Abstract
TLX is an orphan nuclear receptor (also called NR2E1) that regulates the expression of target genes by functioning as a constitutive transrepressor. The physiological significance of TLX in the cytodifferentiation of neural cells in the brain is known. However, the corepressors supporting the transrepressive function of TLX have yet to be identified. In this report, Y79 retinoblastoma cells were subjected to biochemical techniques to purify proteins that interact with TLX, and we identified LSD1 (also called KDM1), which appears to form a complex with CoREST and histone deacetylase 1. LSD1 interacted with TLX directly through its SWIRM and amine oxidase domains. LSD1 potentiated the transrepressive function of TLX through its histone demethylase activity as determined by a luciferase assay using a genomically integrated reporter gene. LSD1 and TLX were recruited to a TLX-binding site in the PTEN gene promoter, accompanied by the demethylation of H3K4me2 and deacetylation of H3. Knockdown of either TLX or LSD1 derepressed expression of the endogenous PTEN gene and inhibited cell proliferation of Y79 cells. Thus, the present study suggests that LSD1 is a prime corepressor for TLX.
Nuclear receptors (NRs) are transcriptional regulators that play pivotal roles in a variety of key metabolic and developmental processes (26). NRs constitute a gene superfamily, and NR protein structure is divided into several functional domains. One of the well-characterized domains is a highly conserved DNA-binding domain (DBD), and the other is a moderately conserved ligand-binding domain (LBD). The NR gene superfamily includes both steroid/thyroid hormone receptors and vitamin A/D receptors. The transcriptional function of NRs is regulated by the binding of specific ligands. In addition to the ligand-dependent NRs, there is a subfamily of so-called orphan NRs, the ligands of which have not yet been characterized (10). In the absence of ligand, orphan NRs constitutively activate or suppress transcription (20, 49, 53).
Ligand-dependent transcriptional control by NRs requires a number of positive or negative coregulatory multiprotein complexes, in addition to basic transcription factors (25, 35, 36, 52). These coregulator complexes can be classified into two groups. The first group comprises ATP-dependent chromatin-remodeling complexes (such as the SWI/SNF complex), which reorganize nucleosomal arrays and potentiate the promoter accessibility of NRs (6, 18, 23, 33). The other group consists of histone modifier complexes, which covalently modify histone tails by acetylation, methylation, ubiquitination, or phosphorylation. These modifications lead to transcriptional repression or activation within the chromatin (2, 17, 42). Of these modifications, methylation of histone lysines is generally regarded as the most significant histone modification, as it triggers alterations in chromatin structure (27, 37). Methylation of H3K9 leads to chromatin silencing, while H3K4 methylation enhances chromatin activity. A number of histone methyltransferases, e.g., SETDB1 (also called KMT1E), and demethylases, e.g., LSD1, have been reported to coregulate ligand-dependent transcriptional controls by NRs (9, 28, 44, 50, 51). In contrast, little is known about coregulators and coregulator complexes supporting the constitutive functions of orphan NRs (8, 13).
TLX belongs to the NR2E orphan NR subfamily and is believed to act as a constitutive transcriptional repressor (55). In the mouse, expression of TLX appears at embryonic day 8.5 and is expressed in the neuroepithelium of the developing central nervous system, including the retina and optic stalk (30, 55). TLX-null mice suffer reduced brain size, thin cerebral cortices, and visual impairment (29, 54). TLX also plays an essential role in maintaining the undifferentiated state of adult neural stem cells in the mouse forebrain (38, 40). Despite the important physiological roles played by TLX in vivo, the molecular mechanisms underlying the transrepressive function of TLX remain elusive (43, 56).
To address this issue, we biochemically purified a TLX corepressor complex from Y79 retinoblastoma cells and identified an LSD1 complex. LSD1 interacted with TLX and possessed corepressor activity for TLX in a chromatin context. Moreover, LSD1 was recruited with TLX to the TLX-targeted promoter of the PTEN gene. Data also showed that LSD1 was required for transrepression of the PTEN gene by TLX. Thus, we conclude that a constitutive-repressor-type NR requires a histone-inactivating demethylase as a corepressor.
MATERIALS AND METHODS
Plasmids and antibodies.
The expression plasmid for full-length TLX cDNA was cloned from a cDNA library of HeLa cells and inserted into a pcDNA3 vector (Invitrogen, Carlsbad, CA). The expression plasmid for the TLX LBD or DBD was cloned into pGEX4T-1 (GE Healthcare, Buckinghamshire, England) or pM vectors (Clontech, Mountain View, CA). The expression plasmids for LSD1 and LSD1ΔAO were described previously (28). The expression plasmid for LSD1 K661A was generated from a pCMX-FLAG LSD1 vector. For in vitro translation of LSD1 mutants, each cDNA was cloned into a pcDNA3 vector. For short hairpin RNA (shRNA) expression plasmids, oligo(DNA) was inserted into a pSUPER.retro.puro vector (Oligoengine, Seattle, WA). The following target sequences were used. For human TLX shRNA, 5′-GTCAACATGAACAAAGACG-3′; for human LSD1 shRNA, 5′-CGGACAAGCTGTTCCTAAA-3′ (28); for Escherichia coli LacZ shRNA, 5′-GCCCATCTACACCAACGTAAC-3′. SMARTpool small interfering RNAs (siRNAs) for each factor were purchased from Dharmacon (Lafayette, CO). Anti-human TLX and human PNR antibodies were purchased from PPMX (Tokyo, Japan). Anti-human LSD1, human CoREST, acetyl histone H3, H3K4me2, H3K4me3, and H3K9me2 antibodies were purchased from Upstate Biotechnology (Lake Placid, NY). Anti-human histone deacetylase 1 (HDAC1) antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-atrophin 1 antibody was purchased from Abcam (Cambridge, United Kingdom). Anti-Ret-CoR antibody was described previously (45).
Cell culture and transfection.
