analysis within the Abd-B region of the bithorax complex: Fab-7 (23), Mcp (29), and Fab-8 (4). All these boundaries display the insulator function, i.e., they are capable of suppressing reporter gene expression when placed between an enhancer and a promoter in a transgenic insulator assay (4, 22, 24, 33, 59, 60, 70, 71). This finding requires explanation as to how the iab enhancers can interact with the Abd-B promoters across insulators such as Fab-7 and Fab-8.
Two models were proposed to explain how iab enhancers flanked by insulators can interact with the proper promoter. The first model is based on special elements called promoter-targeting sequences (PTS elements) that were found to adjoin both Fab-7 and Fab-8 boundary elements (11, 69). In transgenic reporters, PTS elements were shown to allow distal enhancers to bypass intervening insulators (37-39). It was proposed that PTS elements facilitate proper interaction between iab enhancers and promoters in the Abd-B locus (39). Recently, a 255-bp element, named promoter-tethering element, was found near the Abd-B promoter. This element is supposedly capable of selectively recruiting iab enhancers to the promoter (2).
The second model proposes that one of the main roles of boundaries in Abd-B is to bring iab enhancers into close proximity to the promoter (42). This model is based on the observation that the Fab-7 boundary interacts with a region near the Abd-B promoter in vivo (12). It was also shown that this interaction is absolutely dependent on the presence of the Fab-7 boundary element. It appears likely that the boundaries, PTS, and promoter-tethering element cooperate in organizing proper interactions between the enhancers and promoters in the Abd-B locus.
Recently binding sites for the Drosophila melanogaster homolog of vertebrate insulator protein CTCF (dCTCF) were identified in the bithorax complex (27, 49, 50). In vertebrates, almost all insulator elements studied are associated with the binding of CTCF, a DNA-binding protein that contains 11 zinc fingers (66). The dCTCF binding sites were found in the Fab-8 insulator (27, 50) and near one of the four Abd-B promoters, designated A (27). At the same time, the Fab-7 boundary is devoid of the dCTCF binding sites.
The dCTCF protein was suggested to be the key protein involved in organization of chromatin domains in the bithorax complex (27, 49). In mammals, CTCF supports long-distance interactions (66), but there is no evidence that the same activity is also characteristic of dCTCF, which shows homology with its vertebrate counterpart only in the zinc finger domain (50). Our goal in this study was to analyze functional interactions between the Fab-7 and Fab-8 boundaries and between them and the region upstream of Abd-B promoter A and to test the role of dCTCF in supporting such distant interactions. To this end, we used GAL4/white, taking into account that the yeast GAL4 activator cannot activate the white promoter across the yellow gene (33).
As a result, we have found that dCTCF binding sites are essential for pairing between two Fab-8 insulators. The functional interaction between the Fab-7 and Fab-8 boundaries requires the presence of a PTS element adjacent to the Fab-8 insulator. Both the Fab-7 and Fab-8 boundaries interact with the 408-bp region containing a dCTCF site upstream of the Abd-B A promoter. Thus, transcriptional factors bound to the boundaries (such as dCTCF) may ensure long-distance communication between the iab enhancers and Abd-B promoters.
MATERIALS AND METHODS
Plasmid construction.
The 5-kb BamHI-BglII fragment (yc) containing the yellow coding region (20) was subcloned into CaSpeR2 (C2-yc). The 3-kb SalI-BamHI fragment containing the yellow regulatory region (yr) was subcloned into BamHI-plus-XhoI-cleaved pGEM7 (yr plasmid). The pCaSpew15(+RI) plasmid was constructed by inserting an additional EcoRI site at bp +3291 of the mini-white gene in the pCaSpew15 plasmid. An insulator located at the 3′ side of the mini-white gene (mw insulator) was deleted from pCaSpew15(+RI) by digestion with EcoRI to produce the pCaSpeRΔ700 plasmid. The BamHI-BglII fragment of the yellow coding region was cloned into pCaSpeRΔ700 (C2-yc).
Fragments PTS/F8 (nucleotides 63683 to 64582 within the DS07696 sequence of the Abd-B gene [44] [reference no. L07835]), F8 (63683 to 64291), PTS (64292 to 64916), F8469 (63683 to 64151), F8254 (64038 to 64291), PTS/F8337 (64038 to 64374), Fab-7 (83647 to 84504), ACTCF (48350 to 48758), and unnamed Abd-B A promoter fragments (45591 to 47193 and 47496 to 48562) were obtained by PCR amplification and sequenced to verify the results. The PCR-amplified fragments (X or Y) were cloned between either two frt [frt(X)] or two lox [lox(Y)] sites. Ten binding sites for GAL4 (G4) were ligated into the yr plasmid cleaved by NcoI and Eco47III (G4-Δyr).
To mutate both dCTCF binding sites in the F8 fragment (F8m), oligonucleotides carrying the desired mutant sequences (5′ AAGGAAAGCACCAACACAAATTTAAATTATCCGAC 3′ and 5′ CCTAGTTCTACATTACCAAGGTCTAGATTTACTGC 3′) were used to amplify PCR products. The resulting DNA fragment was sequenced to confirm that the intended mutant sequences had been introduced and that other PCR-induced mutations were absent.
The synthetic dCTCF binding region was created by concatamerization of oligonucleotides containing the 20-bp binding site GGCCAGGTGGCGCTGCAAGG (64 205) of the natural Fab-8 insulator (27). Two pairs of single-stranded 27-bp oligonucleotides (corresponding to the sense and antisense strands) were synthesized so as to contain overhangs for either BamHI or BglII. The sequences of the oligonucleotides were 5′ CTGCAGCGCCACCTGGCCTTGGAGATC 3′ and 5′ TCCCAAGGCCAGGTGGCGCTGCAGGATC 3′. The desired concatemers were isolated, purified, and cloned into the plasmid. The resulting DNA fragment was verified by sequencing.
