Abstract
Mediator is a conserved multisubunit complex that acts as a functional interface between regulatory transcription factors and the general RNA polymerase II initiation apparatus. MED1 is a pivotal component of the complex that binds to nuclear receptors and a broad array of other gene-specific activators. Paradoxically, MED1 is found in only a fraction of the total cellular Mediator complexes, and the mechanisms regulating its binding to the core complex remain unclear. Here, we report that phosphorylation of MED1 by mitogen-activated protein kinase-extracellular signal-regulated kinase (MAPK-ERK) promotes its association with Mediator. We show that MED1 directly binds to the MED7 subunit and that ERK phosphorylation of MED1 enhances this interaction. Interestingly, we found that both thyroid and steroid hormones stimulate MED1 phosphorylation in vivo and that MED1 phosphorylation is required for its nuclear hormone receptor coactivator activity. Finally, we show that MED1 phosphorylation by ERK enhances thyroid hormone receptor-dependent transcription in vitro. Our findings suggest that ERK phosphorylation of MED1 is a regulatory mechanism that promotes MED1 association with Mediator and, as such, may facilitate a novel feed-forward action of nuclear hormones.
Mediator is a conserved multisubunit protein complex that plays a key coregulatory role in eukaryotic transcription by RNA polymerase II (Pol II) (15). The complex is thought to function as a molecular bridge between enhancer-bound activators and components of the basal transcription apparatus, thus promoting the assembly and activation of the RNA Pol II preinitiation complex at core gene promoters (21, 31). In mammals, Mediator is comprised of at least 30 subunits arranged into four subcomplexes termed the head, middle, tail, and Cdk8-containing modules (2, 4). Although Mediator interacts with a diverse array of DNA-binding transcription factors (18), the human complex was originally isolated by virtue of its ability to bind nuclear hormone receptors (NRs) in the presence of cognate ligand and was subsequently shown to be required for NR-dependent transcriptional activation (reviewed in reference 1). A single subunit of the complex, MED1 (formerly termed PBP, TRAP220, and DRIP205), directly targets Mediator to ligand-activated NRs via two signature LXXLL motifs that serve as NR binding surfaces (1, 29). Also, MED1 serves as a direct binding target for a number of other regulatory transcription factors that are essential for cell growth and development, including the GATA family of proteins (5, 11, 30), p53 (10), BRCA-1 (36), and GABPα (35).
Despite its functional importance as a binding target for a diverse array of transcriptional activators, MED1 is only variably associated with the Mediator complex (20, 34), and the specific subunits that tether it to the larger assemblage of proteins are poorly defined. Furthermore, the molecular mechanisms regulating MED1 entry into, and exit from, the core Mediator complex remain unclear. A recent study found that MED1 exists in only a subpopulation of human Mediator complexes (less than 20% of the total) that are tightly associated with near-stoichiometric levels of Pol II (41). Human Mediator complexes lacking MED1 are still capable of facilitating activator-dependent transcription (20, 34) and are similar in subunit composition to complexes containing MED1. Electron microscopy suggests that when MED1 is associated with Mediator, it assumes a relatively peripheral location near the middle module (2, 33, 34). These data thus suggest that MED1 is only tangentially associated with Mediator and likely serves specialized functions. Interestingly, we recently reported that human MED1 is phosphorylated by the mitogen-activated protein kinase (MAPK)-extracellular signal-regulated kinase (ERK) in vivo at two sites (threonines 1032 and 1457) (26). A similar finding was reported for murine MED1 (24). Notably, ERK phosphorylation of both human and murine MED1 proteins was found to enhance its intrinsic NR transcriptional coactivation activity. Nonetheless, whether MED1 phosphorylation by MAPK-ERK can influence its association with the core Mediator complex has not been addressed.
In the present study, we show that steroid and thyroid hormones facilitate MED1 phosphorylation in vivo via extranuclear activation of MAPK-ERK. Significantly, we found that MED1 phosphorylation by MAPK-ERK enhances its association with the Mediator complex but has no significant effect on its association with NRs. Using in vitro and in vivo protein-protein interaction assays, we show that MED1 directly binds to another Mediator subunit located in the middle module termed MED7 and that MAPK-ERK phosphorylation of MED1 enhances this interaction. Finally, using a thyroid hormone receptor (TR)-dependent in vitro transcription assay reconstituted with recombinant MED1, we found that prephosphorylation of the MED1 protein with MAPK-ERK significantly enhances transcription. Taken together, our data suggest that MAPK-ERK phosphorylation of MED1 may be a regulatory mechanism that promotes its association with the Mediator complex. Furthermore, our findings are consistent with a feed-forward action of thyroid and steroid hormones in which cognate ligand activates both NRs and NR coactivators.
MATERIALS AND METHODS
Plasmids.
The pSG5-HA-MED1-wt, -ERK mutant (T1032A and T1457A), -CΔ454, -CΔ690, and -CΔ918 expression vectors and the pVL1393-HA-MED1 baculovirus vector were described previously (26, 27). The pBK-CMV-FLAG-TRα expression vector and MMTV-Luc, 2 × TRE-tk-Luc, and (ERE)2-tk-Luc luciferase reporter genes were all described previously (26, 37, 40), as were the GST-ERα and GST-TRα plasmids (13, 39). The MED14 cDNA was from H. Yoshikawa (Johns Hopkins University), the MED21 cDNA was from R. Young (Massachusetts Institute of Technology), the MED26 cDNA was from J. Conaway (Stowers Institute), and the MED7 cDNA was from L. Myers (Dartmouth). GST-MED7-full length, -CΔ60, and CΔ120 were generated by PCR and cloned into the pGEX-2TK vector (Amersham). The MED4 and MED31 cDNAs were purchased from the American Type Culture Collection (ATCC).