Y79 cells were obtained from the ATCC (no. HTB-18) and cultured in RPMI 1640 medium (Wako, Osaka, Japan) supplemented with 10% fetal bovine serum and antibiotics. The cells were grown at 37°C in 5% CO2. For mass cultures, Y79 cells were cultured in spinner flasks under the same conditions and nuclear extracts were prepared by a modification of the method of Dignam et al. (4). For transfection, we used Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
We maintained 293F cells in Dulbecco's modified Eagle's medium (Nissui Pharmaceutical, Tokyo, Japan) supplemented with 10% fetal bovine serum and antibiotics. For establishment of 293F-pGL4.31 cells, 293F cells were grown in 10-cm dishes and transfected with 10 μg of pGL4.31 vector (Promega, Madison, WI) by standard calcium-phosphate methods. After 48 h, cells were selected with 200 μg/ml hygromycin (Invitrogen) and cloned by cloning ring. From 12 hygromycin-resistant clones, we selected appropriate clones by luciferase reporter activity using a luciferase assay kit (Promega) (15).
Immunoprecipitation and glutathione S-transferase (GST) pull-down assays.
Y79 cell nuclear extracts (10 mg each) were immunoprecipitated with each indicated antibody for 1 h and then transferred to protein G Sepharose beads (GE Healthcare) at 4°C. The immunoprecipitates were washed and boiled with Laemmli sample buffer and then subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting with the indicated antibodies.
For GST pull-down assays, LSD1 and its mutants were translated in vitro using TNT Quick Coupled Transcription/Translation Systems (Promega) and incubated with GST-fused TLX mutants immobilized on glutathione-Sepharose beads for 1 h at room temperature. The beads were washed and boiled with Laemmli sample buffer, subjected to SDS-PAGE, and assessed by autoradiography.
Chromatin immunoprecipitation.
Soluble chromatin from Y79 cells was immunoprecipitated using an acetyl-histone H4 immunoprecipitation assay kit (Upstate Biotechnology) with antibodies against the indicated proteins. Specific primer pairs were designed to amplify the promoter region of the human PTEN proximal region (5′-AATCAGCTCTCTCACGGTGACAGG-3′ and 5′-CTCTACATCGACCTATTCTGCGCC-3′) and distal region (5′-GTGGTCCAGCCCAAGCTTCTTTAC-3′ and 5′-GGAGTGCAGTAGCACAATCTCAGC-3′) and the β-actin promoter (5′-GTGTGGTCCTGCGACTTCTAAGTG-3′ and 5′-TCCTAGGTGTGGACATCTCTTGGG-3′). The PCR products were visualized on 1.5% agarose/Tris-acetate-EDTA gels.
RNA isolation, cDNA synthesis, and quantitative reverse transcription-PCR (qPCR).
Total RNA was extracted with Trizol (Invitrogen), and cDNA was synthesized using SuperscriptIII reverse transcriptase (Invitrogen). Reverse transcription of 2 μg total RNA was carried out with 0.2 μg oligo(dT) 15 primer for 50 min at 50°C, and the PCR was performed using a GeneAmp PCR system 9700 (Applied Biosystems, Foster City, CA). The primer sequences for the human genes were as follows: for human TLX, forward, 5′-GTGCTTACAAGCAGGCATGAACCA-3′, and reverse, 5′-AGCTCATTCCATGCCTCTTCCAGC-3′; for human NCoR, forward, 5′-TGAAACACCTAGCGATGCTATTGA-3′, and reverse, 5′-GGTAGGATCATTTTCCGCTTGA-3′; for human SMRT, forward, 5′-GGTCAAGTCCAAGAAGCAAGAGAT-3′, and reverse, 5′-GCTTCTATAGGTCATAAGGCCTGTTC-3′; for human atrophin 1, forward, 5′-TACCCAGCTGGAACTCTCCCTAAC-3′, and reverse, 5′-TGACTGTAGTAGTCCTCCTGGGCA-3′. RNA levels were normalized using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an internal standard.
qPCR was performed using Sybr Premix Ex Taq (Takara, Tokyo, Japan) with the ABI Prism 7900HT (Applied Biosystems) according to the manufacturer's instructions. qPCR primer sets for PTEN and GAPDH were purchased from Takara. Experimental samples were matched to a standard curve generated by amplifying serially diluted products using the same PCR protocol. To correct for variability in RNA recovery and efficiency of reverse transcription, GAPDH cDNA was amplified and quantified in each cDNA preparation.
Purification and separation of the LSD1-CoREST complex.
Y79 nuclear extracts (150 mg) were applied to GST- or GST-TLX LBD columns and washed extensively with BC100 buffer (20 mM HEPES [pH 7.6], 100 mM KCl, 0.2 mM EDTA, 10% glycerol, 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM dithiothreitol). Bound proteins were eluted from the column by incubation with 15 mM glutathione in BC100 for 30 min at room temperature. For fractionation on glycerol density gradients, eluted proteins were layered on top of 13-ml linear 10% to 40% glycerol gradients in BC100 and centrifuged for 16 h at 4°C at 40,000 rpm in an SW40 rotor (Beckman Coulter, Fullerton, CA) (7, 34). Protein standards included β-globulin (158 kDa) and thyroglobulin (667 kDa). Samples were applied to 2% to 15% gradient gels (Daiichi Pure Chemicals, Tokyo, Japan).
Luciferase assay.
293F-pGL4.31 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. At 40% to 50% confluence, the cells were transfected with the indicated plasmids using the standard calcium-phosphate method in 12-well trays. The total amount of cDNA was adjusted by supplementing it with empty vector. Luciferase activities were determined using the dual Luciferase Reporter Assay System (Promega). Renilla luciferase was used as a reference to normalize transfection efficiencies in all experiments. All values are means ± standard deviations from at least three independent experiments.
Retroviral production and infection.
shTLX-, shLSD1-, and shLacZ-expressing retroviruses were generated using pSUPER.retro.puro. vectors (Oligoengine) and PLAT-A cells (31). The PLAT-A cells were kindly provided by Toshio Kitamura (University of Tokyo). The Y79 cells were infected by incubating them with the retrovirus in the presence of 5 μg/ml hexadimethrine bromide (Sigma-Aldrich, St. Louis, MO). After 24 h of incubation, we selected cells expressing shRNA by treatment with 10 μg/ml puromycin (Sigma-Aldrich).
RESULTS
Identification of LSD1 and CoREST as TLX interactants.
