Abstract
JunB, a member of the AP-1 family of dimeric transcription factors, is best known as a cell proliferation inhibitor, a senescence inducer, and a tumor suppressor, although it also has been attributed a cell division-promoting activity. Its effects on the cell cycle have been studied mostly in G1 and S phases, whereas its role in G2 and M phases still is elusive. Using cell synchronization experiments, we show that JunB levels, which are high in S phase, drop during mid- to late G2 phase due to accelerated phosphorylation-dependent degradation by the proteasome. The forced expression of an ectopic JunB protein in late G2 phase indicates that JunB decay is necessary for the subsequent reduction of cyclin A2 levels in prometaphase, the latter event being essential for proper mitosis. Consistently, abnormal JunB expression in late G2 phase entails a variety of mitotic defects. As these aberrations may cause genetic instability, our findings contrast with the acknowledged tumor suppressor activity of JunB and reveal a mechanism by which the deregulation of JunB might contribute to tumorigenesis.
AP-1 is a collection of dimeric transcription factors that bind to the so-called AP1/TRE and CRE DNA motifs. Its best-studied components are the members of the Fos (c-Fos, FosB, Fra-1, and Fra-2) and Jun (c-Jun, JunB, and JunD) families, which bind to DNA owing to a basic motif (DNA-binding domain [DBD]) and dimerize via an adjacent leucine zipper (LZ) domain. AP-1 dimers act positively or negatively on transcription depending on their composition, the target gene, the cell context, and the signals received from the environment (12, 19, 32, 36, 43, 53, 54).
AP-1/TRE and CRE motifs are found in many genes. Hence, AP-1 regulates many fundamental cell processes, including proliferation, differentiation, apoptosis, and responses to stresses, and it is essential for many physiological functions at the whole-organism level. AP-1 also is implicated in various pathologies, notably tumorigenesis, via multiple effects on cell fate (12, 19, 32, 36, 43, 53, 54). Although certain AP-1 proteins can be oncogenic on their own in certain situations, the major contribution of AP-1 to tumorigenesis is as an effector of upstream oncogenic events. For example, the expression of the various Fos and Jun members is altered by mutated Ras, which is instrumental for cell transformation (19, 36, 43). Consistently, deregulated Fos and Jun protein expression is associated with a number of human neoplasias (44). Finally, although AP-1 is best known as a tumorigenesis promoter, some of its components display oncosuppressor activity in certain circumstances, as illustrated by c-Fos (19) and JunB (see below).
Much attention has been paid to cell division control by AP-1. In particular, Fos and Jun proteins regulate the expression of key cell cycle regulators, such as cyclins and cyclin-dependent kinase inhibitors (cki), and the transcriptional control of the latter genes largely depends on changes in the levels of the various Fos and Jun proteins themselves (19, 32, 36, 53, 54). Depending on the condition of their expression and/or the extracellular cues, the Fos and Jun proteins can manifest diverse, and sometimes opposite, functions. For example, when quiescent cells are stimulated by mitogens, c-Fos and c-Jun exert positive effects on cell division, notably via the induction of the cyclin D1 gene in G1 (2, 4, 31). However, they act as effectors of apoptosis in other situations (53).
With respect to cell cycle control, there is evidence that JunB exerts a dual function: even though it is best known as a cell division inhibitor (4, 48), a senescence inducer (48), and a tumor suppressor, at least in the myeloid lineage (47, 49, 55), it also can show cell division-promoting activity. Thus, its expression, which is very low in quiescent cells, is rapidly and transiently induced by mitogenic stimuli during the G0/G1 transition before returning to an intermediate level (38, 39, 41), with both of these events being instrumental for progression toward S phase (40). Rapid progression through S phase depends on JunB, the expression of which increases at the G1/S transition, to positively regulate the transcription of the cyclin A2 gene (Ccna2) (3). However, contrasting with the latter proliferation-stimulation functions, sustained JunB accumulation throughout G1 in response to antiproliferative signals leads to cell cycle arrest via the induction of the p16INK4αcki gene (48) and the down-regulation of that of cyclin D1 (4). Consequently, cells are blocked before they can enter S phase and this can be followed by senescence (48). Finally, JunB levels are low in mitotic and cycling cells traversing early G1 (4). In contrast, c-Jun levels remain constant during the same period of time with, however, a progressive increase in its transcriptional activity during early G1 (4). As JunB represses and c-Jun activates the cyclin D1 promoter, low levels of JunB in mitotic and early G1 phases provide an impetus for progression through G1 toward the S phase, as this permits a temporal increase in cyclin D1 transcription (4).
Notably, JunB appears in an electrophoretically retarded form in mitotic cells. This retardation is suppressed either by phosphatase treatment or by the point mutation of three residues (S23, T150, and S186) into nonphosphorylatable alanines in the mouse JunB (4). Interestingly, these residues are located in S/T-P motifs that are potential target sites for cyclin-containing cdk complexes. Moreover, JunB coimmunoprecipitates with cdk1 from cell extracts and is phosphorylated by cdk1/cyclin B1 complexes on these residues in vitro (4). Consequently, Bakiri et al.'s work raised an interesting hypothesis: cdk1/cyclin B1 complexes would phosphorylate JunB on these residues during mitosis to destabilize it during this specific period of the cell cycle in order to ensure low levels at the onset of the following G1 phase and, thereby, to permit another round of division (4). With the initial aim of testing this possibility and also because JunB function in G2 has not been studied in detail, we have investigated JunB's fate in cell synchronization experiments.
We show here that JunB levels are high in S phase and drop abruptly by mid-/late G2 due to phosphorylation-dependent proteasomal degradation. Interestingly, our data indicate that this JunB disappearance is necessary for the physiological reduction of the abundance of cyclin A2 protein in early mitosis. This most probably occurs via a direct transcriptional effect on the Ccna2 gene. Consistent with the fact that cyclin A2 degradation in prometaphase (just after nuclear envelope breakdown [NEBD]) is an essential event for proper progression through later stages of mitosis (16, 29), the overexpression of JunB in late G2 phase entails mitotic defects reminiscent of those caused by abnormal cyclin A2 accumulation in early M phase. Thus, our work reveals a heretofore unsuspected role for JunB down-regulation in the preparation of mitosis. Moreover, as the perturbation of mitosis may cause genetic instability and facilitate tumorigenesis, our findings contrast with the acknowledged tumor suppressor activity of JunB, since they point to a potential mechanism by which JunB contributes to tumorigenesis.
MATERIALS AND METHODS
Immunoblot analysis.
Protein extract preparation, electrophoresis, and immunoblotting conditions were described previously (7). Final immunodetections were carried out with donkey horseradish peroxidase-conjugated anti-rabbit and anti-mouse immunoglobulin antisera from Santa Cruz and the ECL Western blotting detection reagents from Perkin Elmer. The JunB monoclonal antibody was a kind gift from M. Yaniv. The cyclin A2 monoclonal antibody (CY-A1) was from Sigma Chemical Co., and the cyclin B1 polyclonal antibody (H-20) was from Santa Cruz Biotechnology. The polyclonal anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) rabbit antiserum was made in-house. The anti-phospho-T150 JunB and anti-phospho-S186-JunB antibodies were obtained after the immunization of rabbits with the CHKMNHVTpPNVS and CTNLSSYSpASAS peptides, respectively, coupled to keyhole limpet hemocyanin. They were produced and affinity chromatography purified by Eurogentec (Belgium). The purification procedure involved two successive steps using phosphorylated and nonphosphorylated forms of the above-described peptides. The specificity of the purified antibodies was demonstrated by blocking phosphopeptide recognition by the phosphorylated peptides, but not by the nonphosphorylated ones, in enzyme-linked immunosorbent assays and by blocking the recognition of phospho-JunB by phosphopeptides in immunoblotting experiments using extracts of nocodazole-arrested HeLa cells (not shown).
Expression vectors.
Wild-type JunB (JunBwt) and JunB3A open reading frames (4) were cloned in the pCDNA3 vector (Clontech) to give the PM799 and PM835 plasmids, respectively. DNA-binding-deficient (JunBΔDBD; PM1202) and dimerization-deficient (JunBVAV; PM1204) JunB mutants were generated from JunBwt in pCDNA3 using standard PCR-based techniques with the QuikChange multi-site-directed mutagenesis kit from Stratagene and subsequently verified by nucleotide sequencing. In JunBΔDBD, the DNA-binding domain was entirely removed. In JunBVAV, the last three leucines of the LZ were mutated to valine, alanine, and valine to abolish dimerization. The tetracycline (Tc)-repressible vectors PM1100, PM1101, and PM1102 are presented in Fig. 3C. They are based on the pTRE2 plasmid from Clontech and on the encephalomyocarditis virus internal ribosome entry site (IRES). All cloning details are available on request. pCDNA3, pEGFP, and pEYFP are from Clontech.
FIG. 3.
Phosphorylation-dependent degradation of JunB. (A) Phosphorylation of T150 and S186 in mitotic cells. HeLa and UTA6 cells were treated with nocodazole for 16 h, and mitotic cells were purified by shake-off. Total JunB, phospho-T150-JunB, and phospho-S186-JunB levels were compared by immunoblotting in mitotic (M) and adherent cycling (Adh) cells, using similar amounts of total cell proteins. (B) Phosphorylation of T150 and S186 in thymidine/thymidine-synchronized cells. UTA6 cells were synchronized and released in the cell cycle in the presence of nocodazole as described in the legend to Fig. 1D for kinetic immunoblotting analysis (except that the aphidicolin block was replaced by a second thymidine block, which did not result in any difference in the efficacy of arrest in G1/S). (C) Tc-regulatable bicistronic expression vectors. The vectors expressing EGFP and either JunB (PM1101) or JunB3A (PM1103) were based on the PM1100 expression vector, which only expresses EGFP downstream of the IRES. All plasmids were stably transfected in UTA6 cells in the presence of Tc. (D) JunB expression in thymidine/thymidine-synchronized UTA6 cells expressing PM1100, PM1101, and PM1103. G1/S-synchronized cells were released in the cycle in the absence of Tc but in the presence of nocodazole, as shown in Fig. 1D. The full transcriptional activation of the PM plasmids occurred within 4 h. Direct EGFP fluorescence analysis showed that most cells expressed the transgene upon the removal of Tc. GAPDH was used as an invariant control in immunoblot assays (not shown). The presented luminogram exposures were selected to best show the variations of endogenous and ectopic JunB proteins. (E) Densitometric analysis of the expression of wild-type and mutant JunB proteins. The graph showing the variations of endogenous and ectopic JunB proteins is not deduced from densitometer scanning of the luminograms presented in panel D but from less-exposed luminograms, i.e., luminograms exposed in the linear range of autoradiography film response. The standardization of quantification experiments allowed us to deduce that ectopic JunBwt and JunB3A are expressed approximately fourfold more than endogenous JunB during the period extending from 4 to 12 h after release in the cell cycle. (F) Expression of phospho-T150- and phospho-S186-JunB in synchronized UTA6 cells expressing JunBwt and JunB3A. UTA6 cells transfected with PM1100, PM1101, and PM1103 were synchronized as described for panel D for immunoblot analysis with the antiserum specific for phospho-T150- and phospho-S186-JunB.