Antibodies and Western blotting.
Antibodies against MED1, MED12, CDK8, cyclin C, MED7, MED6, MED17, MED26, RPB1, and TFIIB and anti-goat immunoglobulin G were from Santa Cruz Biotechnology. Rabbit polyclonal antibodies against MED24 and MED1 (used in immunodepletion assays) were described previously (40). Antibodies against phosphothreonine and hemagglutinin (HA) were from Roche. Antibodies against ERK and phospho-ERK were from Cell Signaling. Western blotting was performed as described previously (26).
Cell culture, hormones, and inhibitors.
HeLa, COS-7, and MCF-7 cells were from the ATCC. HeLa-TR and HeLa-AR were generated by stably transfecting HeLa cells with TRα- or androgen receptor (AR)-expressing retrovirus as described previously (9). Cells were routinely cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS). DHT, E2, and T3 were from Sigma. EGF was from Invitrogen. U0126 was from Alexis Biochemicals.
Recombinant baculovirus protein expression in Sf9 cells.
Baculovirus-expressed full-length (FL) HA-MED1 (bv-HA-MED1), human retinoid X receptor α (RXRα), and human TRα were described previously (9, 37). Baculovirus expressing the HA-MED1-ERK mutant (T1042A; T1457A) and the C-terminal MED1 deletions (pAcSG2-MED1-CΔ454 and pAcSG2-MED1-CΔ918) were generated by transfecting the constructs, along with linearized baculovirus DNA (BaculoGold; Pharmingen), into Sf9 cells. Subsequent infection, expression, and purification of recombinant proteins was done as described in detail previously (9).
In vitro kinase assay.
Fifty nanograms of purified baculovirus-expressed MED1 protein (bv-MED1) was incubated with 2 μCi [γ-32P]ATP or nonradioactive ATP (0.1 mM) plus 1 ng MAPK-ERK1 (Upstate) as described previously (26).
Immunoprecipitation of endogenous phosphorylated MED1.
Cells (2 × 107) were cultured in serum-free DMEM for 48 h and then pulsed with 32Pi (0.125 mCi/ml) in serum- and phosphate-free DMEM for 30 min with or without epidermal growth factor (EGF) (50 ng/ml), triiodothyronine (T3) (10−7 M), estradiol (E2) (10−7 M), or dihydrotestosterone (DHT) (10−7 M) as indicated. In assays involving MCF-7 cells, phenol red-free DMEM was used. Anti-MED1 immunoprecipitation was then carried out as described previously (26).
Transient transfections, immunoprecipitation of ectopic MED1, and luciferase assays.
Transient transfections were carried out in 6- or 12-well plates using Lipofectamine (Invitrogen). In assays involving MCF-7 cells, phenol red-free DMEM was used. For immunoprecipitation of 32P-labeled ectopic HA-MED1, 8 × 105 cells were transfected with 4 μg of pSG5-HA-MED1-wt or -ERK mutant. The cells were serum starved for 24 h and then pulsed with 32Pi (0.125 mCi/ml) in serum- and phosphate-free medium for 30 min with or without EGF (50 ng/ml) or hormones (T3, E2, or DHT, all at 10−7 M) as specified. Anti-HA immunoprecipitation was carried out as described above. For transcription assays, 1 × 105 HeLa-AR or MCF-7 cells were transfected with MMTV-Luc (0.5 μg) or (ERE)2-tk-Luc (0.5 μg), respectively, along with 250 ng of either pSG5-HA-MED1-wt or -ERK mutant or empty vector. HeLa cells (1 × 105) were transfected with pBK-CMV-FLAG-TRα (1 μg) and 2 × TRE-tk-Luc (0.3 μg), along with 250 ng of either pSG5-HA-MED1-wt or -ERK mutant or empty vector. The cells were then cultured in charcoal/dextran-stripped FBS (Gemini) for 24 h with or without DHT, E2, or T3 (all at 10−7 M) as indicated. Luciferase activity was measured using a luminometer and normalized using a pSV-β-galactosidase internal control.
In vitro protein-protein interaction studies.
Glutathione S-transferase (GST) and GST fusion proteins were expressed and purified as described previously (29). MED1 and other Mediator subunits were [35S]methionine labeled in vitro using a kit (TNT; Promega Corp.). GST pull-down assays with 35S-labeled or baculovirus-expressed proteins were carried out as previously outlined (29, 37). For experiments involving MED1 phosphorylation, bv-MED1 (50 ng) was incubated in kinase buffer containing or lacking ERK1 for 30 min (as described above); the entire kinase reaction mixtures were then added to the binding reactions. Anti-HA pull-down assays were carried out essentially as described previously (35).
In vivo protein-protein interaction studies.
