Abstract
FANCJ mutations are associated with breast cancer and genetically linked to the bone marrow disease Fanconi anemia (FA). The genomic instability of FA-J mutant cells suggests that FANCJ helicase functions in the replicational stress response. A putative helicase with sequence similarity to FANCJ in Caenorhabditis elegans (DOG-1) and mouse (RTEL) is required for poly(G) tract maintenance, suggesting its involvement in the resolution of alternate DNA structures that impede replication. Under physiological conditions, guanine-rich sequences spontaneously assemble into four-stranded structures (G quadruplexes [G4]) that influence genomic stability. FANCJ unwound G4 DNA substrates in an ATPase-dependent manner. FANCJ G4 unwinding is specific since another superfamily 2 helicase, RECQ1, failed to unwind all G4 substrates tested under conditions in which the helicase unwound duplex DNA. Replication protein A stimulated FANCJ G4 unwinding, whereas the mismatch repair complex MSH2/MSH6 inhibited this activity. FANCJ-depleted cells treated with the G4-interactive compound telomestatin displayed impaired proliferation and elevated levels of apoptosis and DNA damage compared to small interfering RNA control cells, suggesting that G4 DNA is a physiological substrate of FANCJ. Although the FA pathway has been classically described in terms of interstrand cross-link (ICL) repair, the cellular defects associated with FANCJ mutation extend beyond the reduced ability to repair ICLs and involve other types of DNA structural roadblocks to replication.
The identification of FANCJ mutations in early-onset breast cancer patients (3, 34) and Fanconi anemia (FA) group J patients (22, 23, 25) implicates FANCJ as a tumor suppressor caretaker that ensures genomic stability. FANCJ interacts with the tumor suppressor BRCA1 (3) and is a DNA helicase that catalytically unwinds duplex DNA in an ATP hydrolysis-dependent manner (2, 14). FA-J cells are hypersensitive to interstrand cross-linking (ICL) agents (1, 25); untreated FA-J cells exhibit diminished BRCA1 foci, and the cells show delayed formation of ionizing radiation-induced BRCA1 foci (31). In response to DNA damage or replicational stress, FANCJ colocalizes with the single-stranded DNA (ssDNA) binding protein replication protein A (RPA), which serves as an auxiliary factor for the unwinding function of FANCJ (15). FANCJ interacts with the mismatch repair complex MutLα, and this interaction is required for the correction of the ICL response in FA-J cells (32). The activation of FANCJ helicase activity is required for timely progression through S phase of the cell cycle (20); however, the precise functions of the FANCJ helicase in S-phase progression remain to be understood.
Although a role for FANCJ in DNA replication has been proposed previously, definitive evidence for a role of the FANCJ helicase in preventing genomic instability is lacking. One source of genomic instability is alternate DNA structures that may impede the replication fork. Guanine-rich nucleic acids have the potential to form G-quadruplex (G4) DNA stabilized by Hoogsteen hydrogen bonding between guanine residues, and G4 DNA influences gene expression and genomic stability (for a review, see reference 26). In the human genome, the number of distinct sites with the potential to form G4 DNA is estimated to be more than 300,000 (9). Key cellular processes are identified with repetitive G-rich chromosomal regions, where the regulated formation of G4 DNA may contribute to biological functions such as immunoglobulin gene rearrangement (21), promoter activation (38), and telomere maintenance (29, 49).
Interestingly, evidence suggests that a helicase-like protein with sequence similarity to the FANCJ helicase domain in nematodes has a role in guanine-rich-DNA metabolism. Nematodes with mutations in dog-1 (deletions of guanine-rich DNA) show germ line as well as somatic deletions in genes containing guanine-rich DNA (4). It was proposed previously that the putative DOG-1 helicase has a role in the resolution of alternate G4 DNA structures that impede replication. Recent genetic evidence indicates that DOG-1 is the Caenorhabditis elegans FANCJ homologue, based on the sensitivity of dog-1 mutants to ICL agents and the epistatic relationship of dog-1 to fcd-1 (encoding C. elegans FANCD2 [CeFANCD2]) (47).
To better understand the roles of FANCJ in DNA metabolism, we have investigated the potential ability of FANCJ to unwind G4 DNA substrates. Purified FANCJ helicase was found to efficiently unwind a variety of G4 DNA structures in a reaction that was dependent on intrinsic FANCJ ATP hydrolysis and regulated by DNA repair factors through distinct mechanisms. The naturally occurring molecule telomestatin (TMS), which selectively binds G4 structures and interferes with normal telomere metabolism, potently inhibited FANCJ G4 unwinding activity. TMS inhibited proliferation and induced DNA damage, accompanied by apoptosis, to a significantly greater extent among FANCJ-depleted cells than among control cells, providing further evidence that FANCJ is likely to have an important role in unwinding G4 DNA structures that potentially impede replication and cause chromosomal instability.
MATERIALS AND METHODS
Proteins.
Baculovirus encoding FANCJ with a C-terminal FLAG tag, kindly provided by S. Cantor (University of Massachusetts Medical School, Worcester, MA), was used to infect High Five insect cells, and the recombinant FANCJ protein was purified as described previously (2). Baculovirus encoding RECQ1 with a histidine tag, kindly provided by A. Vindigni (International Centre for Genetic Engineering and Biotechnology, Trieste, Italy), was used to infect Sf9 insect cells, and the recombinant RECQ1 protein was purified as described previously (35). Human RPA purified from HeLa cells was provided by M. Kenny (Albert Einstein Cancer Center, Bronx, NY). Recombinant His-tagged MSH2/MSH6 was kindly provided by T. Wilson (University of Maryland, Baltimore, MD). Purified recombinant Bloom's syndrome protein (BLM) was kindly provided by I. Hickson and L. Wu (Cancer Research UK Laboratories). A representative gel with the purified recombinant proteins used in this study is shown in Fig. S1 in the supplemental material.
DNA substrates.
Polyacrylamide gel electrophoresis-purified oligonucleotides used for the preparation of DNA substrates were purchased from Loftstrand Labs (Gaithersburg, MD) and are listed in Table 1. The forked-duplex DNA substrate was prepared from the DC26 and TSTEM25 oligonucleotides as described previously (5). G4 DNA and G2′ DNA substrates were prepared from the oligonucleotides indicated in Table 1 as described previously (28, 43, 46).
TABLE 1.
Substrates used in this study
The guanine residues that compose the G4 structures in the oligonucleotides are underlined.
Helicase assays.
Helicase assay reaction mixtures (20 μl) contained 40 mM Tris-HCl (pH 7.4), 25 mM KCl, 5 mM MgCl2, 2 mM dithiothreitol, 2% glycerol, 100 ng of bovine serum albumin/μl, 2 mM ATP, 10 fmol of the specified tetraplex or duplex DNA substrate (0.5 nM), and the concentrations of FANCJ indicated in the figures. Helicase reactions were initiated by the addition of FANCJ, and the reaction mixtures were incubated at 30°C for 15 min unless otherwise indicated. G4 helicase reactions were terminated by the addition of 20 μl of stop buffer (74% glycerol, 0.01% xylene cyanol, 0.01% bromophenol blue, 10 mM KCl, 20 mM EDTA). Forked-duplex helicase reactions were terminated as described previously (15). Reaction products were resolved on nondenaturing 8 and 12% (19:1 acrylamide-bisacrylamide) polyacrylamide gels for the G4 and forked-duplex substrates, respectively, and quantitated as described previously (15).
G4 DNA binding ligands.
N-Methyl mesoporphyrin IX (NMM) and meso-tetra(N-methyl-4-pyridyl) porphine tetratosylate (T4) were from Frontier Scientific, Inc. (Logan, UT). TMS was purified as described previously (37). Compounds were dissolved in dimethyl sulfoxide.
Cell lines.
HeLa, U2OS, and FA-A (PD220) fibroblasts were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 1% penicillin-streptomycin, and 1% l-glutamine at 37°C in 5% CO2. FA-D2 (PD20) fibroblasts were grown in RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 15% fetal bovine serum, 1% penicillin-streptomycin, and 1% l-glutamine at 37°C in 5% CO2.
FANCJ silencing in HeLa and U2OS cells.
The sequence of FANCJ small interfering RNA (siRNA) was 5′ GUACAGUACCCCACCUUAU 3′ (25), and that of the control siRNA was 5′ AGUUACUCAGCCAAGAACGA 3′ (20). siRNA transfections were done using Lipofectamine 2000 according to the protocol of the manufacturer (Invitrogen). Cells were plated to 40% confluence in 10-cm dishes 24 h prior to transfection. siRNA (0.6 nmol) was mixed with 30 μl of Lipofectamine 2000 in 3 ml of Opti-MEM (Invitrogen). The mixture was added to cells which were subsequently incubated for 6 h. After 24 h, a second transfection was performed similarly. Seventy-two hours after the initial transfection, cells were harvested or treated as indicated below. Cell lysate preparation and Western blot analyses were performed as described previously (15).
Cell proliferation studies.