All constructs were made by using the same general scheme. A fragment flanked by frt sites [frt(X)] was inserted in the direct or reverse orientation into the G4-Δyr plasmid cleaved by KpnI [G4-Δyr-frt(X)]. A fragment flanked by lox sites [lox(Y)]) was cloned into C2-yc between the yellow and white genes [C2-lox(Y)-yc]. Next, G4-Δyr-frt(X) fragments were cloned into the corresponding C2-lox(Y)-yc plasmids.
Generation and analysis of transgenic lines.
The construct and P25.7wc plasmid were injected into yacw1118 preblastoderm (30). The resultant flies were crossed with yacw1118 flies, and the transgenic progeny were identified by their eye color. Chromosome localization of various transgene insertions was determined by crossing the transformants with the yacw1118 balancer stock containing dominant markers, In(2RL),CyO for chromosome 2 and In(3LR)TM3, Sb for chromosome 3.
The lines with DNA fragment excisions were obtained by crossing the transposon-bearing flies with the Flp (w1118; S2CyO, hsFLP, ISA/Sco; +) or Cre (yw; Cyo, P[w+, cre]/Sco; +) recombinase-expressing lines. The Cre recombinase induces 100% excisions in the next generation. The high level of FLP recombinase (almost 90% efficiency) was produced by daily heat shock treatment for 2 h during the first 3 days after hatching. All excisions were confirmed by PCR analysis with the pairs of primers flanking the −893 insertion site (5′ ATCCAGTTGATTTTCAGGGACCA 3′ and 5′ TTGGCAGGTGATTTTGAGCATAC 3′) relative to the yellow transcription start site and the insertion site between the yellow and white genes (5′ TTTTCTTGAGCGGAAAAAGCGGA 3′ and 5′ ATCTACATTCTCCAAAAAAGGGT 3′). Details of the crosses used for genetic analysis and the excision of functional elements are available upon request.
To induce GAL4 expression, we used the modified yw1118; P[w−, tubGAL4]117/TM3, Sb line (Bloomington Center no. 5138), in which the marker mini-white gene was deleted as described previously (33).
The white phenotype was determined from eye pigmentation in adult flies. Wild-type white expression determined the bright red eye color; in the absence of white expression, the eyes were white. Intermediate levels of white expression (in increasing order) were reflected in the eye color, ranging from pale yellow to yellow, dark yellow, orange, dark orange, and finally brown or brownish red.
Electrophoretic mobility shift assay.
For the purpose of synthesizing dCTCF in vitro, the cDNA of dCTCF (kindly provided by J. Zhou) was subcloned into the pET 23a plasmid (Novagen). The dCTCF protein was synthesized in vitro in the TNT-coupled transcription/translation reticulocyte lysate (Promega) from a T7 promoter. In vitro-translated protein (6 μl) was added to 25 fmol of a radioactively labeled DNA probe in a 20-μl final volume of binding reaction in a phosphate-buffered saline buffer also containing 5 mM MgCl2, 0.1 mM ZnCl2, 1 mM dithiothreitol, 0.1% Nonidet P-40, and 10% glycerol. Binding reactions were incubated at room temperature for 30 min and then resolved on a 5% nondenaturing polyacrylamide gel at 5 V/cm using 0.5× Tris-borate-EDTA buffer.
RESULTS
Pairing of Fab-8 boundaries facilitates long-distance stimulation of the white promoter by the GAL4 activator.
Previously we demonstrated the pairing between two copies of Mcp or Fab-7 insulators that facilitated distant communication between an enhancer and a promoter. If this property is common to all insulators in the Abd-B regulatory region, it should be also characteristic of the Fab-8 boundary (Fig. 1A). The complete boundary (here designated PTS/F8) consists of two functionally distinct elements: the Fab-8 insulator (F8) and the promoter targeting sequence (PTS), which has an anti-insulator activity, allowing an enhancer to activate its promoter over the intervening insulator (69).
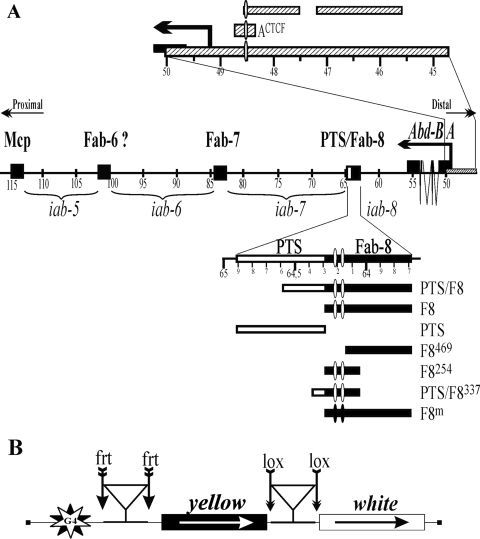
FIG. 1.
Schemes of the distal part of the bithorax complex and the construct for testing the interaction between regulatory elements of the Abd-B locus. (A) The Abd-B gene and part of its 3′ cis-regulatory region. The horizontal line represents the DNA sequence of the bithorax marked off in kilobases (44). The only class A Abd-B transcript that is required for morphogenesis in PS10 to PS13 is drawn above the DNA line. Arrows marked “Proximal” and “Distal” point toward the centromere and the telomere, respectively. Horizontal brackets below the DNA indicate the extents of iab-5, iab-6, iab-7, and part of iab-8. Positions of the boundaries are indicated by filled squares. The promoter A region, marked off in kilobases, is indicated above the DNA line. The DNA fragments tested are shown as hatched boxes. The Fab-8 boundary is drawn below the DNA line. The PTS and Fab-8 insulator are indicated by white and black rectangles, respectively. The fragments taken for analysis are shown below the Fab-8 boundary. White and black ovals represent functional and mutant binding sites for dCTCF. (B) Reductive scheme of transgenic construct used to examine the interaction between regulatory elements at a distance. The yellow and white genes are shown as boxes, with arrows indicating the direction of their transcription. Downward arrows indicate the target sites of the Flp recombinase (frt) or Cre recombinase (lox); the same sites in construct names are denoted by parentheses. The GAL4 binding sites (indicated as G4) are at a distance of approximately 5 kb from the white promoter. Triangles indicate positions of elements tested for interaction.
