Abstract
Exogenous fibroblast growth factor 1 (FGF1) signals through activation of transmembrane FGF receptors (FGFRs) but may also regulate cellular processes after translocation to the cytosol and nucleus of target cells. Translocation of FGF1 occurs across the limiting membrane of intracellular vesicles and is a regulated process that depends on the C-terminal tail of the FGFR. Here, we report that translocation of FGF1 requires activity of the α isoform of p38 mitogen-activated protein kinase (MAPK). FGF1 translocation was inhibited after chemical inhibition of p38 MAPK or after small interfering RNA knockdown of p38α. Translocation was increased after stimulation of p38 MAPK with anisomycin, mannitol, or H2O2. The activity level of p38 MAPK was not found to affect endocytosis or intracellular sorting of FGF1/FGFR1. Instead, we found that p38 MAPK regulates FGF1 translocation by phosphorylation of FGFR1 at Ser777. The FGFR1 mutation S777A abolished FGF1 translocation, while phospho-mimetic mutations of Ser777 to Asp or Glu allowed translocation to take place and bypassed the requirement for active p38 MAPK. Ser777 in FGFR1 was directly phosphorylated by p38α in a cell-free system. These data demonstrate a crucial role for p38α MAPK in the regulated translocation of exogenous FGF1 into the cytosol/nucleus, and they reveal a specific role for p38α MAPK-mediated serine phosphorylation of FGFR1.
Fibroblast growth factor 1 (FGF1) belongs to a family of heparin binding polypeptide growth factors encoded by 22 genes in mice and humans (20). Most FGFs transmit signals to cells by binding and through activation of a family of high-affinity, tyrosine kinase FGF receptors (FGFR1 to -4) (7). FGF1 and FGF2 may, in addition, be translocated from the extracellular space into the cytosol and nucleus of target cells (37, 39, 46, 58). Translocated FGF1 and FGF2, in particular nuclear FGF1 and FGF2, have been reported to be involved in regulating processes such as rRNA synthesis and cell growth (17-19, 21, 36, 44, 45, 52, 54, 56, 61).
The translocation of exogenous FGF1 or FGF2 into the cytosol and nucleus is a highly regulated process that requires phosphatidylinositol 3-kinase (PI3K) activity (23) and active hsp90 (52) and is strictly dependent on binding of FGF to either FGFR1 or FGFR4 (47). Furthermore, translocation was found to be cell cycle dependent (3, 31, 63), it can be stimulated by serum deprivation of cells (1, 3, 18, 25, 31, 32, 55, 63), and it occurs after a several-hour delay compared to the endocytic uptake of FGF (31, 47). The nuclear trafficking of FGF1 is also tightly regulated by two nuclear localization sequences (19, 51), a nuclear export sequence (36), and by phosphorylation of FGF1 at Ser130 by protein kinase Cδ (PKCδ) (57).
The actual translocation of FGF across cellular membranes appears to occur in early endosomes, as it was found to depend on the electrical potential across vesicular membranes (31, 32). Extensive unfolding of the growth factor is not required for the translocation to occur (53). It is not known exactly how FGF crosses the vesicular membrane and to what extent this event involves accessory proteins in addition to FGFR. Previously, we identified certain amino acid residues localized in the C-terminal tail region of FGFR that are crucial for the FGF1 translocation (47).
p38 mitogen-activated protein kinase (MAPK) is a conserved member of the MAPK family and participates in a variety of biological processes. Originally, it was described as a kinase mainly involved in inflammatory and stress-induced responses, often activating proapoptotic stimuli, but it is also involved in regulating growth and differentiation of cells in response to a wide range of growth factors and cytokines (42, 62). More recently it has been shown that p38 MAPK signaling also functions in tumor suppression (15), and it can regulate endocytosis and intracellular sorting. p38 MAPK contributes to regulation of endocytosis by phosphorylation of components of the endocytic machinery, such as GDI-Rab5 (5), EEA1 (8, 28), and rabenosyn 5 (28). Furthermore, it was shown that p38 MAPK can regulate epidermal growth factor receptor (EGFR) internalization (49, 59, 65) and downregulation (9).
Four isoforms of p38 MAPK have been identified in mammals: p38α, -β, -γ, and -δ. p38α and p38β are ubiquitously expressed, whereas p38γ is most prominent in muscle and p38δ is most prominent in lungs and kidneys. p38 MAPK is activated through the sequential activation of MAPK kinase kinases and MAPK kinases (MKKs). MKK3 and MKK6 are known to directly activate p38 MAPK by phosphorylating a Thr-Gly-Tyr dual phosphorylation motif in the activation loop of the p38 kinase subdomain VIII. It has also been shown that the p38α isoform can be activated by alternative mechanisms which involve stimulation of p38α autophosphorylation of its dual phosphorylation motif through interaction with TAB1 (12, 13, 22) or Zap 70 (43). Active p38 MAPK phosphorylates target substrates on serines and threonines, in particular its downstream substrate, MK2/MAPKAP kinase 2, which further activates various substrates, including HSP27, CREB, transcriptional factor ATF1, SRF, and eukaryotic initiation factor 4E (42, 62). p38 MAPK can be activated by FGF/FGFR and has been shown to be a crucial second messenger in regulation of various cellular responses to FGF, such as proliferation, migration, and growth inhibition (2, 11, 27, 29, 30, 34, 40).
In the present study we investigated the role of p38 MAPK in the translocation of exogenous FGF1 into the cytosol and nucleus of cells. We demonstrate that FGF1 translocation requires activity of the α isoform of p38 MAPK and that it can be enhanced upon stimulation of the p38 MAPK activity by chemical stress. We also show that phosphorylation of FGFR1 at Ser777 is required for the translocation of FGF1 and that this phosphorylation is regulated by p38α.
MATERIALS AND METHODS
Materials.
[35S]methionine, [33P]phosphate, and [γ-33P]ATP were obtained from Amersham Biosciences. 125I was from Perkin-Elmer. Heparin-Sepharose was from GE Healthcare Bio-Sciences AB. Complete protease inhibitor cocktail was from Roche Diagnostics. Phosphatase inhibitor cocktail, leptomycin B (LMB), trypsin, mannitol, thapsigargin, digitonin, anisomycin, and 12-O-tetradecanoylphorbol-13-acetate (TPA) were from Sigma-Aldrich. Leupeptin was from the Peptide Institute. PD169316, SB203580, SU5402, SB202474, LY294002, rottlerin, bafilomycin A1, and Gö6976 were from Calbiochem. Antibodies against p38 MAPK (mouse), ERK1/2 (rabbit), p38β MAPK (goat), and MK2 and phospho-MK2 were from Cell Signaling Technology. Anti-p38α MAPK antibody (rabbit) was from BD Transduction Laboratories, and anti-phospho-p38 MAPK antibody (mouse) was from BD Biosciences Pharmingen. Anti-FGF1 antibody (goat), anti-PKCδ (goat) antibody, anti-Sumo-1C19 (goat) antibody, and anti-PKCδ antibody blocking peptide (sc937) were from Santa Cruz Biotechnology. Anti-lamin A (mouse) antibody was from Abcam. Anticalreticulin (rabbit) antibody was from Stressgen. Anti-myelin basic protein (MBP; rabbit) antibody was from AbD Serotec, and MBP was from Sigma-Aldrich. Mouse anti-LAMP-1 antibody was obtained from the Developmental Studies Hybridoma Bank at the University of Iowa. Anti-Rab5 (rabbit) antibody was a gift from Harald Stenmark. Anti-mouse, -rabbit, and -goat horseradish peroxidase-linked and Cy2-labeled anti-mouse secondary antibodies were from Jackson ImmunoResearch Laboratories. Recombinant FGF1 and in vitro-transcribed [35S]methionine-labeled FGF1 (35S-FGF1) were produced as described previously (54). Cy3-FGF1 was made by labeling of FGF1 with Cy3-maleimide (Amersham Biosciences) following the manufacturer's procedure.
Cell cultures.
NIH 3T3 cells, BJ cells, and COS-1 cells were grown in Quantum 333 medium (PAA Laboratories GmbH) supplemented with 100 U/ml penicillin and 100 μg/ml streptomycin, and for NIH 3T3 cells the medium was also supplemented with 2% bovine serum (Gibco). HeLa cells were grown in Dulbecco's modified Eagle's medium (DMEM; Gibco) containing 10% fetal calf serum (FCS; PAA Laboratories GmbH). Cells were seeded into tissue culture plates the day before the start of the experiments.
Plasmids.
The pCDNA3 plasmids encoding full-length human FGFR1 and FGFR4 were described previously (47). The mutations S777A, S777D, S777E, and S777N were introduced into FGFR1 by QuikChange site-directed mutagenesis (Stratagene) and verified by sequencing. The receptors were expressed in COS-1 cells or HeLa cells by transient transfection using FuGENE 6 transfection reagent (Roche).
In vivo phosphorylation of FGF1.
NIH 3T3 cells, or BJ cells treated exactly as NIH 3T3 cells, were first starved for 24 h in DMEM without serum. COS-1 cells were first transfected with the FGFR1 gene of interest by incubation with FuGENE transfection mixture for 6 h. Then, the cells were incubated overnight in phosphate-free DMEM supplemented with 25 μCi/ml [33P]phosphate. After labeling with [33P]phosphate the cells were treated with 10 U/ml heparin and 100 ng/ml recombinant FGF1 for 6 h. The cells were then washed with HEPES medium containing 10 U/ml heparin and once with a high-salt/low-pH buffer (2 M NaCl in 20 mM sodium acetate, pH 4.0) to remove cell surface-bound FGF1 before lysis in lysis buffer (0.1 M NaCl, 10 mM Na2HPO4, 1% Triton X-100, 1 mM EDTA) supplemented with protease and phosphatase inhibitor cocktails. The cell lysate was scraped off the plates. For NIH 3T3 cells, in most cases, the total cell lysate was analyzed for phosphorylated FGF1. In this case the cell lysate was sonicated and the insoluble fraction removed by centrifugation. In some cases the NIH 3T3 cell lysate was fractionated into a nuclear and a cytoplasmic fraction. In this case the lysate was centrifuged at 720 × g for 15 min at 4°C, and the supernatant was centrifuged again for 5 min at 15,800 × g and designated the cytoplasmic (cytosol plus membrane) fraction. The first pellet was washed twice by resuspension in lysis buffer and centrifugation at 720 × g for 15 min at 4°C and then sonicated and centrifuged for 5 min at 15,800 × g. The supernatant after the last centrifugation was designated the nuclear fraction. For COS-1 cells, the cell lysate was centrifuged and only the soluble, cytoplasmic fraction was analyzed further. All fractions were incubated for 2 h at 4°C with heparin-Sepharose to bind the growth factor. The heparin-Sepharose was washed with lysis buffer and treated with 2 μg/ml tosylsulfonyl phenylalanyl chloromethyl ketone-treated trypsin in HEPES medium for 30 min at room temperature. The trypsinization removes most phosphorylated proteins other than FGF1, which is exceptionally resistant to trypsin digestion when bound to heparin. The digestion was terminated by washing with lysis buffer supplemented with protease inhibitors. The bound protein was eluted in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer and subjected to SDS-PAGE and electroblotting onto an Immobilon-P membrane (Millipore). The membrane was first dried and exposed to fluorography to detect 33P-phosphorylated FGF1. Thereafter, the total amount of FGF1 on the same membrane was detected with anti-FGF1 antibody and horseradish peroxidase-coupled secondary antibody.