Seventy-two hours after the first siRNA transfection, cells were treated with TMS (5 μM) or left untreated for 2 h and subsequently washed with phosphate-buffered saline (PBS). To analyze relative growth rates, 5 × 104 cells were seeded in triplicate into a 12-well plate and the total cell numbers at the times indicated in a figure were determined using a Coulter counter (Beckman, Fullerton, CA). The effect of TMS on cellular metabolic activity was determined by a WST-1 assay (Roche). Cells were plated onto 96-well plates at a density of 5 × 103 per well. Ten microliters of WST-1 per well was added to the cells, and the plates were incubated at 37°C for 2 h. The absorbance at 450 nm at the times indicated in a figure was measured using a microplate reader (Bio-Rad). For the quantitative determination of the DNA synthesis rate, mitogenic activity was determined by measuring bromodeoxyuridine (BrdU) incorporation by a BrdU cell proliferation enzyme-linked immunosorbent assay (ELISA; Roche). Cells (5 × 103) were seeded in quadruplicate into 96-well plates and allowed to adhere for 16 h. Cells were then labeled for 3 h with BrdU, and DNA synthesis was measured with the ELISA BrdU assay according to the manufacturer's instructions. Absorbance values correlate directly to the amount of newly synthesized DNA and the number of proliferating cells in the culture. The absorbance at 370 nm and 450 nm was measured with a microplate reader (Bio-Rad). All cell proliferation assays were performed at least four times.
Analysis of cellular apoptosis.
HeLa or U2OS cells were treated with either the control or FANCJ siRNA duplexes for 72 h as described above. To assay apoptosis induction after TMS treatment, equal numbers of siRNA-treated cells were subcultured in complete medium containing 5 μM TMS or no drug for 2 h and then allowed a recovery period in complete medium for 12 h. Subsequently, the cytoplasmic histone-associated DNA fragments, which are indicative of ongoing apoptosis, were quantitatively measured using the cell death detection ELISAPLUS photometric enzyme immunoassay (Roche).
Immunofluorescence studies.
Cells were treated with siRNA as described above. For drug treatments, TMS (5 μM) was added to the cells for 2 h and the cells were washed twice with PBS, returned to complete medium for 2 h, washed, and fixed with paraformaldehyde (3.7%) at room temperature for 15 min. Fixed cells were washed four times with PBS and treated with 0.5% Triton solution (Sigma) at room temperature for 3 min. Cells were then washed four times with PBS and blocked with 10% goat serum (Sigma) overnight at 4°C. Indirect immunostaining was performed by first incubating cells with a mouse anti-γ-H2AX monoclonal antibody (1:500; Upstate) for 1 h at 37°C. Following four washes in PBS containing 0.1% Tween 20, cells were incubated with Alexa Fluor 488-conjugated goat anti-mouse immunoglobulin G (1:400; Invitrogen) for 1 h at 37°C. Cells were washed four times with PBS containing 0.1% Tween 20 and coated with Prolong Gold antifade reagent containing DAPI (4′,6′-diamidino-2-phenylindole; Invitrogen). Coverslips were placed on chamber slides, and cells were cured at room temperature in the dark for 24 h. Immunofluorescence analyses were performed with a Zeiss LSM 510 META inverted Axiovert 200 M laser scan microscope with a Plan-Apochromat 63× 1.4-numerical-aperature oil immersion differential interference contrast objective lens. Images were captured with a charge-coupled device camera and analyzed using the LSM Browser software package.
RESULTS
FANCJ efficiently unwinds G4 DNA in an ATP hydrolysis-dependent manner.
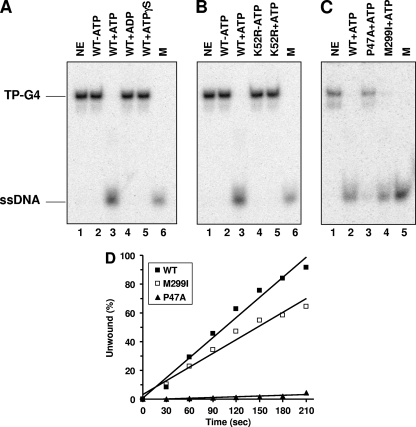
To better understand the DNA targets FANCJ acts upon to perform its cellular functions, we tested the ability of purified FANCJ recombinant protein to unwind a variety of G-rich tetraplex structures. We began by testing the ability of FANCJ helicase to unwind a well-characterized four-stranded parallel G4 DNA substrate (TP-G4) derived from a mouse immunoglobulin sequence. FANCJ unwound the TP-G4 substrate in the presence of ATP, as evidenced by the product of the helicase reaction (Fig. 1A, lane 3), which comigrated with the unannealed control radiolabeled oligonucleotide (lane 6). FANCJ failed to unwind the G4 substrate in the absence of ATP (lane 2) or in the presence of ADP (lane 4) or ATPγS (lane 5), indicating that FANCJ G4 unwinding requires the hydrolysis of nucleoside triphosphate. FANCJ bound the G4 substrate in the presence of ATPγS or in the absence of a nucleotide (data not shown), indicating that G4 binding by FANCJ is not nucleotide dependent.
FIG. 1.
FANCJ efficiently unwinds G4 DNA in an ATP hydrolysis-dependent manner. (A) G4 unwinding assays were performed by incubating 4.8 nM wild-type (WT) FANCJ with 0.5 nM TP-G4 DNA substrate at 30°C for 15 min under standard helicase assay conditions as described in Materials and Methods. Lane 1, no-enzyme control (NE); lanes 2 to 5, FANCJ G4 helicase reaction mixtures with or without the indicated nucleotide (2 mM); lane 6, 32P-labeled single-stranded oligonucleotide marker (M). (B) Catalytically inactive FANCJ mutant protein K52R fails to unwind G4 DNA. Lane 1, no-enzyme control; lanes 2 and 3, wild-type FANCJ (4.8 nM) in the absence or presence of 2 mM ATP; lanes 4 and 5, K52R (4.8 nM) in the absence or presence of 2 mM ATP; lane 6, marker. (C) Wild-type FANCJ (4.8 nM) or variant forms (with the mutation P47A or M299I; 4.8 nM) tested for G4 unwinding in the presence of 2 mM ATP. Lane 1, no-enzyme control; lane 2, wild-type FANCJ; lane 3, FANCJ P47A mutant protein (P47A); lane 4, FANCJ M299I mutant protein (M299I); lane 5, marker. (D) Kinetic analyses of DNA unwinding of the G4-TP substrate by wild-type FANCJ or FANCJ polymorphic variants. Wild-type FANCJ (0.3 nM) or an associated variant (FANCJ M299I or P47A mutant protein) was incubated with the 0.5 nM TP-G4 DNA substrate under standard helicase reaction conditions at 30°C for the indicated times. A quantitative analysis of results from FANCJ helicase assays is shown.
To determine if G4 unwinding was dependent on ATP hydrolysis intrinsic to FANCJ, we tested an engineered FANCJ ATPase-dead mutant protein (K52R) purified in a manner identical to that of wild-type FANCJ, which was shown previously to be completely inactive as a helicase on M13- or oligonucleotide-based duplex DNA substrates (2, 14), for G4 unwinding activity. The K52R mutant protein failed to resolve the G4 substrate (Fig. 1B, lane 5), confirming that FANCJ G4 unwinding activity is dependent on FANCJ ATP hydrolysis.
Naturally occurring mutations (P47A and M299I) in the conserved helicase domain of FANCJ previously identified in breast cancer patients lacking BRCA1 or BRCA2 mutations (3) were evaluated for their effects on FANCJ G4 unwinding activity. The P47A mutant protein, which has a 12-fold reduction in the kcat for DNA-dependent ATP hydrolysis and an 80-fold reduction in helicase activity on a forked, 19-bp duplex substrate compared to wild-type FANCJ (13), unwound only 5% of the G4 substrate compared to the 100% unwound by wild-type FANCJ at a 4.8 nM protein concentration (Fig. 1C, lane 3 versus lane 2). The M299I mutant protein (lane 4) retained a level of G4 helicase activity similar to that of wild-type FANCJ, a result that is consistent with the ability of FANCJ to unwind duplex DNA substrates efficiently (13). Kinetic analyses of the G4-unwinding reactions confirmed that the wild-type and M299I FANCJ proteins unwound the TP-G4 substrate in a time-dependent manner but that the P47A mutant protein displayed a 50-fold reduction in the rate of unwinding of the G4 substrate compared to the rate of wild-type FANCJ (Fig. 1D).
We next examined G4 helicase activity as a function of FANCJ concentration. FANCJ unwound the G4 DNA and forked-duplex substrates (0.5 nM) in a protein concentration-dependent manner (see Fig. S2A and B in the supplemental material). Comparable percentages of the 19-bp forked-duplex and G4 substrates were unwound by FANCJ protein concentrations up to 0.3 nM; however, FANCJ concentrations of 0.6 nM and greater unwound significantly more of the G4 substrate than of the forked duplex (see Fig. S2C in the supplemental material).