To test for distant interactions between regulatory elements, we relied on our previous finding that the yeast GAL4 activator bound to sites located upstream of the yellow gene fails to stimulate the white promoter placed downstream of the yellow 3′ end (33). In the test constructs (Fig. 1B), 10 GAL4 binding sites (G4) were inserted at −893 relative to the yellow transcription start site. As a result, the distance between the white gene and the GAL4 binding sites was almost 5 kb.
To examine the functional interaction between two regulatory elements, one element flanked by FRT sites (21) was inserted near G4, and the other, flanked by LOX sites (61), was inserted near the mini-white promoter. The presence of the FRT and LOX sites allowed us to delete the DNA fragments tested and to compare stimulation of transcription by GAL4 in transgenic lines before deletion of the regulatory elements and after it (control).
Initially we studied whether the interaction between two PTS/F8 boundaries could facilitate white stimulation by GAL4 across the yellow gene. The PTS/F8 boundaries were inserted in either the opposite (Fig. 2A) or the same (Fig. 2B) orientation relative to each other. To express the GAL4 protein, we used the transgenic line carrying the GAL4 gene under control of the ubiquitous tubulin promoter (33). In transgenic lines carrying two boundaries inserted in opposite orientations (Fig. 1A), GAL4 strongly induced white expression: flies had brown to red eyes in more than half of transgenic lines (10 out of 16). When PTS/F8 elements were deleted from transgenic lines, GAL4 lost the ability to stimulate white expression: a slight increase in eye pigmentation was observed in only 3 out of 16 transgenic lines. Thus, the interaction between the PTS/F8 insulators allows GAL4 activator to stimulate transcription of the white gene.
FIG. 2.
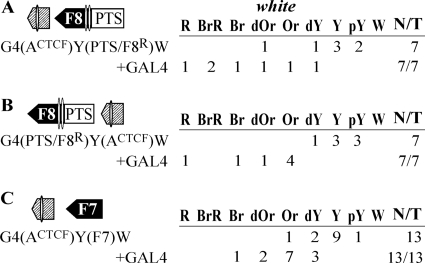
Experimental evidence that interacting elements facilitate stimulation of white by a distantly located GAL4 activator. The Fab-8 insulator (F8) and PTS are shown as black and white boxes, respectively, with apexes indicating their orientation in constructs. A superscript “R” indicates that the corresponding element is inserted in the reverse orientation relative to the white gene in the construct. “+GAL4” indicates that eye phenotypes in transgenic lines were examined after induction of GAL4 expression. The “white” column shows the numbers of transgenic lines with different levels of white pigmentation in the eyes. Wild-type white expression determined the bright red eye color (R); in the absence of white expression, the eyes were white (W). Intermediate levels of pigmentation, with the eye color ranging from pale yellow (pY) through yellow (Y), dark yellow (dY), orange (Or), dark orange (dOr), and brown (Br) to brownish red (BrR), reflect the increasing levels of white expression. N is the number of lines in which flies acquired a new w phenotype upon induction of GAL4 or deletion (Δ) of the specified DNA fragment; T is the total number of lines examined for each particular construct. Other designations are as in Fig. 1.
When the PTS/F8 elements were in the same orientation (Fig. 2B), we observed much weaker stimulation of white by GAL4: 10 out of 20 transgenic lines had an orange to dark-orange eye color. However, GAL4 almost completely lost the ability to stimulate white transcription when both PTS/F8 elements were deleted. These results suggest that the interacting boundaries inserted in the same orientation bring together GAL4 and the white promoter on the one hand but do not permit strong stimulation of white by GAL4 on the other hand. Recently we observed a similar orientation-dependent interaction between two Mcp insulators (33).
Next, we tested whether PTS and Fab-8 have the same ability to interact in pairs in an orientation-dependent manner. Two Fab-8 insulators inserted in opposite orientations (Fig. 2C) allowed strong stimulation of white by GAL4: flies in 11 out of 16 transgenic lines tested acquired brown to red eye pigmentation. When Fab-8 insulators were placed in the same orientation (Fig. 2D), GAL4 only weakly stimulated white expression. Thus, two Fab-8 insulators interact in an orientation-dependent manner.
PTSs were inserted in opposite orientations (Fig. 2E) and in the same orientation (Fig. 2F), but, irrespective of their mutual orientation, they provided only weak white activation. In transgenic lines with both constructs, GAL4 increased eye pigmentation only to a dark-yellow to orange color. The failure of two PTS elements to provide for strong white stimulation by GAL4 might be explained by weak interaction between these elements.
To simplify further presentation of the results, we designated white stimulation by GAL4 “strong,” “moderate,” or “weak” when flies from more than half of tested transgenic lines acquired brown to red, dark-orange to orange, or orange to dark-yellow eye pigmentation, respectively.
dCTCF binding sites are essential for interaction between the Fab-8 boundaries.
Previously (50) two closely spaced binding sites for the dCTCF protein were identified in the Fab-8 insulator (Fig. 1A and 3). To test their role in the long-distance interaction between the Fab-8 insulators, we divided the insulator into two overlapping 469-bp (F8469) and 254-bp (F8254) fragments (Fig. 1A). Both dCTCF binding sites were located in the 254-bp fragment.