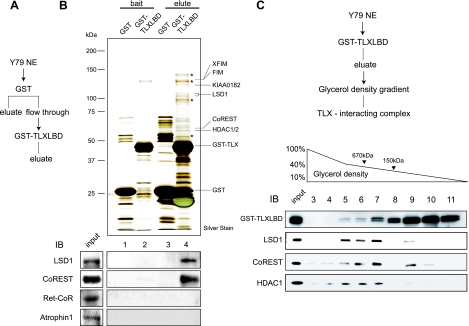
To identify the components of TLX's coregulator complex, Y79 retinoblastoma cell nuclear extracts were incubated with a chimeric TLX LBD (amino acids 109 to 385) fused to GST at its N terminus. Proteins that associated with TLX LBD were eluted with glutathione (Fig. 1A), separated by SDS-PAGE, and then subjected to silver staining (Fig. 1B, top). Matrix-assisted laser desorption ionization-time of flight mass spectrometric analyses of the bands revealed the presence of LSD1, CoREST, and HDAC1/2. Supporting these mass spectrometric analyses, we detected LSD1 and CoREST by Western blotting of the bands. We were unable to detect either Ret-CoR, which had been reported as a corepressor for PNR (also known as NR2E3), or atrophin 1, reportedly a corepressor for TLX (Fig. 1B, bottom). Furthermore, the eluted proteins were separated on 10% to 40% glycerol gradients and then analyzed by Western blotting with specific antibodies (Fig. 1C). Three of the polypeptides listed above migrated together with GST-TLX LBD in fractions 5 to 7, suggesting that they formed a complex. The smeared pattern of GST-TLX LBD may imply the presence of other TLX-interacting complexes in addition to LSD1/CoREST/HDAC1 (Fig. 1C).
FIG. 1.
Purification and identification of TLX interactants. (A) Schematic diagram of the purification of the TLX-associated complex. Y79 cell nuclear extract was loaded on a column as described in Materials and Methods. Bound proteins were eluted with 15 mM glutathione in BC100. NE, nuclear extracts. (B) Identification of TLX-interacting proteins. In the upper image, eluted proteins were subjected to SDS-PAGE, followed by silver staining. Molecular masses of marker proteins are indicated on the left side. Matrix-assisted laser desorption ionization-time of flight mass spectrometric analysis of the indicated proteins is shown on the right side. The asterisks indicate background peptides. The lower image shows Western blots (IB) using the indicated antibodies. (C) LSD1 forms a multiprotein complex with CoREST and HDAC1. The purification scheme is shown in the upper image. The eluted proteins were fractionated by a 10% to 40% glycerol density gradient. Western blot analysis of glycerol density gradient fractions using the indicated antibodies is shown in the lower image.
Interaction between TLX and LSD1 in vivo and in vitro.
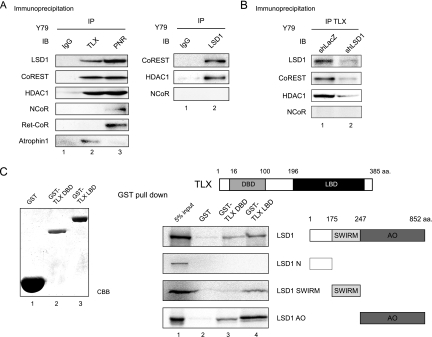
To confirm the interaction between TLX and the biochemically identified factors, we performed immunoprecipitation assays using nuclear extracts of Y79 cells. Immunoblot analysis revealed that LSD1, CoREST, or HDAC1 was coimmunoprecipitated with TLX from Y79 cells (Fig. 2A, left). Furthermore, CoREST and HDAC1 were coimmunoprecipitated with LSD1 (Fig. 2A, right), suggesting these factors formed a multiprotein complex. We could also detect TLX in the LSD1 immunoprecipitates when we overexpressed TLX (data not shown). Knockdown by LSD1 RNA interference (RNAi) decreased the amounts of CoREST and HDAC1 in the TLX immunoprecipitates (Fig. 2B). Thus, it appears that LSD1 mediates the interaction of TLX with the CoREST complex. Interestingly, these factors were also detected in an anti-PNR immunoprecipitate (Fig. 2A, left). Taken together, these data suggest that the LSD1 complex can interact with the NR2E orphan NR subfamily.
FIG. 2.
LSD1 associates with TLX in Y79 cells and in vitro. (A) Immunoprecipitation of nuclear extracts of Y79 cells, followed by Western blot (IB) analysis using anti-TLX, anti-PNR, and anti-LSD1 antibodies. Mouse immunoglobulin G (IgG) immunoprecipitate was used as a negative control. (B) Knockdown by LSD1 RNAi. Immunoprecipitation of nuclear extracts from control Y79 cells (shLacZ) and LSD1-depleted Y79 cells (shLSD1), followed by Western blot analysis using anti-TLX antibody. The confirmation of knockdown is shown in Fig. 4B. (C) In vitro interaction between TLX and LSD1. GST pull-down assays were performed with S35-labeled LSD1 or its deletion mutants and bacterially expressed GST-TLX DBD and GST-TLX LBD fusion proteins. GST protein was used as a negative control. aa., amino acids.
To further verify physical interaction between TLX and LSD1, we performed in vitro GST pull-down assays. GST-fused TLX mutants were incubated with in vitro-translated 35S-labeled LSD-1 deletion mutants (N terminus domain [NTD], SWIRM domain, or amine oxidase [AO] domain). As shown in Fig. 2C, full-length LSD1, as well as the SWIRM and AO domains, interacted with TLX LBD. TLX DBD was capable of associating only with the LSD1 AO domain. Altogether, it is likely that LSD1 interacts directly with TLX via both the SWIRM and AO domains.
LSD1 mediates the transrepressive function of TLX.