In addition to FANCJ, two other superfamily 2 (SF2) helicases, Werner syndrome protein (WRN) and BLM, were reported previously to unwind a variety of G4 DNA structures, including the TP-G4 substrate (10, 43). To determine if G4 DNA-unwinding activity is a general property of all SF2 helicases, we tested human RECQ1 helicase, an enzyme that we have previously shown to unwind a variety of duplex DNA structures, including a 3′ ssDNA substrate, a forked duplex, a three-stranded D loop, and a four-stranded synthetic Holliday junction (35). RECQ1 was unable to unwind all tetraplex DNA substrates tested, including TP-G4 and a G4 substrate derived from the human telomeric sequence, as well as G4 substrates with a 15- or 25-nucleotide (nt) 3′ tail, under conditions in which the enzyme efficiently unwound a forked-duplex substrate (see Fig. S3 in the supplemental material). These results suggest that G4 DNA unwinding by FANCJ is specific and not a general feature of SF2 DNA helicases, including a member of the RecQ helicase family.
FANCJ unwinds a two-stranded G2′ tetraplex.
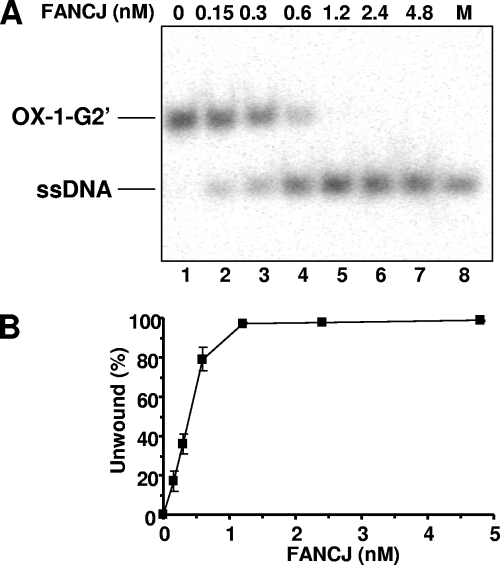
An alternative form of G-G-paired DNA, designated G2′, can form when hairpin dimers of two antiparallel strands form Hoogsteen hydrogen bonds (42). We examined the ability of FANCJ to unwind G2′ tetraplex structures formed by double T4G4 repeats derived from the Oxytricha telomeric sequence. FANCJ unwound the 5′-tailed G2′ tetraplex (OX-1-G2′) in a protein concentration-dependent manner (Fig. 2A). The percentages of the substrate unwound at given FANCJ protein concentrations were comparable for the two-stranded G2′ tetraplex substrate (Fig. 2B) and the four-stranded parallel TP-G4 tetraplex (see Fig. S2C in the supplemental material).
FIG. 2.
FANCJ unwinds two-stranded OX-1-G2′ tetraplex DNA. The indicated FANCJ concentrations (lanes 2 to 7) were incubated with 0.5 nM OX-1-G2′ at 30°C for 15 min under standard helicase assay conditions. Lane 1, no-enzyme control; lane 8, 32P-labeled single-stranded oligonucleotide marker (M). (B) Quantitative analysis of FANCJ helicase activity on OX-1-G2′. The data shown are the means of results from three independent experiments, with standard deviations (SD) indicated by error bars.
FANCJ G4 helicase activity is differentially modulated by DNA repair factors. (i) RPA stimulates FANCJ G4 helicase activity.
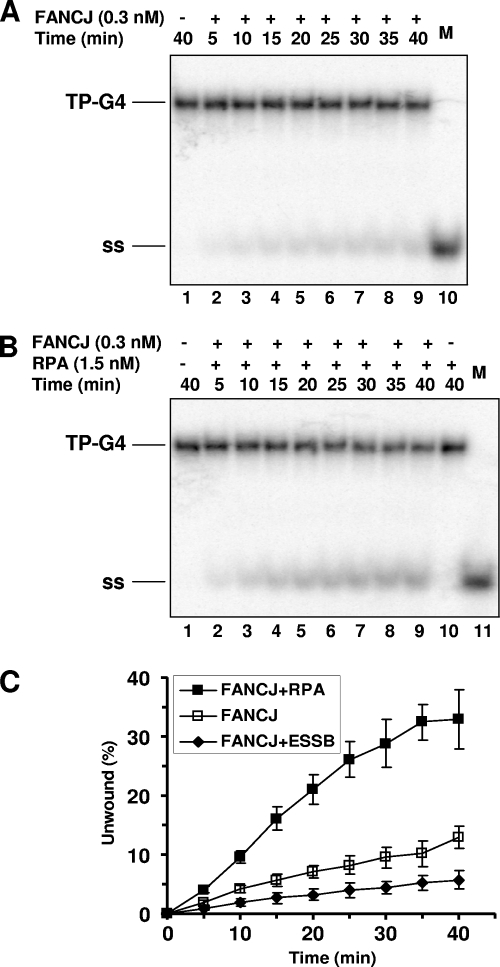
Recently, we reported that FANCJ associates with RPA in a DNA damage-inducible manner and that, through the protein interaction, RPA stimulates FANCJ helicase to better unwind duplex DNA substrates (15). These findings identified RPA as the first regulatory partner of FANCJ and suggested that RPA may be important for the helicase to more efficiently unwind DNA repair intermediates or alternate DNA structures to maintain genomic stability. To further explore the functional interaction between FANCJ and RPA, we performed kinetic experiments to determine if RPA affects FANCJ G4 helicase activity. A low concentration of FANCJ (0.3 nM) unwound the TP-G4 substrate in a time-dependent manner, achieving a level of unwound G4 substrate of 10% by the end of a 40-min time course (Fig. 3A and C). In the presence of 1.5 nM RPA, FANCJ G4 unwinding was stimulated throughout the time course (5 to 40 min) (Fig. 3B and C). A linear regression analysis of the kinetic data demonstrated a threefold increase in the rate of FANCJ G4 helicase activity when RPA was present. In control experiments, RPA alone did not destabilize the G4 substrate (Fig. 3B, lane 10; also data not shown). The same concentration (1.5 nM) of Escherichia coli ssDNA binding protein failed to stimulate FANCJ G4 helicase activity and in fact moderately inhibited G4 unwinding by FANCJ (Fig. 3C). The protection-of-telomeres protein POT1 also did not stimulate (or inhibit) FANCJ G4 helicase activity (data not shown). Importantly, the results demonstrate that RPA stimulates FANCJ G4 unwinding in a specific manner.
FIG. 3.
RPA stimulates FANCJ unwinding of G4 DNA. (A and B) FANCJ protein (0.3 nM) was incubated with 0.5 nM TP-G4 substrate in the absence (A) or the presence (B) of 1.5 nM RPA heterotrimer under standard helicase reaction conditions at 30°C for the indicated times. M, 32P-labeled single-stranded oligonucleotide marker; +, present; −, absent; ss, ssDNA. (C) Quantitative analysis of FANCJ G4 helicase activity in the absence or presence of RPA or E. coli ssDNA binding protein (ESSB). The data shown are the averages of results from at least three independent experiments, with SD indicated by error bars.
(ii) MSH2/MSH6 inhibits FANCJ G4 helicase activity.
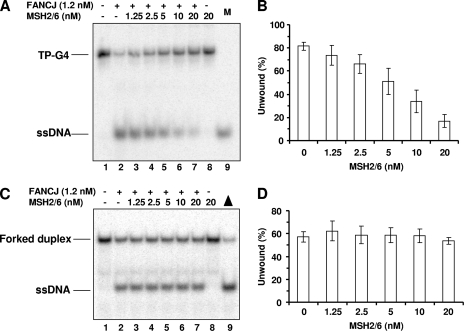
Recently, it was reported that the mismatch repair heterodimer MutSα (MSH2/MSH6) binds with high affinity to G4 DNA formed upon the transcription of the switch regions and that the interaction of MutSα with the switch regions in switching B cells promotes DNA synapsis and recombination (21). We performed experiments to determine if MutSα regulates FANCJ G4 unwinding. MSH2/MSH6 inhibited FANCJ unwinding of the TP-G4 substrate in a protein concentration-dependent manner (Fig. 4A and B). At 5 nM MSH2/MSH6, FANCJ G4 helicase activity was reduced by ∼30%. Higher MSH2/MSH6 concentrations (10 and 20 nM) further reduced G4 unwinding by 60 and 80%, respectively. MSH2/MSH6 inhibition of FANCJ helicase activity was restricted to G4 structures, since no inhibition of FANCJ unwinding of a forked-duplex substrate by MSH2/MSH6 was observed (Fig. 4C and D). MSH2/MSH6 was able to stably bind the G4 substrate under previously described conditions (21), as evidenced by the results of a gel shift analysis (data not shown), suggesting that MSH2/MSH6 interfered with FANCJ G4 helicase activity by preferentially binding the G4 substrate over a forked substrate with a homoduplex region.
FIG. 4.