Cell culture and transfections.
HeLa and U2OS-derived UTA6 (20) cells were cultured in fetal calf serum-containing Dulbecco's modified Eagle's medium (DMEM). UTA6 cells were cotransfected with pTRE2-based plasmids and the hygromycin resistance-conferring pCMVHygro plasmid using the Fugene 6 transfection agent (Roche). Stable UTA6 cell clones were selected in the presence of 150 μg/ml of hygromycin B and of 1 μg/ml Tc and were tested individually for JunB and EGFP coinduction in the absence of Tc before use. A volume of 7.5 × 104 UTA6 cells/ml was seeded in a 6-well culture plate. After 24 h, cells were transfected with 2 μg of PM799 and PM835 or 10 μg of PM1202 and PM1204 using the Fugene 6 transfection reagent according to the manufacturer's protocol (Roche). Cells were collected 48 h after transfection.
Cell synchronization.
For G1/S synchronization, 6 × 105 HeLa cells were routinely seeded into 10-cm-diameter culture dishes. Twenty-four hours later, 2.5 mM thymidine was added for 16 h. Cells were further cultured in thymidine-free medium for 12 h and then in the presence of 5 μg/ml aphidicolin for another 12 h (thymidine/aphidicolin block) and were released in the cycle by washing out aphidicolin. They were subsequently cultured in standard culture medium containing 0.04 μg/ml nocodazole when necessary. Due to the toxicity of aphidicolin for UTA6 cells, cell populations derived from this cell line were subjected to longer culture times in the presence (24 h) and in the absence (12 h) of thymidine and again in the presence of thymidine (24 h) (thymidine/thymidine block). Mitotic cells were collected by shake-off after 16 h in the presence of 0.04 μg/ml nocodazole, washed twice, and replated in nocodazole-free medium for subsequent culture.
Flow cytometry.
For flow cytometry, cells were (i) washed once in ice-cold phosphate-buffered saline (PBS), (ii) fixed in 70% ethanol at −20°C overnight, (iii) resuspended in 50 μg/ml RNase A-containing and 50 μM propidium iodide-containing PBS, and (iv) incubated for 1 h before the quantification of propidium iodide fluorescence and/or cell numbering using the FACSCalibur flow cytometer (Becton Dickinson). The cell distribution in the cell cycle was determined with the Cellquest software (Becton Dickinson) after gating out cell debris signals.
Protein half-life measurements.
For JunB half-life measurements, G1/S-synchronized cells released in the cycle were given cycloheximide (CHX; 50 μg/ml), alone or in combination with MG132 (25 μM), at various time points before kinetic immunoblot analysis. The densitometer analysis of luminograms was carried out using the ImageQuant system (Amersham).
Fluorescence microscopy and time-lapse analysis.
For the detection of JunB and phospho-S10 histone H3, cells were fixed in 4% paraformaldehyde at room temperature for 30 min, washed with 0.1% Triton X-100-containing PBS, and incubated in the blocking buffer (PBS containing 2% bovine serum albumin and 0.1% Triton X-100) for 15 min. Primary and secondary antibodies contained in the blocking buffer were successively added to cells for 1 h. After five washes in 0.1% Triton X-100-containing PBS, slides were mounted in Vectashields in the presence of DAPI (Vectalabs). JunB was detected using the JunB mouse monoclonal antibody and phospho-S10-histone H3 with antiserum from Cell Signaling Technology. For β-tubulin analysis, cells were fixed at room temperature in ice-cold methanol for 5 min and processed as described above using the T-0198 antiserum from Sigma. All secondary antibodies (Molecular Probes) were conjugated with Alexa 568. Microscopic examination was performed using the Leica DM 6000 B device using ×40 and ×63 objectives. Time-lapse differential interface contrast (DIC) fluorescence microscopy was carried out as described previously (13, 27). G1/S-synchronized cells released from the aphidicolin block were transfected 1 h later with either pCDNA3 plus pEYFP or the PM799 JunB expression vector plus pEYFP. Images were captured every 5 min with a Leica DMIRBE microscope equipped with a PentaMax camera (Princeton Instruments) and a PowerWave computer (PowerComputing) running the IP Lab Spectrum imaging software (Scanalytics Inc.). A ×40 oil objective was used. DIC images were used to determine mitotic phases and were converted to PICT format for exportation into the Adobe Photoshop program.
qRT-PCR.
For quantitative reverse transcription-PCR (qRT-PCR), total RNA was extracted using the Illustra kit (Amersham). Two micrograms of DNase-treated RNA was reverse transcribed using random hexanucleotide primers (3 μg), deoxynucleoside triphosphates (0.5 mM), dithiothreitol (10 mM), RNase Out (20 U), and the SuperScript III reverse transcriptase (100 U) from Invitrogen according to the supplier's specifications. cDNAs were amplified using the Sybr green PCR master mix from Applied Biosystems. Amplification products were detected by real-time PCR using the Gene Amp 5700 sequence detector system according to the manufacturer's specifications (Applied Biosystems). Triplicate reactions were carried out (10 min at 95°C, followed by 40 cycles of 15 min at 95°C and 60 min at 60°C). Primers were designed to span two exons and were selected using Primer Express 2.0 software (Applied Biosystems) to obtain products with lengths ranging from 100 to 400 bp. RT-PCR data were calculated by measuring the average cycle threshold (CT) for the mRNA of interest (Ccna2) and were normalized to the values for the housekeeping gene GAPDH or β-actin. The formula 2−ΔCT was used to express the normalized values. The range given for Ccna2 relative to that of the housekeeping gene was determined by evaluating the expression of 2−ΔCT with 2−ΔCT + s and 2−ΔCT − s, where s is the standard deviation of the 2−ΔCT value (user bulletin no. 2 for the ABI PRISM 7700 sequence detection system, 11 December 1997 [updated October 2001]). 5′-ATCAGTTATTGCTGGAGCTGCCT-3′ and 5′-TTCGTATTAATGATTCAGGCCAGCT-3′ were used as forward and reverse primers for cyclin A2, 5′-CTGGTGGCCTCTCTCTACACG-3′ and 5′-CCCGCGGGGGTAAAAGTACTG-3′ were used as forward and reverse primers for JunB, 5′-ACCAACTGGGACGATATGGAGAAGA-3′ and 5′-CGCACGATTTCCCTCTCAGC-3′ were used as forward and reverse primers for β-actin, and 5′-CATCTTCCAGGAGCGAGATC-3′ and 5′-GTTCACACCCATGACGAACAT-3′ were used as forward and reverse primers for GAPDH.
RNA interference experiments.
UTA6 cells stably transfected with either the control (PM1100) or the JunB3A (PM1103) expression vector were plated in fetal calf serum-containing DMEM in the presence of Tc at a density of 7.5 × 104 cells/ml in a 6-well culture plate. Twenty-four hours later, Tc was removed from the culture and cells were transfected with the amounts of short interfering RNA (siRNA) indicated in the figures, using Oligofectamine (Invitrogen) as a transfection reagent according to the manufacturer's protocol. The siRNAs were from Dharmacon Inc. (Lafayette, CO). The reference of the control siRNA is D-001210-01-05. Human Ccna2 mRNA was targeted using ON-Targetplus SMART pool L-003205-00-0005.
RESULTS
Decrease of JunB levels in G2.
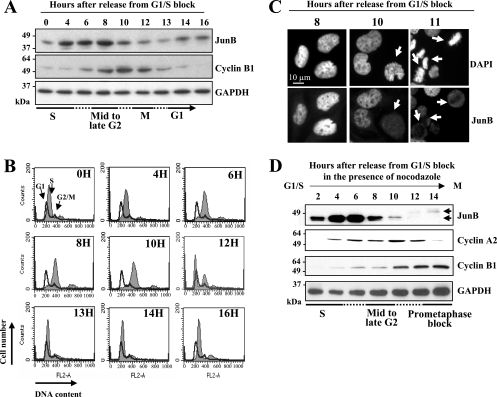
We first asked when JunB disappears during the period extending from S to the following G1 phase. For this, we reversibly arrested human HeLa cells at the onset of S phase (G1/S arrest) by sequential thymidine/aphidicolin blocks. After release from the aphidicolin block, JunB and cyclin B1 levels were monitored by immunoblotting, with GAPDH as a control (Fig. 1A). In this set of experiments, cyclin B1 was used as a mitotic marker, as its destruction, which is required for exit from mitosis, begins in metaphase (13). Progression through the cell cycle was assayed by flow cytometry analysis of the DNA content (Fig. 1B). Under these conditions, cells underwent mitosis between 11 and 13 h after release from the G1/S arrest. JunB levels increased during early S phase, remained high during late S and early G2 phases, and dropped between 8 and 10 h after release from the G1/S-phase block, i.e., at least 2 h before the onset of cyclin B1 destruction (Fig. 1A). As prophase and prometaphase last approximately 20 min and 20 to 40 min, respectively, in HeLa cells, this indicated that JunB levels were rapidly reduced in mid- to late G2 phase. The reduction in JunB levels was confirmed by immunofluorescence analysis. Residual JunB protein distributed homogenously in mitotic cells and was excluded from condensed chromosomes, whereas the protein was exclusively nuclear with nucleolar exclusions during interphase (Fig. 1C).
FIG. 1.