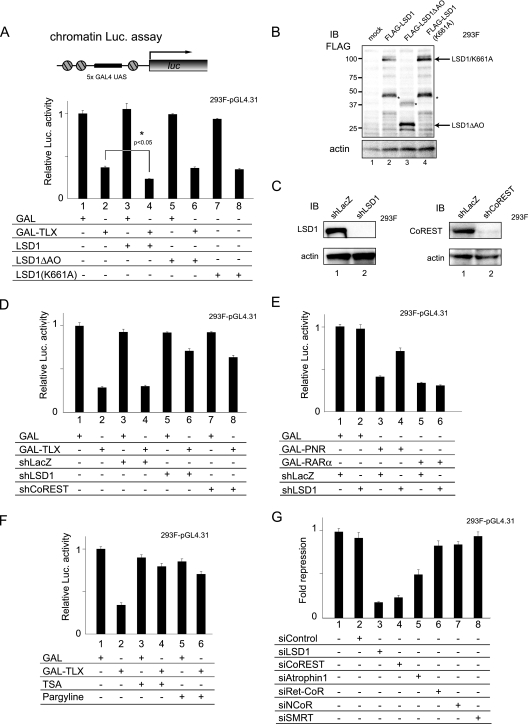
Next, we investigated the significance of the physical interaction between LSD1 and TLX. A reporter gene assay was developed to determine whether LSD1 acted as a corepressor with TLX through demethylation of histones. We generated a 293F cell line stably integrating a pGL4.31 reporter gene containing five GAL4 upstream activation sequence sites in the luciferase gene promoter (chromatin luciferase assay) (Fig. 3A, top). As shown in Fig. 3A, LSD1 potentiated the transrepressive activity of TLX (Fig. 3A, compare lanes 2 and 4). However, LSD1 mutants lacking the demethylase activities (LSD1ΔAO and LSD1 K661A) (Fig. 3B) lost their corepressor activities for TLX, implying that LSD1 potentiates the transrepressive activity of TLX through its histone demethylase activity. The transrepression activities of TLX and PNR, but not unliganded-RARα (NR1B1), were abolished by RNAi for LSD1 (Fig. 3D and E), and transrepression by TLX was also abolished by RNAi for CoREST (Fig. 3D). Furthermore, we tested two inhibitors of specific histone-modifying enzymes, trichostatin A (TSA) (an HDAC inhibitor), and pargyline (a monoamine oxidase-type histone demethylase inhibitor). As expected, transcriptional repression by TLX was partially relieved by TSA and pargyline (Fig. 3F, compare lanes 2, 4, and 6).
FIG. 3.
LSD1 is a TLX corepressor. (A) Corepressor activity of LSD1 with TLX in chromatin. 293F cells, which have a stably integrated pGL4.31 reporter gene, were transiently transfected with pM or pM TLX LBD vector (200 ng each) and with 400 ng of pcDNA empty vector, pCMX-FLAG LSD1, LSD1ΔAO, or LSD1(K661A) (chromatin luciferase [Luc] assay). The P value (bar 4 versus bar 2) was calculated by Student's t test (n = 3). UAS, upstream activation sequence. The error bars indicate standard deviations. (B) Confirmation of the expression level of each LSD1 mutant. Actin was used as a loading control. The asterisks denote nonspecific peptides. IB, immunoblot. (C) Confirmation of knockdown by shRNA. 293F-pGL4.31 cells were transfected with shRNA vector and examined for expression of LSD1 and CoREST by Western blotting. (D) Knockdown of LSD1 or CoREST by RNAi and its effect on the transrepression activity of TLX. A chromatin luciferase assay was performed using 293F-pGL4.31 reporter cells. shRNA expression vectors for LSD1 or CoREST were used at 400 ng. shLacZ expression vector was used as a negative control. (E) Knockdown of LSD1 and its effect on transrepression activity of PNR. Unliganded-RARα was used as a negative control (F) Inhibition of the histone-modifying enzyme and its effect on the transrepression activity of TLX. A chromatin luciferase assay was performed using 293F cells-pGL4.31 following 18 h of treatment with 100 nM TSA, a histone deacetylase inhibitor, or 3 mM pargyline, a monoamine oxidase-type histone demethylase inhibitor. (G) Knockdown of other corepressors by siRNAs and its effect on the transrepression activity of TLX. The repression of GAL-TLX LBD was determined relative to GAL activity.
We used siRNA knockdown assays to examine the effects of other known NR corepressors on the transrepression activity of TLX (see Fig. S1A in the supplemental material). As shown in Fig. 3G, neither NCoR/SMRT RNAi nor Ret-CoR RNAi affected the transrepressive function of TLX. However, RNAi for atrophin 1 weakly impaired TLX function, consistent with previous reports (48, 56).
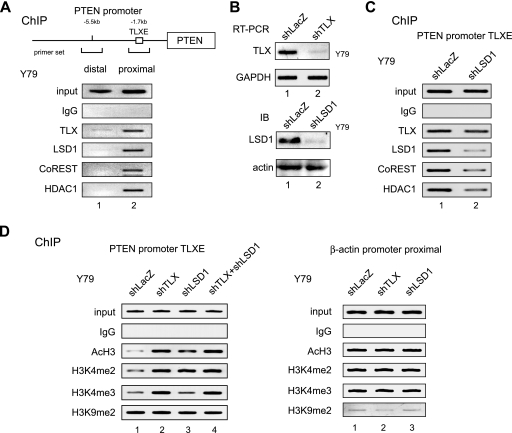
LSD1 and TLX are recruited to the PTEN promoter in Y79 cells.
To confirm that LSD1 functioned as a corepressor in TLX-mediated transrepression of endogenous genes, we conducted chromatin immunoprecipitation (ChIP) using Y79 cells (Fig. 4). In the human PTEN promoter, a TLX-binding site (TLXE) is located in a proximal region that is 1.7 kb upstream from the transcription initiation site (56). Using antibodies shown in the figure, the immunoprecipitated chromatins were subjected to PCR using primers corresponding to the proximal region and a distal region 5.5 kb upstream, which was used as a negative control. As shown in Fig. 4A, LSD1, CoREST, HDAC1, and TLX were all detectable in the proximal region, but not in the distal region, of the PTEN promoter. Knockdown of LSD1 decreased the amounts of CoREST and HDAC1 in the PTEN promoter, supporting the notion that LSD1 mediates the interaction of TLX with the CoREST complex (Fig. 4B and C). Next, we used ChIP analysis to determine if the recruited corepressors modified histones in the PTEN promoter. Knockdown of either TLX or LSD1 attenuated demethylation of H3K4me2 without affecting H3K9 methylation (Fig. 4D). Knockdown of TLX or LSD1 also impaired histone H3 deacetylation. When both factors were knocked down simultaneously, no additive effects in histone modifications were observed (Fig. 4D). Therefore, it appears that LSD1 demethylates H3K4me2 at the proximal region of the PTEN promoter through its association with TLX.
FIG. 4.