COS-7 cells (5 × 105) were transfected with 3 μg of either pSG5-HA-MED1-wt or pSG5-HA-MED1-ERK mutant, together with pCIN4-MED7 (50 ng). For MED1 deletion mutant analyses, pSG5-HA-MED1Δ454 (0.25 μg), pSG5-HA-MED1Δ690 (0.5 μg), or pSG5-HA-MED1Δ918 (1.25 μg) was transfected, along with pCIN4-MED7 (50 ng). Twenty-four hours posttransfection, the cells were serum starved for an additional 24 h and then treated with EGF (50 ng/ml) or U0126 (50 μM) for 1 h or untreated. Whole-cell extract was prepared by using cell lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM EDTA, 0.5% NP-40, 1 μg/ml aprotinin, 1 μg/ml leupeptin, 0.2 mM phenylmethylsulfonyl fluoride). Equal amounts of cell lysate were incubated with 5 μl packed anti-HA agarose beads (Sigma) at 4°C overnight and then washed three times with cell lysis buffer. The anti-HA-bound proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and detected by Western blotting using anti-MED7 antibodies.
Immunoaffinity purification of Mediator complexes.
HeLa and HeLa-AR cells were cultured in 3-liter spinner flasks, whereas MCF-7 cells were cultured in 15-cm culture dishes containing phenol red-free DMEM; 2 × 109 cells were then serum starved for 24 h. The HeLa cells were then treated with or without EGF (50 ng/ml) for 1 h, whereas HeLa-AR and MCF-7 cells were treated with 10−7 M DHT or E2, respectively, for 6 h. Nuclear extracts were prepared as described previously (9). Affinity-purified goat anti-MED1 antibodies (Santa Cruz) or rabbit polyclonal antibodies against MED1 (40) were conjugated to protein G-agarose as detailed previously (21); preimmune antibodies from the same rabbits were used as mock controls. Equal amounts of nuclear extract were incubated with mock or anti-MED1 beads overnight at 4°C. The immunocomplexes were washed twice with BC-300/NP-40 (20 mM Tris-HCl [pH 7.9], 20% glycerol, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride, 5 mM β-mercaptoethanol, 300 mM KCl, 0.1% NP-40) and once with BC-100/NP-40 (same as BC300/NP-40 except 100 mM KCl), eluted with 0.1 M glycine, pH 3.0, and immediately neutralized with 1 M Tris-HCl, pH 8.0. The eluates were normalized for equivalent MED1 levels via anti-MED1 immunoblotting and subsequently reprobed with antibodies against phosphothreonine and other Mediator subunits.
MED1 immunodepletion and in vitro transcription assay.
HeLa cell nuclear extract was prepared as described previously (9) and then semidepleted of MED1 by passing it through a column prepared with rabbit polyclonal antibodies against MED1 (40) coupled to protein G-agarose as described previously (21). Preimmune antibodies from the same rabbits were utilized to prepare mock control columns. Transcription was measured in vitro by incubating the TRE3Δ53 (see below) G-free cassette template (50 ng) in MED1-depleted (or preimmune mock-depleted) HeLa cell nuclear extract (50 μg), together with purified baculovirus-expressed TRα (20 ng), RXRα (20 ng), and 10−7 M T3 as described previously (9). Fifty nanograms of purified bv-MED1-wt, MED1-ERK mutant, MED1Δ454, or MED1Δ918 was added to the transcription reactions where indicated. For experiments involving MED1 phosphorylation, the bv-MED1 protein (50 ng) was preincubated in kinase buffer containing or lacking purified ERK1 for 30 min (as described above); the entire kinase reaction mixture was then added to the transcription reaction mixture.
RESULTS
Steroid and thyroid hormones stimulate MED1 phosphorylation via activated MAPK-ERK signaling.
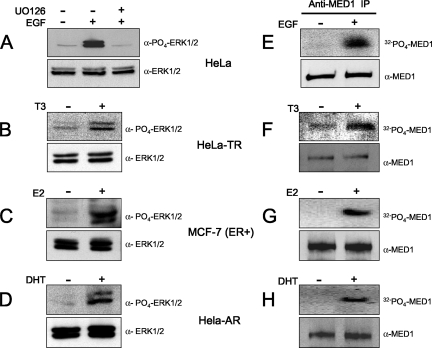
In addition to their classic role as activators of NR-dependent transcription, nuclear hormones perform extranuclear actions, including stimulation of phosphorylation cascades (3, 8). Indeed, gonadal steroid hormones (estrogens, progestins, and androgens) are rapid and effective stimulators of the Src/Ras/Raf signaling pathway (23) that ultimately leads to MAPK-ERK activation (reviewed in references 16 and 17). Similarly, thyroid hormones (T3 and T4) are potent stimulators of activated MAPK-ERK (19), although the initial upstream signaling events stimulated by T3/T4 are believed to be different from those activated by steroid hormones (7). Given that MED1 is a direct substrate for activated ERK, as well as a binding target for ligand-activated NRs, we asked whether steroid or thyroid hormones could facilitate MED1 phosphosphorylation. For these studies, we utilized HeLa cells stably expressing human TRα or AR (HeLa-TR and HeLa-AR, respectively) or human breast cancer MCF-7 cells that endogenously express estrogen receptor α (ER). Consistent with earlier reports, we found that T3, E2, and DHT are all capable of activating ERK in cells expressing their cognate NRs (Fig. 1B to D).
FIG. 1.