Decrease of JunB levels in G2. (A to C) Variations of levels of JunB in synchronized HeLa cells. Cells were G1/S synchronized and sampled at various times after release in the cell cycle for the immunoblot analysis of JunB, cyclin B1, and GAPDH (A); flow cytometry assay after propidium iodide labeling (B); and immunofluorescence analysis with anti-JunB antibodies after the fixation and staining of DNA with DAPI (C). For panel B, the thin-line profile corresponds to control asynchronous cells. The left peak, the right peak, and the zone in between correspond to G1, G2/M, and S cells, respectively. The solid profile corresponds to synchronized cells. Under the conditions used, the first M-to-G1 transition events are detected 10 to 11 h after release from the G1/S block and the last ones are detected at between 12 and 13 h. In panel C, the arrows show cells with low levels of JunB (time, 10 h) and condensed chromosomes (time, 11 h). (D) JunB decays before cyclin A2. Immunoblotting and synchronization experiments were carried out as described for panel A, except that nocodazole was added to the culture medium when aphidicolin was removed. Due to the presence of nocodazole, (i) cells could not progress beyond metaphase, explaining cyclin B1 accumulation (which cannot occur in the cells used for panel A), and (ii) JunB decay is clearer than that seen in panel A, because most cells escaping thymidine/aphidicolin synchronization are blocked in prometaphase (see the text).
We next positioned JunB decay with respect to the onset of cyclin A2 degradation, which starts at NEBD (16, 29), i.e., before the destruction of cyclin B1. In these experiments, HeLa cells were released from G1/S arrest in the presence of nocodazole to trap cells in mitosis. This experimental procedure change was intended to give a more accurate idea of the JunB disappearance profile in late G2, as the JunB decay occurring in the synchronized cells is partially obscured by the JunB content of the 10 to 20% of cells that always escape the G1/S block in classical thymidine/aphidicolin synchronization experiments. In contrast, in the presence of nocodazole, a significant proportion of the nonsynchronized cells are blocked in prometaphase (i.e., in a low-JunB-content state), which minimizes biases in the immunoblotting assays of JunB's fate in thymidine/aphidicolin-synchronized cells. As shown in Fig. 1D, cyclin B1, the degradation of which requires the inactivation of the spindle assembly checkpoint (13), was stable from 10 h until the end of the experiment, indicating efficient mitotic arrest by nocodazole (compare the kinetics of expression in Fig. 1A and D), whereas JunB was degraded approximately 2 to 4 h before cyclin A2, i.e., by mid- to late G2. These results also confirmed the previous observation (4) that the remnant of JunB in mitosis is subjected to phosphorylation, as visualized by retarded electrophoretic mobility (Fig. 1D) (see the JunB phosphosite analysis below).
Accelerated proteasomal degradation of JunB in mid-/late G2.
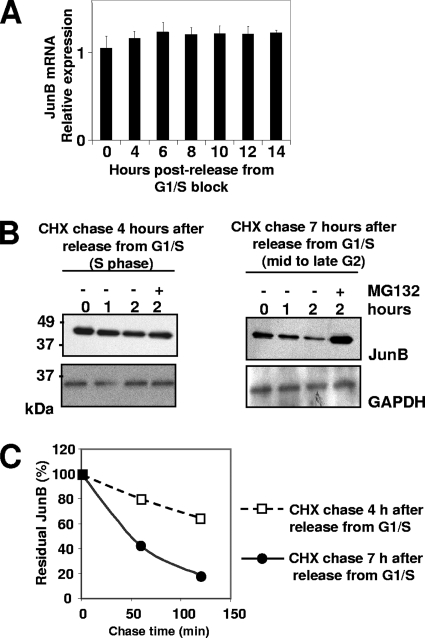
We next addressed the mechanisms underlying JunB decay in mid-/late G2. qRT-PCR revealed no change in JunB RNA levels from S phase to the next G1 phase (Fig. 2A). Thus, JunB disappearance is not due to transcriptional down-regulation. We also compared JunB half-lives in late S to those in late G2. Translation was inhibited by CHX either 4 or 7 h after G1/S-arrested HeLa cells were released in the cycle, and subsequently JunB levels were monitored by immunoblotting (Fig. 2B). An approximately 30% decay of JunB was seen after 2 h in late S, whereas most of it was gone after the same time in late G2 phase. Luminogram quantification indicated a 40- to 50-min half-life for JunB in late G2 and a >2.5-h half-life in S phase. This did not rule out a possible reduction of JunB translation in mid-/late G2 but clearly indicated a major contribution of protein destabilization to reduced JunB levels. We then asked whether JunB degradation was dependent on the proteasome, which is the main intracellular proteolytic machinery (24), as the proteasomal degradation of JunB already has been described by others (22, 25, 28). This was indeed the case, as JunB was stabilized in late G2 in the presence of the proteasome inhibitor MG132 (Fig. 2B, right).
FIG. 2.
Accelerated proteasomal degradation of JunB in G2. (A) JunB mRNA levels in synchronized HeLa cells. HeLa cells were G1/S synchronized and released in the cell cycle as described in the legend to Fig. 1A. Total RNA was prepared at various time points, and JunB mRNA was assayed by qRT-PCR as described in Materials and Methods, using β-actin mRNA as a normalization standard. The data are presented in arbitrary units and are the averages from three experiments. Bars correspond to standard deviations. (B) G2 destabilization of JunB. HeLa cells were synchronized as described in the legend to Fig. 1D. CHX was added at the indicated times in the absence or presence of MG132. JunB was analyzed by immunoblotting using GAPDH as an invariant reference. (C) JunB half-life in S and G2. The graph showing JunB decay is deduced from the densitometer scanning of appropriately exposed luminograms corresponding to the experiment presented in panel B.
Phosphorylation-dependent degradation of JunB in mid-/late G2.
Bakiri et al. (4) have shown that, in transfected 293 human kidney epithelial cells, the combined mutations of S23, T150, and S186 into nonphosphorylatable alanines in the mouse JunB protein (JunB3A mutant) lead both to the suppression of JunB electrophoretic retardation and to higher accumulation in nocodazole-arrested cells. We therefore asked whether these phosphorylations could be instrumental for destabilization in mid-/late G2 phase in a two-step approach.
First, we addressed endogenous JunB phosphorylation in synchronized human cells. To this end, we developed anti-phosphopeptide antibodies specifically recognizing JunB phosphorylated on either T150 or S186. S23 was not considered, as it is not conserved in human cells. HeLa and UTA6 (a subclone of human osteosarcoma U2OS cells; see below) cells were arrested in mitosis by nocodazole treatment, and their total and T150- and S186-phosphorylated JunB contents were compared to those of cycling cells by immunoblotting. Total JunB levels were severalfold lower in mitotic cells than in cycling cells, whereas the reverse was observed for T150 and S186 phosphorylation signals (Fig. 3A). This formally demonstrated the increased phosphorylation of T150 and S186 in mitotic cells, which was, thus far, inferred from indirect arguments. We next positioned the JunB phosphorylation onset in thymidine/aphidicolin-synchronized UTA6 cells. As shown in Fig. 3B, endogenous JunB levels dropped between 12 and 16 h, which is later than that for HeLa cells due to their longer doubling time (not shown). Consistently with our previous data for HeLa cells (Fig. 1D), the disappearance of JunB preceded that of cyclin A2, which occurred by 16 to 18 h after release in the cell cycle. Little or no phospho-T150 JunB and phospho-S186-JunB could be detected during S phase. However, the T150 and S186 phospho-JunB/total JunB ratios started to increase by the time the protein started to decay and reached a maximal level in mitosis. Taken together, these data indicated that the phosphorylation of T150 and S186 begins in G2 and continues at least during the first part of mitosis.
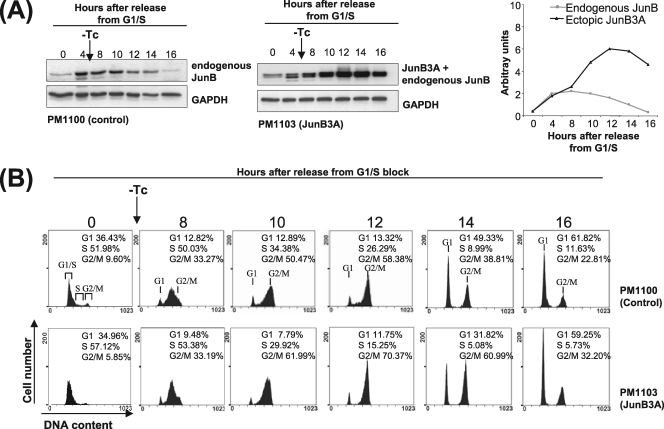
We next asked whether the alteration of JunB phosphorylation sites would suppress destabilization in mid-/late G2. The stable expression of JunB being incompatible with long-term cell proliferation (48), we resorted to a Tc-inducible expression system in the UTA6 subclone (20) of U2OS cells to address the destabilization of ectopic wild-type and mutant JunB proteins. JunBwt and JunB3A were cloned in a Tc-responsive bicistronic vector with the enhanced green fluorescent protein (EGFP) under the control of an IRES (Fig. 3C). This facilitated the selection of stably transfected Tc-responsive clones. Cells were synchronized at G1/S in the presence of Tc and subsequently were released in the presence of nocodazole after Tc was washed out. Under these conditions, it took 4 to 8 h to detect maximal amounts of both the endogenous and the two ectopic JunB proteins. The two ectopic proteins accumulated to similar levels, i.e., approximately fourfold more than that of the endogenous protein at these time points (Fig. 3D and E). Both endogenous JunB and ectopic wild-type JunB levels decreased with similar kinetics in mid-/late G2 (Fig. 3D and E), i.e., between 12 and 16 h after release from G1/S. In contrast, the nonphosphorylatable JunB3A levels hardly decreased during the time course (Fig. 3D and E). This supported the idea that the phosphorylation of T150 and/or S186 (and possibly S23 in the case of mouse JunB) is important for the destabilization of JunB in late G2 and, maybe, the beginning of mitosis. Additionally, we assessed the level of phospho-S186-JunB in synchronized control and transfected UTA6 cells. Phospho-S186-JunB levels were higher in JunBwt-expressing cells than in control and JunB3A-expressing cells, which showed similar signals (Fig. 3F). This was consistent with the fact that JunB3A is not phosphorylatable on S186 and was indicative of the specificity of the anti-phospho-S186-JunB antibody we developed. Similar data were obtained in the case of T150 (Fig. 3F).
Thus, our data indicate that JunB undergoes accelerated degradation in mid-/late G2 and destabilization involves the phosphorylation of T150 and/or S186.