The MSH2/MSH6 complex inhibits FANCJ G4 helicase activity. (A and C) FANCJ (1.2 nM) was incubated with 0.5 nM TP-G4 (A) or forked duplex (C) in the presence of the indicated MSH2/MSH6 concentrations under standard helicase reaction conditions at 30°C for 15 min. Lanes 9, 32P-labeled single-stranded oligonucleotide marker (M) or heat-denatured substrate control (triangle); +, present; −, absent. (B and D) Quantitative analyses of results presented in panels A and C, respectively. The data shown are the means of results from three independent experiments, with SD indicated by error bars.
Characterization of FANCJ helicase activity on human G4 substrates.
Motifs for the formation of G4 DNA structures are widely dispersed in eukaryotic genomes and can be found in telomeres, gene promoter elements, and recombination hot spots (39). Of particular interest from a biological standpoint are the G4 structures that form from human telomeric repeats (TTAGGG) and triplet repeats such as CGG that are known to be unstable in fragile X syndrome. We therefore tested FANCJ for the unwinding of four-stranded G4 substrates derived from the human telomere and triplet repeat sequences.
FANCJ unwinds human telomeric G4 structures.
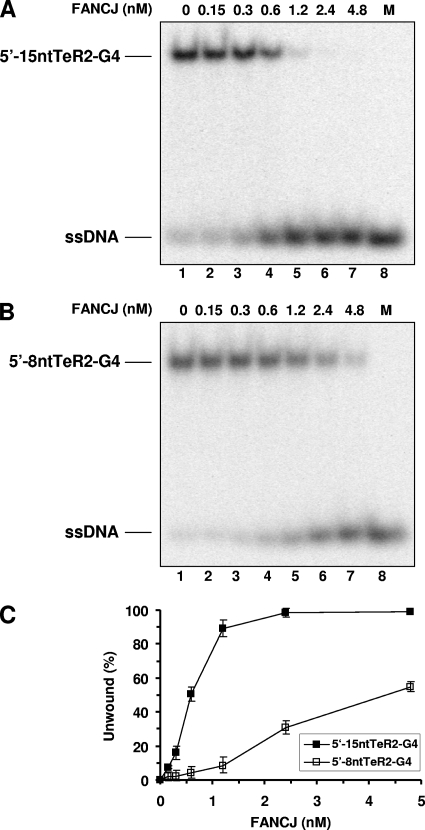
For the human telomeric sequence, we tested FANCJ unwinding of four-stranded parallel G4 substrates with either a 15-nt 5′ ssDNA tail (5′-15ntTeR2-G4) or an 8-nt 5′ ssDNA tail (5′-8ntTeR2-G4). This testing was done to determine if the length of the 5′ ssDNA tail may affect the ability of FANCJ to unwind the telomeric G4 structure, since a previous study demonstrated that FANCJ unwinding of substrates with simple 5′ tails or forked duplexes is significantly increased when the 5′ tail lengths are increased from 5 to 20 nt (14). As shown in Fig. 5, FANCJ unwound the two telomeric G4 substrates in a protein concentration-dependent manner; however, the G4 substrate with a 15-nt 5′ tail was unwound much more efficiently by FANCJ than the substrate with an 8-nt 5′ tail. For example, at 1.2 nM FANCJ, ∼90% of the 5′-15ntTeR2-G4 substrate was unwound, compared with <10% of the 5′-8ntTeR2-G4 substrate. At the highest FANCJ concentration tested (4.8 nM), ∼55% of the 5′-8ntTeR2-G4 substrate was unwound. These results demonstrate that the length of the 5′ ssDNA tail adjacent to the G4 structure dramatically affected the ability of FANCJ to unwind the human telomeric G4 substrate.
FIG. 5.
FANCJ unwinds human telomeric G4 DNA. (A and B) The indicated FANCJ concentrations (lanes 2 to 7) were incubated with 0.5 nM 5′-15ntTeR2-G4 (A) or 5′-8ntTeR2-G4 (B) at 30°C for 15 min under standard helicase assay conditions. Lane 1, no-extract control; lane 8, 32P-labeled single-stranded oligonucleotide marker (M). (C) Quantitative analysis of FANCJ helicase activity on 5′-15ntTeR2-G4 and 5′-8ntTeR2-G4. The data shown are the means of results from three independent experiments, with SD indicated by error bars.
FANCJ unwinds fragile X triplet repeat G4 structures.
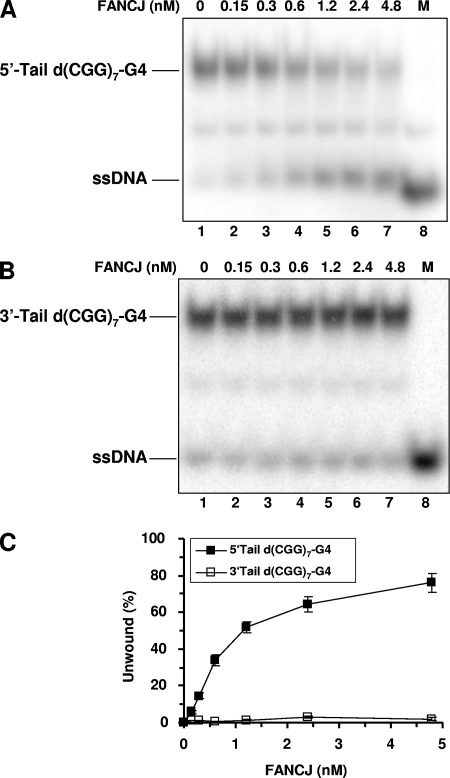
The enhanced ability of FANCJ to unwind the G4 structure with a longer 5′ ssDNA tail is consistent with the demonstrated 5′-to-3′ directionality of FANCJ unwinding of duplex DNA substrates, as defined by the classic M13 DNA directionality substrate (2) or oligonucleotide-based substrates (14). However, since there has been some controversy over the DNA substrate and/or tail polarity requirements for WRN helicase activity on G4 substrates (10, 28), we examined this issue carefully by testing FANCJ helicase activity on four-stranded parallel G4 substrates in which the (CGG)7 triplet repeat sequence was flanked by either an 8-nt 5′ ssDNA tail [5′-tailed d(CGG)7-G4] or an 8-nt 3′ ssDNA tail [3′-tailed d(CGG)7-G4]. The results from these experiments, shown in Fig. 6, demonstrate that FANCJ efficiently unwound the 5′-tailed d(CGG)7-G4 substrate but that little to no activity was observed for the 3′-tailed d(CGG)7-G4 substrate. From these findings, we conclude that G4 unwinding by FANCJ requires a 5′ ssDNA tail adjacent to the G4 structure. Presumably, the 5′ ssDNA tail provides a loading dock for FANCJ to unwind the G4 structure. This substrate requirement distinguishes FANCJ from the WRN and BLM enzymes, which are reported to require a 3′ ssDNA tail to unwind G4 substrates (28, 43).
FIG. 6.
FANCJ requires a 5′ ssDNA tail to unwind a G4 substrate derived from a human triplet repeat sequence. (A and B) The indicated FANCJ concentrations (lanes 2 to 7) were incubated with 0.5 nM 5′-tailed d(CGG)7-G4 (A) or 3′-tailed d(CGG)7-G4 (B) at 30°C for 15 min under standard helicase assay conditions. Lane 1, no-extract control; lane 8, 32P-labeled single-stranded oligonucleotide marker (M). (C) Quantitative analysis of FANCJ helicase activities on 5′-tailed d(CGG)7-G4 and 3′-tailed d(CGG)7-G4. The data shown are the means of results from three independent experiments, with SD indicated by error bars.
The G4-interactive molecule TMS potently inhibits FANCJ G4 helicase activity.
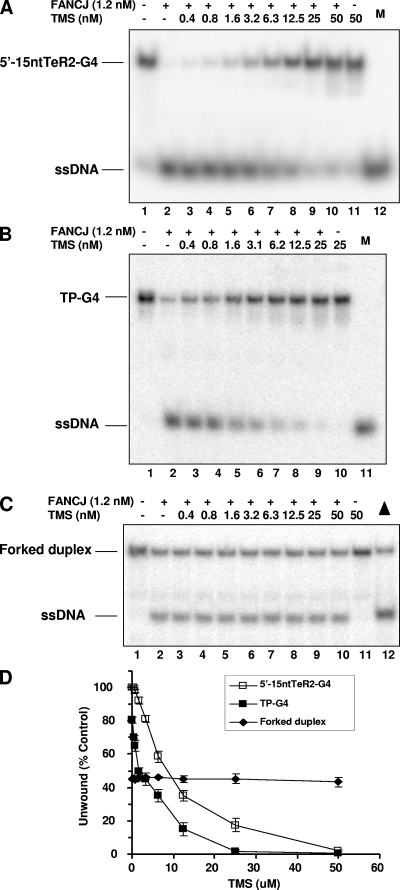
TMS, a natural product isolated from Streptomyces anulatus 3533-SV4, was reported previously to interact selectively with the G4 DNA structure formed by the human telomere sequence (36). Although TMS inhibits telomerase activity (37), its effect on the helicase-catalyzed unwinding of a G4 substrate had not been characterized previously. Therefore, we assayed the effect of TMS on G4 unwinding by FANCJ. TMS inhibited FANCJ unwinding of G4 substrates derived from the human telomeric sequence (Fig. 7A) or an immunoglobulin sequence (Fig. 7B) in a drug concentration-dependent manner. However, TMS inhibition of FANCJ helicase activity was specific to G4 DNA structures, since little or no effect of the drug on FANCJ unwinding of a forked-duplex substrate (Fig. 7C and D) or triplex DNA (data not shown) was observed. TMS inhibited FANCJ helicase activity on all telomeric and nontelomeric four-stranded G4 and two-stranded G2′ tetraplex substrates tested, and the 50% inhibitory concentrations (IC50s) of TMS were all determined to be approximately 6 nM (for TP-G4, 6.2 nM; for 5′-15ntTeR2-G4, 6.5 nM; for 5′-8ntTeR2-G4, 6.1 nM; and for OX-1-G2′, 6.2 nM; the IC50 for the forked duplex was not determined). Thus, TMS potently inhibited FANCJ unwinding activity on a variety of G4 DNA structures but did not affect FANCJ unwinding of duplex DNA, consistent with the compound's ability to selectively bind G4 DNA structures (19, 36).