Digitonin cell fractionation.
NIH 3T3 or BJ cells were serum starved for 24 h, then incubated with 35S-FGF1 and 10 U/ml heparin for 6 h at 37°C, and then washed with high-salt/low-pH buffer to remove FGF1 bound to the cell surface. The cells were then washed with phosphate-buffered saline (PBS), and 20 μg/ml digitonin was added to permeabilize the cells. The cells were kept at 25°C for 5 min and then on ice for an additional 30 min to allow the cytosol to diffuse into the buffer. The buffer was recovered and designated the cytosolic fraction. The remainder of the cells were lysed with lysis buffer, scraped from the plastic, and centrifuged at 15,800 × g for 15 min. The supernatant was designated the membrane fraction, and the pellet was designated the nuclear fraction. The nuclear fraction was resuspended in PBS, sonicated, and centrifuged to remove undissolved material. FGF1 from all fractions was adsorbed to heparin-Sepharose beads. Then, the beads were washed with lysis buffer and the FGF1 was subsequently eluted and analyzed by SDS-PAGE and fluorography.
Assay for PKCδ activity.
Serum-starved NIH 3T3 cells were pretreated for 30 min at 37°C with or without 10 μM SB203580, 10 μM anisomycin, 10 μM PD 169316, 1 μM Gö6976, or 5 μM rottlerin and then treated for 30 min at 37°C with 20 nM TPA. The cells were washed, lysed, and sonicated in lysis buffer containing phosphatase and protease inhibitor cocktails. Then, immunoprecipitation was performed with 2 μg of antibody against PKCδ or Sumo-1 (control) and protein A-Sepharose, in one case in the presence of 3 μg of anti-PKCδ antibody blocking peptide. The immunoprecipitates were incubated in 25 μl of kinase buffer (25 mM HEPES, pH 7.0, 20 mM MgCl2, 150 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol, and phosphatase and protease inhibitor cocktails) containing 20 nM TPA for 30 min at 37°C with 2.5 μg of MBP and 30 μCi of [γ-33P]ATP in the absence or presence of 10 μM SB203580, 10 μM anisomycin, 10 μM PD169316, 1 μM Gö6976, or 5 μM rottlerin. Finally, the samples were centrifuged, and collected supernatants were subjected to SDS-PAGE, fluorography, and immunoblotting.
siRNA design and transfection.
Three small interfering RNAs (siRNAs) targeting mouse p38α mRNA, two siRNAs targeting human p38α mRNA, and two siRNAs targeting human p38β mRNA were designed. All constructs were checked in silico for target mRNA specificity by BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST/), and their nucleotide composition was fitted as closely as possible to the criteria depicted in reference 41. Sequences against mouse p38α mRNA were GAACGUUGUUUCCUGGUACTT (α1), GUAUAUACAUUCGGCUGACAU (α2), and GAUGCUCGUUUUGGACUCAG (α3). Sequences against human p38α (α1 and α2) and p38β mRNA (β1 and β2) were the same as previously published (50). Constructs were ordered as high-performance liquid chromatography purified from MWG Biotech with TT overhangs at the 3′ end of each strand. A negative control siRNA was obtained from Eurogentec. For siRNA transfection studies, NIH 3T3 cells or BJ cells (1 × 105 cells/ml) were seeded in the absence of penicillin and streptomycin. After 24 h the cells were transfected with 75 nM of the indicated siRNA using Oligofectamine transfection reagent (Invitrogen) according to the procedure given by the company. Four hours after transfection, 10% FCS, 100 U/ml penicillin, and 100 U/ml streptomycin were added to the cells, and the cells were left for 72 h.
Receptor binding assay.
Binding of FGF1 to cell surface receptors was performed essentially as published previously (60). Confluent NIH 3T3 cells were washed with ice-cold binding buffer (HEPES medium, 10 U/ml heparin), incubated with inhibitors for 30 min, and then incubated with increasing concentrations of 125I-labeled FGF1 (125I-FGF1) in the absence or presence of different inhibitors for 3 h at 4°C. Subsequently, the cells were washed twice with ice-cold binding buffer and once with ice-cold PBS, followed by the addition of ice-cold 1 M NaCl in PBS to dissociate the growth factor from heparan sulfate proteoglycans at the cell surface. After washing, the cells were lysed in 0.1 M KOH, and the solubilized radioactivity, representing the cell surface FGFR-bound 125I-FGF1, was measured using a gamma counter.
Endocytosis of 35S-FGF1.
NIH 3T3 cells (2 × 105/ml) were preincubated in the absence or presence of 20 μM anisomycin for 15 min at 37°C. Next, the cells were incubated with 5 ng/ml of 35S-FGF1 and 10 U/ml heparin at 4°C for 2 h. Then, the cells were washed extensively with HEPES medium containing 10 U/ml heparin. To measure binding to cell surface receptors, the cells were washed with 1 M NaCl in PBS and lysed at this stage. To measure endocytosis, the cells were incubated further for 0, 15, 30, or 60 min at 37°C in the presence or absence of 20 μM anisomycin. Next, the cells were washed twice with high-salt/low-pH buffer and once with HEPES medium and lysed. 35S-FGF1 was extracted from the cell lysates by binding to heparin-Sepharose and subjected to SDS-PAGE and fluorography.
Microscopy.
HeLa cells were transiently transfected with FGFR1 or FGFR4. Cy3-FGF1 (100 ng/ml) was bound to the cell surface by incubation at 4°C in HEPES medium in the presence of heparin (50 U/ml) and in the presence or absence of SB203580 (10 μM) or anisomycin (10 μM). The cells were then washed with PBS and incubated further for 2 h at 37°C in DMEM with 10% FCS and 0.3 mM leupeptin and in the presence or absence of SB203580 or anisomycin. The cells were then fixed with Formalin solution (10%; Sigma-Aldrich), permeabilized with 0.05% saponin, and stained with mouse anti-LAMP-1 antibody and Cy2-labeled anti-mouse antibody. The cells were examined with a Zeiss LSM Duo confocal microscope. Images were prepared with the Zeiss LSM image browser (version 3.2.0.115) and CorelDraw11. Images of randomly chosen cells were examined. The proportion of red structures, indicating internalized Cy3-FGF1, which colocalized with LAMP-1-positive (green) structures, was calculated. The means and standard deviations were calculated from 15 cells in each case.
In vitro phosphorylation of the FGFR1 C-terminal tail.
In vitro phosphorylation experiments were performed with recombinant human p38α MAPK and its inactive form (R&D Systems). Recombinant fusion proteins consisting of the 68 most C-terminal amino acids from FGFR1 fused to the C-terminal end of glutathione S-transferase (GST) were produced in bacteria and purified with glutathione-Sepharose (Amersham Biosciences) according to standard procedures. One μg of the fusion proteins was incubated with p38α kinase and 40 μCi/ml [γ-33P]ATP in reaction buffer (25 mM HEPES, pH 7.5, 20 mM MgCl2, 1 mM Na2MO3, 20 mM sodium β-glycerophosphate, 1 mM dithiothreitol, 5 mM EGTA) at 30°C for 30 min in the presence or absence of SB203580. The reaction was stopped by trichloroacetic acid precipitation (30 min on ice). Then, the samples were centrifuged, washed twice with cold acetone, and resuspended in sample buffer. The proteins were analyzed by SDS-PAGE, electroblotting, and autoradiography, and then the membrane was stained with Coomassie blue.
RESULTS
p38 MAPK activity is required for translocation of exogenous FGF1 into cells.
To test for the transport of FGF1 from the cell surface and into the cytosol and nucleus, we have developed an in vivo FGF1 phosphorylation assay which has been extensively described and validated in earlier studies (23, 25, 31, 32, 36, 47, 52, 57). In this assay the translocation of exogenously added FGF1 into the cytosol and nucleus of the cells is monitored by phosphorylation of the growth factor. FGF1 contains only one functional phosphorylation site, a PKC site at Ser130. PKC is only found in the cytosol and nucleus, and therefore, phosphorylation of externally added FGF1 can be taken as evidence that the growth factor has reached the cytosol or the nucleus.
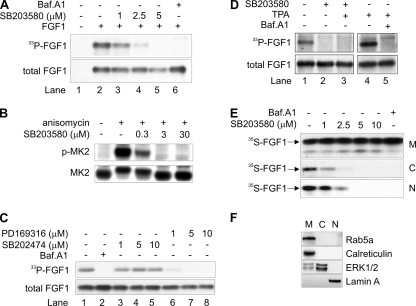
To test if p38 MAPK activity is required for translocation of exogenous FGF1 to the cytosol and nucleus, we first studied the effect of a specific low-molecular-weight inhibitor of p38 MAPK, SB203580 (6, 35), on in vivo phosphorylation of FGF1. NIH 3T3 cells, which are known to express FGFR1, were first allowed to accumulate [33P]phosphate, then incubated with FGF1 for 6 h, and then the intracellularly accumulated FGF1 was extracted from total cell lysates as described in Materials and Methods, run on an SDS-PAGE gel, and electroblotted onto a membrane. FGF1 that was radiolabeled by in vivo phosphorylation (33P-FGF1, representing FGF1 translocated to cytosol/nucleus) was visualized by fluorography as shown in Fig. 1A, upper panel. This shows that phosphorylation of FGF1 was reduced upon treatment with 2.5 μM of SB203580 (lane 4) and was completely abolished at 5 μM (lane 5). No significant effect was observed on the total cellular uptake of FGF1 (total FGF1, largely derived from endosomes), which was visualized by anti-FGF1 immunodetection (Fig. 1A, lower panel) on the same membrane as shown in the upper panel.