LSD1 and CoREST are recruited to TLX in the endogenous PTEN gene promoter. (A) ChIP analysis of the core components of the TLX-interacting complex on the human PTEN promoter using anti-TLX, anti-LSD1, anti-CoREST, and anti-HDAC1 antibodies. The region distal from the PTEN promoter was used as a negative control. IgG, immunoglobulin G. (B) Confirmation of RNAi specific for TLX and LSD1. Y79 cells were infected with each shRNA-expressing retrovirus and examined for expression of TLX and LSD1 by reverse transcription-PCR or Western blotting. GAPDH and actin were used as negative controls. (C) The effect of LSD1 knockdown for recruitment of the TLX-interacting complex. ChIP analysis was performed using LSD1-depleted Y79 cells. (D) The effect of TLX and LSD1 RNAi on histone tail modification. ChIP analysis was performed using the indicated antibodies. shLacZ and the β-actin promoter constituted the negative controls. A double-knockdown experiment was performed by transfecting shTLX-expressing Y79 cells with the shLSD1 expression vector.
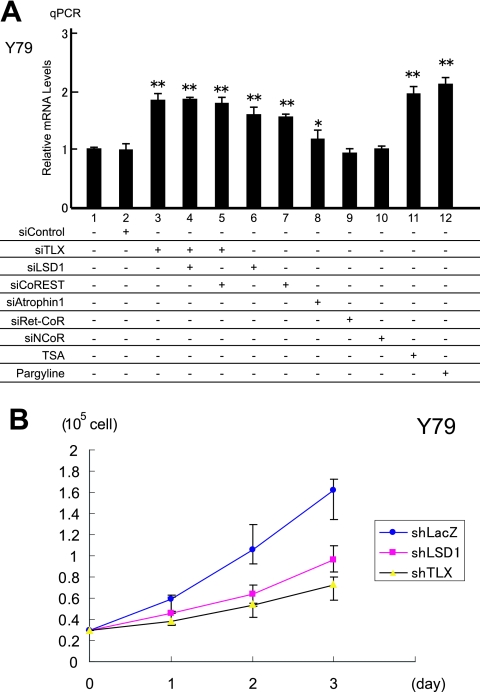
Finally, we used qPCR analysis to verify that TLX and LSD1 had transrepressive activities for endogenous expression of the PTEN gene in Y79 cells. Consistent with the present findings, PTEN mRNA was up-regulated by knockdown of either TLX, LSD1, or CoREST (Fig. 5A; see Fig. S1B in the supplemental material). PTEN gene expression was also increased by treatments with either TSA or pargyline (Fig. 5A). As anticipated, double knockdown of TLX and LSD1 failed to enhance PTEN expression, suggesting that both factors are in the same pathway. RNAi for NCoR, as well as for Ret-CoR, failed to derepress PTEN expression, while atrophin 1 knockdown weakly increased expression. These data suggest that LSD1 is a prime corepressor of TLX.
FIG. 5.
TLX and LSD1 regulate endogenous PTEN gene expression and proliferation of Y79 cells. (A) Increased expression of the PTEN gene was assessed by qPCR. Y79 cells were treated with various siRNAs or enzyme inhibitors as indicated. The expression levels of the PTEN gene were normalized to the endogenous expression of the GAPDH gene. *, P < 0.05, and **, P < 0.01 versus siControl (n = 3). (B) Cell proliferation assays for Y79 cells and effects of RNAi for TLX or LSD1. Y79 cells were infected with retrovirus expressing each shRNA. shLacZ was used as a negative control. The error bars indicate standard deviations.
PTEN is one of the most frequently mutated tumor suppressor genes in cancer cells, and it is well established that PTEN protein negatively regulates cell proliferation (5). Thus, we explored the effects of TLX and LSD1 RNAi on cell proliferation. Following RNAi treatment, 3 days of cultivation clearly revealed that the Y79 cell proliferation rate had slowed due to the decrease in either LSD1 or TLX (Fig. 5B). Thus, it appears that cell proliferation controlled by PTEN is subject to regulation by TLX and its prime corepressor, LSD1 (Fig. 6).
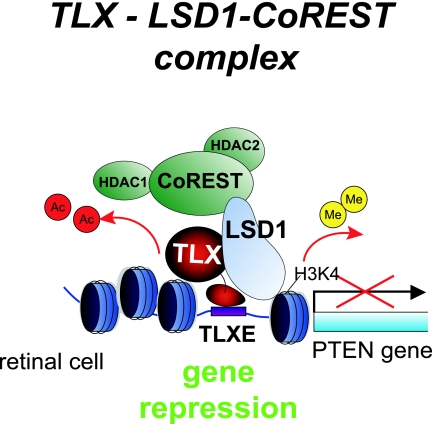
FIG. 6.
Schematic diagram of an LSD1 corepressor complex for TLX. In Y79 cells, the LSD1 complex is recruited to TLX bound to the target gene promoter, exhibiting transrepression activity by its histone demethylase and deacetylase activities.
DISCUSSION
It is well documented that transrepression by ligand-free NRs requires HDAC corepressor complexes containing NCoR/SMRT (1, 12, 14, 32). However, the NCoR/SMRT complex was not effective for the constitutive repressive function of TLX (Fig. 3G) and PNR (14, 45). These findings prompted us to search for other corepressor complexes for these orphan NRs. In our recent study, we reported that a Ret-CoR protein complex acted as a novel corepressor for PNR specifically in retinal development. In the present report, we analyzed nuclear extracts from Y79 retinoblastoma cells and identified LSD1 as a corepressor for TLX and PNR (Fig. 1B). As described in the previous paper, we suppose that the Ret-CoR corepressor complex is formed in a specific phase of the cell cycle and that it functions in a specific stage of development (45). We hypothesize that though both LSD1 and Ret-CoR can work as corepressors for PNR, the functional cellular context for each factor may be different.
Among biochemically identified interactants like KIAA0182, FIM (Znf198), and XFIM (Znf261), LSD1 appeared to form a complex with CoREST and HDAC1, since these three factors were detected in the same region of glycerol gradients (Fig. 1C, fractions 5 to 7). As the other three factors were also recently identified in LSD1 complexes, our purified complex may be the same as or closely related to those LSD1 complexes (22, 41).