Steroid and thyroid hormones stimulate MED1 phosphorylation via activated MAPK-ERK signaling. (A to D) HeLa, HeLa-TR, MCF-7, and HeLa-AR cells were serum starved and then treated with EGF, U0126, T3, E2, or DHT (+) or untreated (−), as indicated (see Materials and Methods). The whole-cell lysate was then probed by immunoblotting for ERK1/2 expression (bottom) and ERK1/2 phosphorylation (top). (E to H) HeLa, HeLa-TR, MCF-7, and HeLa-AR cells were serum starved and then pulsed with 32Pi with or without hormones, as indicated. Parallel reactions lacking 32Pi were performed side by side. Native MED1 was immunoprecipitated with anti-MED1 antibodies and resolved by SDS-PAGE. Reactions containing 32Pi were detected by autoradiography (top), while reactions lacking 32Pi were detected by anti-MED1 immunoblotting (bottom).
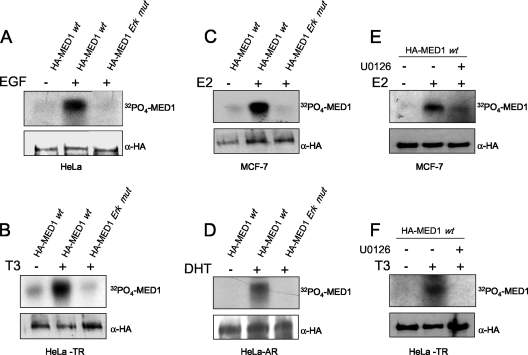
To investigate whether thyroid or steroid hormone activation of ERK could in turn facilitate endogenous MED1 phosphorylation, we cultured HeLa-TR, MCF-7, or HeLa-AR cells with or without cognate ligand (T3, E2, or DHT, respectively) and then pulsed the cells with 32Pi. Native MED1 was then precipitated with anti-MED1 antibodies and subjected to autoradiography. Remarkably, we found that T3, E2, and DHT were all capable of triggering MED1 phosphorylation to levels comparable to that observed in HeLa cells stimulated with EGF, a well-documented and potent activator of ERK (27) (compare Fig. 1E and F to H). Furthermore, we found that when HeLa-TR, MCF-7, or HeLa-AR cells were transiently transfected with a wild-type HA-tagged MED1 expression vector, stimulation with cognate ligand again led to phosphorylation of the ectopic HA-tagged MED1 to levels comparable to that observed in EGF-stimulated HeLa cells (Fig. 2A to D). Conversely, when the cells were transfected with a MED1 expression vector in which the two ERK phosphorylation sites were mutated (T1032A and T1457A) (26), no significant thyroid-, steroid-, or EGF-induced MED1 phosphorylation was observed. Moreover, thyroid and steroid hormone induction of MED1 phosphorylation was blocked upon addition of U0126, a specific chemical inhibitor of ERK1/2 activation (Fig. 2E and F). Intriguingly, these findings reveal a novel molecular action of thyroid and steroid hormones: phosphorylation of the Mediator subunit MED1 via extranuclear activation of ERK.
FIG. 2.
Steroid and thyroid hormones stimulate phosphorylation of ectopic MED1. HeLa, HeLa-TR, MCF-7, and HeLa-AR cells were transfected with (+) pSG5-HA-MED1-wt (wild type) (HA-MED1 wt) or pSG5-HA-ERK mutant (HA-MED1 Erk mut), as indicated at the top of each panel. The cells were serum starved and then pulsed with 32Pi with or without specific hormones or U0126, as indicated (see Materials and Methods). Parallel reactions lacking 32Pi were performed side by side. HA-MED1 was immunoprecipitated with anti-HA antibodies and resolved by SDS-PAGE. Reactions containing 32Pi were detected by autoradiography (top), while reactions lacking 32Pi were detected by anti-HA immunoblotting (bottom).
MED1 phosphorylation is required for NR coactivator activity, but not for NR binding.
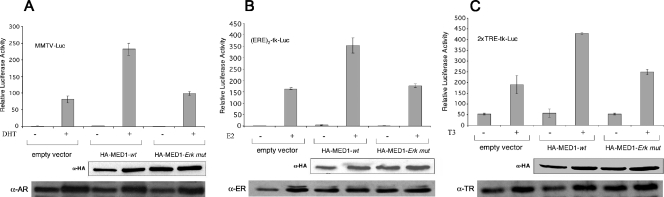
To explore the functional significance of MED1 phosphorylation, we compared the NR coactivator activity of wild-type MED1 to that of the phosphorylation-defective MED1 mutant. In agreement with previous studies (20, 26, 37, 42), transient expression of wild-type MED1 robustly enhanced AR-, ER-, and TR-dependent transcription in the presence of their cognate ligands (Fig. 3A to C). By contrast, transient transfection of the MED1-ERK mutant conferred only a modest coactivation. While we previously reported that ERK phosphorylation of MED1 increases protein stability (26), the decrease in NR coactivator activity observed with the MED1-ERK mutant cannot be attributed to decreased protein levels, since expression of the mutant was comparable to that of the wild-type protein (Fig. 3A to C, bottom).
FIG. 3.