FIG. 1.
Inhibition of p38 MAPK inhibits translocation of FGF1 to the cytosol and nucleus. (A) In vivo phosphorylation of exogenously added FGF1 in NIH 3T3 cells was analyzed in the absence or presence of micromolar concentrations of SB203580 or 10 nM bafilomycin A1 (Baf.A1), as indicated. In vivo-phosphorylated FGF1 (33P-FGF1) was detected by fluorography, and the total cellular uptake of FGF1 (total FGF1) was detected by anti-FGF1 immunodetection. (B) NIH 3T3 cells were incubated with or without 5 μM anisomycin in the absence or presence of micromolar concentrations of SB203580 as indicated. Total cell lysates were analyzed by Western blotting using an anti-phospho-MK2-specific antibody (p-MK2) and an anti-total MK2 antibody. (C) The in vivo phosphorylation of FGF1 was analyzed in the presence of micromolar concentrations of PD169316 or SB202474 or 10 nM bafilomycin A1 as indicated. (D) In vivo phosphorylation of FGF1 was analyzed in the absence or presence of 5 μM SB203580, 20 nM TPA, or 10 nM bafilomycin A1 as indicated. (E) NIH 3T3 cells were incubated with in vitro-labeled 35S-FGF1 and heparin for 6 h in the absence or presence of micromolar concentrations of SB203580 or 10 nM bafilomycin A1 as indicated. The cells were fractionated into membrane (M), cytosolic (C), and nuclear (N) fractions. FGF1 was extracted from each fraction by binding to heparin-Sepharose and analyzed by SDS-PAGE and fluorography. (C and N fractions were exposed to film four times longer than the M fractions.) (F) The subcellular fractions obtained by fractionation as for panel E were tested for the presence of the membrane-associated proteins Rab5a and calreticulin, the cytosolic protein ERK1/2, and the nuclear protein lamin A by specific antibodies.
Phosphorylation of FGF1 was also blocked by bafilomycin A1 (Fig. 1A, lane 6), which prevents FGF1 translocation due to inhibition of the vesicular proton pump required to generate the vesicular membrane potential, as shown previously (31, 32). Bafilomycin A1 in the following experiments was used as a negative control.
Three micromolar SB203580 was sufficient to completely block the activity of p38 MAPK, as shown in Fig. 1B, where NIH 3T3 cells were treated with anisomycin, a well-known efficient stimulator of p38 MAPK (4, 64), and with increasing concentrations of SB203580. The activity of p38 MAPK in total cell lysates was measured by the level of phosphorylated MK2, a substrate of p38 MAPK.
We also tested if another inhibitor of p38 MAPK, PD169316, and an inactive analogue of SB203580, SB202474, were able to inhibit FGF1 phosphorylation. As demonstrated in Fig. 1C, SB202474 had no inhibitory effect (lanes 3 to 5), while PD169316 reduced phosphorylation of FGF1 already at 1 μM (lanes 6 to 8).
In order to test the possibility that inhibition of p38 MAPK affects only phosphorylation of the translocated FGF1 and not the translocation process as such, we studied phosphorylation of FGF1 in cells that were treated with SB203580 and TPA (Fig. 1D). TPA strongly stimulates the activity of cytosolic and nuclear PKCδ, which is the enzyme that phosphorylates FGF1 (57) (Fig. 1D, lane 4). However, the TPA treatment did not overcome the inhibitory effect of SB203580 in the FGF1 phosphorylation assay (lane 3), indicating that SB203580 inhibits the translocation of FGF1 to the cytosol and nucleus.
Further evidence for this was obtained in experiments where translocation of FGF1 to the cytosol and nucleus was studied by cell fractionation. We employed a digitonin-based fractionation assay (52, 53, 57) where translocation of exogenously added, in vitro-labeled FGF1 (35S-FGF1) to the cytosol and nucleus is monitored in a manner unrelated to the phosphorylation status of FGF1. NIH 3T3 cells were incubated with 35S-FGF1 for 6 h and then fractionated into a cytosolic, a nuclear, and a membrane fraction as described in Materials and Methods. The membrane fraction includes growth factor present in intracellular vesicles (endosomes) and can here be considered a loading control, as the largest part of the cell-associated FGF1 is present in the endosomes at the time of cell lysis. In the absence of inhibitors, labeled growth factor was detected in all three fractions (Fig. 1E). Also in this assay, translocation of FGF1 to the cytosol and nucleus was blocked by bafilomycin A1. Upon treatment of the cells with increasing concentrations of SB203580, the amount of 35S-FGF1 in the nuclear and cytosolic fractions, but not in the membrane fraction, decreased in a concentration-dependent manner and was undetectable at 5 μM SB203580. Thus, treatment of cells with the p38 MAPK inhibitor SB203580 clearly inhibits the translocation of FGF1 to the cytosol and nucleus. As shown in Fig. 1F, the cytosolic and nuclear fractions obtained by the digitonin fractionation assay were devoid of calreticulin and Rab5a, which are marker proteins for the membrane fraction of cells.
Increased FGF1 translocation at increased cellular activity of p38 MAPK.
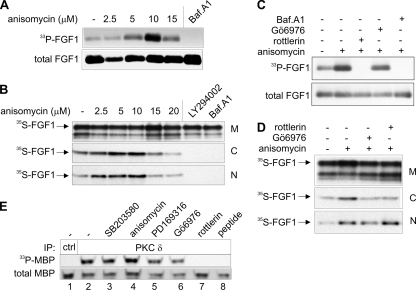
Since inhibition of p38 MAPK blocked translocation of FGF1 into cells, we tested if increased p38 MAPK activity, stimulated by anisomycin, would increase translocation. As shown in Fig. 2A, increasing amounts of anisomycin increased the amount of in vivo-phosphorylated FGF1. Furthermore, in a digitonin fractionation experiment, more 35S-FGF1 was detected in the nuclear and the cytosolic fractions of cells treated with anisomycin than in control cells (Fig. 2B). Quantitation of the amount of cytosolic and nuclear 35S-FGF1 by phosphorimager analysis of several repeated experiments showed that 10 μM anisomycin increased FGF1 translocation by two- to fourfold. At higher concentrations of anisomycin (15 μM and 20 μM) the enhancing effect on FGF1 translocation was not observed, probably due to the toxicity of the compound, which inhibits protein synthesis. FGF1 translocation was completely inhibited by the PI3K inhibitor LY294002, as reported previously (23), and by bafilomycin A1. These results indicate that an increased activity of p38 MAPK in the cells is able to increase the translocation of FGF1 into the cytosol and nucleus.
FIG. 2.
Activation of p38 MAPK by anisomycin enhances translocation of FGF1. (A) In vivo phosphorylation of exogenously added FGF1 in NIH 3T3 cells was analyzed in the presence of increasing micromolar concentrations of anisomycin or 10 nM bafilomycin A1 (Baf.A1) as indicated. In vivo phosphorylated FGF1 (33P-FGF1) was detected by fluorography, and the total cellular uptake of FGF1 (total FGF1) was detected by anti-FGF1 immunodetection. (B) NIH 3T3 cells were incubated with in vitro-labeled 35S-FGF1 and heparin for 6 h in the presence of increasing concentrations of anisomycin as indicated. The cells were fractionated into membrane (M), cytosolic (C), and nuclear (N) fractions. FGF1 was extracted from each fraction by binding to heparin-Sepharose and analyzed by SDS-PAGE and fluorography. (C and N fractions were exposed to film four times longer than the M fractions.) (C) In vivo phosphorylation of FGF1 was analyzed in the absence or presence of 10 μM anisomycin, 1 μM Gö6976, 5 μM rottlerin, or 10 nM bafilomycin A1 as indicated. (D) NIH 3T3 cells were treated as explained for panel B but in the absence or presence of 10 μM anisomycin, 1 μM Gö6976, or 5 μM rottlerin as indicated. (E) The activity of PKCδ in the presence of various inhibitors was tested in vitro. Lysates of NIH 3T3 were subjected to immunoprecipitation (IP) using anti-Sumo-1 antibody (ctrl; lane 1) or anti-PKCδ antibody (lanes 2 to 7) in the absence (lane1 to 6) or presence (lane 7) of anti-PKCδ antibody-blocking peptide. The immunoprecipitated material was incubated in kinase buffer with MBP and [γ-33P]ATP, in the presence or absence of 10 μM SB203580, 10 μM anisomycin, 10 μM PD169316, 1 μM Gö6976, or 5 μM rottlerin, and thereafter the samples were analyzed by SDS-PAGE, fluorography (33P-MBP), and Western blotting using anti-MBP antibody (total MBP).
We tested if the translocated FGF1 was modified by PKCδ, as previously reported (57), also during treatment with anisomycin. Figure 2C shows that FGF1 was not phosphorylated during treatment of the cells with 10 μM anisomycin and 5 μM rottlerin, a specific inhibitor of PKCδ (14). On the other hand, in the presence of 10 μM anisomycin and 1 μM Gö6976, a specific inhibitor of PKCα (33), the phosphorylation of FGF1 was not inhibited. Using the digitonin fractionation assay we found that the tested PKC inhibitors did not significantly reduce translocation of 35S-FGF1 (Fig. 2D). This indicates that FGF1 is phosphorylated only by PKCδ, even during hyperactivation of p38 MAPK. Furthermore, as shown in Fig. 2E, in vitro phosphorylation of the PKCδ substrate MBP by PKCδ immunoprecipitated from TPA-stimulated cells was not inhibited by SB203580, PD169316, or Gö6976, indicating that the PKCδ activity and FGF1 in vivo phosphorylation should be unaffected by these compounds.
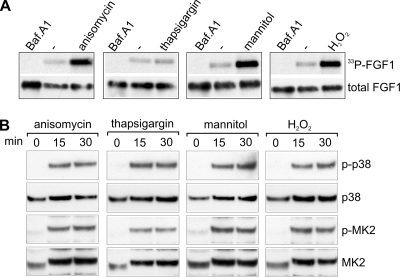
Previously, it was shown that p38α activation by thapsigargin requires MKK3/6, whereas anisomycin, sorbitol (hyperosmolarity), and peroxyhydrogen can activate p38α also by an MKK3/6-independent pathway (22). To test if the increased FGF1 translocation obtained with increased p38 MAPK activity correlates with a specific activation pathway for p38 MAPK, we carried out the FGF1 phosphorylation assay in the presence of anisomycin, thapsigargin, mannitol (hyperosmolarity), and H2O2. As shown in Fig. 3A, anisomycin, mannitol, and H2O2 clearly increased the amount of phosphorylated FGF1, while in the case of thapsigargin there was no significant increase. Figure 3B demonstrates that each of these compounds was able to induce hyperphosphorylation of p38 MAPK and phosphorylation of its downstream substrate, MK2. Thus, activation of p38 MAPK by different types of chemical stress that can activate p38 MAPK by an MKK3/6-independent pathway enhances FGF1 translocation.