FIG. 7.
TMS potently inhibits FANCJ G4 helicase. (A to C) FANCJ (1.2 nM) was incubated with 0.5 nM 5′-15ntTeR2-G4 (A), TP-G4 (B), or a forked duplex (C) in the presence of the indicated TMS concentrations under standard helicase reaction conditions. M, 32P-labeled single-stranded oligonucleotide marker; −, absent; +, present; triangle, heat-denatured substrate control. (D) Quantitative analyses of FANCJ helicase activity on 5′-15ntTeR2-G4, TP-G4, or a forked duplex. The data shown are the means of results from three independent experiments, with SD indicated by error bars.
We next sought to determine if the effect of TMS on unwinding activity was specific to FANCJ or if another G4-unwinding helicase may be similarly inhibited. Therefore, we examined the effect of TMS on DNA unwinding catalyzed by BLM helicase. TMS effectively inhibited BLM G4-unwinding in a dose-dependent manner (see Fig. S4A and C in the supplemental material), resulting in an IC50 of 6.0 nM, very similar to that observed for FANCJ helicase on the G4 substrates. TMS failed to inhibit BLM helicase activity on a forked-duplex substrate (see Fig. S4B and C in the supplemental material), indicating that the effect of TMS on BLM helicase was specific to G4 DNA unwinding. From these results, we conclude that the inhibition of FANCJ or BLM G4 helicase activity by TMS is substrate specific.
The ability of TMS to potently inhibit FANCJ G4 helicase raised the question of whether other G4-interactive compounds might similarly affect FANCJ activity. Previously, the porphyrin compound NMM was reported to be a specific inhibitor of G4 DNA unwinding by BLM and the Saccharomyces cerevisiae RecQ helicase Sgs1 (16), and the cationic porphyrin T4 was found to inhibit BLM and WRN helicase activities on both G4 and duplex DNA substrates (24). We therefore tested the abilities of NMM and T4 to inhibit FANCJ unwinding of G4 and duplex DNA substrates. NMM inhibited FANCJ unwinding of the TP-G4 substrate in a dose-dependent manner (see Fig. S5A in the supplemental material); however, significantly higher concentrations of NMM (IC50, ∼25,000 nM) than of TMS (IC50, ∼6 nM) were required (see Table S1 in the supplemental material). Moreover, NMM also inhibited FANCJ unwinding of the forked-duplex substrate at high concentrations (see Fig. S5A and Table S1 in the supplemental material), indicating that the effect was not specific to G4 DNA. T4 inhibited FANCJ unwinding of both the G4 and duplex DNA substrates (see Fig. S5B in the supplemental material), resulting in IC50s of 100 and 160 nM, respectively (see Table S1 in the supplemental material). Thus, T4 or NMM inhibition of FANCJ helicase activity was neither substrate specific nor potent compared to the effect of TMS.
FANCJ depletion sensitizes cells to the antiproliferative effects of TMS.
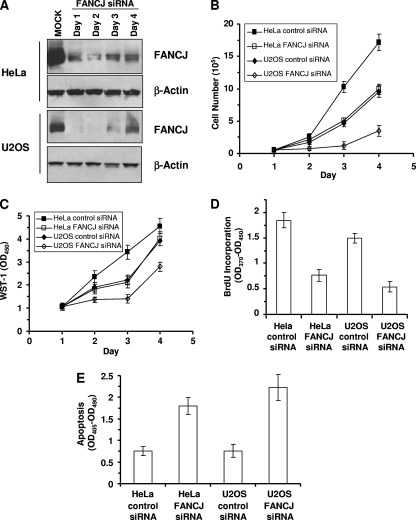
To investigate a potential role of FANCJ in cellular G4 DNA metabolism, we asked if FANCJ-depleted cells were sensitive to the G4-stabilizing compound TMS. Telomerase-positive (HeLa) and telomerase-negative (U2OS) human cell lines were evaluated for TMS sensitivity as a function of FANCJ status. siRNA depletion of FANCJ in HeLa cells resulted in decreases of FANCJ expression by ≥90% (days 1 and 2), 84% (day 3), and 75% (day 4) compared to that in the control cells, as observed by Western blotting (Fig. 8A). In U2OS cells treated with FANCJ siRNA, FANCJ levels were decreased by ≥98% (days 1 and 2), 86% (day 3), and 74% (day 4) (Fig. 8A). To determine the proliferative survival of FANCJ-depleted cells, equivalent numbers of cells either untreated or exposed to 5 μM TMS for 2 h were plated and the cells were counted sequentially for 4 days by using a Coulter counter. TMS-treated HeLa cells that had been transfected with FANCJ siRNA displayed approximately twofold reductions in numbers compared to those of the siRNA control cells on days 3 and 4 (Fig. 8B). TMS-treated U2OS cells depleted of FANCJ also displayed reduced growth, with a fivefold difference compared to siRNA control cells on days 3 and 4 (Fig. 8B). Only a marginal difference in growth between FANCJ-depleted HeLa or U2OS cells and siRNA control cells in the absence of TMS treatment was observed (data not shown). TMS significantly impaired the proliferation of FANCJ-depleted HeLa and U2OS cells (Fig. 8C), whereas there was little to no difference in metabolic activity between untreated FANCJ-depleted and control cells, as observed using the WST-1 assay (data not shown). The measured decrease in the metabolic activity of FANCJ-depleted cells treated with TMS is consistent with the decrease in cell number determined by the Coulter counter. HeLa or U2OS cells depleted of FANCJ were sensitive to the ICL agent mitomycin C (MMC) (see Fig. S6A in the supplemental material). The cotreatment of HeLa cells with 1 μM MMC and 5 μM TMS resulted in an additive inhibition of cell growth (see Fig. S6B in the supplemental material). An additive effect of MMC and TMS on cell growth was also observed for U2OS cells (data not shown).
FIG. 8.
FANCJ depletion sensitizes cells to TMS. (A) Whole-cell extracts from HeLa or U2OS cells transfected with control or FANCJ siRNA were probed by Western blotting with antibodies against FANCJ or actin (as a loading control). (B) The proliferation of control or FANCJ siRNA-treated HeLa or U2OS cells was assessed by Coulter counting of the total populations of cells at the indicated time points. (C) WST-1 assay of the proliferation of control cells and cells transfected with FANCJ siRNA. OD450, optical density at 450 nm. (D) Colorimetric BrdU cell proliferation ELISA of control or FANCJ siRNA-treated HeLa or U2OS cells. (E) Effect of FANCJ depletion on TMS-induced apoptosis. HeLa or U2OS cells were siRNA treated for 72 h, and cells were incubated with 5 μM TMS for 2 h, washed, and allowed to recover in complete medium for 12 h. Enrichment with the cytoplasmic histone-associated DNA fragments is indicative of apoptosis among cells with the indicated siRNA oligonucleotides. The data shown are the means and SD of results from three independent experiments.
The impaired proliferation of FANCJ-depleted cells exposed to TMS raised the question of whether cell lines deficient in other FA genes would be sensitive to TMS. Previously, Youds et al. reported that dog-1 mutants, but not mutants with alterations in CeFANCD2, CeBRCA1, or CeBLM or in a number of activators or regulators of the FA pathway (e.g., ALT-1 and CLK-2) or associated FA components (e.g., BRC-2 and MLH-1), exhibit G tract deletions (47, 48). To address the importance of FA gene products other than FANCJ for TMS resistance in human cells, we examined the TMS sensitivities of FA-A and FA-D2 cell lines. The results from these experiments demonstrated that neither the FA-A nor the FA-D2 cell line was sensitive to TMS (see Fig. S7 in the supplemental material), suggesting that genomic instability induced by TMS-stabilized G4 structures is of the greatest consequence in FA group J patients.