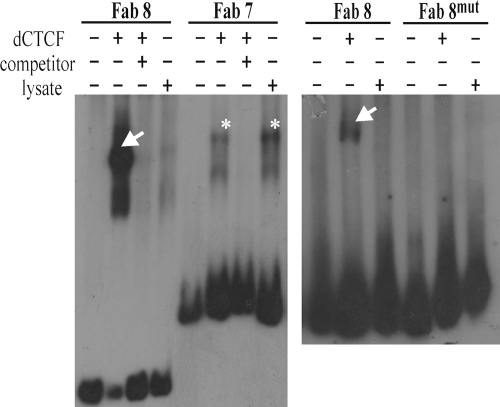
FIG. 3.
Electrophoretic mobility shift assays. Radioactively labeled Fab-7, Fab-8, and Fab-8m DNA fragments used as probes were incubated with the in vitro-synthesized dCTCF protein in the presence of competitors (unlabeled Fab-7 or Fab-8 fragment added in excess) or without them and subjected to electrophoresis in 5% polyacrylamide (see Materials and Methods). One shifted band (indicated by an arrow) presumably corresponds to a protein-DNA complex formed by dCTCF with a binding site. Asterisks indicate nonspecific binding of lysate to the Fab-7 DNA.
The fragments tested, F8469 (Fig. 4A) and F8254 (Fig. 4B), were inserted only in opposite orientations. We observed strong white stimulation by GAL4 in transgenic lines carrying the 254-bp fragments (Fig. 4B), which indicated that the dCTCF-containing fragment contributed to the long-distance interaction between Fab-8 boundaries. In contrast, only weak signs of GAL4-mediated stimulation were detected in transgenic lines with the 469-bp fragment (Fig. 4A). This is additional evidence for the main role of a dCTCF-containing fragment in the interaction between the Fab-8 insulators.
FIG. 4.
Role of dCTCF binding sites in the functional interaction between the Fab-8 boundaries. For designations, see the legends for Fig. 1 and 2.
To corroborate the role of dCTCF in the distant interaction between the Fab-8 insulators, we made mutations in both dCTCF binding sites (F8m). Electrophoretic mobility shift assay results showed that the dCTCF protein bound to the Fab-8 fragment but not to the F8m fragment (Fig. 3). The F8m fragments were inserted in opposite orientations (Fig. 4C). In transgenic lines, GAL4 only weakly stimulated white expression, indicating that there was no interaction between the F8m fragments.
These results suggest that dCTCF either directly participates in the distant interaction between the Fab-8 insulators or facilitates the binding of a protein complex involved in this process. To discriminate between these possibilities, we made a DNA fragment containing four consensus binding sites for the dCTCF protein (the CTCF fragment), and two such fragments were inserted in opposite orientations (Fig. 4D). In transgenic lines, the presence of the dCTCF sites provided for strong activation of white expression by GAL4. These results show that dCTCF may be directly involved in the distant interaction between the Fab-8 insulators.
Demonstration of functional interaction between Fab-7 and Fab-8 boundaries.
It was shown recently that dCTCF does not bind to Fab-7 (27). The results of electrophoretic mobility shift assay confirmed this conclusion (Fig. 3). We found that pairing between the 858-bp Fab-7 boundaries (Fig. 1A) facilitated interaction between the white promoter and the eye-specific enhancer (54). To confirm the pairing between Fab-7 insulators, we inserted two Fab-7 insulators in either the same or opposite orientations near the GAL4 binding sites and the white promoter (Fig. 5). GAL4 strongly stimulated white expression in both series of transgenic lines, supporting our previous observation (54) that the functional interaction between the Fab-7 insulators is not dependent on their relative orientation.
FIG. 5.
Testing the functional interaction between the Fab-7 boundaries. For designations, see the legends for Fig. 1 and 2.
Next, we tested whether the Fab-7 and PTS/F8 boundaries are able to interact with each other. When these boundaries were inserted in opposite orientations, we observed strong activation of white by GAL4 (Fig. 6A); when Fab-7 and PTS/F8 were inserted in the same orientation, GAL4-mediated stimulation was at a moderate level (Fig. 6B). These results suggest that the boundaries flanking the iab-7 enhancer can interact in an orientation-dependent manner.
FIG. 6.
Testing of the functional interaction between the Fab-7 and Fab-8 boundaries. For designations, see the legends for Fig. 1 and 2.
A question arose as to which part of the PTS/F8 boundary is responsible for the interaction with Fab-7. To test the role of the Fab-8 insulator, Fab-7 and Fab-8 were placed in both orientations relative to each other (Fig. 6C and D). Unexpectedly, GAL4 failed to stimulate white transcription in most of the transgenic lines tested, suggesting the absence of the functional interaction between the Fab-7 and Fab-8 insulators. However, strong stimulation of white transcription by GAL4 was observed in 3 out of 25 transgenic lines in which the deletion of the insulators led to loss of GAL4-mediated activation (data not shown). Thus, in rare genomic positions, the Fab-7 and Fab-8 insulators were capable of functional interaction.
To test for the functional interaction between Fab-7 and PTS, we inserted them only in opposite orientations (Fig. 6E). Once again, GAL4 failed to stimulate white transcription in most of the transgenic lines tested. These results suggest that both parts of the PTS/F8 boundary are required for functional interaction with Fab-7. To confirm this conclusion, we tested a combination of Fab-7 and a PTS/F8337 fragment containing 83 bp from the 3′ end of PTS and 254 bp from the 5′ end of Fab-8, including dCTCF binding sites (Fig. 1A). Strong stimulation of white transcription by GAL4 was observed when PTS/F8337 and Fab-7 were placed in opposite orientations (Fig. 6F). These results suggest that protein(s) bound to the regions close to the boundary between PTS and Fab-8 are essential for the interaction with the Fab-7 insulator.