FIG. 3.
Effect of activation of p38 MAPK by anisomycin, thapsigargin, mannitol, and H2O2 on translocation of FGF1. (A) In vivo phosphorylation of exogenously added FGF1 in NIH 3T3 cells was analyzed in the absence or presence of 10 μM anisomycin, 1 μg/ml thapsigargin, 0.5 M mannitol, 250 μM H2O2, or 10 nM bafilomycin A1 (Baf.A1) as indicated. In vivo-phosphorylated FGF1 (33P-FGF1) was detected by fluorography, and the total cellular uptake of FGF1 (total FGF1) was detected by anti-FGF1 immunodetection. (B) NIH 3T3 cells were treated with 10 μM anisomycin, 1 μg/ml thapsigargin, 0.5 M mannitol, or 250 μM H2O2 for 0, 15, or 30 min as indicated. Cell lysates were analyzed by SDS-PAGE and Western blotting using antibodies specific for phosphorylated p38 MAPK (p-p38), total p38 MAPK (p38), phosphorylated MK2 (p-MK2), and total MK2 as indicated.
Knockdown of p38α by siRNA inhibits FGF1 translocation.
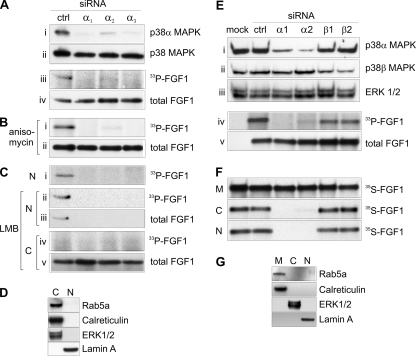
To further test if inhibition of the activity of p38 MAPK is the direct cause of the inhibition of FGF1 translocation by SB203580, and to elucidate which isoform of the p38 MAPK is involved, we specifically knocked down the expression of the α isoform of p38 MAPK in NIH 3T3 cells by three different variants of siRNA (α1, α2, and α3) (see Materials and Methods). A scrambled siRNA pool was used as a control. As demonstrated in Fig. 4, p38α siRNA selectively knocked down expression of p38α, while the total amount of p38 MAPK (all isoforms) was little affected, as determined by immunoblotting using anti-p38α or anti-p38 MAPK antibodies, respectively (Fig. 4A, panels i and ii). We next examined if knockdown of p38α affected the ability of the cells to translocate FGF1 in an FGF1 phosphorylation assay. FGF1 was not phosphorylated in cells treated with p38α siRNA (Fig. 4A, panel iii). No effect on the total cellular uptake of FGF1 was observed for any siRNA treatment (panel iv).
FIG. 4.
siRNA knockdown of p38α inhibits translocation of FGF1. (A to C) NIH 3T3 cells were transfected with control siRNA (ctrl) or three different siRNAs specific for mouse p38α (α1, α2, and α3). (A) Total cell lysates were analyzed for p38α (i) and total p38 MAPK (ii) by Western blotting. In vivo phosphorylation of FGF1 was analyzed in the siRNA-transfected cells. Phosphorylated FGF1 (33P-FGF1) was detected by fluorography (iii), and the total cellular uptake was detected by anti-FGF1 immunodetection (total FGF1 [iv]). (B) In vivo phosphorylation of FGF1 was analyzed in the siRNA-transfected cells in the presence of 10 μM anisomycin. (C) In vivo phosphorylation of FGF1 was analyzed in the siRNA-transfected cells in the absence (i) or presence (ii to v) of LMB, and the cells were fractionated into nuclear (N) and cytoplasmic (C) fractions before extraction of FGF1. (D) Nuclear and cytoplasmic fractions of NIH 3T3 cells obtained by the cellular fractionation method described for panel C were tested for the presence of the cytoplasmic proteins rab5a, calreticulin, and ERK1/2 and the nuclear protein lamin A by specific antibodies. (E and F) BJ cells were mock transfected or transfected with control siRNA (ctrl), two different siRNAs specific for human p38α (α1 and α2), or two different siRNAs specific for human p38β (β1 and β2). (E) The transfected cells were lysed and tested for the amount of p38α (i), p38β (ii), and ERK1/2 (loading control, [iii]) by immunoblotting. Translocation of FGF1 was tested in the transfected cells in an in vivo FGF1 phosphorylation assay. Phosphorylated FGF1 (33P-FGF1) was detected by fluorography (iv), and the total cellular uptake was detected by anti-FGF1 immunodetection (total FGF1 [v]). No FGF1 was added in the first lane, panels iv and v. (F) siRNA-transfected BJ cells were incubated with in vitro-labeled 35S-FGF1 and heparin for 6 h, and then the cells were fractionated into membrane (M), cytosolic (C), and nuclear (N) fractions. FGF1 was extracted from each fraction by binding to heparin-Sepharose and analyzed by SDS-PAGE and fluorography. (C and N fractions were exposed to film four times longer than the M fractions.) (G) The subcellular fractions obtained by fractionation as described for panel F were tested for the presence of the membrane-associated proteins Rab5a and calreticulin, the cytosolic protein ERK1/2, and the nuclear protein lamin A by specific antibodies.
Since anisomycin is an efficient stimulator of p38 MAPK and can enhance FGF1 translocation, we examined if this drug had an effect on phosphorylation of FGF1 in cells with knocked down p38α. As shown in Fig. 4B, even under anisomycin treatment we were unable to detect phosphorylated FGF1 in p38α knockdown cells.
We have previously shown that FGF1 is phosphorylated in the nucleus and then rapidly exported to the cytosol, where it is apparently degraded. However, in the presence of LMB the growth factor is trapped in the nucleus and thereby protected from degradation in the cytosol (57). To test the possibility that phosphorylated FGF1 was not detected in p38α knockdown cells due to rapid transport to the cytosol, we carried out FGF1 phosphorylation experiments in p38α knockdown cells in the presence of LMB and fractionated the cells into cytoplasmic and nuclear fractions. Also under LMB treatment we were unable to detect phosphorylated FGF1 in p38α knockdown cells, while phosphorylated FGF1 was easily detected in the nuclear fraction obtained from control (mock-transfected) cells (Fig. 4C, panels i and ii). By using anti-FGF1, nuclear FGF1 could be detected in mock-treated cells, but not in p38α knockdown cells (panel iii), demonstrating that translocation of FGF1, and not only phosphorylation of the growth factor, is inhibited in the presence of p38α siRNA. With LMB treatment, phosphorylated FGF1 was not detected in the cytoplasmic fraction, as expected (panel iv), while the total amount of FGF1 in the cytoplasmic fraction was similar for all treatments (panel v). Figure 4D shows that the nuclear fraction obtained by the fractionation procedure used in Fig. 4C is not contaminated with marker proteins for the cytoplasmic fraction of cells.
We were not able to successfully measure knockdown of p38β in NIH 3T3 cells, and we therefore carried out siRNA knockdown of p38 MAPK also in BJ cells, a human fibroblast cell line. The BJ cells were found to behave similarly to NIH 3T3 cells with respect to translocation of FGF1 and sensitivity to SB203580 and anisomycin (data not shown). BJ cells were transfected with two different siRNAs against human p38α (α1 and α2), two siRNAs against human p38β (β1 and β2), control siRNA, or mock transfected (no RNA) (Fig. 4E and F). Considerable knockdown of p38α as well as p38β was obtained with the specific siRNAs (Fig. 4E, panels i and ii). Translocation of FGF1 was studied in the siRNA-transfected cells by the in vivo FGF1 phosphorylation method (Fig. 4E, panels iv and v) and by the digitonin fractionation method (Fig. 4F). Knockdown of p38α, but not p38β, clearly inhibited the translocation of FGF1 to the cytosol and nucleus. Figure 4G demonstrates the purity of cytosolic and nuclear fractions obtained by digitonin-based fractionation of the BJ cells.
These experiments indicated that p38α MAPK, and not p38β MAPK, is required for translocation of exogenous FGF1 into the cytosol and nucleus.
Effect of serum starvation and FGF1 stimulation on p38 MAPK activity in NIH 3T3 cells.
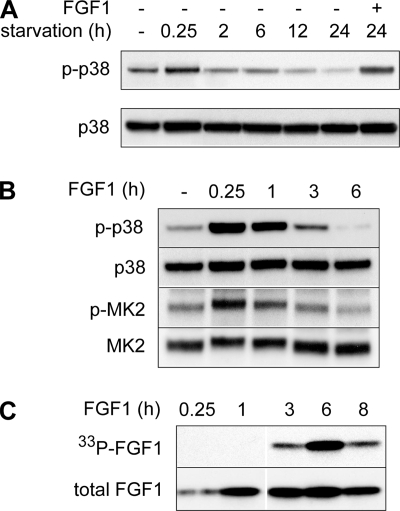
To investigate the level of p38 MAPK activity necessary for FGF1 translocation, we monitored the level of phosphorylated p38 MAPK (p-p38) in NIH 3T3 cells at different time points. To enhance FGF1 translocation in NIH 3T3 cells, we routinely deprive the cells of serum for 24 h prior to stimulation with FGF1 (25, 31). As shown in Fig. 5A, serum deprivation induced a brief increase in p-p38 (at 0.25 h), but thereafter the level of p-p38 declined gradually during the course of serum starvation. After 24 h of serum starvation, the level of p-p38 was considerably lower than the level found in NIH 3T3 cells grown in medium containing serum.
FIG. 5.