To test the effect of TMS on the mitogenic efficiency of FANCJ-depleted cells, we evaluated BrdU incorporation as a measure of DNA synthesis. The results from these experiments, shown in Fig. 8D, demonstrate that FANCJ-depleted HeLa cells or U2OS cells exposed to TMS were impaired in their ability to synthesize DNA compared to control siRNA-treated cells. Little reduction in DNA synthesis was observed for the FANCJ-depleted HeLa or U2OS cells in the absence of TMS treatment. Collectively, these results demonstrate that FANCJ depletion sensitizes telomerase-positive and telomerase-negative cells to the cytotoxic effects of TMS.
Effect of FANCJ depletion on TMS-induced apoptosis.
Programmed cell death by the apoptotic pathway is an important component of the cellular response to stress or DNA damage. The negative effect of FANCJ depletion on cellular proliferation upon TMS exposure raised the possibility that the FANCJ status may affect apoptotic potential. As shown quantitatively in Fig. 8E, FANCJ-depleted HeLa or U2OS cells exposed to 5 μM TMS displayed a significantly (>2-fold) greater level of apoptosis after TMS exposure than siRNA control cells, as measured by cytoplasmic mono- and oligonucleosomes. No difference in apoptosis between untreated control and FANCJ siRNA-treated cells was observed (data not shown). These results suggest that the cellular depletion of FANCJ renders both telomerase-positive and telomerase-negative cells more prone to apoptosis upon TMS exposure.
The depletion of FANCJ elevates TMS-induced DNA damage.
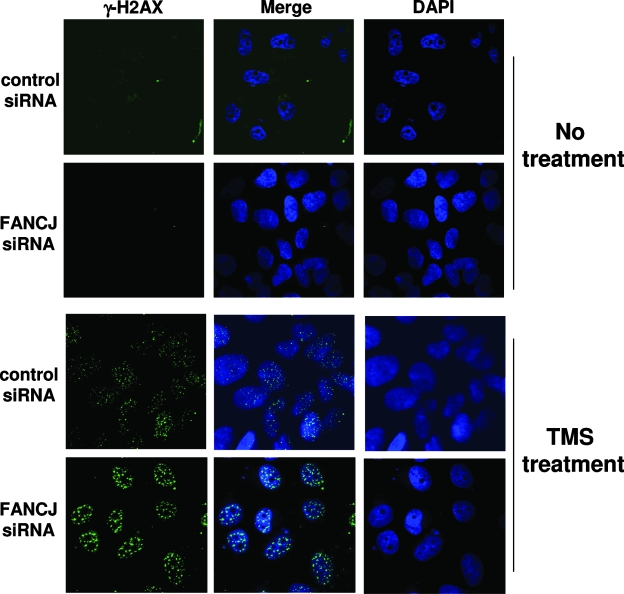
We next asked if FANCJ depletion affected the cellular ability to cope with replicational stress induced by the G4-stabilizing compound TMS. Stalled replication forks can lead to fork collapse and double-strand breaks. A well-known marker for double-strand breaks is γ-H2AX. To assess the importance of FANCJ in preventing or reducing the number of double-strand breaks that arise from TMS treatment, we analyzed γ-H2AX focus formation in control or FANCJ siRNA-transfected cells. In the absence of TMS treatment, FANCJ depletion had no detectable effect on γ-H2AX focus formation (Fig. 9), a result that is consistent with the data in a previous report (31). However, the γ-H2AX foci were significantly more intense in TMS-treated FANCJ-depleted cells than in control siRNA cells (Fig. 9), a result that was confirmed using MCID Core software (InterFocus Imaging Ltd., Cambridge, United Kingdom) to analyze and compare γ-H2AX foci in the two cell lines exposed to TMS. The increased γ-H2AX staining in FANCJ-depleted U2OS cells compared to that in undepleted cells, also observed for HeLa cells (see Fig. S8 in the supplemental material), suggests that FANCJ alleviates the replicative stress induced by TMS, thereby decreasing the extent of double-strand breaks.
FIG. 9.
TMS induces elevated numbers of γ-H2AX foci in FANCJ-depleted cells. FANCJ siRNA-treated or control siRNA-treated U2OS cells were either exposed to 5 μM TMS for 2 h or left unexposed and were washed and allowed to recover in complete medium for 2 h, fixed with formaldehyde, and stained with anti-γ-H2AX antibody or DAPI. The merged picture shows cells stained with anti-γ-H2AX (green) and DAPI (blue).
DISCUSSION
Failure to replicate DNA in a timely and faithful manner can result in mutations, genomic instability, and cellular dysfunction manifested by cell transformation, senescence, or death. To deal with replication-blocking lesions, proteins associated with the DNA replication fork or involved in distinct DNA damage response pathways facilitate fork progression or the accurate repair and restarting of damaged or broken replication forks. Alternate DNA structures such as G4 structures have been implicated in critical biological processes and can interfere with normal cellular DNA transactions such as the advancement of the replication fork during S phase. In this study, we have examined the role of FANCJ in the replicational stress response by evaluating its ability to unwind G4 alternate DNA structures that potentially cause genomic instability. The results of our investigations demonstrate that FANCJ efficiently unwinds a variety of G4 DNA substrates, providing the first biochemical evidence that a helicase outside the classic RecQ family utilizes its ATP hydrolysis function to operate in this capacity. This finding bears significance since it was proposed previously that the peculiar type of genomic instability in C. elegans mutants with alterations in the product of dog-1, a FANCJ homologue that also functions in ICL repair (47), is due to the accumulation of secondary DNA structures in G-rich tracts (4). Although the FA pathway has been classically described in terms of ICL repair, our results suggest that the cellular defects associated with an inactivating mutation in the FANCJ gene extend beyond the reduced ability to repair ICLs and involve other types of DNA structural roadblocks to replication. Recent work shows that other FA proteins function to prevent DNA breaks during normal replication (41).
To probe the functional importance of FANCJ in G4 metabolism in vivo, we examined the effects of TMS, a selective quadruplex binding agent, on cell proliferation and γ-H2AX focus formation as a marker of DNA damage in FANCJ-depleted versus control cells. Although TMS was originally identified as a potent telomerase inhibitor by virtue of its ability to stabilize telomeric quadruplex DNA and inhibit the polymerase reaction (37), results from a number of recent studies suggest that TMS can interfere with telomere capping by additional mechanisms and may also exert cytotoxic effects through its action on nontelomere G4 targets (6, 11, 12, 44). Based on the ability of FANCJ to unwind G4 DNA, we hypothesized that FANCJ-depleted cells would be hypersensitive to the biological effects of TMS. Indeed, this turned out to be the case. TMS was observed to significantly impair cell proliferation and growth and induce an elevated level of apoptosis in FANCJ-depleted cells. The antiproliferative and apoptotic effects of TMS were observed in both telomerase-positive and telomerase-negative cells depleted of FANCJ, suggesting that FANCJ plays an important role in G4 metabolism that is not dependent on the telomerase status. Results from cellular immunofluorescence assays demonstrated that TMS induced significantly more intense γ-H2AX foci in the FANCJ-depleted cells than in the siRNA control cells. Five percent of the γ-H2AX foci colocalized with TRF2, suggesting that the double-strand breaks that occurred after TMS treatment were present in extratelomeric regions and, to a lesser extent, in the telomeres (data not shown). The lack of γ-H2AX foci in untreated FANCJ siRNA-transfected cells suggests that transient G4 structures not stabilized by TMS do not impose strong blocks to replication that would result in double-strand breaks. Thus, the G4-stabilizing effect of TMS adversely affected proliferation and DNA damage accumulation among FANCJ-depleted cells to a greater extent than among the control cells. TMS-stabilized G4 structures that presumably accumulate in FANCJ-depleted cells may lead to stalled replication forks that are repaired by homologous recombination, as evidenced for C. elegans dog-1 mutants (48).
Collectively, the molecular and cellular evidence suggests that G4 DNA represents a physiological substrate acted upon by FANCJ to facilitate replication and/or DNA repair. Consistent with a role of FANCJ in the metabolism of G4 structures stabilized by TMS, endogenous FANCJ is phosphorylated in response to cellular exposure to TMS (data not shown). FANCJ phosphorylation was also observed when cells were subjected to UV (31), suggesting a mechanism for the modulation of FANCJ function during the DNA damage response. The phosphorylation state of FANCJ is also important for functions other than the response to exogenous DNA damage, since FANCJ is silenced during the G1 phase of the cell cycle and is activated through a dephosphorylation event as cells enter S phase (20). In the future, it will be important to assess the importance of FANCJ phosphorylation and other posttranslational modifications in modulating the molecular functions of FANCJ during replication or DNA repair. In addition, the cross talk of FANCJ and other DNA repair factors such as RPA and MSH2/MSH6 plays an important role in regulating FANCJ helicase function. The strong colocalization of FANCJ and RPA in response to nucleotide depletion by hydroxyurea (15) suggests that the two proteins collaborate in situations of replicational stress, a condition that arises when the replication fork is impeded by a G4 structure. The observation that the stimulation of FANCJ G4 helicase activity by RPA is specific suggests that the physical interaction between FANCJ and RPA (15) plays a role in the functional interaction. The critical importance of helicase-RPA physical interaction for the stimulation of DNA unwinding was demonstrated previously for the WRN and BLM helicases on duplex DNA substrates (8).