MED1 phosphorylation is required for NR coactivator activity. (A and B) HeLa-AR or MCF-7 cells were transfected with MMTV-Luc or (ERE)2-tk-Luc, respectively, along with pSG5-HA-MED1-wt (HA-MED1-wt) or -ERK mutant (HA-MED1-Erk mut) or empty vector, as shown. (C) HeLa cells were transfected with pBK-CMV-FLAG-TRα and 2 × TRE-tk-Luc, along with pSG5-HA-MED1-wt or -ERK mutant or empty vector as shown. For panels A to C, the cells were subsequently cultured in charcoal/dextran-stripped FBS with or without DHT, E2, or T3, as indicated. Whole-cell lysate was assayed for luciferase activity and for MED1 and NR expression via immunoblotting (bottom). All luciferase assays were performed in triplicate and normalized against β-galactosidase (pSV-βgal) expression. The results are presented as the mean ± standard error.
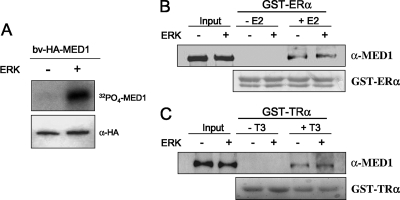
To investigate whether MED1 phosphorylation affects its NR binding activity, GST-tagged ER and TR pull-down assays were carried out with bv-HA-MED1 (see Fig. 6G). As shown in Fig. 4A, purified bv-HA-MED1 is effectively phosphorylated by purified ERK in vitro, thus confirming that MED1 is a direct substrate. Nonetheless, prephosphorylation of bv-HA-MED1 by ERK in vitro had no significant affect on ligand-dependent binding to either ER or TR (Fig. 4B and C). Given that only a portion of the recombinant MED1 protein is presumably phosphorylated by ERK in vitro, we cannot completely rule out phosphorylation-dependent binding affects. However, our data are consistent with an earlier report showing that ERK phosphorylation of murine MED1 enhanced transcription by the peroxisome proliferator-activated receptor (PPAR) yet conferred no enhanced ligand-dependent MED1 binding to PPAR (24). Taken together, the data suggest that ERK phosphorylation of MED1 facilitates a mechanism important for NR coactivator activity yet distinct from NR binding.
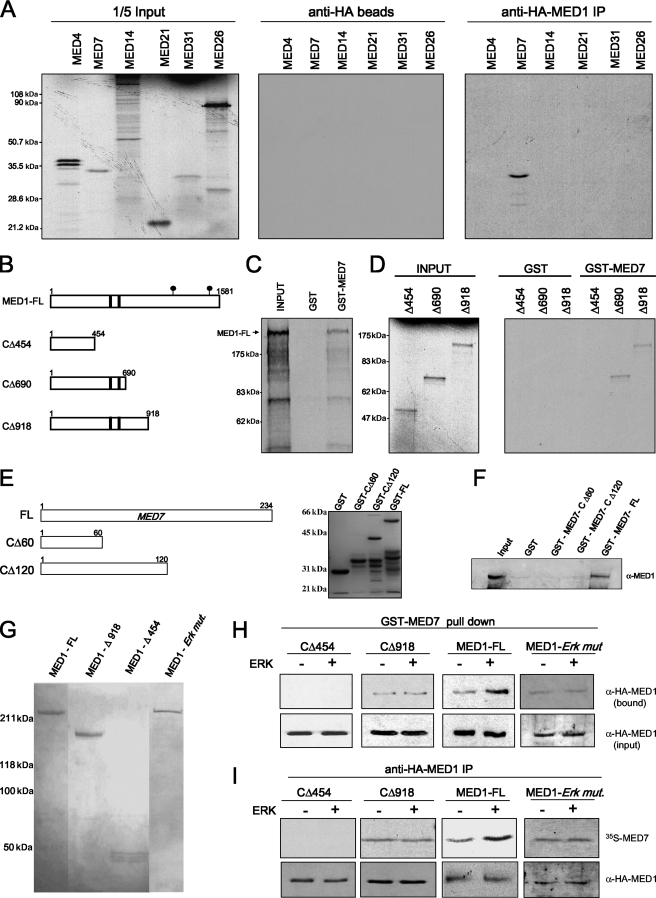
FIG. 6.
MED1 interacts with MED7 in vitro. (A) [35S]methionine-labeled Mediator subunits were incubated with bv-FL-HA-MED1. HA pull-down was carried out, and the bound proteins were detected by autoradiography. IP, immunoprecipitation. (B) Schematic representation of FL-MED1 and MED1 C-terminal deletion mutants. The black bars indicate LXXLL motifs; the black circles indicate ERK phosphorylation sites. (C and D) GST or GST-MED7 was incubated with [35S]methionine-labeled FL-MED1 (MED1-FL) (C) or C-terminal deletion mutants (D). GST pull-down assays were carried out, and the bound proteins were detected by autoradiography. (E) Schematic representation and corresponding Coomassie blue-stained gel showing each GST-MED7 fusion protein. (F) GST-MED7 FL, GST-MED7-Δ1 or -Δ2, or GST alone was incubated with FL-bv-HA-MED1. GST pull-down assays were carried out, and the bound proteins were detected via anti-MED1 immunoblotting. (G) Purified FL-bv-HA-MED1 (MED1 - FL) and -HA-MED1-ERK mutant (MED1 - Erk mut.) and C-terminal MED1 deletion mutants stained with Coomassie blue (panel B shows a schematic representation). (H and I) FL or truncated bv-HA-MED1 proteins were preincubated in kinase buffer containing (+) or lacking (−) ERK1 and then incubated with GST-MED7 (H) or [35S]methionine-labeled MED7 (I). In panel H, GST pull-down assays were carried out, and bound MED1 protein was detected via anti-HA immunoblotting. In panel I, HA pull-down assays were carried out, and bound MED7 protein was blotted to a membrane and detected by autoradiography; the same blot was stripped and reprobed with anti-HA antibodies to determine precipitated HA-MED1 levels.