Effect of serum starvation and FGF1 stimulation on the activity of p38 MAPK. (A) NIH 3T3 cells were incubated for the indicated length of time in serum-free medium. In one case the cells were starved for 24 h and thereafter stimulated with FGF1 and heparin for 15 min. Total cell lysates were analyzed for phosphorylated p38 MAPK (p-p38) and total p38 MAPK by Western blotting. (B) NIH 3T3 cells were serum starved for 24 h and then stimulated with FGF1 and heparin for the indicated periods of time. Total cell lysates were analyzed for phosphorylated p38 MAPK (p-p38), total p38 MAPK (p38), phosphorylated MK2 (p-MK2), and total MK2 by Western blotting, as indicated. (C) In vivo phosphorylation of exogenously added FGF1 in NIH 3T3 cells was analyzed after incubation of the cells with FGF1 for the indicated length of time. In vivo-phosphorylated FGF1 (33P-FGF1) was detected by fluorography, and the total cellular uptake of FGF1 (total FGF1) was detected by anti-FGF1 immunodetection.
Treatment of serum-starved NIH 3T3 cells with FGF1 stimulated phosphorylation of p38 MAPK (Fig. 5A and B), conceivably through activation of FGFR1 as described elsewhere (11, 60). However, the FGF1-induced increase in p-p38, as well as phosphorylation of the p38 MAPK substrate MK2, declined to a relatively low level after 3 h (Fig. 5B). Translocation of FGF1 into the cytosol and nucleus is known to be a rather slow process and peaks approximately 6 h after addition of FGF1 to the cells (Fig. 5C), as shown previously (31, 47, 53). At this time point the total cellular activity level of p-p38 was very low (Fig. 5B), indicating that a highly activated state of p38 MAPK is not required during the FGF1 translocation process.
Lack of regulation of the endosomal sorting of endocytosed FGF1 by p38 MAPK.
Most likely, FGF1 is translocated from endosomes (32). Since p38 MAPK has been shown to regulate components of the endocytic machinery as well as EGFR endocytosis, we investigated if p38 MAPK regulates the endosomal uptake and intracellular sorting of FGF1.
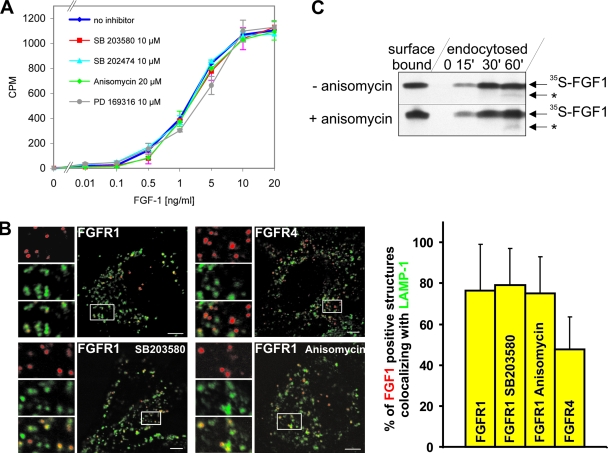
By measuring binding of 125I-labeled FGF1 to the surface of NIH 3T3 cells that were pretreated with 10 μM SB203580, 10 μM SB202474, 20 μM anisomycin, or 10 μM PD169316, we found that none of these compounds had any effect on the apparent number of binding sites for FGF1 available at the cell surface (Fig. 6A).
FIG. 6.
Effect of activation and inhibition of p38 MAPK on endocytosis and intracellular sorting of FGF1. (A) NIH 3T3 cells were incubated with SB203580, SB202474, anisomycin, or PD169316 and increasing concentrations of 125I-labeled FGF1 for 3 h at 4°C. The cells were washed with 1 M NaCl and lysed, and the solubilized radioactivity, representing the cell surface FGFR-bound 125I-FGF1, was measured (reported as counts per minute [CPM]). (B) HeLa cells transfected with FGFR1 or FGFR4 were incubated with Cy3-FGF1 and heparin in the presence or absence of SB203580 (10 μM) or anisomycin (10 μM) for 2 h at 37°C in DMEM with 0.3 mM leupeptin. The cells were fixed, stained for LAMP-1 (Cy2), and examined by confocal microscopy. Bar, 5 μm. Images shown are merged images (red plus green) where a section of each image has been magnified and shown to the left as red, green, and merged images separately. The proportion of Cy3-positive structures that colocalized with Cy2-positive structures (indicated by yellow in the merged images) was calculated and presented as the mean from 15 cells in each case. Error bars indicate the standard deviations. (C) NIH 3T3 cells were preincubated with or without 20 μM anisomycin for 15 min at 37°C and then incubated with 35S-FGF1 and heparin at 4°C for 2 h. The cells were washed with 1 M NaCl and lysed to measure surface-bound FGF1 or, to measure endocytosis, incubated further for 0, 15, 30, or 60 min at 37°C in the presence and absence of 20 μM anisomycin. 35S-FGF1 was extracted from cell lysates and subjected to SDS-PAGE and fluorography. *, partially degraded FGF1.
Previously, we have shown that after endocytosis of FGF1/FGFR1, FGFR1 follows mainly the endocytic route to lysosomes, while FGFR4 is to a larger extent sorted to the endosomal recycling compartment (16). We investigated the trafficking of Cy3-FGF1 in cells expressing FGFR1 in the presence of 10 μM SB203580 or 10 μM anisomycin and compared it to the normal trafficking of FGFR1 and FGFR4. HeLa cells transfected with FGFR1 or FGFR4 were incubated with Cy3-FGF1, first for 1 h on ice and then for 2 h at 37°C, and then they were fixed and stained for LAMP-1 as a marker for the lysosomal pathway. Then, the colocalization between LAMP-1 and Cy3-FGF1 in vesicular structures was quantitated as described in Materials and Methods. As shown in Fig. 6B, there was no significant difference in the colocalization with LAMP-1 for Cy3-FGF1 endocytosed as a complex with FGFR1 in the absence or presence of SB203580 or anisomycin. In contrast, Cy3-FGF1 endocytosed as a complex with FGFR4 showed much less colocalization with LAMP-1.
Anisomycin has been reported to stimulate endosomal uptake of EGFR (49, 65). We therefore measured the uptake and intracellular stability of in vitro-labeled 35S-FGF1 in the presence of anisomycin. NIH 3T3 cells were preincubated in the absence and presence of anisomycin for 15 min at 37°C. Then, 35S-FGF1 was bound to NIH 3T3 cells at 4°C and its endocytosis was measured after 15, 30, or 60 min in the presence or absence of 20 μM anisomycin at 37°C. As shown in Fig. 6C, anisomycin did not significantly change the efficiency of cell surface binding, endocytic uptake, or the intracellular degradation of 35S-FGF1 (Fig. 6C).
Thus, varying the activity level of p38 MAPK does not seem to alter significantly the endosomal uptake and intracellular sorting pattern of FGF1/FGFR1.
FGF1 translocation depends on p38 MAPK-regulated phosphorylation of Ser777 in FGFR1.
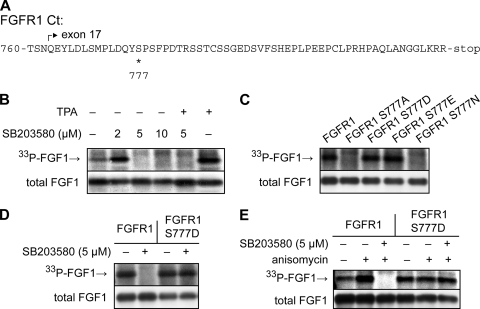
We have previously shown that in COS-1 cells the translocation of FGF1 is crucially dependent on the C-terminal tail (Ct) of the FGFR (47), which is composed of 59 amino acids and encoded by exon 17 in FGFR1. This Ct is rich in phosphorylatable amino acid residues, particularly serines (Fig. 7A), and we therefore investigated the possibility that the function of this Ct is regulated by phosphorylation. We constructed several FGFR1 mutants that were expressed in COS-1 cells.
FIG. 7.
Effects of inhibition of p38 MAPK and mutations in FGFR1 on translocation of FGF1 in COS-1 cells. (A) Amino acid sequence of the Ct of FGFR1. The start of exon 17 is marked by an arrow, and Ser777 is marked by an asterisk. (B) In vivo phosphorylation of exogenously added FGF1 in COS-1 cells transfected to express FGFR1 was analyzed in the absence or presence of TPA (20 nM) or micromolar concentrations of SB203580. In vivo-phosphorylated FGF1 (33P-FGF1) was detected by fluorography, and the total cellular uptake of FGF1 (total FGF1) was detected by anti-FGF1 immunodetection. (C) COS-1 cells were transfected with wild-type FGFR1 or with FGFR1 with point mutations, as indicated, and then the in vivo phosphorylation of FGF1 (33P-FGF1) and the total cellular uptake (total FGF1) were analyzed. (D and E) COS-1 cells were transfected with wild-type FGFR1 or FGFR1 S777D, and then the in vivo phosphorylation of FGF1 (33P-FGF1) and the total cellular uptake (total FGF1) was analyzed in the absence or presence of 5 μM SB203580 (D) and in the absence or presence of 5 μM anisomycin and 5 μM SB203580 as indicated (E).
We first tested if translocation of FGF1 in COS-1 cells expressing FGFR1 required p38 MAPK activity, as observed in NIH 3T3 cells. As shown in Fig. 7B, phosphorylation of FGF1 was completely inhibited by 5 μM of SB203580, and this inhibition could not be overcome by stimulation of PKC with TPA, indicating that SB203580 abolished the translocation of FGF1.
Comparison of the FGFR1 Ct to known consensus sites for phosphorylation suggested that Ser777 could be a MAPK phosphorylation site. When Ser777 was mutated to alanine (FGFR1 S777A), the ability of the receptor to mediate FGF1 translocation was abolished (Fig. 7C), possibly because a phosphate group at the 777 position is required. To test this, we mutated the 777 residue to aspartic acid and glutamic acid, which by their acidic charge may mimic a phospho group, and to asparagine, which is not charged but has a similar structure as aspartic acid. We found that FGFR1 S777D and FGFR1 S777E, but not FGFR1 S777N, mediated FGF1 translocation (Fig. 7C), suggesting that phosphorylated Ser777 in FGFR1 is required for FGF1 translocation. The lower panel in Fig. 7C shows that the various receptors were similarly expressed and functional in endocytosis of FGF1, as the total cellular uptake of FGF1 (total FGF1), which is largely endocytosed FGF1, was similar.