By immunoprecipitation, we could detect interaction between TLX and atrophin 1 (Fig. 2A), which had been identified as a corepressor for TLX in a yeast-two hybrid screening (48, 56). However, we were unable to detect atrophin 1 in our purification even by Western blotting using specific antibody (Fig. 1B). We attribute this difference to the biochemical approach we used for TLX corepressor purification. This method is strongly dependent on the concentrations of interactants and the strengths of association between given bait proteins and putative corepressors. Furthermore, in Fig. 3G, siatrophin 1 weakly decreased the transrepression function of TLX. Thus, we conclude that atrophin 1 also works as a corepressor for TLX under our experimental conditions. However, we believe that the prime corepressor for TLX is LSD1, at least in Y79. Thus, we have a view that, like the other NRs, TLX interacts with a number of corepressor complexes to mediate its cell-type-specific function (25, 35, 36). According to this idea, the corepressor activity of atrophin 1 for TLX might depend on physiological conditions, like Ret-CoR.
In the present study, we provide the first evidence that LSD1 acts as a corepressor for a constitutively repressive type of orphan NR. LSD1 was identified as the first histone lysine demethylase capable of demethylating H3K4me2 and H3K4me1 in vitro (39). In vivo, LSD1 demethylates either H3K4me2/me1 or H3K9me2/me1 in a context-dependent manner and, accordingly, acts as a transcriptional repressor or activator, respectively (11, 22, 28, 41). In a murine model of pituitary development, Wang et al. recently showed that LSD1 was required for both repression and activation of developmental genes (47).
As anticipated, knockdown of LSD1 impaired TLX-induced demethylation of H3K4me2, but not H3K4me3, in the target gene promoter. In contrast, demethylation of both H3K4me2 and H3K4me3 was abrogated by TLX RNAi. As LSD1 is unable to demethylate H3K4me3 due to the inherent chemistry of the flavin-containing amine oxidases, it is likely that TLX associates with a trimethylated-histone demethylase for its transrepressive function (46). In this respect, it would be interesting to determine if JARID family members have corepressor activity for TLX, since this family is potent at demethylating H3K4me3 (3, 16, 19, 21).
TLX is capable of maintaining an undifferentiated state of neural stem cells (38, 40). Conceivably, TLX could transrepress expression of differentiation-related genes, such as GFAP and S100β (38, 56). More recently, it was reported that TLX targets signal transduction factors, including the PTEN tumor suppressor gene, and cell cycle-related factors, such as p21, a cyclin-dependent kinase inhibitor (24, 43, 56). In this report, we provide evidence that both TLX and LSD1 are indispensable for Y79 retinoblastoma proliferation through their regulation of PTEN expression (Fig. 5B). From these findings, we speculate that TLX modulates tumorigenesis or tumor progression by modulating the expression of genes controlling cell proliferation.
It is likely that DNA-binding transcription factors require highly regulated exchange of coregulator complexes for their transcriptional controls. From the present study, it appears that TLX is capable of interacting with a variety of coregulator complexes, such as the LSD1 complex and the atrophin 1 complex, presumably depending on physiological conditions, including the developmental stage and/or cell cycle phase. Further biochemical analyses to identify other TLX coregulators are essential to understand the molecular mechanism of TLX function.
Supplementary Material
Acknowledgments
We thank T. Kitamura for the PLAT-A cells. We also thank all the members of our laboratory for their help and discussion, especially Ken-ichi Takeyama, Ichiro Takada, Sally Fujiyama, Fumiaki Ohtake, Ryoji Fujiki, Atsushi Baba, and Maiko Okada for helpful discussions. We also express our appreciation for the technical assistance provided by Yuko Fukuda, Ikuko Yamaoka, Kei Iwasaki, and Chie Yoshida, and we thank Kazuyo Motoi and Hiroko Yamazaki for their preparation of the manuscript.
This work was supported in part by priority areas from the Ministry of Education, Culture, Sports, Science and Technology (to H.K. and S.K.). This work was also supported by a research fellowship for young scientists from the Japan Society for the Promotion of Science (to A.Y.).
Footnotes
Published ahead of print on 7 April 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alenghat, T., J. Yu, and M. A. Lazar. 2006. The N-CoR complex enables chromatin remodeler SNF2H to enhance repression by thyroid hormone receptor. EMBO J. 253966-3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bernstein, B. E., A. Meissner, and E. S. Lander. 2007. The mammalian epigenome. Cell 128669-681. [DOI] [PubMed] [Google Scholar]
- 3.Christensen, J., K. Agger, P. A. Cloos, D. Pasini, S. Rose, L. Sennels, J. Rappsilber, K. H. Hansen, A. E. Salcini, and K. Helin. 2007. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell 1281063-1076. [DOI] [PubMed] [Google Scholar]
- 4.Dignam, J. D., R. M. Lebovitz, and R. G. Roeder. 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 111475-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engelman, J. A., J. Luo, and L. C. Cantley. 2006. The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat. Rev. Genet. 7606-619. [DOI] [PubMed] [Google Scholar]