PTS/F8 and Fab-7 boundaries functionally interact with the upstream region of the Abd-B A promoter.
A dCTCF binding site was found near promoter A of the Abd-B gene (27). We tested if the 370-bp regulatory element including the CTCF site (ACTCF) can functionally interact with the PTS/F8 and Fab-7 boundaries (Fig. 1A).
To test for the functional interaction between PTS/F8 and ACTCF, we inserted the ACTCF fragment either near the GAL4 binding sites (Fig. 7A) or at the white promoter (Fig. 7B). In both series of transgenic lines, GAL4 effectively stimulated white expression, suggesting that PTS/F8 functionally interacts with ACTCF.
FIG. 7.
Testing of the functional interaction between the boundaries and promoter A of the Abd-B gene. For designations, see the legends for Fig. 1 and 2.
We also combined ACTCF and Fab-7 (Fig. 7C). In the transgenic lines, GAL4 induced white expression at a moderate level, which was indicative of the functional interaction between the Fab-7 insulator and the ACTCF fragment. Thus, the ACTCF region functionally interacts with both boundaries.
Previous transvection experiments with Abd-B alleles (62) provided evidence for the presence of a long tethering region upstream of the Abd-B A promoter that is essential for long-distance communication between the iab enhancers and the promoter. Thus, the question arose as to whether other regions upstream of the Abd-B A promoter are capable of interacting with the boundaries.
We tested two DNA fragments, 1,603 and 1,067 bp in size (Fig. 1A). Transgenic lines were obtained in which either PTS/F8 or Fab-7 was inserted in the same or opposite orientation relative to the test fragments of the Abd-B promoter. No stimulation of mini-white transcription by the GAL4 activator was observed in these experiments (data not shown). These results suggest that only the ACTCF fragment containing a dCTCF binding site is capable of functional interaction with the Fab-7 and PTS/F8 boundaries.
DISCUSSION
Previously we found that the relative orientation of Mcp elements defines the mode of loop formation that either allows or blocks stimulation of the white promoter by the GAL4 activator (33). Here we have demonstrated that two PTS/F8 boundaries or Fab-8 insulators alone are also capable of orientation-dependent interaction. When these elements are located in opposite orientations, the loop configuration is favorable for communication between regulatory elements located beyond the loop. The loop formed by two insulators located in the same orientation juxtaposes two elements located within and beyond the loop, which leads to partial isolation of the GAL4 binding sites and the white promoter placed on the opposite sides of the insulators.
The orientation-dependent interaction may be accounted for by at least two proteins bound to the insulator that are involved in specific protein-protein interactions. In the case of a Fab-8 insulator, we demonstrated that dCTCF is likely to be directly involved in pairing between two insulators. Since mutated Fab-8 insulators devoid of dCTCF binding sites proved to be incapable of interacting with each other, we hypothesize that dCTCF facilitates the binding of a certain as yet unidentified protein (or proteins) that, in combination with dCTCF, accounts for orientation-dependent interaction between the Fab-8 insulators. Functional interactions between the Fab-7 boundary devoid of dCTCF binding sites and PTS/F8 or the upstream Abd-B A promoter region are also evidence for the existence of unidentified proteins that support organization of distance interactions in the Abd-B locus.
Recently it was shown that in the repressed state of the bithorax complex, all of its major regulatory elements binding PcG proteins, including PREs with adjacent boundaries and core promoters, interact at a distance, giving rise to a topologically complex structure (34). The question arises as to what proteins are important for such interactions. All PREs tested in the above study (34) are flanked by boundaries, suggesting that all these regulatory elements may be involved in long-distance interactions. As shown previously, the Fab-7 (3) or Mcp (51, 65) boundaries including PREs can support physical association between even transposons located on different chromosomes. One of relevant models proposes that PcG proteins are capable of supporting highly specific long-distance interactions between transposons (3, 34). However, it is known that many PcG complexes with similar properties can bind to Drosophila chromosomes (58), which leaves open the question as to how such protein complexes can ensure a high specificity of interactions between distantly located transposons. Moreover, there is no experimental evidence that PREs without additional regulatory elements can support long-distance interactions. In contrast, there are many proven cases showing that insulator proteins are involved in physical association between distant chromosomal regions. For example, the interaction between gypsy insulators can support activation of the yellow promoter by enhancers separated by many megabases (31). The Mod(mdg4)-67.2 and Su(Hw) proteins bound to the gypsy insulator are essential for such long-distance interactions. In mammals, the interaction of the imprinting control region on chromosome 7 with the Wsb1/Nf1 locus on chromosome 11 depends on the presence of the CTCF protein (40). In vivo interaction between Fab-7 and the Abd-B promoter is absolutely dependent on the presence of the Fab-7 insulator (12). Finally, we have demonstrated here the functional interaction between the Fab-7 and Fab-8 boundaries and the Abd-B promoter. These results support the model (26, 33, 42) that transcriptional factors bound to boundaries can facilitate enhancer-promoter interactions in the bithorax complex. Further studies are necessary for identifying new proteins involved in long-distance interactions and for elucidating the mechanisms that allow interactions either between proper active enhancers and promoters or between only silenced enhancers and promoters.
Acknowledgments
We are grateful to J. Zhou for his gift of the dCTCF clone.
This study was supported by the Russian Foundation for Basic Research (project no. N 06-04-48360), the Molecular and Cell Biology Program of the Russian Academy of Sciences, and the International Research Scholar Award from the Howard Hughes Medical Institute (to P.G.).
Footnotes
Published ahead of print on 21 April 2008.