If p38 MAPK regulated FGF1 translocation through phosphorylation of Ser777 in FGFR1, one would expect that translocation mediated by FGFR1 S777D, which mimics a receptor that is constitutively phosphorylated at the 777 position, would not require p38 MAPK activity. Indeed, we found that translocation of FGF1 by FGFR1 S777D was not inhibited by 5 μM SB203580 (Fig. 7D). This suggests that p38 MAPK-regulated phosphorylation of FGFR1 at Ser777 is an important event in the regulation of translocation of FGF1 into cells. Whereas anisomycin could enhance FGF1 translocation by wild-type FGFR1 also in COS-1 cells, we did not observe a similar increase in FGF1 translocation when FGFR1 S777D-expressing cells were stimulated with anisomycin (Fig. 7E). This is consistent with the idea that the enhancing effect of anisomycin on FGF1 translocation by FGFR1 is primarily due to enhanced p38 MAPK activity and enhanced phosphorylation of Ser777.
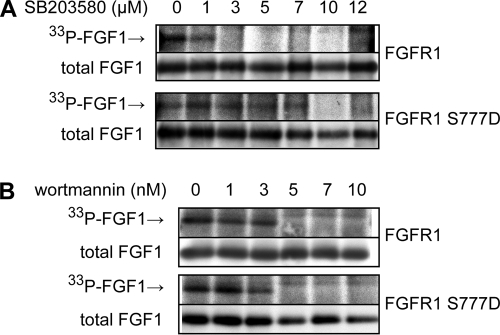
To investigate the sensitivity of FGFR1 and FGFR1 S777D to SB203580 in more detail, we performed an FGF1 phosphorylation experiment where the concentration of SB203580 was carefully titrated. Whereas translocation of FGF1 by FGFR1 was strongly inhibited at 3 μM of SB203580, translocation of FGF1 by FGFR1 S777D was unaffected by up to 7 μM of SB203580 (Fig. 8A). At 10 μM SB203580, however, translocation mediated by FGFR1 S777D was also inhibited. In contrast, FGFR1 and FGFR1 S777D exhibited equal sensitivity to wortmannin (Fig. 8B), a compound that inhibits the activity of PI3K, which was previously found to be required for FGF1 translocation (23).
FIG. 8.
Inhibitory effect of increasing concentrations of SB203580 and wortmannin on translocation of FGF1 in COS-1 cells expressing wild-type FGFR1 or FGFR1 S777D. COS-1 cells were transfected with wild-type FGFR1 or FGFR1 S777D, and then the in vivo phosphorylation of FGF1 was analyzed in the presence of increasing concentrations of SB203580 (A) or wortmannin (B) as indicated. In vivo-phosphorylated FGF1 (33P-FGF1) was detected by fluorography, and the total cellular uptake of FGF1 (total FGF1) was detected by anti-FGF1 immunodetection.
A possible reason that translocation of FGFR1 S777D is inhibited by the higher concentrations of SB203580 is that at the higher concentrations the inhibitor may be less selective and also inhibit other kinases that play a role in FGF1 translocation (26).
In vitro phosphorylation of Ser777 in FGFR1 by p38α.
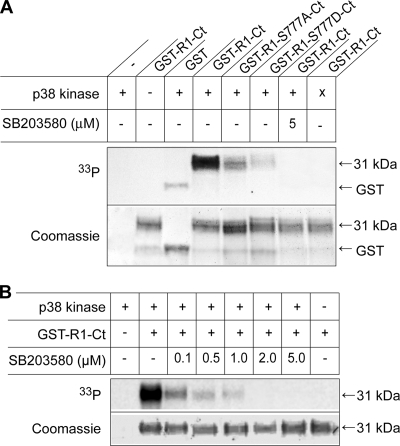
In order to elucidate whether p38 MAPK is able to directly phosphorylate FGFR1 at Ser777, we carried out an in vitro phosphorylation assay, using pure recombinant p38α, [γ-33P]ATP, and recombinant Ct from FGFR1 fused to GST as substrates. The Ct of wild-type FGFR1 could clearly be phosphorylated by a recombinant active form of p38α, whereas the Ct of FGFR1 S777A, as well as that of FGFR1 S777D, was only phosphorylated to about the background level obtained with GST alone (Fig. 9A). Phosphorylation of the wild-type Ct was abolished in the presence of 5 μM SB 203580 or when an inactive form of p38α was added instead of the active p38α.
FIG. 9.
In vitro phosphorylation of the FGFR1 Ct by recombinant p38α. (A) Recombinant, active (+), or inactive (x) p38α kinase and recombinant fusion proteins of GST and the Ct of wild-type FGFR1 (GST-R1-Ct), FGFR1 S777A (GST-R1-S777A-Ct), or FGFR1 S777D (GST-R1-S777D-Ct) were incubated with [γ-33P]ATP in reaction buffer at 30°C for 30 min in the presence or absence of 5 μM SB203580. The proteins were analyzed by SDS-PAGE, electroblotting, and autoradiography (upper panel), and then the membrane was stained with Coomassie (lower panel). The 31-kDa band indicates the fusion proteins. (B) In vitro phosphorylation of GST-R1-Ct was performed as for panel A, but in the presence of increasing concentrations of SB203580, as indicated.
As shown in Fig. 9B, there is a dose-dependent inhibitory effect of SB203580 on the in vitro phosphorylation of the wild-type FGFR1 Ct. Already at 0.1 μM SB203580 the phosphorylation was reduced, while it was completely inhibited by 2 to 5 μM SB203580, which is in agreement with results from in vivo experiments. These data indicate that p38α can directly phosphorylate FGFR1 at Ser777 and, furthermore, Ser777 appears to be the only site in the Ct of FGFR1 that is phosphorylated by p38α.
DISCUSSION
We report here that translocation of exogenous FGF1 to the cytosol and nucleus requires active p38 MAPK. The requirement for p38 MAPK activity is, at least partly, due to the requirement for p38 MAPK-mediated phosphorylation of FGFR1 at Ser777, demonstrating for the first time a specific role for serine phosphorylation of FGFR1.
As shown previously, the translocation of FGF1 across membranes of intracellular vesicles is a tightly regulated process that depends on special features of FGFR (47) as well as several additional cellular factors (3, 23, 31, 52, 63). Here, we found that translocation of exogenous FGF1 to the cytosol and the nucleus is completely inhibited by low concentrations of two low-molecular-weight inhibitors of p38 MAPK, SB203580 and PD169316, providing evidence that the activity of this kinase is essential for the translocation to occur. A requirement for p38 MAPK was confirmed and extended by the specific knockdown of the α isoform of p38 MAPK by siRNA in two different cell lines, which also reduced FGF1 translocation to below a detectable level. This showed that the α isoform of p38 MAPK is strictly required for FGF1 translocation.
Since translocation of FGF1 is known to be a highly regulated process, we investigated whether FGF1 translocation correlated with the cellular activity of p38 MAPK. We found that chemically induced hyperactivation of p38 MAPK by anisomycin, mannitol, or H2O2 increased the translocation of FGF1 to the cytosol and nucleus severalfold. This suggests that cells can respond to stress conditions that activate p38 MAPK by increasing the translocation of FGF1, possibly pointing toward an important biological role for translocated FGF1. Thapsigargin was also able to enhance the p38 MAPK activity but did not increase FGF1 translocation. This could be due to negative side effects of thapsigargin. Another possible explanation is that the stress-induced increase in FGF1 translocation is related to the mode of activation of p38 MAPK, as the activators anisomycin, sorbitol, and hydrogen peroxide, but not thapsigargin, were previously found to be able to activate p38 MAPK in an MKK3/6-independent manner (22).
In the absence of stress activation, we found that the cellular activity of p38 MAPK was at a low level during the time interval for most efficient FGF1 translocation (3 to 8 h after addition of FGF1). Thus, it appears that a low level of p38 MAPK activity in the cells, possibly reflecting a basal activity required for certain housekeeping processes, is sufficient to support FGF1 translocation. Although stimulation of the cells with FGF1 induces an initial rise in p38 MAPK activity (0 to 3 h), this activation is probably not important, since we have previously shown that FGF1 translocation can be facilitated by kinase-dead mutants of FGFR (24, 47), and FGF1 translocation can occur in the presence of chemical inhibition of the FGFR1 kinase activity (unpublished results), both representing situations where FGFR-induced phosphorylation cascades, including activation of MKKs and thereby p38 MAPK, are absent. Previously, also, it was shown that in the absence of stress, a low activity level of p38 MAPK could regulate endocytosis of a cell surface receptor (28).
Although a low cellular activity level of p38 MAPK may be sufficient for FGF1 translocation, it is possible that FGF1 translocation depends on variable properties of p38 MAPK that are not reflected in its total cellular activity, for instance, activation or a recruitment of p38 MAPK at specific subcellular sites. Importantly, p38 MAPK has been shown to regulate general components of the endocytic machinery, such as EEA1, GDI-rab5, and rabenosyn 5, all of which play roles in the functioning of early endosomes (5, 8, 28). In this study, we did not find any evidence that the cellular activity level of p38 MAPK affected the endocytosis or the major intracellular transport routes of endocytosed FGF1/FGFR1. Nevertheless, it cannot be excluded that p38 MAPK regulates features of endosomes that are important for FGF1 translocation. Notably, the translocation of FGF1 probably occurs from endosomal vesicles (32). Thus, it is conceivable that the p38 MAPK-mediated phosphorylation of FGFR1 also occurs at the endosome, prior to, or during, FGF1 translocation.
The intracellular part of FGFRs consists of a juxtamembrane region of about 75 amino acids, a split kinase domain encompassing approximately 290 amino acids, and a C-terminal tail region downstream of the kinase of 49 to 59 amino acids. In a previous study we showed that whereas the entire kinase domain of FGFR1 was dispensable for FGF1 translocation, the C-terminal tail region was crucial. By mutational analysis we also identified Met771 and His798 within this domain as important for the facilitation of FGF1 translocation (47). In the present study we have provided evidence that also Ser777 in the C-terminal tail of FGFR1 is crucial for FGF1 translocation. The first half of the C-terminal tail of FGFR1 includes 11 Ser/Thr residues as well as two tyrosines. These amino acids are quite well-conserved between the four FGFR isoforms. Among these Ser/Thr residues, we found Ser777 to be of particular interest, as its context (DQYS777PSF) made it a putative MAPK phosphorylation site. When Ser777 was mutated to alanine, translocation of FGF1 was abolished, while when Ser777 was mutated to an acidic amino acid, aspartic acid or glutamic acid, which may mimic a constitutively phosphorylated serine, FGF1 translocation was efficient. Importantly, translocation of FGF1 mediated by the FGFR1 S777D mutant was not dependent on p38 MAPK activity. This suggests that the role of p38 MAPK in FGF1 translocation is to regulate phosphorylation of Ser777 in FGFR1. Using recombinant p38α protein and recombinant C-terminal tail regions from FGFR1 in in vitro reactions, we found that p38α could phosphorylate the C-terminal tail from wild-type FGFR1 but not the C-terminal tail from FGFR1 S777A or FGFR1 S777D. This suggests that Ser777 is a direct phosphorylation site for p38α and, also, that Ser777 is the only p38α phosphorylation site in the C-terminal tail of FGFR1. These results highlight the importance of the C-terminal tail region of FGFR1 and indicate that this domain is able to regulate receptor functions in response to cell signaling events.