- 6.Fujiki, R., M. S. Kim, Y. Sasaki, K. Yoshimura, H. Kitagawa, and S. Kato. 2005. Ligand-induced transrepression by VDR through association of WSTF with acetylated histones. EMBO J. 243881-3894. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 7.Fukuda, T., K. Yamagata, S. Fujiyama, T. Matsumoto, I. Koshida, K. Yoshimura, M. Mihara, M. Naitou, H. Endoh, T. Nakamura, C. Akimoto, Y. Yamamoto, T. Katagiri, C. Foulds, S. Takezawa, H. Kitagawa, K. Takeyama, B. W. O'Malley, and S. Kato. 2007. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nat. Cell Biol. 9604-611. [DOI] [PubMed] [Google Scholar]
- 8.Gaillard, S., L. L. Grasfeder, C. L. Haeffele, E. K. Lobenhofer, T. M. Chu, R. Wolfinger, D. Kazmin, T. R. Koves, D. M. Muoio, C. Y. Chang, and D. P. McDonnell. 2006. Receptor-selective coactivators as tools to define the biology of specific receptor-coactivator pairs. Mol. Cell 24797-803. [DOI] [PubMed] [Google Scholar]
- 9.Garcia-Bassets, I., Y. S. Kwon, F. Telese, G. G. Prefontaine, K. R. Hutt, C. S. Cheng, B. G. Ju, K. A. Ohgi, J. Wang, L. Escoubet-Lozach, D. W. Rose, C. K. Glass, X. D. Fu, and M. G. Rosenfeld. 2007. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 128505-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giguere, V. 1999. Orphan nuclear receptors: from gene to function. Endocr. Rev. 20689-725. [DOI] [PubMed] [Google Scholar]
- 11.Hakimi, M. A., Y. Dong, W. S. Lane, D. W. Speicher, and R. Shiekhattar. 2003. A candidate X-linked mental retardation gene is a component of a new family of histone deacetylase-containing complexes. J. Biol. Chem. 2787234-7239. [DOI] [PubMed] [Google Scholar]
- 12.Heinzel, T., R. M. Lavinsky, T. M. Mullen, M. Soderstrom, C. D. Laherty, J. Torchia, W. M. Yang, G. Brard, S. D. Ngo, J. R. Davie, E. Seto, R. N. Eisenman, D. W. Rose, C. K. Glass, and M. G. Rosenfeld. 1997. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature 38743-48. [DOI] [PubMed] [Google Scholar]
- 13.Hermanson, O., C. K. Glass, and M. G. Rosenfeld. 2002. Nuclear receptor coregulators: multiple modes of modification. Trends Endocrinol. Metab. 1355-60. [DOI] [PubMed] [Google Scholar]
- 14.Hu, X., and M. A. Lazar. 2000. Transcriptional repression by nuclear hormone receptors. Trends Endocrinol. Metab. 116-10. [DOI] [PubMed] [Google Scholar]
- 15.Ishizuka, T., and M. A. Lazar. 2003. The N-CoR/histone deacetylase 3 complex is required for repression by thyroid hormone receptor. Mol. Cell. Biol. 235122-5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwase, S., F. Lan, P. Bayliss, L. de la Torre-Ubieta, M. Huarte, H. H. Qi, J. R. Whetstine, A. Bonni, T. M. Roberts, and Y. Shi. 2007. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell 1281077-1088. [DOI] [PubMed] [Google Scholar]
- 17.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 18.Kitagawa, H., R. Fujiki, K. Yoshimura, Y. Mezaki, Y. Uematsu, D. Matsui, S. Ogawa, K. Unno, M. Okubo, A. Tokita, T. Nakagawa, T. Ito, Y. Ishimi, H. Nagasawa, T. Matsumoto, J. Yanagisawa, and S. Kato. 2003. The chromatin-remodeling complex WINAC targets a nuclear receptor to promoters and is impaired in Williams syndrome. Cell 113905-917. [DOI] [PubMed] [Google Scholar]
- 19.Klose, R. J., Q. Yan, Z. Tothova, K. Yamane, H. Erdjument-Bromage, P. Tempst, D. G. Gilliland, Y. Zhang, and W. G. Kaelin, Jr. 2007. The retinoblastoma binding protein RBP2 is an H3K4 demethylase. Cell 128889-900. [DOI] [PubMed] [Google Scholar]
- 20.Lee, M. B., L. A. Lebedeva, M. Suzawa, S. A. Wadekar, M. Desclozeaux, and H. A. Ingraham. 2005. The DEAD-box protein DP103 (Ddx20 or Gemin-3) represses orphan nuclear receptor activity via SUMO modification. Mol. Cell. Biol. 251879-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, M. G., J. Norman, A. Shilatifard, and R. Shiekhattar. 2007. Physical and functional association of a trimethyl H3K4 demethylase and Ring6a/MBLR, a polycomb-like protein. Cell 128877-887. [DOI] [PubMed] [Google Scholar]
- 22.Lee, M. G., C. Wynder, N. Cooch, and R. Shiekhattar. 2005. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature 437432-435. [DOI] [PubMed] [Google Scholar]
- 23.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 24.Li, W., G. Sun, S. Yang, Q. Qu, K. Nakashima, and Y. Shi. 2007. Nuclear receptor TLX regulates cell cycle progression in neural stem cells of the developing brain. Mol. Endocrinol. 2256-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lonard, D. M., and W. B. O'Malley. 2007. Nuclear receptor coregulators: judges, juries, and executioners of cellular regulation. Mol. Cell 27691-700. [DOI] [PubMed] [Google Scholar]
- 26.Mangelsdorf, D. J., C. Thummel, M. Beato, P. Herrlich, G. Schutz, K. Umesono, B. Blumberg, P. Kastner, M. Mark, P. Chambon, and R. M. Evans. 1995. The nuclear receptor superfamily: the second decade. Cell 83835-839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martin, C., and Y. Zhang. 2005. The diverse functions of histone lysine methylation. Nat. Rev. Mol. Cell Biol. 6838-849. [DOI] [PubMed] [Google Scholar]
- 28.Metzger, E., M. Wissmann, N. Yin, J. M. Muller, R. Schneider, A. H. Peters, T. Gunther, R. Buettner, and R. Schule. 2005. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437436-439. [DOI] [PubMed] [Google Scholar]
- 29.Monaghan, A. P., D. Bock, P. Gass, A. Schwager, D. P. Wolfer, H. P. Lipp, and G. Schutz. 1997. Defective limbic system in mice lacking the tailless gene. Nature 390515-517. [DOI] [PubMed] [Google Scholar]
- 30.Monaghan, A. P., E. Grau, D. Bock, and G. Schutz. 1995. The mouse homolog of the orphan nuclear receptor tailless is expressed in the developing forebrain. Development 121839-853. [DOI] [PubMed] [Google Scholar]
- 31.Morita, S., T. Kojima, and T. Kitamura. 2000. Plat-E: an efficient and stable system for transient packaging of retroviruses. Gene Ther. 71063-1066. [DOI] [PubMed] [Google Scholar]