Several reports have provided evidence that small molecules which stabilize G4 structures cause replicative senescence after long-term exposure to tumor cell cultures (18, 33). Among them, the natural product TMS is one of the most potent and selective telomeric G4 ligands. TMS impairs the conformation and length of the telomeric G overhang, an effect that is thought to be more relevant than double-stranded telomere erosion as a marker for TMS cellular activity (44). TMS treatment of human cancer cell lines results in the dissociation of TRF2 from telomeres, impairs cell proliferation, and results in telomere dysfunction, leading to anaphase bridge formation and apoptosis (44). TMS also inhibits POT1 binding to the telomeric G overhang in vitro and induces POT1 dissociation from telomeres in telomerase-positive cells (11). The pleiotropic effects of TMS on telomere dynamics are proposed to revolve around its stabilizing effect on the G4 structure at the telomeric end. Although there is no evidence for a direct role of FANCJ in telomere biology, it is plausible that FANCJ G4 helicase activity is important in telomere maintenance. The potent and specific inhibition of the unwinding of G4 DNA by FANCJ or a related helicase (e.g., BLM or WRN) by TMS may be an underlying cause of the cytotoxic effects of TMS. In this regard, it will be of interest to determine the molecular basis of genomic instability in organisms with deficiencies in genes (rtel, chl1, and Ddx11) that encode proteins with sequence similarity in the helicase core to FANCJ. Rtel (Regulator of telomere length) knockout mice die between days 10 and 11.5, and embryonic stem cells from these knockout mice show telomere loss and display many chromosome breaks and fusions upon differentiation (7), suggesting a role of RTEL in the maintenance of telomere length or capping. The sister chromatid cohesion defects in Saccharomyces cerevisiae chl1 mutants (27, 40), Ddx11 mutant mice (17), and ChlR1-depleted human cells (30), as well as the telomere shortening in human cells containing a mutation in the DDX11 gene encoding ChlR1 (45), may be a consequence of aberrant mitotic chromosome structures due to genomic sequences that form alternate DNA arrangements such as G4 structures. Our results suggest that FANCJ preserves genomic stability by directly unwinding DNA roadblocks such as G4 structures that destabilize or impede the replication fork.
Supplementary Material
Acknowledgments
This work was supported by the Intramural Research program of the National Institute on Aging, NIH, and the Fanconi Anemia Research Fund (R.M.B.).
We acknowledge S. Cantor (University of Massachusetts Medical Center, Worcester, MA) for the FANCJ baculovirus and L. Wu and I. Hickson (Cancer Research UK Laboratories) for generously providing recombinant BLM. We thank M. Kenny (Albert Einstein Cancer Center, Bronx, NY) for the human RPA and T. Wilson (University of Maryland, Baltimore, MD) for the purified MSH2/MSH6 complex. We thank Ming Lei (University of Michigan, Ann Arbor, MI) for kindly providing the recombinant POT1. We are grateful to N. Maizels's group (University of Washington, Seattle, WA) for technical advice on G4 DNA substrate preparation. We acknowledge the Fanconi Anemia Research Fund for FA-A and FA-D2 cell lines.
Footnotes
Published ahead of print on 21 April 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Bridge, W. L., C. J. Vandenberg, R. J. Franklin, and K. Hiom. 2005. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat. Genet. 37953-957. [DOI] [PubMed] [Google Scholar]
- 2.Cantor, S., R. Drapkin, F. Zhang, Y. Lin, J. Han, S. Pamidi, and D. M. Livingston. 2004. The BRCA1-associated protein BACH1 is a DNA helicase targeted by clinically relevant inactivating mutations. Proc. Natl. Acad. Sci. USA 1012357-2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cantor, S. B., D. W. Bell, S. Ganesan, E. M. Kass, R. Drapkin, S. Grossman, D. C. Wahrer, D. C. Sgroi, W. S. Lane, D. A. Haber, and D. M. Livingston. 2001. BACH1, a novel helicase-like protein, interacts directly with BRCA1 and contributes to its DNA repair function. Cell 105149-160. [DOI] [PubMed] [Google Scholar]
- 4.Cheung, I., M. Schertzer, A. Rose, and P. M. Lansdorp. 2002. Disruption of dog-1 in Caenorhabditis elegans triggers deletions upstream of guanine-rich DNA. Nat. Genet. 31405-409. [DOI] [PubMed] [Google Scholar]
- 5.Choudhary, S., J. A. Sommers, and R. M. Brosh, Jr. 2004. Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of Werner syndrome protein. J. Biol. Chem. 27934603-34613. [DOI] [PubMed] [Google Scholar]
- 6.De Cian, A., G. Cristofari, P. Reichenbach, E. De Lemos, D. Monchaud, M. P. Teulade-Fichou, K. Shin-ya, L. Lacroix, J. Lingner, and J. L. Mergny. 2007. Reevaluation of telomerase inhibition by quadruplex ligands and their mechanisms of action. Proc. Natl. Acad. Sci. USA 10417347-17352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding, H., M. Schertzer, X. Wu, M. Gertsenstein, S. Selig, M. Kammori, R. Pourvali, S. Poon, I. Vulto, E. Chavez, P. P. Tam, A. Nagy, and P. M. Lansdorp. 2004. Regulation of murine telomere length by Rtel: an essential gene encoding a helicase-like protein. Cell 117873-886. [DOI] [PubMed] [Google Scholar]
- 8.Doherty, K. M., J. A. Sommers, M. D. Gray, J. W. Lee, C. von Kobbe, N. H. Thoma, R. P. Kureekattil, M. K. Kenny, and R. M. Brosh, Jr. 2005. Physical and functional mapping of the replication protein A interaction domain of the Werner and Bloom syndrome helicases. J. Biol. Chem. 28029494-29505. [DOI] [PubMed] [Google Scholar]
- 9.Eddy, J., and N. Maizels. 2006. Gene function correlates with potential for G4 DNA formation in the human genome. Nucleic Acids Res. 343887-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fry, M., and L. A. Loeb. 1999. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 27412797-12802. [DOI] [PubMed] [Google Scholar]
- 11.Gomez, D., M. F. O'Donohue, T. Wenner, C. Douarre, J. Macadre, P. Koebel, M. J. Giraud-Panis, H. Kaplan, A. Kolkes, K. Shin-ya, and J. F. Riou. 2006. The G-quadruplex ligand telomestatin inhibits POT1 binding to telomeric sequences in vitro and induces GFP-POT1 dissociation from telomeres in human cells. Cancer Res. 666908-6912. [DOI] [PubMed] [Google Scholar]
- 12.Gomez, D., T. Wenner, B. Brassart, C. Douarre, M. F. O'Donohue, K. El, V. K. Shin-ya, H. Morjani, C. Trentesaux, and J. F. Riou. 2006. Telomestatin-induced telomere uncapping is modulated by POT1 through G-overhang extension in HT1080 human tumor cells. J. Biol. Chem. 28138721-38729. [DOI] [PubMed] [Google Scholar]
- 13.Gupta, R., S. Sharma, K. M. Doherty, J. A. Sommers, S. B. Cantor, and R. M. Brosh, Jr. 2006. Inhibition of BACH1 (FANCJ) helicase by backbone discontinuity is overcome by increased motor ATPase or length of loading strand. Nucleic Acids Res. 346673-6683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta, R., S. Sharma, J. A. Sommers, Z. Jin, S. B. Cantor, and R. M. Brosh, Jr. 2005. Analysis of the DNA substrate specificity of the human BACH1 helicase associated with breast cancer. J. Biol. Chem. 28025450-25460. [DOI] [PubMed] [Google Scholar]
- 15.Gupta, R., S. Sharma, J. A. Sommers, M. K. Kenny, S. B. Cantor, and R. M. Brosh, Jr. 2007. FANCJ (BACH1) helicase forms DNA damage inducible foci with replication protein A and interacts physically and functionally with the single-stranded DNA binding protein. Blood 1102390-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huber, M. D., D. C. Lee, and N. Maizels. 2002. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 303954-3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Inoue, A., T. Li, S. K. Roby, M. B. Valentine, M. Inoue, K. Boyd, V. J. Kidd, and J. M. Lahti. 2007. Loss of ChlR1 helicase in mouse causes lethality due to the accumulation of aneuploid cells generated by cohesion defects and placental malformation. Cell Cycle 61646-1654. [DOI] [PubMed] [Google Scholar]