REFERENCES
- 1.Akbari, O. S., A. Bousum, E. Bae, and R. A. Drewell. 2006. Unraveling cis-regulatory mechanisms at the Abdominal-A and Abdominal-B genes in the Drosophila bithorax complex. Dev. Biol. 293294-304. [DOI] [PubMed] [Google Scholar]
- 2.Akbari, O. S., E. Bae, H. Johnsen, A. Villaluz, D. Wong, and R. A. Drewell. 2008. A novel promoter-tethering element regulates enhancer-driven gene expression at the bithorax complex in the Drosophila embryo. Development 135123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bantignies, F., C. Grimaud, S. Lavrov, M. Gabut, and G. Cavalli. 2003. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 172406-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barges, S., J. Mihaly, M. Galloni, K. Hagstrom, M. Muller, G. Shanower, P. Schedl, H. Gyurkovics, and F. Karch. 2000. The Fab-8 boundary defines the distal limit of the bithorax complex iab-7 domain and insulates iab-7 from initiation elements and a PRE in the adjacent iab-8 domain. Development 127779-790. [DOI] [PubMed] [Google Scholar]
- 5.Boulet, A. M., A. Lloyd, and S. Sakonju. 1991. Molecular definition of the morphogenetic and regulatory functions and the cis-regulatory elements of the Drosophila Abd-B homeotic gene. Development 111393-405. [DOI] [PubMed] [Google Scholar]
- 6.Brasset, E., and C. Vaury. 2005. Insulators and fundamental components of the eukaryotic genomes. Heredity 94571-576. [DOI] [PubMed] [Google Scholar]
- 7.Busturia, A., and M. Bienz. 1993. Silencers in abdominal-B, a homeotic Drosophila gene. EMBO J. 121415-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Busturia, A., C. D. Wightman, and S. Sakonju. 1997. A silencer is required for maintenance of transcriptional repression throughout Drosophila development. Development 1244343-4350. [DOI] [PubMed] [Google Scholar]
- 9.Celniker, S. E., S. Sharma, D. J. Keelan, and E. B. Lewis. 1990. The molecular genetics of the bithorax complex of Drosophila: cis-regulation in the Abdominal-B domain. EMBO J. 94227-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chambeyron, S., and W. A. Bickmore. 2004. Does looping and clustering in the nucleus regulate gene expression? Curr. Opin. Cell Biol. 16256-262. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Q., L. Lin, S. Smith, Q. Lin, and J. Zhou. 2005. Multiple promoter targeting sequences exist in Abdominal-B to regulate long-range gene activation. Dev. Biol. 286629-636. [DOI] [PubMed] [Google Scholar]
- 12.Cleard, F., Y. Moshkin, F. Karch, and R. K. Maeda. 2006. Probing long-distance regulatory interactions in the Drosophila melanogaster bithorax complex using Dam identification. Nat. Genet. 38931-935. [DOI] [PubMed] [Google Scholar]
- 13.Comet, I., E. Savitskaya, B. Schuettengruber, N. Negre, S. Lavrov, A. Parshikov, F. Juge, E. Gracheva, P. Georgiev, and G. Cavalli. 2006. PRE-mediated bypass of two Su(Hw) insulators targets PcG proteins to a downstream promoter. Dev. Cell 11117-124. [DOI] [PubMed] [Google Scholar]
- 14.de Laat, W., and F. Grosveld. 2003. Spatial organization of gene expression: the active chromatin hub. Chromosome Res. 11447-459. [DOI] [PubMed] [Google Scholar]
- 15.Dillon, N., and P. Sabbattini. 2000. Functional gene expression domains: defining functional unit of eukaryotic gene regulation. BioEssays 22657-665. [DOI] [PubMed] [Google Scholar]
- 16.Dorsett, D. 1999. Distant liaisons: long range enhancer-promoter interactions in Drosophila. Curr. Opin. Genet. 9505-514. [DOI] [PubMed] [Google Scholar]
- 17.Duncan, I. 1987. The bithorax complex. Annu. Rev. Genet. 21285-319. [DOI] [PubMed] [Google Scholar]
- 18.Fraser, P., and W. Bickmore. 2007. Nuclear organization of the genome and the potential for gene regulation. Nature 447413-417. [DOI] [PubMed] [Google Scholar]
- 19.Galloni, M., H. Gyurkovics, P. Schedl, and F. Karch. 1993. The bluetail transposon: evidence for independent cis-regulatory domains and domain boundaries in the bithorax complex. EMBO J. 121087-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geyer, P. K., and V. G. Corces. 1987. Separate regulatory elements are responsible for the complex pattern of tissue-specific and developmental transcription of the yellow locus in Drosophila melanogaster. Genes Dev. 1996-1004. [DOI] [PubMed] [Google Scholar]
- 21.Golic, K. G., and S. Lindquist. 1989. The FLP recombinase of yeast catalyzes site-specific recombination in the Drosophila genome. Cell 59499-509. [DOI] [PubMed] [Google Scholar]
- 22.Gruzdeva, N., O. Kyrchanova, A. Parshikov, A. Kullyev, and P. Georgiev. 2005. The Mcp element from the bithorax complex contains an insulator that is capable of pairwise interactions and can facilitate enhancer-promoter communication. Mol. Cell. Biol. 253682-3689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gyurkovics, H., J. Gausz, J. Kummer, and F. Karch. 1990. A new homeotic mutation in the Drosophila bithorax complex removes a boundary separating two domains of regulation. EMBO J. 92579-2585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagstrom, K., M. Muller, and P. Schedl. 1996. Fab-7 functions as a chromatin domain boundary to ensure proper segment specification by the Drosophila bithorax complex. Genes Dev. 103202-3215. [DOI] [PubMed] [Google Scholar]