FIG. 4.
MED1 phosphorylation is not required for NR binding. (A) ERK phosphorylation of MED1 in vitro. Purified bv-HA-MED1 was incubated with [γ-32P]ATP in the presence (+) or absence (−) of ERK1, as indicated. The reactions were resolved by SDS-PAGE and exposed for autoradiography. As a loading control, parallel reactions lacking [γ-32P]ATP were probed by immunoblotting with anti-HA antibodies. (B and C) bv-HA-MED1 was incubated with nonisotopic ATP in kinase buffer containing (+) or lacking (−) purified ERK1 (as in panel A) and then incubated with GST-ER or GST-TR with or without ligand, as indicated. Bound protein was detected by immunoblotting using anti-MED1 antibodies; 50% of the input was loaded in the first two lanes as a control. As a loading control for equivalent GST fusion protein, the nitrocellulose membranes were stained with Ponceau S, thus revealing the GST-ER and GST-TR polypeptides in each reaction (bottom).
MED1 phosphorylation promotes its association with the Mediator complex.
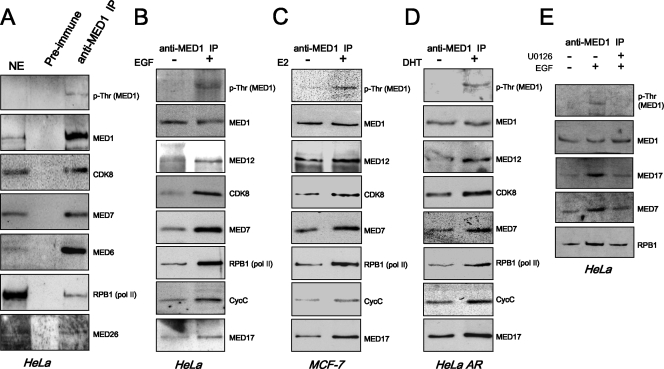
We next asked whether MED1 phosphorylation might influence its association with the Mediator complex. Toward that end, nuclear extracts from HeLa cells cultured in FBS were prepared and MED1-containing Mediator complexes were immunoprecipitated using anti-MED1 antibodies cross-linked to protein G-agarose beads. Interestingly, immunoblotting of the isolated complexes revealed low but detectable levels of threonine-phosphorylated MED1 (Fig. 5A). Moreover, and consistent with a recent study (41), the MED1-immunoprecipitated Mediator complexes were clearly enriched with RNA Pol II, as evidenced by the presence of its largest subunit, RPB1 (Fig. 5A).
FIG. 5.
MED1 phosphorylation promotes its association with the Mediator complex. (A) Isolation of native Mediator complexes containing phosphorylated MED1 and RNA Pol II. Nuclear extract prepared from HeLa cells cultured in FBS was incubated with anti-MED1 or preimmune antibodies. The bound complexes were then eluted and probed by immunoblotting with the antibodies indicated on the right of the panels. HeLa nuclear extract (NE) (10 μg) was loaded as a control. IP, immunoprecipitation. (B to E) HeLa, MCF-7, and HeLa-AR cells were serum starved and then stimulated with (+) or without (−) EGF, E2, DHT, or U0126, as indicated. Nuclear extracts were then prepared and incubated with anti-MED1 antibodies. The bound complexes were first normalized for equivalent MED1 concentrations via anti-MED1 immunoblotting. The normalized blots were then stripped and reprobed for RNA Pol II and other Mediator subunits, as indicated. For panels A to D, the immunoblot signal generated by the anti-phosphothreonine antibody precisely overlapped the band generated by the anti-MED1 antibody on the same blot.
To investigate more closely whether MED1 phosphorylation affects its association with Mediator, HeLa cells were serum starved for 24 h and then stimulated with or without EGF. Nuclear extracts were then prepared, and MED1-containing Mediator complexes were precipitated with anti-MED1 antibodies. Immunoprecipitates from the EGF-stimulated and unstimulated extracts were first normalized for equivalent MED1 levels by immunoblotting and then reprobed with antibodies against other Mediator subunits. Remarkably, we found that MED1 isolated from EGF-stimulated cells was more highly associated (about two- to threefold) with various components of the Mediator complex (MED7, MED17, MED12, CDK8, and CycC) and RNA Pol II (RPB1) than was the MED1 protein isolated from serum-starved HeLa cells (Fig. 5B). Similarly, when MED1 was immunoprecipitated from MCF-7 or HeLa-AR cells stimulated with E2 or DHT, respectively, we again observed that the phosphorylated MED1 protein was more highly associated with components of the Mediator complex and RNA Pol II (Fig. 5C and D). Moreover, when HeLa cells were costimulated with EGF and U0126, MED1 association with Mediator decreased (Fig. 5E), thus confirming that MED1 phosphorylation via ERK promotes its interaction with Mediator. In sum, these findings suggest that MED1 phosphorylation may serve to stabilize or enhance its association with the Mediator complex.