Abnormal expression of JunB alters mitosis.
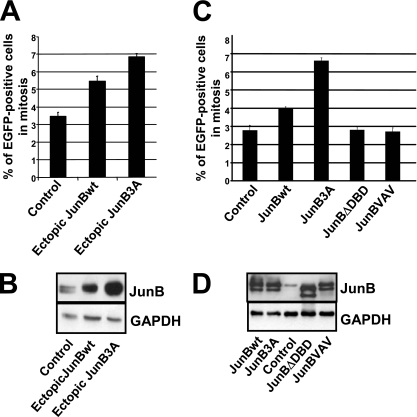
It was important to address whether the JunB decrease in mid-/late G2 phase is required for further progression through the cell cycle and, in particular, mitosis. With this aim, we first asked whether the overexpression of JunB in asynchronous cells could alter the fraction of mitotic cells, as this would indicate changes in the duration of mitosis. In a first series of experiments, Tc was removed from cultures of asynchronous UTA6 cells stably transfected with the regulatable bicistronic vectors for EGFP and either JunBwt or JunB3A and fluorescent mitotic cells were scored 48 h later. The induction of ectopic JunB proteins caused a 1.5- to 2-fold increase in the fraction of mitotic cells. The effect of JunB3A was stronger than that of JunBwt (Fig. 4A and B), possibly due to a higher accumulation level (see below). In a second series of experiments, asynchronous UTA6 cells were transiently transfected with cytomegalovirus (CMV) promoter-based plasmids constitutively expressing not only JunBwt and JunB3A but also a dimerization-deficient variant mutated in the LZ domain (JunBVAV) or a DNA-binding-deficient variant deleted of the DBD (JunBΔDBD). Consistent with the experiments presented in Fig. 4A, JunBwt and JunB3A led to the higher accumulation of mitotic cells, whereas JunBVAV and JunBΔDBD had no effect (Fig. 4C), despite the fact that their levels of accumulation were similar to those of JunBwt and JunB3A in transfected cells (Fig. 4D). Thus, abnormal JunB expression can impinge on mitosis with a stronger effect for JunB3A than for JunBwt, whereas that of the transcription-deficient mutant has no effect.
FIG. 4.
Effect of ectopic JunB on mitosis. (A) Cell accumulation in mitosis upon ectopic JunB expression in asynchronous UTA6 cells stably transfected with PM1100, PM1101, and PM1103. Tc was removed from the culture medium of asynchronous UTA6-derived cells stably transfected with PM1100 (control), PM1101 (JunBwt), and PM1103 (JunB3A) vectors (described in the legend to Fig. 3C). Fluorescent mitotic cells were scored 48 h later. At least 200 cells were counted in each experiment. The data are the means from three experiments. Error bars indicate the standard deviations. (B) Immunoblot analysis of endogenous JunB (control), ectopic JunBwt, and ectopic JunB3A in cells used for panel A. The immunoblotting analysis was performed 48 h after the removal of Tc from the culture medium. (C) UTA6 cell accumulation in mitosis upon transient transfection with plasmids expressing different JunB mutants. UTA6 cells were transiently transfected with a CMV promoter-based plasmid (pCDNA3) expressing either JunBwt, JunB3A, JunBΔDBD (DNA-binding-deficient mutant), or JunBVAV (LZ-deficient mutant). Mitotic cell analysis was carried out 48 h later as described for panel A. (D) Immunoblot analysis of transiently transfected UTA6 cells. The immunoblot analysis of transfected UTA6 cells was carried out 48 h after transfection.
To establish a more direct link between the aberrant timing of JunB expression and the alteration of mitosis, we resorted again to our stably transfected UTA6 cells, the progression of which through the cell cycle was monitored by flow cytometry after release from a G1/S arrest. In a first set of experiments, thymidine and Tc were removed at the same time, which corresponded to the conditions of expression of ectopic JunBwt and JunB3A proteins presented in Fig. 3D and E. A typical experiment is presented in Fig. 5. Because of the time required for the induction of the ectopic JunB proteins, no effect on the cell cycle was expected during the initial part of S phase. However, the cells overexpressing JunBwt and JunB3A reached G2/M faster than control cells due to faster progression through late S. This was consistent with work by others, who showed that JunB positively influences progression through S phase (3). Thereafter, both JunBwt- and JunB3A-expressing cells showed slower progression through G2 phase and/or mitosis, as judged from the smaller fraction of cells reentering G1 phase. Thus, the aberrant expression of JunB clearly extended the duration of mitosis in this experiment. However, it was not clear if this was due to aberrant timing and/or the expression level of ectopic JunB, as those were expressed to particularly high levels during both S and G2. To answer this question, UTA6 cells stably transfected with the JunB3A expression plasmid were G1/S arrested and released in the cell cycle and Tc was removed 6 h later to delay JunB3A induction. Under this condition, JunB3A became detectable by 10 h after release in the cell cycle (Fig. 6A). As in the experiments shown in Fig. 5, flow cytometry analysis showed that progression through mitosis was slowed down in JunB3A-expressing cells (Fig. 6B). This was consistent with the idea that the aberrant expression of JunB in G2 rather than overexpression at earlier time points is responsible for this effect (also see below).
FIG. 5.
Extended mitosis duration in stably transfected UTA6 cells upon early induction of JunBwt and JunB3A expression vector. UTA6 cells transfected with PM1100, PM1101, and PM1103 (described in the legend to Fig. 3C) were G1/S synchronized in the presence of Tc and then released in the cycle in its absence. They were harvested at different times and propidium iodide stained for flow cytometry analysis. The values are the averages from two independent synchronization experiments. Kinetics of induction of JunBwt and JunB3A are presented in Fig. 3D and E.
FIG. 6.
Extended mitosis duration in stably transfected UTA6 cells upon the late induction of JunB3A expression vector. UTA6 cells stably transfected with either the control (PM1100) or the inducible JunB3A expression vector (PM1103) (described in the legend to Fig. 3C) were G1/S synchronized and released in the cell cycle in the presence of Tc. Tc was removed 6 h later to allow for PM1100 and PM1103 induction. (A) Immunoblot analysis. Control and JunB3A-expressing cells were harvested at different time points for JunB and GAPDH content analysis. (B) Progression through the cell cycle. Progression through the cell cycle was monitored by flow cytometry analysis after propidium iodide staining. The values are the averages from two independent synchronization experiments.
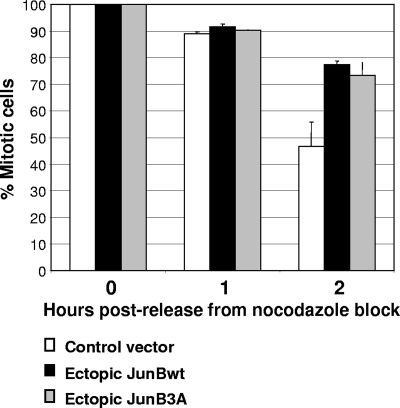
We next asked whether the G2 phase and/or mitosis was affected by deregulated JunB expression. G1/S-arrested HeLa cells were released into the cell cycle by the removal of aphidicolin. At the same time, they were transfected with either an empty CMV promoter-based plasmid or one expressing JunBwt along with one for yellow fluorescent protein (YFP) to identify transfected cells. Because HeLa cells are easily amenable to transfection, this technique allows the substantial expression of ectopic proteins from the next G2 phase onwards. Progression through mitosis was monitored by time-lapse videomicroscopy. Consistent with den Elzen and Pines (16), the time from the completion of NEBD to anaphase onset in HeLa cells, either those not transfected or those transfected with a void expression vector, was 25 to 35 min. In contrast, in the presence of ectopic JunB this time was extended in 90% of the analyzed cells, taking from 40 to 180 min. Typical experiments are presented in Fig. 7. The times between NEBD completion and anaphase were 38 and 106 min for control and ectopic JunB-expressing cells, respectively. To address whether the duration of mitosis could be impaired by abnormal JunB expression, we also resorted to our stable UTA6 transfectants. These cells were blocked in prometaphase by nocodazole treatment after the induction of either JunBwt or JunB3A, mitotic cells were collected by shake-off, and the kinetics of passage in G1 were monitored by the flow cytometry analysis of DNA content after the removal of nocodazole. Whereas almost 50% of control cells had entered G1 after 2 h, only approximately 25% of JunBwt- and JunB3A-expressing cells, respectively, had done so (Fig. 8). This confirmed that aberrant JunB expression entails delayed progression through mitosis. Thus, abnormal JunB expression prior to mitosis can alter mitosis.
FIG. 7.
Time-lapse analysis of the delay in prometaphase-anaphase transition induced by ectopic JunB expression. HeLa cells were G1/S synchronized. One hour after release in the cycle, they were transfected with either pCDNA3 or a pCDNA3-based JunB expression plasmid (PM799) in the presence of the pEYFP plasmid. The latter plasmid, which encodes the fluorescent EYFP protein, served to identify transfected cells. Progression through mitosis was monitored by time-lapse DIC microscopy. Images were taken at 3-min intervals. In the photogram, a representative cell overexpressing JunB is compared to a typical control cell. Pictures were selected to show the prophase (P), prometaphase (P/M), metaphase (M), anaphase (A), and telophase (T). Twenty-eight individual cells were observed in three independent experiments.
FIG. 8.
Extended mitosis in UTA6 cells ectopically expressing JunB and JunB3A. Tc was removed from the culture medium of UTA6-derived cells stably transfected with PM1100 (control), PM1101 (JunB), and PM1103 (JunB3A) vectors (described in the legend to Fig. 3C). Thirty-six hours later, nocodazole was added for 14 h. Mitotic cells were collected by shake-off and replated in the absence of nocodazole. 4N and 2N DNA contents were analyzed by flow cytometry 1 and 2 h later. The results of three independent experiments are plotted as the percentages of mitotic cells. Bars correspond to standard deviations.
Abnormal expression of JunB induces mitotic abnormalities.