- 32.Nagy, L., H. Y. Kao, D. Chakravarti, R. J. Lin, C. A. Hassig, D. E. Ayer, S. L. Schreiber, and R. M. Evans. 1997. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell 89373-380. [DOI] [PubMed] [Google Scholar]
- 33.Narlikar, G. J., H. Y. Fan, and R. E. Kingston. 2002. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108475-487. [DOI] [PubMed] [Google Scholar]
- 34.Ohtake, F., A. Baba, I. Takada, M. Okada, K. Iwasaki, H. Miki, S. Takahashi, A. Kouzmenko, K. Nohara, T. Chiba, Y. Fujii-Kuriyama, and S. Kato. 2007. Dioxin receptor is a ligand-dependent E3 ubiquitin ligase. Nature 446562-566. [DOI] [PubMed] [Google Scholar]
- 35.Perissi, V., and M. G. Rosenfeld. 2005. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol. 6542-554. [DOI] [PubMed] [Google Scholar]
- 36.Rosenfeld, M. G., V. V. Lunyak, and C. K. Glass. 2006. Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev. 201405-1428. [DOI] [PubMed] [Google Scholar]
- 37.Ruthenburg, A. J., C. D. Allis, and J. Wysocka. 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol. Cell 2515-30. [DOI] [PubMed] [Google Scholar]
- 38.Shi, Y., D. C. Lie, P. Taupin, K. Nakashima, J. Ray, R. T. Yu, F. H. Gage, and R. M. Evans. 2004. Expression and function of orphan nuclear receptor TLX in adult neural stem cells. Nature 42778-83. [DOI] [PubMed] [Google Scholar]
- 39.Shi, Y., F. Lan, C. Matson, P. Mulligan, J. R. Whetstine, P. A. Cole, and R. A. Casero. 2004. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119941-953. [DOI] [PubMed] [Google Scholar]
- 40.Shi, Y., G. Sun, C. Zhao, and R. Stewart. 2007. Neural stem cell self-renewal. Crit. Rev. Oncol. Hematol. 6543-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shi, Y. J., C. Matson, F. Lan, S. Iwase, T. Baba, and Y. Shi. 2005. Regulation of LSD1 histone demethylase activity by its associated factors. Mol. Cell 19857-864. [DOI] [PubMed] [Google Scholar]
- 42.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 43.Sun, G., R. T. Yu, R. M. Evans, and Y. Shi. 2007. Orphan nuclear receptor TLX recruits histone deacetylases to repress transcription and regulate neural stem cell proliferation. Proc. Natl. Acad. Sci. USA 10415282-15287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takada, I., M. Mihara, M. Suzawa, F. Ohtake, S. Kobayashi, M. Igarashi, M. Y. Youn, K. Takeyama, T. Nakamura, Y. Mezaki, S. Takezawa, Y. Yogiashi, H. Kitagawa, G. Yamada, S. Takada, Y. Minami, H. Shibuya, K. Matsumoto, and S. Kato. 2007. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat. Cell Biol. 91273-1285. [DOI] [PubMed] [Google Scholar]
- 45.Takezawa, S., A. Yokoyama, M. Okada, R. Fujiki, A. Iriyama, Y. Yanagi, H. Ito, I. Takada, M. Kishimoto, A. Miyajima, K. Takeyama, K. Umesono, H. Kitagawa, and S. Kato. 2007. A cell cycle-dependent co-repressor mediates photoreceptor cell-specific nuclear receptor function. EMBO J. 26764-774. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 46.Tsukada, Y., J. Fang, H. Erdjument-Bromage, M. E. Warren, C. H. Borchers, P. Tempst, and Y. Zhang. 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439811-816. [DOI] [PubMed] [Google Scholar]
- 47.Wang, J., K. Scully, X. Zhu, L. Cai, J. Zhang, G. G. Prefontaine, A. Krones, K. A. Ohgi, P. Zhu, I. Garcia-Bassets, F. Liu, H. Taylor, J. Lozach, F. L. Jayes, K. S. Korach, C. K. Glass, X. D. Fu, and M. G. Rosenfeld. 2007. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 446882-887. [DOI] [PubMed] [Google Scholar]
- 48.Wang, L., H. Rajan, J. L. Pitman, M. McKeown, and C. C. Tsai. 2006. Histone deacetylase-associating Atrophin proteins are nuclear receptor corepressors. Genes Dev. 20525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang, Z., G. Benoit, J. Liu, S. Prasad, P. Aarnisalo, X. Liu, H. Xu, N. P. Walker, and T. Perlmann. 2003. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 423555-560. [DOI] [PubMed] [Google Scholar]
- 50.Wissmann, M., N. Yin, J. M. Muller, H. Greschik, B. D. Fodor, T. Jenuwein, C. Vogler, R. Schneider, T. Gunther, R. Buettner, E. Metzger, and R. Schule. 2007. Cooperative demethylation by JMJD2C and LSD1 promotes androgen receptor-dependent gene expression. Nat. Cell Biol. 9347-353. [DOI] [PubMed] [Google Scholar]
- 51.Yamane, K., C. Toumazou, Y. Tsukada, H. Erdjument-Bromage, P. Tempst, J. Wong, and Y. Zhang. 2006. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125483-495. [DOI] [PubMed] [Google Scholar]
- 52.Yanagisawa, J., H. Kitagawa, M. Yanagida, O. Wada, S. Ogawa, M. Nakagomi, H. Oishi, Y. Yamamoto, H. Nagasawa, S. B. McMahon, M. D. Cole, L. Tora, N. Takahashi, and S. Kato. 2002. Nuclear receptor function requires a TFTC-type histone acetyl transferase complex. Mol. Cell 9553-562. [DOI] [PubMed] [Google Scholar]
- 53.You, L. R., F. J. Lin, C. T. Lee, F. J. DeMayo, M. J. Tsai, and S. Y. Tsai. 2005. Suppression of Notch signalling by the COUP-TFII transcription factor regulates vein identity. Nature 43598-104. [DOI] [PubMed] [Google Scholar]
- 54.Young, K. A., M. L. Berry, C. L. Mahaffey, J. R. Saionz, N. L. Hawes, B. Chang, Q. Y. Zheng, R. S. Smith, R. T. Bronson, R. J. Nelson, and E. M. Simpson. 2002. Fierce: a new mouse deletion of Nr2e1; violent behaviour and ocular abnormalities are background-dependent. Behav. Brain Res. 132145-158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, R. T., M. McKeown, R. M. Evans, and K. Umesono. 1994. Relationship between Drosophila gap gene tailless and a vertebrate nuclear receptor Tlx. Nature 370375-379. [DOI] [PubMed] [Google Scholar]
- 56.Zhang, C. L., Y. Zou, R. T. Yu, F. H. Gage, and R. M. Evans. 2006. Nuclear receptor TLX prevents retinal dystrophy and recruits the corepressor atrophin1. Genes Dev. 201308-1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.