- 25.Hagstrom, K., M. Muller, and P. Schedl. 1997. A Polycomb and GAGA dependent silencer adjoin the Fab-7 boundary in the Drosophila bithorax complex. Genetics 1461365-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hogga, I., J. Mihaly, S. Barges, and F. Karch. 2001. Replacement of Fab-7 by the gypsy or scs insulator disrupts long-distance regulatory interactions in the Abd-B gene of the bithorax complex. Mol. Cell 81145-1151. [DOI] [PubMed] [Google Scholar]
- 27.Holohan, E. E., C. Kwong, B. Adryan, M. Bartkuhn, M. Herold, R. Renkawitz, S. Russel, and R. White. 2007. CTCF genomic binding sites in Drosophila and the organization of the bithorax complex. PLOS Genet. 3e112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karch, F., B. Weiffenbach, M. Peifer, W. Bender, I. Duncan, S. Celniker, M. Crosby, and E. B. Lewis. 1985. The abdominal region of the bithorax complex. Cell 4381-96. [DOI] [PubMed] [Google Scholar]
- 29.Karch, F., M. Galloni, L. Sipos, J. Gausz, H. Gyurkovics, and P. Schedl. 1994. Mcp and Fab-7: molecular analysis of putative boundaries of cis-regulatory domains in the bithorax complex of Drosophila melanogaster. Nucleic Acids Res. 223138-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karess, R. E., and G. M. Rubin. 1984. Analysis of P transposable element functions in Drosophila. Cell 38135-146. [DOI] [PubMed] [Google Scholar]
- 31.Kravchenko, E., E. Savitskaya, O. Kravchuk, A. Parshikov, P. Georgiev, and M. Savitsky. 2005. Pairing between gypsy insulators facilitates the enhancer action in trans throughout the Drosophila genome. Mol. Cell. Biol. 259283-9291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuhn, E. J., and P. K. Geyer. 2003. Genomic insulators: connecting properties to mechanism. Curr. Opin. Cell Biol. 15259-265. [DOI] [PubMed] [Google Scholar]
- 33.Kyrchanova, O., S. Toshchakov, A. Parshikov, and P. Georgiev. 2007. Study of the functional interaction between Mcp insulators from the Drosophila bithorax complex: effects of insulator pairing on enhancer-promoter communication. Mol. Cell. Biol. 273035-3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lanzuolo, C., V. Roure, J. Dekker, F. Bantignies, and V. Orlando. 2007. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat. Cell Biol. 91167-1174. [DOI] [PubMed] [Google Scholar]
- 35.Levine, M. R., and R. Tjian. 2003. Transcription regulation and animal diversity. Nature 424147-151. [DOI] [PubMed] [Google Scholar]
- 36.Lewis, E. B. 1978. A gene complex controlling segmentation in Drosophila. Nature 276565-570. [DOI] [PubMed] [Google Scholar]
- 37.Lin, Q., D. Wu, and J. Zhou. 2003. The promoter targeting sequence facilitates and restricts a distant enhancer to a single promoter in the Drosophila embryo. Development 130519-526. [DOI] [PubMed] [Google Scholar]
- 38.Lin, Q., Q. Chen, L. Lin, and J. Zhou. 2004. The promoter targeting sequence mediates epigenetically heritable transcription memory. Genes Dev. 182639-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, Q., Q. Chen, L. Lin, S. Smith, and J. Zhou. 2007. Promoter targeting sequence mediates enhancer interference in the Drosophila embryo. Proc. Natl. Acad. Sci. USA 1043237-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling, J. Q., T. Li, J. F. Hu, T. H. Vu, H. L. Chen, X. W. Qiu, A. M. Cherry, and A. R. Hoffman. 2006. CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312269-272. [DOI] [PubMed] [Google Scholar]
- 41.Maeda, R. K., and F. Karch. 2006. The ABC of the BX-C: the bithorax complex explained. Development 1331413-1422. [DOI] [PubMed] [Google Scholar]
- 42.Maeda, R. K., and F. Karch. 2007. Making connections: boundaries and insulators in Drosophila. Curr. Opin. Genet. Dev. 17394-399. [DOI] [PubMed] [Google Scholar]
- 43.Maksimenko, O. G., D. A. Chetverina, and P. G. Georgiev. 2006. Insulators of higher eukaryotes: properties, mechanisms of action, and role in transcription regulation. Genetika 421029-1044. [PubMed] [Google Scholar]
- 44.Martin, C. H., C. A. Mayeda, C. A. Davis, C. L. Ericsson, J. D. Knafels, D. R. Mathog, S. E. Celniker, E. B. Lewis, and M. J. Palazzolo. 1995. Complete sequence of the bithorax complex of Drosophila. Proc. Natl. Acad. Sci. USA 928398-8402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCall, K., M. B. O'Connor, and W. Bender. 1994. Enhancer traps in the Drosophila bithorax complex mark parasegmental domains. Genetics 138387-399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mihaly, J., I. Hogga, J. Gausz, H. Gyurkovics, and F. Karch. 1997. In situ dissection of the Fab-7 region of the bithorax complex into a chromatin domain boundary and a Polycomb-response element. Development 1241809-1820. [DOI] [PubMed] [Google Scholar]
- 47.Mihaly, J., I. Hogga, S. Barges, M. Galloni, R. K. Mishra, K. Hagstrom, M. Muller, P. Schedl, L. Sipos, J. Gausz, H. Gyurkovics, and F. Karch. 1998. Chromatin domain boundaries in the bithorax complex. Cell Mol. Life Sci. 5460-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mihaly, J., S. Barges, L. Sipos, R. Maeda, F. Cleard, I. Hogga, W. Bender, H. Gyurkovics, and F. Karch. 2006. Dissecting the regulatory landscape of the Abd-B gene of the bithorax complex. Development 1332983-2993. [DOI] [PubMed] [Google Scholar]