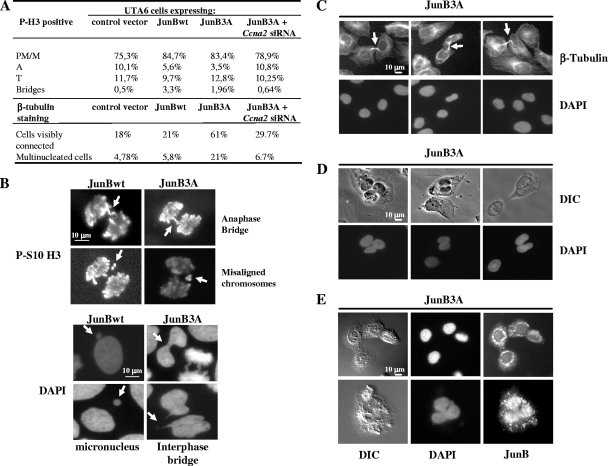
We next asked whether, in addition to lengthening mitosis, aberrant JunB expression could induce mitotic abnormalities. First, the stage distribution and the morphology of mitotic cells, visualized using antibodies directed against histone H3 phosphorylated on S10 (30) and DAPI staining, were examined under the microscope 48 h after the removal of Tc from cultures of our stable UTA6 transfectants. The fractions of cells in prophase, prometaphase, and metaphase in JunBwt- and JunB3A-expressing cells were higher than those in control cells (Fig. 9A). In contrast, the number of anaphase cells was slightly reduced in JunBwt- and JunB3A-expressing cells (Fig. 9A). This confirmed that predominantly early mitosis is affected. Interestingly, a significant fraction of JunBwt-expressing anaphase cells and, to a lesser extent, those expressing JunB3A showed misaligned chromosomes, chromatin bridges, and micronuclei, as visualized by DAPI staining and in immunofluorescence assays with the anti-phospho-S10-histone H3 antibody (Fig. 9B), whereas these abnormalities were rarely seen in control cells. However, the completion of cytokinesis was more strongly affected in JunB3A-expressing cells (Fig. 9A), which remained connected by a cytoplasmic bridge detectable upon β-tubulin labeling (Fig. 9C). Confirming the inhibitory effect of JunB3A on cytokinesis, the fraction of cells containing more than one nucleus 48 h after Tc removal was larger than those of control and JunBwt-expressing cells (Fig. 9A). The majority (90%) of multinucleated cells harbored two nuclei of normal and similar sizes (Fig. 9D), which is consistent with the idea of abortive cytokinesis rather than nuclear fragmentation or improper chromosome segregation. We cannot, however, eliminate the possibility of the regression of the cleavage furrow after initial formation. Intriguingly, the cell-to-cell JunB3A distribution was heterogeneous in all multinucleated cells, with strong perinuclear accumulation and cytoplasmic foci (Fig. 9E) in addition to its more classical nuclear localization (Fig. 1C).
FIG. 9.
Late-mitosis defects induced by JunB and RNA interference experiments targeting Ccna2 mRNA. Tc was removed from the culture medium of asynchronous UTA6-derived cells stably transfected with PM100 (control), PM1101 (JunB), and PM1103 (JunB3A) vectors (described in the legend to Fig. 3C). Microscopic analyses were carried out 48 h later. Mitotic cells were identified by being DAPI stained and labeled with an antibody directed against S10-phosphorylated histone H3. In the case of RNA interference experiments, Tc was removed from the medium and cells were transfected with control (not shown) and Ccna2-targeting siRNA (Fig. 12). (A) Distribution among the mitotic stages and frequencies of mitotic defects. PM/M, prometaphase/metaphase; A, anaphase; T, telophase. (B) Chromosomal abnormalities. DAPI-stained cells were analyzed by indirect immunofluorescence with an anti-phospho-S10 histone H3 antibody. Arrows indicate anaphase bridges, misaligned chromosomes, interphase bridges, and micronuclei. (C, D, and E) Cytokinesis phenotypes. DAPI-stained cells were analyzed by indirect immunofluorescence with antitubulin (C) and anti-JunB (E) antibodies or by DIC (D). (C) The daughter cells remain connected by cytoplasmic bridges indicated by arrows. (D) Multinucleation induced by JunB3A. (E) JunB3A localization in the nucleus, the cytoplasm, and intercellular bridges formed by daughter cells.
Thus, aberrant JunB expression can induce a variety of mitotic abnormalities, including those at late stages.
Dysregulation of cyclin A2 expression by ectopic JunB in late G2.
Andrecht et al. have shown that Ccna2 is a direct transcription target of JunB in S phase (3; also see Discussion). Moreover, den Elzen and Pines have reported that the overexpression of cyclin A2 in early mitosis, via the microinjection of an expression vector in thymidine/aphidicolin-synchronized cells released in the cell cycle, is sufficient to delay anaphase onset (16) in a manner that is very reminiscent of the mitotic dysfunctioning generated by ectopic JunB (Fig. 7). We therefore wondered whether aberrant JunB expression in late G2 could alter cyclin A2 expression. In a first step, we probed for both cyclin A2 and cyclin B1 in the protein extracts of the synchronization experiments presented in Fig. 3D and 5, in which ectopic JunBwt and JunB3A levels were severalfold higher than that of endogenous JunB. No striking difference was observed for cyclin B1 compared to that of control cells, indicating that cyclin B1 expression is independent of JunB. In contrast, higher levels of cyclin A2 were observed in JunBwt- and JunB3A-expressing cells than in control cells for the whole duration of the experiment, including mitosis (Fig. 10A and B). We then probed for cyclin A2 in extracts from the synchronized JunB3A-expressing UTA6 cells presented in Fig. 6A, in which JunB3A expression (i) was induced later and (ii) reached a level comparable to that of endogenous JunB, but (iii) did not decay in late G2 and mitosis. Interestingly, cyclin A2 was expressed to physiological levels during the time course of the experiments, except at the latest time points tested, at which point it did not decay, in contrast to what occurred in control cells. Taken together with the observations of Andrecht et al. (3) and den Elzen and Pines (16), these data are consistent with the idea that mitotic abnormalities in cells abnormally expressing JunB (Fig. 4 to 9) are, at least in part, due to the deregulated accumulation of cyclin A2.
FIG. 10.
Alteration of cyclin A2 expression by JunB. (A) Cyclin A2 and cyclin B1 levels upon the early induction of JunB and JunB3A in stably transfected UTA6 cells released from G1/S arrest. Protein extracts from the synchronized cells presented in Fig. 3D and E were probed for cyclins A2 and B1. Luminograms were selected to best show the variations in cyclin A2 levels. (B) Variations in cyclin A2 abundance. Variations were deduced by the densitometer scanning of appropriately exposed luminograms corresponding to the analysis presented in panel A. (C) Cyclin A2 levels upon the late induction of JunB3A in stably transfected UTA6 cells in thymidine/aphidicolin synchronization experiments. Protein extracts from the synchronized cells presented in Fig. 6 were probed for cyclin A2 abundance.
We next addressed whether cyclin A2 deregulation was transcriptional or posttranscriptional in ectopic JunB-expressing cells. To this end, we compared by qRT-PCR the cyclin A2 mRNA levels of synchronized UTA6 cells expressing JunB3A, from which Tc was removed at the same time that cells were allowed to reenter the cell cycle, to those of control cells (Fig. 11). In control cells, cyclin A2 mRNA expression increased 1.5-fold between 4 and 10 h after release from the G1/S block in the absence of Tc. Thereafter, it decreased when cells reached mitosis, reflecting the normal regulation of the Ccna2 gene (51). In contrast, in Jun3A-expressing cells, the Ccna2 mRNA level increased by fourfold between 4 and 10 h after release and did not significantly decrease thereafter. As the Ccna2 gene is a transcriptional target for JunB (3), higher Ccna2 mRNA levels were consistent with both the high level of ectopic JunB found in these cells (Fig. 3B) and the fact that mitotic abnormalities are dependent on the transcriptional activity of JunB (Fig. 4C and D).
FIG. 11.
Ccna2 mRNA expression in stably transfected JunB3A-expressing UTA6 cells after release from the G1/S arrest. UTA6 cells stably transfected with either the control or the JunB3A-expressing plasmid (described in the legend to Fig. 3C) were G1/S synchronized and released in nocodazole-containing medium in the absence of Tc. Relative cyclin A2 and GAPDH mRNA levels were assayed by qRT-PCR. The data are the averages from three experiments. Bars correspond to standard deviations.
JunB-induced mitotic abnormalities are dependent on cyclin A2 deregulation.
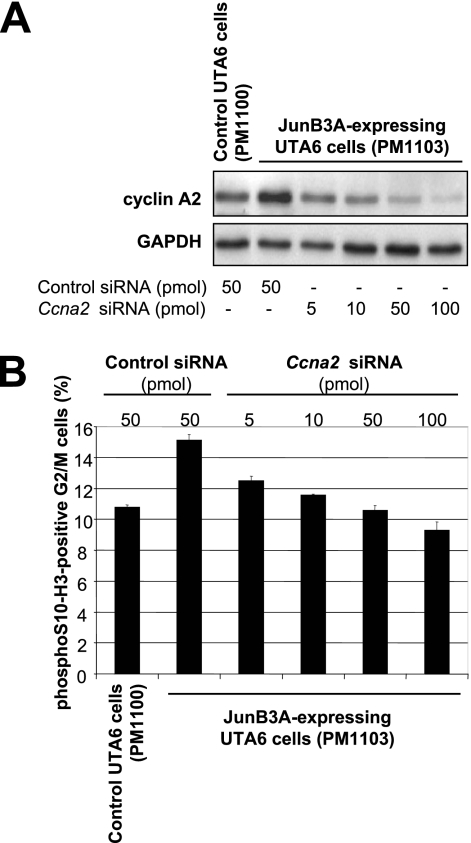
Lastly, we investigated whether the JunB-induced deregulation of cyclin A2 was essential for mitosis alteration. To this end, JunB3A was induced in asynchronous UTA6 cells, which were transfected with increasing amounts of an siRNA mixture targeting Ccna2 mRNA. Immunoblot analysis showed a dose-dependent effect on the reduction of cyclin A2 abundance, whereas a control siRNA had no effect (Fig. 12A). Moreover, a decrease in the fraction of mitotic cells was associated with the reduction in cyclin A2 abundance, as assayed by flow cytometry analysis of G2/M cells positive for phospho-S10 histone H3 48 h after JunB induction (Fig. 12B). Importantly, the reduction of cyclin A2 levels to that in control cells (Fig. 12B) was sufficient to reduce significantly the fraction of mitotic cells. This fraction was, however, larger than that of control cells, suggesting that JunB targets other genes to perturb mitosis. We also analyzed, under the microscope, the stage distribution and the morphology of Ccna2 siRNA-transfected mitotic cells. Their distributions among the various mitotic stages were restored to values close to those of control cells (Fig. 9A). Moreover, the numbers of both multinucleated cells and cells remaining connected by a bridge were dramatically reduced compared to those of JunB3A-expressing cells (Fig. 9A). Thus, the deregulation of cyclin A2 accumulation is essential for mitotic abnormalities induced by deregulated JunB expression.