MED1 directly binds to the MED7 subunit of the Mediator complex.
The identity of the specific human Mediator subunit(s) that tethers MED1 to the core Mediator complex remains unknown. Electron microscopy of purified Saccharomyces cerevisiae and human Mediator complexes suggests that MED1 is peripherally associated with the middle module (2, 33, 34). Supporting this notion, an in vitro binding study found that yeast MED1 can bind two yeast subunits located in the middle module, MED4 and MED7 (14), whereas a yeast two-hybrid assay found that yeast MED1 can bind as many as four subunits in the middle module (MED4, MED5, MED7, and MED9) and possibly one subunit in the tail module (MED14) (12).
Using the yeast binding studies as a starting point, we set out to identify human Mediator subunits that directly bind human MED1 and presumably help tether the protein to the core complex. Given that MED4, MED7, and MED14 are conserved in humans, we [35S]methionine radiolabeled human MED4, MED7, and MED14 polypeptides in vitro and tested them for binding to FL bv-HA-MED1 using an HA pull-down assay. Additionally, three other human Mediator subunits, MED21, MED26, and MED31, all believed to reside in the middle module (2), were tested for MED1 binding in parallel. Interestingly, and consistent with the earlier yeast studies, we found that human MED1 specifically bound to MED7 (Fig. 6A). To map the MED7-binding domain of MED1, we 35S labeled three MED1 deletions (CΔ454, CΔ690, and CΔ918) and tested them for binding with GST-MED7. Only CΔ454 failed to bind (Fig. 6B to D), thus revealing a minimal MED7-binding domain between MED1 amino acids 454 and 690. To map the MED1-binding domain of MED7, two additional GST-MED7 deletion mutants (CΔ60 and CΔ120) were tested for binding to FL bv-HA-MED1. However, only GST-FL-MED7 bound MED1 (Fig. 6E and F), thus indicating that the MED1-binding domain resides between MED7 amino acids 120 and 234. These results are in agreement with the yeast studies that mapped the MED1-binding domain of yeast MED7 to its C terminus (12). Importantly, these findings indicate that the MED1-MED7 interaction first reported in yeast is conserved in humans.
Phosphorylation of MED1 enhances its binding to MED7.
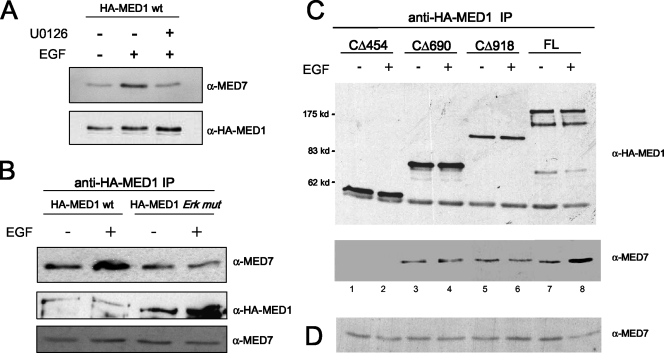
Given our results suggesting that MED1 phosphorylation promotes its association with Mediator (Fig. 5), we next investigated whether MED1 phosphorylation influences its interaction with MED7. To address this issue, we utilized FL-bv-HA-MED1, demonstrated earlier to be efficiently phosphorylated by ERK in vitro (Fig. 4A), as well as a phosphorylation-deficient FL bv-HA-MED1-ERK mutant (T1032A; T1457A). Additionally, we expressed via baculovirus two HA-tagged MED1 deletion mutants (CΔ918 and CΔ454), both lacking ERK phosphorylation sites (Fig. 6B). Accordingly, the FL and truncated MED1 baculovirus-expressed proteins (Fig. 6G) were preincubated in kinase reactions containing or lacking ERK and subsequently tested for binding to GST-MED7. As expected, CΔ454 failed to bind MED7, whereas CΔ918, FL-MED1, and the MED1-ERK mutant bound MED7, even in the absence of ERK phosphorylation (Fig. 6H). These results are consistent with the presence of a minimal MED7-binding domain (amino acids 454 to 690) in the CΔ918 and FL-MED1 polypeptides. Interestingly, preincubation of FL-MED1 with ERK increased MED7 binding ∼2- to 3-fold, whereas preincubation of CΔ918 or the MED1-ERK mutant with ERK had no significant effect on MED7 binding (Fig. 6H). Reciprocally using an anti-HA pull-down assay, we found that CΔ918, FL-MED1, and the MED1-ERK mutant, but not the CΔ454 mutant, bound to 35S-radiolabeled MED7 in the absence of ERK exposure (Fig. 6I). Once again, preincubation of FL-MED1 with ERK increased MED7 binding ∼2-fold, whereas preincubation of the phosphorylation-deficient CΔ918 or MED1-ERK mutant with ERK had no significant effect on MED7 binding (Fig. 6I).