- 18.Izbicka, E., R. T. Wheelhouse, E. Raymond, K. K. Davidson, R. A. Lawrence, D. Sun, B. E. Windle, L. H. Hurley, and D. D. Von Hoff. 1999. Effects of cationic porphyrins as G-quadruplex interactive agents in human tumor cells. Cancer Res. 59639-644. [PubMed] [Google Scholar]
- 19.Kim, M. Y., H. Vankayalapati, K. Shin-ya, K. Wierzba, and L. H. Hurley. 2002. Telomestatin, a potent telomerase inhibitor that interacts quite specifically with the human telomeric intramolecular G-quadruplex. J. Am. Chem. Soc. 1242098-2099. [DOI] [PubMed] [Google Scholar]
- 20.Kumaraswamy, E., and R. Shiekhattar. 2007. Activation of BRCA1/BRCA2-associated helicase BACH1 is required for timely progression through S phase. Mol. Cell. Biol. 276733-6741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Larson, E. D., M. L. Duquette, W. J. Cummings, R. J. Streiff, and N. Maizels. 2005. MutSα binds to and promotes synapsis of transcriptionally activated immunoglobulin switch regions. Curr. Biol. 15470-474. [DOI] [PubMed] [Google Scholar]
- 22.Levitus, M., Q. Waisfisz, B. C. Godthelp, Y. de Vries, S. Hussain, W. W. Wiegant, E. Elghalbzouri-Maghrani, J. Steltenpool, M. A. Rooimans, G. Pals, F. Arwert, C. G. Mathew, M. Z. Zdzienicka, K. Hiom, J. P. de Winter, and H. Joenje. 2005. The DNA helicase BRIP1 is defective in Fanconi anemia complementation group J. Nat. Genet. 37934-935. [DOI] [PubMed] [Google Scholar]
- 23.Levran, O., C. Attwooll, R. T. Henry, K. L. Milton, K. Neveling, P. Rio, S. D. Batish, R. Kalb, E. Velleuer, S. Barral, J. Ott, J. Petrini, D. Schindler, H. Hanenberg, and A. D. Auerbach. 2005. The BRCA1-interacting helicase BRIP1 is deficient in Fanconi anemia. Nat. Genet. 37931-933. [DOI] [PubMed] [Google Scholar]
- 24.Li, J. L., R. J. Harrison, A. P. Reszka, R. M. Brosh, Jr., V. A. Bohr, S. Neidle, and I. D. Hickson. 2001. Inhibition of the Bloom's and Werner's syndrome helicases by G-quadruplex interacting ligands. Biochemistry 4015194-15202. [DOI] [PubMed] [Google Scholar]
- 25.Litman, R., M. Peng, Z. Jin, F. Zhang, J. Zhang, S. Powell, P. R. Andreassen, and S. B. Cantor. 2005. BACH1 is critical for homologous recombination and appears to be the Fanconi anemia gene product FANCJ. Cancer Cell 8255-265. [DOI] [PubMed] [Google Scholar]
- 26.Maizels, N. 2006. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 131055-1059. [DOI] [PubMed] [Google Scholar]
- 27.Mayer, M. L., I. Pot, M. Chang, H. Xu, V. Aneliunas, T. Kwok, R. Newitt, R. Aebersold, C. Boone, G. W. Brown, and P. Hieter. 2004. Identification of protein complexes required for efficient sister chromatid cohesion. Mol. Biol. Cell 151736-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mohaghegh, P., J. K. Karow, J. R. Brosh, Jr., V. A. Bohr, and I. D. Hickson. 2001. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 292843-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oganesian, L., I. K. Moon, T. M. Bryan, and M. B. Jarstfer. 2006. Extension of G-quadruplex DNA by ciliate telomerase. EMBO J. 251148-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parish, J. L., J. Rosa, X. Wang, J. M. Lahti, S. J. Doxsey, and E. J. Androphy. 2006. The DNA helicase ChlR1 is required for sister chromatid cohesion in mammalian cells. J. Cell Sci. 1194857-4865. [DOI] [PubMed] [Google Scholar]
- 31.Peng, M., R. Litman, Z. Jin, G. Fong, and S. B. Cantor. 2006. BACH1 is a DNA repair protein supporting BRCA1 damage response. Oncogene 252245-2253. [DOI] [PubMed] [Google Scholar]
- 32.Peng, M., R. Litman, J. Xie, S. Sharma, R. M. Brosh, Jr., and S. B. Cantor. 2007. The FANCJ/MutLα interaction is required for correction of the cross-link response in FA-J. cells. EMBO J. 263238-3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Riou, J. F., L. Guittat, P. Mailliet, A. Laoui, E. Renou, O. Petitgenet, F. Megnin-Chanet, C. Helene, and J. L. Mergny. 2002. Cell senescence and telomere shortening induced by a new series of specific G-quadruplex DNA ligands. Proc. Natl. Acad. Sci. USA 992672-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Seal, S., D. Thompson, A. Renwick, A. Elliott, P. Kelly, R. Barfoot, T. Chagtai, H. Jayatilake, M. Ahmed, K. Spanova, B. North, L. McGuffog, D. G. Evans, D. Eccles, D. F. Easton, M. R. Stratton, and N. Rahman. 2006. Truncating mutations in the Fanconi anemia J gene BRIP1 are low-penetrance breast cancer susceptibility alleles. Nat. Genet. 381239-1241. [DOI] [PubMed] [Google Scholar]
- 35.Sharma, S., J. A. Sommers, S. Choudhary, J. K. Faulkner, S. Cui, L. Andreoli, L. Muzzolini, A. Vindigni, and R. M. Brosh, Jr. 2005. Biochemical analysis of the DNA unwinding and strand annealing activities catalyzed by human RECQ1. J. Biol. Chem. 28028072-28084. [DOI] [PubMed] [Google Scholar]
- 36.Shin-ya, K. 2004. Telomerase inhibitor, telomestatin, a specific mechanism to interact with telomere structure. Nippon Rinsho 621277-1282. (In Japanese.) [PubMed] [Google Scholar]
- 37.Shin-ya, K., K. Wierzba, K. Matsuo, T. Ohtani, Y. Yamada, K. Furihata, Y. Hayakawa, and H. Seto. 2001. Telomestatin, a novel telomerase inhibitor from Streptomyces anulatus. J. Am. Chem. Soc. 1231262-1263. [DOI] [PubMed] [Google Scholar]
- 38.Siddiqui-Jain, A., C. L. Grand, D. J. Bearss, and L. H. Hurley. 2002. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 9911593-11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simonsson, T. 2001. G-quadruplex DNA structures: variations on a theme. Biol. Chem. 382621-628. [DOI] [PubMed] [Google Scholar]
- 40.Skibbens, R. V. 2004. Chl1p, a DNA helicase-like protein in budding yeast, functions in sister-chromatid cohesion. Genetics 16633-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sobeck, A., S. Stone, V. Costanzo, B. de Graaf, T. Reuter, J. de Winter, M. Wallisch, Y. Akkari, S. Olson, W. Wang, H. Joenje, J. L. Christian, P. J. Lupardus, K. A. Cimprich, J. Gautier, and M. E. Hoatlin. 2006. Fanconi anemia proteins are required to prevent accumulation of replication-associated DNA double-strand breaks. Mol. Cell. Biol. 26425-437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sun, H., R. J. Bennett, and N. Maizels. 1999. The Saccharomyces cerevisiae Sgs1 helicase efficiently unwinds G-G paired DNAs. Nucleic Acids Res. 271978-1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sun, H., J. K. Karow, I. D. Hickson, and N. Maizels. 1998. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 27327587-27592. [DOI] [PubMed] [Google Scholar]
- 44.Tahara, H., K. Shin-ya, H. Seimiya, H. Yamada, T. Tsuruo, and T. Ide. 2006. G-Quadruplex stabilization by telomestatin induces TRF2 protein dissociation from telomeres and anaphase bridge formation accompanied by loss of the 3′ telomeric overhang in cancer cells. Oncogene 251955-1966. [DOI] [PubMed] [Google Scholar]
- 45.Vasa-Nicotera, M., S. Brouilette, M. Mangino, J. R. Thompson, P. Braund, J. R. Clemitson, A. Mason, C. L. Bodycote, S. M. Raleigh, E. Louis, and N. J. Samani. 2005. Mapping of a major locus that determines telomere length in humans. Am. J. Hum. Genet. 76147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vaughn, J. P., S. D. Creacy, E. D. Routh, C. Joyner-Butt, G. S. Jenkins, S. Pauli, Y. Nagamine, and S. A. Akman. 2005. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 28038117-38120. [DOI] [PubMed] [Google Scholar]
- 47.Youds, J. L., L. J. Barber, J. D. Ward, S. J. Collis, N. J. O'Neil, S. J. Boulton, and A. M. Rose. 2008. DOG-1 is the Caenorhabditis elegans BRIP1/FANCJ homologue and functions in interstrand cross-link repair. Mol. Cell. Biol. 281470-1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Youds, J. L., N. J. O'Neil, and A. M. Rose. 2006. Homologous recombination is required for genome stability in the absence of DOG-1 in Caenorhabditis elegans. Genetics 173697-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zahler, A. M., J. R. Williamson, T. R. Cech, and D. M. Prescott. 1991. Inhibition of telomerase by G-quartet DNA structures. Nature 350718-720. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.