FIG. 12.
Effect of Ccna2 siRNA on UTA6 cells expressing JunB3A. Asynchronous UTA6-derived cells stably transfected with either PM1100 (control) or PM1103 (JunB3A) were transfected with either a control siRNA or increasing amounts of an anti-Ccna2 siRNA pool as indicated on the figure, and Tc was removed from the medium. Cells were collected 48 h later for further analysis, as described in the legend to Fig. 9A. (A) Immunoblot analysis of cyclin A2 and GAPDH levels. (B) Percentage of mitotic cells with respect to the total amount of G2/M cells. Cells were analyzed by flow cytometry for DNA content and phospho-S10 histone H3 fluorescence associated with mitotic cells. The values are the averages from three independent experiments. Bars correspond to standard deviations.
DISCUSSION
We report that the JunB level abruptly decreases by mid-/late G2 due to accelerated phosphorylation-dependent proteasomal degradation. Moreover, forced JunB expression in late G2 delays the transcriptional repression of Ccna2 and induces delayed progression through mitosis as well as a number of mitotic abnormalities. RNA interference experiments targeting cyclin A2 partially reversed the phenotype of JunB-overexpressing cells, pointing to an essential effector role of cyclin A2 in the JunB-induced perturbation of mitosis.
JunB degradation in mid-/late G2.
Bakiri et al. (4) have reported that a variety of mouse and human mitotic cells express low levels of JunB. As (i) the mouse JunB is phosphorylated in vitro by cdk1/cyclin B1 complexes on S23, T150, and/or S186, (ii) the mutation of these residues into nonphosphorylatable alanines entails the high accumulation of JunB in mitotic human and mouse cells, and (iii) JunB coimmunoprecipitates with cdk1 from metaphasic cells (4), it was logically proposed that the reason for low mitotic JunB levels was accelerated degradation during mitosis due to cdk1-mediated phosphorylation. Our data support another scenario, in which low mitotic JunB levels result from protein destabilization by mid-/late G2 phase, i.e., before mitosis. There are several pieces of evidence for this. Firstly, thymidine/aphidicolin-based cell synchronization experiments indicated that JunB is expressed at high levels during S and the first part of G2 and undergoes rapid decay by mid-/late G2 (Fig. 1). Secondly, the absence of variations in JunB mRNA levels (Fig. 2A) excluded transcriptional mechanisms for late-G2 down-regulation. Rather, turnover analysis demonstrated the accelerated degradation of JunB in mid-/late G2 (Fig. 2B). Moreover, pharmacological inhibition pointed to a role for the proteasome in this process (Fig. 2A). Thirdly, JunB (even phosphorylated, as visualized by its electrophoretic shift and by immunoblotting using antisera specifically recognizing phospho-T150- and phospho-S186-JunB) levels did not decrease upon the prolongation of the nocodazole-induced prometaphase arrest and we were unable to detect any degradation of JunB from (at least) prometaphase to exit from mitosis in synchronization experiments in which HeLa cells were released from a nocodazole block (R. Farràs and M. Piechaczyk, unpublished data). Interestingly, in vitro degradation experiments further supported these data, as cell extracts from G1/S-arrested HeLa cells released in the cycle for 8 to 9 h efficiently degraded a JunB protein produced in the reticulocyte lysate, whereas extracts prepared at earlier or later times or from prometaphase-blocked cells could not (Farràs and Piechaczyk, unpublished). Importantly, JunB degradation in these cell-free assays also was inhibited by proteasome inhibitors. It is important to underline that the techniques used during the course of this work, even though they allow the defining of the time at which JunB is destabilized, did not allow us to precisely delineate the whole period of JunB instability. At this stage of investigation, we cannot, therefore, exclude the possibility that JunB may be degraded actively until the early phases of mitosis before the restoration of a slower degradation rate from prometaphase on.
In most instances, proteasomal degradation has been associated with the prior ubiquitylation of the substrate protein (24). Such a mechanism has been described already for JunB in various transfection assays and upon the activation of mouse helper T cells (22, 25, 28). In these T cells, the E3 ligase involved is the HECT protein Itch (22, 28). In preliminary synchronization experiments (Farràs and Piechaczyk, unpublished), we have detected a slight increase in JunB-ubiquitin conjugates at the time JunB gets destabilized by mid-/late G2, which is suggestive, but not demonstrative, of the ubiquitin-dependent degradation of JunB under these conditions. We also have seen that, in HeLa cells, AIP4, the human homolog of Itch, (i) is expressed in all phases of the cell cycle, (ii) can interact with JunB, and (iii) promotes JunB ubiquitylation in asynchronous cell cotransfection assays (Farràs and Piechaczyk, unpublished). However, the RNA interference-mediated reduction of its levels by at least 90% led to JunB stabilization in neither S- nor G2-synchronized HeLa cells (Farràs and Piechaczyk, unpublished). Although we cannot formally rule out the possible that a functional redundancy with other E3s masked a role for AIP4 or that residual AIP4 was sufficient for JunB destruction, this finding argues for the involvement of another E3, if ubiquitylation actually is instrumental for the degradation of JunB in mid-/late G2. In fact, this would not be surprising, as several proteins, including c-Jun (23, 28, 46, 59), now have been demonstrated to be ubiquitylatable by various E3s. However, it is important to underline that ubiquitylation may serve purposes other than the addressing of substrates to the proteasome (15, 33, 45, 52) and that other AP-1 proteins, albeit ubiquitylatable, can be degraded by the proteasome independently of their ubiquitylation either in vitro (c-Jun [35]) or in vivo (c-Fos and Fra-1 [5, 7]). The formal demonstration of the ubiquitylation-dependent degradation of JunB in mid-/late G2 therefore will await the identification of the relevant JunB E3(s) complemented by functional assays.
Two lines of evidence strongly suggest that accelerated JunB degradation in mid-/late G2 is phosphorylation dependent. Firstly, the ratios of phospho-T150-JunB/total JunB and phospho-S186-JunB/total JunB start to increase by the time the JunB level begins to drop in mid-/late G2, as assayed by immunoblotting with specific antibodies (Fig. 3). Secondly, the mouse JunB3A mutant, in which S23, T153, and S186 were mutated into nonphosphorylatable alanines, shows higher stability than JunBwt in late G2 (Fig. 3). Although our work does not disqualify a possible contribution of S23 phosphorylation to the G2 destabilization of mouse JunB, it clearly suggests a role for the phosphorylation of the T150 and/or S186 residue conserved among species. However, further work is necessary to estimate the relative effects of each one of these residues on JunB-accelerated destruction. It also will be important to investigate the possibility that still-to-be-identified phosphorylations cooperate with those of T150 and S186 to destabilize JunB. An important question is which kinase is responsible for the phosphorylation of these two residues. As described by Bakiri et al. (4), we confirmed that JunB, but not JunB3A, is phosphorylated by cdk1/cyclin B1 in vitro (V. Baldin, unpublished data). However, the latter kinase is not a good candidate, as it is activated abruptly only at the end of G2 and shows maximal activity during mitosis (26). Moreover, cyclin B is predominantly cytoplasmic during interphase (50), whereas JunB is essentially nuclear (Fig. 1D). Another possibility is that, instead of cdk1/cyclin B1, cdk1/cyclin A2 phosphorylates JunB. This would provide a regulatory loop in which the phosphorylating complex would induce the degradation of a factor acting positively on the transcription of the gene encoding its regulatory subunits at a time the latter must disappear. However, we have not been able to demonstrate the phosphorylation of S23, T150, and/or S186 in an in vitro assay involving in vitro-translated JunB and baculovirus-produced cdk1/cyclin A2 (not shown). Whether an ancillary factor cooperating with, or another kinase activated by, cdk1/cyclin A2 is involved in triggering G2 JunB degradation still deserves investigation. Finally, it is of note that JunB is phosphorylated in mitotic cells when its degradation is slowed down. This raises the interesting possibility of another role for the phosphorylation of T150 and/or S186 (and possibly S23 in the mouse) in mitosis. It would, for example, be worth determining whether they also could down-regulate JunB transcriptional activity, which would functionally inactivate the remnant of JunB to prevent possible perturbations of this particular phase of the cell cycle.
Dysregulation of Ccna2 by JunB.
Consistent with the role of cyclin A2 as a regulatory partner of cdk2 and cdk1 during S and G2/M, Ccna2 is periodically transcribed with low levels of expression in G0 and G1. However, at what time repression is established has not been investigated. Our work suggests that this occurs as soon as mid-/late G2.
The precise molecular mechanisms by which Ccna2 transcription is regulated are far from being understood. Various transcriptional cofactors, including the RASSF1 oncosuppressor protein (1), the HMG2A architectural transcription factor (57), and the SWI/SNF chromatin-remodeling complex (14), are involved in this process. Through binding at several sites, various transcription factors, including E2F and Rb family members, p120E4F, c-Jun, JunB, Fra-1, ATF2, and CREB, also have been implicated, although a number of functional interpretations are still debated (1, 3, 10, 14). With regard to the implication of AP-1, band shift and luciferase reporter gene assays initially have suggested that a CRE motif residing approximately 80 nucleotides upstream of the Ccna2 initiation transcription site is responsive to JunB in S phase (3). More recent chromatin immunoprecipitation experiments showed that JunB actually can bind to this DNA element in Ras-transformed rat thyroid cells traversing G2 (10), which we confirmed in exponentially growing HeLa cells (Farràs and Piechaczyk, unpublished). However, in this work, the authors also identified four upstream AP-1/TRE sites localized within the −397/−569 region (10). Three of these sites could bind JunB- and Fra-1-containing AP-1 dimers more efficiently than the CRE. As in Ras-transformed cells both Fra-1 and JunB levels are dramatically increased compared to those of their normal cell counterparts (11, 42, 58), it is important to investigate the physiological regulation of Ccna2 by JunB during G2 in nontransformed cells. Further work involving extensive chromatin immunoprecipitation experiments and the functional analyses of the whole Ccna2 promoter will be necessary to determine which of the possible AP-1 binding sites are involved in the physiological regulation of Ccna2 in S and G2 and in the delay in Ccna2 repression when JunB is aberrantly expressed in late G2. This study certainly will be complicated by the fact that various other transcription factors also have been proposed to target the CRE (1, 6, 17, 21, 34, 37).