Due to the important role of FGFR as a tyrosine kinase, the phosphorylation pattern for Tyr residues in FGFR, and also the role of such phosphorylations, has been mapped and studied quite extensively (7, 10). The role of serine phosphorylations has not received similar attention. The FGFR is, however, quite rich in serines in its intracellular juxtamembrane region as well as in the C-terminal tail region. Although it has been recognized that Ser phosphorylations in FGFR occur (48), the role of such phosphorylations has been elusive. To our knowledge, this report is the first to describe a specific role of a single phospho-serine in FGFR1.
p38 MAPK is emerging as a signaling kinase involved in regulating endosome functions and intracellular sorting. The EGFR has been described to be phosphorylated on several Ser/Thr by p38 MAPK, and this is involved in regulating endosomal sorting of EGFR (59, 65). Recently, it was also found that the retrograde transport of Shiga toxin to the cytosol requires p38 MAPK signaling (50). Genome-wide analyses of kinases and their role in endocytosis have shown that transport along endocytic routes is highly regulated by a number of signaling kinases (38). This report demonstrates a link between the signaling molecule p38α, phosphorylation of FGFR1 at Ser777, and the translocation of exogenous FGF1 across membranes of endosomal vesicles to gain access to cytosol and nucleus.
Acknowledgments
Y.Z. and E.M.H. are predoctoral fellows of the Norwegian Cancer Society. M.Z. is a fellow of the Norwegian Research Council, and her work was supported by the Foundation for Polish Science and Ministry of Science and Higher Education (grant no. N301 419233). This work was supported by the Research Council of Norway, the Novo Nordisk Foundation, the Blix Fund for Promotion of Medical Research, the Rachel and Otto Kr. Bruns Fund, and the Torsteds Fund.
Footnotes
Published ahead of print on 14 April 2008.
REFERENCES
- 1.Baldin, V., A. M. Roman, I. Bosc-Bierne, F. Amalric, and G. Bouche. 1990. Translocation of bFGF to the nucleus is G1 phase cell cycle specific in bovine aortic endothelial cells. EMBO J. 91511-1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boilly, B., A. S. Vercoutter-Edouart, H. Hondermarck, V. Nurcombe, and X. Le Bourhis. 2000. FGF signals for cell proliferation and migration through different pathways. Cytokine Growth Factor Rev. 11295-302. [DOI] [PubMed] [Google Scholar]
- 3.Bouche, G., N. Gas, H. Prats, V. Baldin, J. P. Tauber, J. Teissie, and F. Amalric. 1987. Basic fibroblast growth factor enters the nucleolus and stimulates the transcription of ribosomal genes in ABAE cells undergoing G0-G1 transition. Proc. Natl. Acad. Sci. USA 846770-6774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cano, E., C. A. Hazzalin, and L. C. Mahadevan. 1994. Anisomycin-activated protein kinases p45 and p55 but not mitogen-activated protein kinases ERK-1 and -2 are implicated in the induction of c-fos and c-jun. Mol. Cell. Biol. 147352-7362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavalli, V., F. Vilbois, M. Corti, M. J. Marcote, K. Tamura, M. Karin, S. Arkinstall, and J. Gruenberg. 2001. The stress-induced MAP kinase p38 regulates endocytic trafficking via the GDI:Rab5 complex. Mol. Cell 7421-432. [DOI] [PubMed] [Google Scholar]
- 6.Cuenda, A., J. Rouse, Y. N. Doza, R. Meier, P. Cohen, T. F. Gallagher, P. R. Young, and J. C. Lee. 1995. SB 203580 is a specific inhibitor of a MAP kinase homologue which is stimulated by cellular stresses and interleukin-1. FEBS Lett. 364229-233. [DOI] [PubMed] [Google Scholar]
- 7.Eswarakumar, V. P., I. Lax, and J. Schlessinger. 2005. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 16139-149. [DOI] [PubMed] [Google Scholar]
- 8.Fratti, R. A., J. Chua, and V. Deretic. 2003. Induction of p38 mitogen-activated protein kinase reduces early endosome autoantigen 1 (EEA1) recruitment to phagosomal membranes. J. Biol. Chem. 27846961-46967. [DOI] [PubMed] [Google Scholar]
- 9.Frey, M. R., R. S. Dise, K. L. Edelblum, and D. B. Polk. 2006. p38 kinase regulates epidermal growth factor receptor downregulation and cellular migration. EMBO J. 255683-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furdui, C. M., E. D. Lew, J. Schlessinger, and K. S. Anderson. 2006. Autophosphorylation of FGFR1 kinase is mediated by a sequential and precisely ordered reaction. Mol. Cell 21711-717. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Maya, M., A. A. Anderson, C. E. Kendal, A. V. Kenny, L. C. Edwards-Ingram, A. Holladay, and J. L. Saffell. 2006. Ligand concentration is a driver of divergent signaling and pleiotropic cellular responses to FGF. J. Cell Physiol. 206386-393. [DOI] [PubMed] [Google Scholar]
- 12.Ge, B., H. Gram, P. F. Di, B. Huang, L. New, R. J. Ulevitch, Y. Luo, and J. Han. 2002. MAPKK-independent activation of p38alpha mediated by TAB1-dependent autophosphorylation of p38α. Science 2951291-1294. [DOI] [PubMed] [Google Scholar]
- 13.Ge, B., X. Xiong, Q. Jing, J. L. Mosley, A. Filose, D. Bian, S. Huang, and J. Han. 2003. TAB1β (transforming growth factor-β-activated protein kinase 1-binding protein 1β), a novel splicing variant of TAB1 that interacts with p38α but not TAK1. J. Biol. Chem. 2782286-2293. [DOI] [PubMed] [Google Scholar]
- 14.Gschwendt, M., H. J. Muller, K. Kielbassa, R. Zang, W. Kittstein, G. Rincke, and F. Marks. 1994. Rottlerin, a novel protein kinase inhibitor. Biochem. Biophys. Res. Commun. 19993-98. [DOI] [PubMed] [Google Scholar]
- 15.Han, J., and P. Sun. 2007. The pathways to tumor suppression via route p38. Trends Biochem. Sci. 32364-371. [DOI] [PubMed] [Google Scholar]
- 16.Haugsten, E. M., V. Sorensen, A. Brech, S. Olsnes, and J. Wesche. 2005. Different intracellular trafficking of FGF1 endocytosed by the four homologous FGF receptors. J. Cell Sci. 1183869-3881. [DOI] [PubMed] [Google Scholar]
- 17.Imamura, T., K. Engleka, X. Zhan, Y. Tokita, R. Forough, D. Roeder, A. Jackson, J. A. Maier, T. Hla, and T. Maciag. 1990. Recovery of mitogenic activity of a growth factor mutant with a nuclear translocation sequence. Science 2491567-1570. [DOI] [PubMed] [Google Scholar]
- 18.Imamura, T., S. Oka, T. Tanahashi, and Y. Okita. 1994. Cell cycle-dependent nuclear localization of exogenously added fibroblast growth factor-1 in BALB/c 3T3 and human vascular endothelial cells. Exp. Cell Res. 215363-372. [DOI] [PubMed] [Google Scholar]
- 19.Imamura, T., Y. Tokita, and Y. Mitsui. 1992. Identification of a heparin-binding growth factor-1 nuclear translocation sequence by deletion mutation analysis. J. Biol. Chem. 2675676-5679. [PubMed] [Google Scholar]
- 20.Itoh, N., and D. M. Ornitz. 2004. Evolution of the Fgf and Fgfr gene families. Trends Genet. 20563-569. [DOI] [PubMed] [Google Scholar]
- 21.Jaye, M., R. M. Lyall, R. Mudd, J. Schlessinger, and N. Sarver. 1988. Expression of acidic fibroblast growth factor cDNA confers growth advantage and tumorigenesis to Swiss 3T3 cells. EMBO J. 7963-969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kang, Y. J., A. Seit-Nebi, R. J. Davis, and J. Han. 2006. Multiple activation mechanisms of p38α mitogen-activated protein kinase. J. Biol. Chem. 28126225-26234. [DOI] [PubMed] [Google Scholar]
- 23.Klingenberg, O., A. Wiedlocha, L. Citores, and S. Olsnes. 2000. Requirement of phosphatidylinositol 3-kinase activity for translocation of exogenous aFGF to the cytosol and nucleus. J. Biol. Chem. 27511972-11980. [DOI] [PubMed] [Google Scholar]
- 24.Klingenberg, O., A. Wiedlocha, A. Rapak, D. Khnykin, L. Citores, and S. Olsnes. 2000. Requirement for C-terminal end of fibroblast growth factor receptor 4 in translocation of acidic fibroblast growth factor to cytosol and nucleus. J. Cell Sci. 1131827-1838. [DOI] [PubMed] [Google Scholar]
- 25.Klingenberg, O., A. Wiedlocha, A. Rapak, R. Munoz, P. Falnes, and S. Olsnes. 1998. Inability of the acidic fibroblast growth factor mutant K132E to stimulate DNA synthesis after translocation into cells. J. Biol. Chem. 27311164-11172. [DOI] [PubMed] [Google Scholar]
- 26.Lali, F. V., A. E. Hunt, S. J. Turner, and B. M. Foxwell. 2000. The pyridinyl imidazole inhibitor SB203580 blocks phosphoinositide-dependent protein kinase activity, protein kinase B phosphorylation, and retinoblastoma hyperphosphorylation in interleukin-2-stimulated T cells independently of p38 mitogen-activated protein kinase. J. Biol. Chem. 2757395-7402. [DOI] [PubMed] [Google Scholar]
- 27.Liu, Z., N. Neiss, S. Zhou, D. Henne-Bruns, M. Korc, M. Bachem, and M. Kornmann. 2007. Identification of a fibroblast growth factor receptor 1 splice variant that inhibits pancreatic cancer cell growth. Cancer Res. 672712-2719. [DOI] [PubMed] [Google Scholar]
- 28.Mace, G., M. Miaczynska, M. Zerial, and A. R. Nebreda. 2005. Phosphorylation of EEA1 by p38 MAP kinase regulates mu opioid receptor endocytosis. EMBO J. 243235-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maher, P. 1999. p38 mitogen-activated protein kinase activation is required for fibroblast growth factor-2-stimulated cell proliferation but not differentiation. J. Biol. Chem. 27417491-17498. [DOI] [PubMed] [Google Scholar]