- 49.Mohan, M., M. Bartkuhn, M. Herold, A. Philippen, N. Heinl, I. Bardenhagen, J. Leers, R. A. H. White, R. Renkawitz-Pohl, Harald Saumweber, and R. Renkawitz. 2007. The Drosophila insulator proteins CTCF and CP190 link enhancer blocking to body patterning. EMBO J. 264203-4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moon, H., G. Filippova, D. Loukinov, E. Pugacheva, Q. Chen, S. T. Smith, A. Munhall, B. Grewe, M. Bartkuhn, R. Arnold, L. J. Burke, R. Renkawitz-Pohl, R. Ohlsson, J. Zhou, R. Renkawitz, and V. Lobanenkov. 2005. CTCF is conserved from Drosophila to humans and confers enhancer blocking of the Fab-8 insulator. EMBO Rep. 6165-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Muller, M., K. Hagstrom, H. Gyurkovics, V. Pirrotta, and P. Schedl. 1999. The Mcp element from the Drosophila melanogaster bithorax complex mediates long-distance regulatory interactions. Genetics 1531333-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Petrascheck, M., D. Escher, T. Mahmoudi, C. P. Verrijer, W. Schaffner, and A. Barberis. 2005. DNA looping induced by a transcriptional enhancer in vivo. Nucleic Acids Res. 333743-3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ptashne, M. 1986. Gene regulation by proteins acting nearby and at distance. Nature 322697-701. [DOI] [PubMed] [Google Scholar]
- 54.Rodin, S., O. Kyrchanova, E. Pomerantseva, A. Parshikov, and P. Georgiev. 2007. New properties of Drosophila Fab-7 insulator. Genetics 177113-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sanchez-Herrero, E., I. Vernos, R. Marco, and G. Morato. 1985. Genetic organization of Drosophila bithorax complex. Nature 313108-113. [DOI] [PubMed] [Google Scholar]
- 56.Sanchez-Herrero, E. 1991. Control of the expression of the bithorax complex abdominal-A and Abdominal-B by cis-regulatory regions in Drosophila embryos. Development 111437-448. [DOI] [PubMed] [Google Scholar]
- 57.Savitskaya, E., L. Melnikova, M. Kostuchenko, E. Kravchenko, E. Pomerantseva, T. Boikova, D. Chetverina, A. Parshikov, P. Zobacheva, E. Gracheva, A. Galkin, and P. Georgiev. 2006. Study of long-distance functional interactions between Su(Hw) insulators that can regulate enhancer-promoter communication in Drosophila melanogaster. Mol. Cell. Biol. 26754-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schwartz, Y. B., T. G. Kahn, D. A. Nix, X. Y. Li, R. Bourgon, M. Biggin, and V. Pirrotta. 2006. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat. Genet. 38700-705. [DOI] [PubMed] [Google Scholar]
- 59.Schweinsberg, S., and P. Schedl. 2004. Developmental modulation of Fab-7 boundary function. Development 1314743-4749. [DOI] [PubMed] [Google Scholar]
- 60.Schweinsberg, S., K. Hagstrom, D. Gohl, P. Schedl, R. P. Kumar, R. Mishra, and F. Karch. 2004. The enhancer-blocking activity of the Fab-7 boundary from the Drosophila bithorax complex requires GAGA-factor-binding sites. Genetics 1681371-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Siegal, M. L., and D. L. Hartl. 2000. Application of Cre/loxP in Drosophila. Site-specific recombination and transgene co-placement. Methods Mol. Biol. 136487-495. [DOI] [PubMed] [Google Scholar]
- 62.Sipos, L., J. Mihaly, F. Karch, P. Schedl, J. Gausz, and H. Gyurkovics. 1998. Transvection in the Drosophila Abd-B domain: extensive upstream sequences are involved in anchoring distant cis-regulatory regions to the promoter. Genetics 1491031-1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sipos, L., and H. Gyurkovics. 2005. Long-distance interactions between enhancers and promoters. The case of the Abd-B domain of the Drosophila bithorax complex. FEBS J. 2723253-3259. [DOI] [PubMed] [Google Scholar]
- 64.Valenzuela, L., and R. T. Kamakaka. 2006. Chromatin insulators. Annu. Rev. Genet. 40107-138. [DOI] [PubMed] [Google Scholar]
- 65.Vazquez, J., M. Muller, V. Pirrotta, and J. W. Sedat. 2006. The Mcp element mediates stable long-range chromosome-chromosome interactions in Drosophila. Mol. Biol. Cell 172158-2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wallace, J. A., and G. Felsenfeld. 2007. We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17400-407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.West, A. G., and P. Fraser. 2005. Remote control of gene transcription. Hum. Mol. Genet. 14R101-R111. [DOI] [PubMed] [Google Scholar]
- 68.Zhao, H., and A. Dean. 2005. Organizing the genome: enhancers and insulators. Biochem. Cell Biol. 83516-524. [DOI] [PubMed] [Google Scholar]
- 69.Zhou, J., and M. Levine. 1999. A novel cis-regulatory element, the PTS, mediates an anti-insulator activity in the Drosophila embryo. Cell 99567-575. [DOI] [PubMed] [Google Scholar]
- 70.Zhou, J., S. Barolo, P. Szymanski, and M. Levine. 1996. The Fab-7 element of the bithorax complex attenuates enhancer-promoter interactions in the Drosophila embryo. Genes Dev. 103195-3201. [DOI] [PubMed] [Google Scholar]
- 71.Zhou, J., H. Ashe, C. Burks, and M. Levine. 1999. Characterization of the transvection mediating region of the Abdominal-B locus in Drosophila. Development 1263057-3065. [DOI] [PubMed] [Google Scholar]