Mitotic perturbations generated by aberrant JunB expression.
Taken together, our data show that JunB overexpression in G2 affects mitosis (Fig. 4 to 9). Thus, in addition to ensuring proper progression through the next G1 phase as proposed by Bakiri et al. (4), the reduction in JunB levels is necessary for proper mitosis. It is unlikely that increased amounts of JunB activate the mitotic spindle checkpoint, as JunB-overexpressing cells still could degrade cyclin B1 and proceed through mitosis. In contrast, exogenous JunB delays anaphase onset, slows down exit from mitosis, and generates cytokinesis abnormalities (Fig. 4 to 9). The early mitotic perturbations are reminiscent of those seen upon the simple ectopic expression of cyclin A2 in synchronized PtK1 and HeLa cells (16). Several lines of evidence support the idea that the early mitotic perturbations are contributed by JunB-induced cyclin A2 dysregulation: (i) mitotic abnormalities are dependent on JunB transcriptional activity (Fig. 4C and D), (ii) JunB overexpression in late G2 causes delayed cyclin A2 protein decay, which is linked to the delayed shutoff of the Ccna2 gene (Fig. 3, 6, 10, and 11), and (iii) RNA interference targeting cyclin A2 expression in JunB3A-expressing cells inhibited the perturbation of JunB-induced mitotic abnormalities (Fig. 9A and 12). However, it is of note that, on the one hand, JunB also entails late mitosis abnormalities (such as the formation of multinucleated cells) that were not observed upon overexpression of normal cyclin A2 (16), and on the other hand, the reduction of cyclin A2 levels to physiological levels in the RNA interference experiments presented in Fig. 12 did not totally reverse the mitotic phenotype induced by JunB3A. Therefore, it is possible that the reduction of JunB levels in late G2 also is necessary for the proper regulation of cell functions other than the timely repression of Ccna2. The analysis of the transcriptome controlled by JunB in this specific cell cycle phase will help clarify this point and, possibly, identify genes encoding proteins that are involved in the control of chromosome alignment, anaphase initiation, and the completion of cytokinesis. Finally, as there are significant differences between the mitotic abnormalities generated by ectopic JunB and JunB3A (Fig. 9), it will be necessary to establish whether this is simply due to differences in the levels and/or the timing of expression of these two proteins or whether JunBwt and JunB3A are not functionally equivalent. As already mentioned, it will be interesting to investigate whether the phosphorylation of JunB by cdk1/cyclin B1 complexes in mitosis is responsible for the transcriptional inactivation of JunB.
JunB's implication in cancer is twofold and context dependent. In addition to its well-described cell proliferation inhibition- and senescence-promoting activities (see the introduction), it can act as a tumor suppressor. For example, its expression is down-regulated in human chronic (9, 60) and acute myeloid leukemia (18), and transgenic mice lacking JunB expression in the myeloid lineage develop tumors resembling human chronic myeloid leukemia (48, 49), most probably because decreased amounts of JunB increase the self-renewal capacity of leukemic stem cells (55). Moreover, JunB overexpression inhibits the transformation of B cells by the v-abl oncogene in the mouse (56). Besides this, JunB can contribute to the tumor phenotype. For example, it cooperates with c-Jun in the development of mouse fibrosarcoma (8), and its increased expression seems essential for the pathogenesis of human anaplastic large-cell lymphoma and certain Hodgkin lymphomas through the induction of the CD30 promoter (59). As disrupted passage through mitosis often leads to chromosome missegregation and the production of aneuploid progeny, our work raises the possibility that the overexpression of JunB in late G2 represents a thus far unsuspected oncogenic mechanism. Further work will aim at determining whether such a dysregulation favors genomic rearrangements or instability eventually resulting in tumor outcomes.
Acknowledgments
M.P.'s laboratory is an Equipe Labellisée of the Ligue Nationale contre le Cancer. This work also was supported by grants from the ARC, the European QLP program (contract QLRT-2000-02026), and Fondo de Investigaciones Sanitarias PI05/2139. R.F. was initially supported by fellowships from the ARC and the Human Frontier Science Program at the IGMM. She is now supported by the Fondo de Investigaciones Sanitarias CP03/00106.
We are grateful to M. Yaniv for the gift of the JunB vectors and antibodies and for his support. We also thank J. Pines for fruitful discussions and the critical reading of the manuscript.
Footnotes
Published ahead of print on 7 April 2008.
REFERENCES
- 1.Ahmed-Choudhury, J., A. Agathanggelou, S. L. Fenton, C. Ricketts, G. J. Clark, E. R. Maher, and F. Latif. 2005. Transcriptional regulation of cyclin A2 by RASSF1A through the enhanced binding of p120E4F to the cyclin A2 promoter. Cancer Res. 652690-2697. [DOI] [PubMed] [Google Scholar]
- 2.Albanese, C., J. Johnson, G. Watanabe, N. Eklund, D. Vu, A. Arnold, and R. G. Pestell. 1995. Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J. Biol. Chem. 27023589-23597. [DOI] [PubMed] [Google Scholar]
- 3.Andrecht, S., A. Kolbus, B. Hartenstein, P. Angel, and M. Schorpp-Kistner. 2002. Cell cycle promoting activity of JunB through cyclin A activation. J. Biol. Chem. 27735961-35968. [DOI] [PubMed] [Google Scholar]
- 4.Bakiri, L., D. Lallemand, E. Bossy-Wetzel, and M. Yaniv. 2000. Cell cycle-dependent variations in c-Jun and JunB phosphorylation: a role in the control of cyclin D1 expression. EMBO J. 192056-2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Basbous, J., D. Chalbos, R. Hipskind, I. Jariel-Encontre, and M. Piechaczyk. 2007. Ubiquitin-independent proteasomal degradation of Fra-1 is antagonized by Erk1/2 pathway-mediated phosphorylation of a unique C-terminal destabilizer. Mol. Cell. Biol. 273936-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beier, F., A. C. Taylor, and P. LuValle. 2000. Activating transcription factor 2 is necessary for maximal activity and serum induction of the cyclin A promoter in chondrocytes. J. Biol. Chem. 27512948-12953. [DOI] [PubMed] [Google Scholar]
- 7.Bossis, G., P. Ferrara, C. Acquaviva, I. Jariel-Encontre, and M. Piechaczyk. 2003. c-Fos proto-oncoprotein is degraded by the proteasome independently of its own ubiquitinylation in vivo. Mol. Cell. Biol. 237425-7436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bossy-Wetzel, E., R. Bravo, and D. Hanahan. 1992. Transcription factors junB and c-jun are selectively up-regulated and functionally implicated in fibrosarcoma development. Genes Dev. 62340-2351. [DOI] [PubMed] [Google Scholar]
- 9.Bruchova, H., T. Borovanova, H. Klamova, and R. Brdicka. 2002. Gene expression profiling in chronic myeloid leukemia patients treated with hydroxyurea. Leuk. Lymphoma 431289-1295. [DOI] [PubMed] [Google Scholar]
- 10.Casalino, L., L. Bakiri, F. Talotta, J. B. Weitzman, A. Fusco, M. Yaniv, and P. Verde. 2007. Fra-1 promotes growth and survival in RAS-transformed thyroid cells by controlling cyclin A transcription. EMBO J. 261878-1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Casalino, L., D. De Cesare, and P. Verde. 2003. Accumulation of Fra-1 in ras-transformed cells depends on both transcriptional autoregulation and MEK-dependent posttranslational stabilization. Mol. Cell. Biol. 234401-4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chinenov, Y., and T. K. Kerppola. 2001. Close encounters of many kinds: Fos-Jun interactions that mediate transcription regulatory specificity. Oncogene 202438-2452. [DOI] [PubMed] [Google Scholar]
- 13.Clute, P., and J. Pines. 1999. Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 182-87. [DOI] [PubMed] [Google Scholar]
- 14.Coisy, M., V. Roure, M. Ribot, A. Philips, C. Muchardt, J. M. Blanchard, and J. C. Dantonel. 2004. Cyclin A repression in quiescent cells is associated with chromatin remodeling of its promoter and requires Brahma/SNF2α. Mol. Cell 1543-56. [DOI] [PubMed] [Google Scholar]
- 15.Collins, G. A., and W. P. Tansey. 2006. The proteasome: a utility tool for transcription? Curr. Opin. Genet. Dev. 16197-202. [DOI] [PubMed] [Google Scholar]
- 16.den Elzen, N., and J. Pines. 2001. Cyclin A is destroyed in prometaphase and can delay chromosome alignment and anaphase. J. Cell Biol. 153121-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desdouets, C., G. Matesic, C. A. Molina, N. S. Foulkes, P. Sassone-Corsi, C. Brechot, and J. Sobczak-Thepot. 1995. Cell cycle regulation of cyclin A gene expression by the cyclic AMP-responsive transcription factors CREB and CREM. Mol. Cell. Biol. 153301-3309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dorsam, S. T., C. M. Ferrell, G. P. Dorsam, M. K. Derynck, U. Vijapurkar, D. Khodabakhsh, B. Pau, H. Bernstein, C. M. Haqq, C. Largman, and H. J. Lawrence. 2004. The transcriptome of the leukemogenic homeoprotein HOXA9 in human hematopoietic cells. Blood 1031676-1684. [DOI] [PubMed] [Google Scholar]
- 19.Eferl, R., and E. F. Wagner. 2003. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer 3859-868. [DOI] [PubMed] [Google Scholar]