- 30.Maher, P. 2002. Phorbol esters inhibit fibroblast growth factor-2-stimulated fibroblast proliferation by a p38 MAP kinase dependent pathway. Oncogene 211978-1988. [DOI] [PubMed] [Google Scholar]
- 31.Malecki, J., J. Wesche, C. S. Skjerpen, A. Wiedlocha, and S. Olsnes. 2004. Translocation of FGF-1 and FGF-2 across vesicular membranes occurs during G1-phase by a common mechanism. Mol. Biol. Cell 15801-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malecki, J., A. Wiedlocha, J. Wesche, and S. Olsnes. 2002. Vesicle transmembrane potential is required for translocation to the cytosol of externally added FGF-1. EMBO J. 214480-4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martiny-Baron, G., M. G. Kazanietz, H. Mischak, P. M. Blumberg, G. Kochs, H. Hug, D. Marme, and C. Schachtele. 1993. Selective inhibition of protein kinase C isozymes by the indolocarbazole Go 6976. J. Biol. Chem. 2689194-9197. [PubMed] [Google Scholar]
- 34.Matsumoto, T., I. Turesson, M. Book, P. Gerwins, and L. Claesson-Welsh. 2002. p38 MAP kinase negatively regulates endothelial cell survival, proliferation, and differentiation in FGF-2-stimulated angiogenesis. J. Cell Biol. 156149-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McLaughlin, M. M., S. Kumar, P. C. McDonnell, H. S. Van, J. C. Lee, G. P. Livi, and P. R. Young. 1996. Identification of mitogen-activated protein (MAP) kinase-activated protein kinase-3, a novel substrate of CSBP p38 MAP kinase. J. Biol. Chem. 2718488-8492. [DOI] [PubMed] [Google Scholar]
- 36.Nilsen, T., K. R. Rosendal, V. Sorensen, J. Wesche, S. Olsnes, and A. Wiedlocha. 2007. A nuclear export sequence located on a beta-strand in fibroblast growth factor-1. J. Biol. Chem. 28226245-26256. [DOI] [PubMed] [Google Scholar]
- 37.Olsnes, S., O. Klingenberg, and A. Wiedlocha. 2003. Transport of exogenous growth factors and cytokines to the cytosol and to the nucleus. Physiol. Rev. 83163-182. [DOI] [PubMed] [Google Scholar]
- 38.Pelkmans, L., E. Fava, H. Grabner, M. Hannus, B. Habermann, E. Krausz, and M. Zerial. 2005. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature 43678-86. [DOI] [PubMed] [Google Scholar]
- 39.Planque, N. 2006. Nuclear trafficking of secreted factors and cell-surface receptors: new pathways to regulate cell proliferation and differentiation, and involvement in cancers. Cell Commun. Signal. 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raucci, A., E. Laplantine, A. Mansukhani, and C. Basilico. 2004. Activation of the ERK1/2 and p38 mitogen-activated protein kinase pathways mediates fibroblast growth factor-induced growth arrest of chondrocytes. J. Biol. Chem. 2791747-1756. [DOI] [PubMed] [Google Scholar]
- 41.Reynolds, A., D. Leake, Q. Boese, S. Scaringe, W. S. Marshall, and A. Khvorova. 2004. Rational siRNA design for RNA interference. Nat. Biotechnol. 22326-330. [DOI] [PubMed] [Google Scholar]
- 42.Roux, P. P., and J. Blenis. 2004. ERK and p38 MAPK-activated protein kinases: a family of protein kinases with diverse biological functions. Microbiol. Mol. Biol. Rev. 68320-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salvador, J. M., P. R. Mittelstadt, T. Guszczynski, T. D. Copeland, H. Yamaguchi, E. Appella, A. J. Fornace, Jr., and J. D. Ashwell. 2005. Alternative p38 activation pathway mediated by T cell receptor-proximal tyrosine kinases. Nat. Immunol. 6390-395. [DOI] [PubMed] [Google Scholar]
- 44.Sheng, Z., Y. Liang, C. Y. Lin, L. Comai, and W. J. Chirico. 2005. Direct regulation of rRNA transcription by fibroblast growth factor 2. Mol. Cell. Biol. 259419-9426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Skjerpen, C. S., T. Nilsen, J. Wesche, and S. Olsnes. 2002. Binding of FGF-1 variants to protein kinase CK2 correlates with mitogenicity. EMBO J. 214058-4069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sorensen, V., T. Nilsen, and A. Wiedlocha. 2006. Functional diversity of FGF-2 isoforms by intracellular sorting. Bioessays 28504-514. [DOI] [PubMed] [Google Scholar]
- 47.Sorensen, V., A. Wiedlocha, E. M. Haugsten, D. Khnykin, J. Wesche, and S. Olsnes. 2006. Different abilities of the four FGFRs to mediate FGF-1 translocation are linked to differences in the receptor C-terminal tail. J. Cell Sci. 1194332-4341. [DOI] [PubMed] [Google Scholar]
- 48.Vainikka, S., V. Joukov, P. Klint, and K. Alitalo. 1996. Association of a 85-kDa serine kinase with activated fibroblast growth factor receptor-4. J. Biol. Chem. 2711270-1273. [DOI] [PubMed] [Google Scholar]
- 49.Vergarajauregui, S., M. A. San, and R. Puertollano. 2006. Activation of p38 mitogen-activated protein kinase promotes epidermal growth factor receptor internalization. Traffic 7686-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Walchli, S., S. S. Skanland, T. F. Gregers, S. U. Lauvrak, M. L. Torgersen, M. Ying, S. Kuroda, A. Maturana, and K. Sandvig. 2008. The mitogen-activated protein kinase p38 links Shiga toxin-dependent signaling and trafficking. Mol. Biol. Cell 1995-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wesche, J., J. Malecki, A. Wiedlocha, M. Ehsani, E. Marcinkowska, T. Nilsen, and S. Olsnes. 2005. Two nuclear localization signals required for transport from the cytosol to the nucleus of externally added FGF-1 translocated into cells. Biochemistry 446071-6080. [DOI] [PubMed] [Google Scholar]
- 52.Wesche, J., J. Malecki, A. Wiedlocha, C. S. Skjerpen, P. Claus, and S. Olsnes. 2006. FGF-1 and FGF-2 require the cytosolic chaperone Hsp90 for translocation into the cytosol and the cell nucleus. J. Biol. Chem. 28111405-11412. [DOI] [PubMed] [Google Scholar]
- 53.Wesche, J., A. Wiedlocha, P. O. Falnes, S. Choe, and S. Olsnes. 2000. Externally added aFGF mutants do not require extensive unfolding for transport to the cytosol and the nucleus in NIH/3T3 cells. Biochemistry 3915091-15100. [DOI] [PubMed] [Google Scholar]
- 54.Wiedlocha, A., P. O. Falnes, I. H. Madshus, K. Sandvig, and S. Olsnes. 1994. Dual mode of signal transduction by externally added acidic fibroblast growth factor. Cell 761039-1051. [DOI] [PubMed] [Google Scholar]
- 55.Wiedlocha, A., P. O. Falnes, A. Rapak, O. Klingenberg, R. Munoz, and S. Olsnes. 1995. Translocation of cytosol of exogenous, CAAX-tagged acidic fibroblast growth factor. J. Biol. Chem. 27030680-30685. [DOI] [PubMed] [Google Scholar]
- 56.Wiedlocha, A., P. O. Falnes, A. Rapak, R. Munoz, O. Klingenberg, and S. Olsnes. 1996. Stimulation of proliferation of a human osteosarcoma cell line by exogenous acidic fibroblast growth factor requires both activation of receptor tyrosine kinase and growth factor internalization. Mol. Cell. Biol. 16270-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wiedlocha, A., T. Nilsen, J. Wesche, V. Sorensen, J. Malecki, E. Marcinkowska, and S. Olsnes. 2005. Phosphorylation-regulated nucleocytoplasmic trafficking of internalized fibroblast growth factor-1. Mol. Biol. Cell 16794-810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiedlocha, A., and V. Sorensen. 2004. Signaling, internalization, and intracellular activity of fibroblast growth factor. Curr. Top. Microbiol. Immunol. 28645-79. [DOI] [PubMed] [Google Scholar]
- 59.Winograd-Katz, S. E., and A. Levitzki. 2006. Cisplatin induces PKB/Akt activation and p38(MAPK) phosphorylation of the EGF receptor. Oncogene 257381-7390. [DOI] [PubMed] [Google Scholar]
- 60.Zakrzewska, M., D. Krowarsch, A. Wiedlocha, S. Olsnes, and J. Otlewski. 2005. Highly stable mutants of human fibroblast growth factor-1 exhibit prolonged biological action. J. Mol. Biol. 352860-875. [DOI] [PubMed] [Google Scholar]
- 61.Zalecki, P., C. Radzikowski, S. Olsnes, and A. Wiedlocha. 1998. Modulation by interleukin-2 of cellular response to fibroblast growth factor-1 in F69-3 fibrosarcoma cells. Exp. Cell Res. 24461-70. [DOI] [PubMed] [Google Scholar]
- 62.Zarubin, T., and J. Han. 2005. Activation and signaling of the p38 MAP kinase pathway. Cell Res. 1511-18. [DOI] [PubMed] [Google Scholar]
- 63.Zhan, X., X. Hu, R. Friesel, and T. Maciag. 1993. Long term growth factor exposure and differential tyrosine phosphorylation are required for DNA synthesis in BALB/c 3T3 cells. J. Biol. Chem. 2689611-9620. [PubMed] [Google Scholar]
- 64.Zinck, R., M. A. Cahill, M. Kracht, C. Sachsenmaier, R. A. Hipskind, and A. Nordheim. 1995. Protein synthesis inhibitors reveal differential regulation of mitogen-activated protein kinase and stress-activated protein kinase pathways that converge on Elk-1. Mol. Cell. Biol. 154930-4938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zwang, Y., and Y. Yarden. 2006. p38 MAP kinase mediates stress-induced internalization of EGFR: implications for cancer chemotherapy. EMBO J. 254195-4206. [DOI] [PMC free article] [PubMed] [Google Scholar]