- 20.Englert, C., X. Hou, S. Maheswaran, P. Bennett, C. Ngwu, G. G. Re, A. J. Garvin, M. R. Rosner, and D. A. Haber. 1995. WT1 suppresses synthesis of the epidermal growth factor receptor and induces apoptosis. EMBO J. 144662-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fajas, L., C. Paul, A. Vie, S. Estrach, R. Medema, J. M. Blanchard, C. Sardet, and M. L. Vignais. 2001. Cyclin A is a mediator of p120E4F-dependent cell cycle arrest in G1. Mol. Cell. Biol. 212956-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang, D., C. Elly, B. Gao, N. Fang, Y. Altman, C. Joazeiro, T. Hunter, N. Copeland, N. Jenkins, and Y. C. Liu. 2002. Dysregulation of T lymphocyte function in itchy mice: a role for Itch in TH2 differentiation. Nat. Immunol. 3281-287. [DOI] [PubMed] [Google Scholar]
- 23.Fang, D., and T. K. Kerppola. 2004. Ubiquitin-mediated fluorescence complementation reveals that Jun ubiquitinated by Itch/AIP4 is localized to lysosomes. Proc. Natl. Acad. Sci. USA 10114782-14787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farràs, R., G. Bossis, E. Andermarcher, I. Jariel-Encontre, and M. Piechaczyk. 2005. Mechanisms of delivery of ubiquitylated proteins to the proteasome: new target for anti-cancer therapy? Crit. Rev. Oncol. Hematol. 5431-51. [DOI] [PubMed] [Google Scholar]
- 25.Fuchs, S. Y., B. Xie, V. Adler, V. A. Fried, R. J. Davis, and Z. Ronai. 1997. c-Jun NH2-terminal kinases target the ubiquitination of their associated transcription factors. J. Biol. Chem. 27232163-32168. [DOI] [PubMed] [Google Scholar]
- 26.Fung, T. K., and R. Y. Poon. 2005. A roller coaster ride with the mitotic cyclins. Semin. Cell Dev. Biol. 16335-342. [DOI] [PubMed] [Google Scholar]
- 27.Furuno, N., N. den Elzen, and J. Pines. 1999. Human cyclin A is required for mitosis until mid prophase. J. Cell Biol. 147295-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao, M., T. Labuda, Y. Xia, E. Gallagher, D. Fang, Y. C. Liu, and M. Karin. 2004. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 306271-275. [DOI] [PubMed] [Google Scholar]
- 29.Geley, S., E. Kramer, C. Gieffers, J. Gannon, J. M. Peters, and T. Hunt. 2001. Anaphase-promoting complex/cyclosome-dependent proteolysis of human cyclin A starts at the beginning of mitosis and is not subject to the spindle assembly checkpoint. J. Cell Biol. 153137-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hendzel, M. J., Y. Wei, M. A. Mancini, A. Van Hooser, T. Ranalli, B. R. Brinkley, D. P. Bazett-Jones, and C. D. Allis. 1997. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 106348-360. [DOI] [PubMed] [Google Scholar]
- 31.Herber, B., M. Truss, M. Beato, and R. Muller. 1994. Inducible regulatory elements in the human cyclin D1 promoter. Oncogene 91295-1304. [PubMed] [Google Scholar]
- 32.Hess, J., P. Angel, and M. Schorpp-Kistner. 2004. AP-1 subunits: quarrel and harmony among siblings. J. Cell Sci. 1175965-5973. [DOI] [PubMed] [Google Scholar]
- 33.Huang, T. T., and A. D. D'Andrea. 2006. Regulation of DNA repair by ubiquitylation. Nat. Rev. Mol. Cell Biol. 7323-334. [DOI] [PubMed] [Google Scholar]
- 34.James, C. G., A. Woods, T. M. Underhill, and F. Beier. 2006. The transcription factor ATF3 is upregulated during chondrocyte differentiation and represses cyclin D1 and A gene transcription. BMC Mol. Biol. 730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jariel-Encontre, I., M. Pariat, F. Martin, S. Carillo, C. Salvat, and M. Piechaczyk. 1995. Ubiquitinylation is not an absolute requirement for degradation of c-Jun protein by the 26 S proteasome. J. Biol. Chem. 27011623-11627. [DOI] [PubMed] [Google Scholar]
- 36.Jochum, W., E. Passegue, and E. F. Wagner. 2001. AP-1 in mouse development and tumorigenesis. Oncogene 202401-2412. [DOI] [PubMed] [Google Scholar]
- 37.Katabami, M., H. Donninger, F. Hommura, V. D. Leaner, I. Kinoshita, J. F. Chick, and M. J. Birrer. 2005. Cyclin A is a c-Jun target gene and is necessary for c-Jun-induced anchorage-independent growth in RAT1a cells. J. Biol. Chem. 28016728-16738. [DOI] [PubMed] [Google Scholar]
- 38.Kovary, K., and R. Bravo. 1992. Existence of different Fos/Jun complexes during the G0-to-G1 transition and during exponential growth in mouse fibroblasts: differential role of Fos proteins. Mol. Cell. Biol. 125015-5023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kovary, K., and R. Bravo. 1991. Expression of different Jun and Fos proteins during the G0-to-G1 transition in mouse fibroblasts: in vitro and in vivo associations. Mol. Cell. Biol. 112451-2459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kovary, K., and R. Bravo. 1991. The Jun and Fos protein families are both required for cell cycle progression in fibroblasts. Mol. Cell. Biol. 114466-4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lallemand, D., G. Spyrou, M. Yaniv, and C. M. Pfarr. 1997. Variations in Jun and Fos protein expression and AP-1 activity in cycling, resting and stimulated fibroblasts. Oncogene 14819-830. [DOI] [PubMed] [Google Scholar]
- 42.Mechta, F., D. Lallemand, C. M. Pfarr, and M. Yaniv. 1997. Transformation by ras modifies AP1 composition and activity. Oncogene 14837-847. [DOI] [PubMed] [Google Scholar]
- 43.Mechta-Grigoriou, F., D. Gerald, and M. Yaniv. 2001. The mammalian Jun proteins: redundancy and specificity. Oncogene 202378-2389. [DOI] [PubMed] [Google Scholar]
- 44.Milde-Langosch, K. 2005. The Fos family of transcription factors and their role in tumourigenesis. Eur. J. Cancer 412449-2461. [DOI] [PubMed] [Google Scholar]
- 45.Mukhopadhyay, D., and H. Riezman. 2007. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315201-205. [DOI] [PubMed] [Google Scholar]
- 46.Nateri, A. S., L. Riera-Sans, C. Da Costa, and A. Behrens. 2004. The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 3031374-1378. [DOI] [PubMed] [Google Scholar]
- 47.Passegué, E., W. Jochum, M. Schorpp-Kistner, U. Mohle-Steinlein, and E. F. Wagner. 2001. Chronic myeloid leukemia with increased granulocyte progenitors in mice lacking junB expression in the myeloid lineage. Cell 10421-32. [DOI] [PubMed] [Google Scholar]
- 48.Passegué, E., and E. F. Wagner. 2000. JunB suppresses cell proliferation by transcriptional activation of p16(INK4a) expression. EMBO J. 192969-2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Passegué, E., E. F. Wagner, and I. L. Weissman. 2004. JunB deficiency leads to a myeloproliferative disorder arising from hematopoietic stem cells. Cell 119431-443. [DOI] [PubMed] [Google Scholar]
- 50.Pines, J., and T. Hunter. 1991. Human cyclins A and B1 are differentially located in the cell and undergo cell cycle-dependent nuclear transport. J. Cell Biol. 1151-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pines, J., and T. Hunter. 1989. Isolation of a human cyclin cDNA: evidence for cyclin mRNA and protein regulation in the cell cycle and for interaction with p34cdc2. Cell 58833-846. [DOI] [PubMed] [Google Scholar]
- 52.Reed, S. H., and T. G. Gillette. 2007. Nucleotide excision repair and the ubiquitin proteasome pathway-do all roads lead to Rome? DNA Repair 6149-156. [DOI] [PubMed] [Google Scholar]
- 53.Shaulian, E., and M. Karin. 2002. AP-1 as a regulator of cell life and death. Nat. Cell Biol. 4E131-E136. [DOI] [PubMed] [Google Scholar]
- 54.Shaulian, E., and M. Karin. 2001. AP-1 in cell proliferation and survival. Oncogene 202390-2400. [DOI] [PubMed] [Google Scholar]
- 55.Steidl, U., F. Rosenbauer, R. G. Verhaak, X. Gu, A. Ebralidze, H. H. Otu, S. Klippel, C. Steidl, I. Bruns, D. B. Costa, K. Wagner, M. Aivado, G. Kobbe, P. J. Valk, E. Passegue, T. A. Libermann, R. Delwel, and D. G. Tenen. 2006. Essential role of Jun family transcription factors in PU.1 knockdown-induced leukemic stem cells. Nat. Genet. 381269-1277. [DOI] [PubMed] [Google Scholar]
- 56.Szremska, A. P., L. Kenner, E. Weisz, R. G. Ott, E. Passegue, M. Artwohl, M. Freissmuth, R. Stoxreiter, H. C. Theussl, S. B. Parzer, R. Moriggl, E. F. Wagner, and V. Sexl. 2003. JunB inhibits proliferation and transformation in B-lymphoid cells. Blood 1024159-4165. [DOI] [PubMed] [Google Scholar]
- 57.Tessari, M. A., M. Gostissa, S. Altamura, R. Sgarra, A. Rustighi, C. Salvagno, G. Caretti, C. Imbriano, R. Mantovani, G. Del Sal, V. Giancotti, and G. Manfioletti. 2003. Transcriptional activation of the cyclin A gene by the architectural transcription factor HMGA2. Mol. Cell. Biol. 239104-9116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vallone, D., S. Battista, G. M. Pierantoni, M. Fedele, L. Casalino, M. Santoro, G. Viglietto, A. Fusco, and P. Verde. 1997. Neoplastic transformation of rat thyroid cells requires the junB and fra-1 gene induction which is dependent on the HMGI-C gene product. EMBO J. 165310-5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe, M., M. Sasaki, K. Itoh, M. Higashihara, K. Umezawa, M. E. Kadin, L. J. Abraham, T. Watanabe, and R. Horie. 2005. JunB induced by constitutive CD30-extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase signaling activates the CD30 promoter in anaplastic large cell lymphoma and Reed-Sternberg cells of Hodgkin lymphoma. Cancer Res. 657628-7634. [DOI] [PubMed] [Google Scholar]
- 60.Wertz, I. E., K. M. O'Rourke, Z. Zhang, D. Dornan, D. Arnott, R. J. Deshaies, and V. M. Dixit. 2004. Human de-etiolated-1 regulates c-Jun by assembling a CUL4A ubiquitin ligase. Science 3031371-1374. [DOI] [PubMed] [Google Scholar]