Because the experiment in Fig. 6 was performed in vitro, it was important to further examine whether MED1 phosphorylation affects its interaction with MED7 in vivo. Accordingly, we found that when FL-MED1 and -MED7 were transiently overexpressed in COS cells, the two proteins coimmunoprecipitated more strongly from cells stimulated with EGF (∼3-fold more) than from unstimulated cells (Fig. 7A). Furthermore, the EGF-induced increase in MED1-MED7 binding was not observed in the presence of U0126, a specific inhibitor of ERK activation (Fig. 7A), or when the MED1-ERK mutant was used instead (Fig. 7B). To map the MED7-binding domain of MED1 in vivo, we overexpressed MED1 deletion mutants (CΔ454, CΔ690, and CΔ918) in COS cells, along with MED7, and then performed coimmunoprecipitation assays. In agreement with the in vitro data, we found that only MED1 proteins containing amino acids 454 to 690 bound MED7 (Fig. 7C), and as before, EGF stimulation enhanced MED7 binding to FL-MED1 (Fig. 7C, lanes 7 and 8) but had no significant effect on MED7 binding with the phosphorylation-deficient MED1 deletion mutants (Fig. 7C, lanes 3 to 6). In sum, these data confirm the presence of a minimal MED7-binding domain between amino acid residues 454 and 690 of the MED1 polypeptide. More intriguingly, however, the findings suggest that ERK phosphorylation of MED1 at Thr-1032 and Thr-1457 serves to stabilize and/or enhance the MED1-MED7 interaction. Thus, the increased association of phosphorylated MED1 with Mediator may be accounted for, at least in part, by enhanced or stabilized interactions with MED7.
FIG. 7.
ERK phosphorylation enhances MED1 association with MED7 in vivo. (A and B) COS-7 cells were transfected with pCIN4-MED7 and either pSG5-HA-MED1-wt or pSG5-HA-MED1-ERK mutant (HA-MED1 Erk mut). The cells were serum starved and then treated with EGF or U0126 (+) or untreated (−), as indicated. Whole-cell lysate was then incubated with anti-HA antibodies, and the precipitated proteins were detected by anti-MED7 and anti-HA immunoblotting. Equal amounts of whole-cell extract, taken prior to the anti-HA immunoprecipitation (IP) in panel B, were probed by immunoblotting with anti-MED7 antibodies. (C) COS-7 cells were transfected with pCIN4-MED7, together with either pSG5-HA-MED1 (FL), -Δ454, -Δ690, or -Δ918. The cells were serum starved and then treated with EGF or untreated. Whole-cell lysate was incubated with anti-HA antibodies, and the precipitated proteins were detected by anti-MED7 and anti-HA immunoblotting. (D) Equal amounts of whole-cell extract, taken prior to the anti-HA immunoprecipitation in panel C, were probed by immunoblotting with anti-MED7 antibodies.
Phosphorylated MED1 enhances TR-dependent transcription in vitro.
To investigate more closely the molecular mechanisms by which MED1 functions as a transcriptional coactivator for NRs, we set up an in vitro TR-dependent transcription assay using HeLa cell nuclear extract as a source of basal transcription factors, RNA Pol II, and coregulatory factors. As described previously (9), transcription was measured from a naked DNA reporter plasmid containing three TR binding elements (TREs) inserted upstream of the adenovirus major late minimal promoter (−53 to +10) (TRE3Δ53). Importantly, efficient activation of the TRE3Δ53 template required the addition of purified recombinant human TRα, human RXRα (the heterodimerization partner of TR), and T3 (Fig. 8B, compare lanes 1 and 2).
FIG. 8.
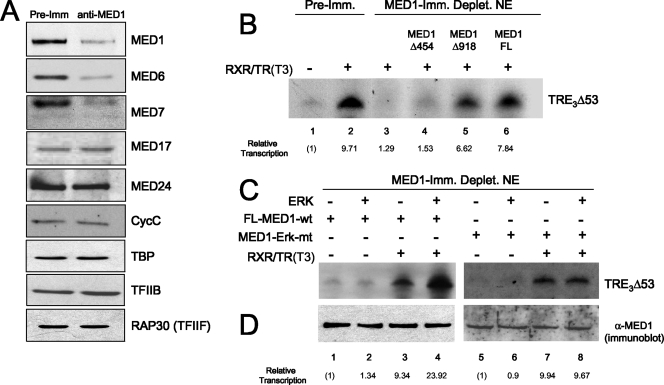
MED1 phosphorylation enhances TR-dependent transcription in vitro. (A) Immunodepletion of MED1 from HeLa cell nuclear extract. Shown is an immunoblot of HeLa nuclear extracts incubated with either anti-MED1 antibodies or preimmune serum (Pre-Imm). The specific antibodies used for immunoblotting are indicated on the right. (B) Purified recombinant MED1 restores TR-mediated transcription in MED1-depleted nuclear extract. Transcription was measured in vitro by incubating the TRE3Δ53 reporter gene in MED1-depleted (or preimmune mock-depleted) (Deplet.) HeLa cell nuclear extracts, together with purified baculovirus-expressed TRα, RXRα, and T3, as described previously (9). Purified bv-FL-MED1 (FL), -MED1-Δ454, or -MED1-Δ918 was added to reactions as indicated. (C) MED1 phosphorylation enhances TR-dependent transcription. In vitro transcription assays were performed as for panel B, except that either FL-MED1 wild type (FL-MED1-wt) or MED1-ERK mutant (MED1-Erk-mt) protein was preincubated in kinase buffer containing (+) or lacking (−) ERK1. The entire mock or true kinase reaction mixtures were then added to the transcription reactions. (D) As a control, parallel ERK (+ or −) kinase reactions were set up side by side and assayed via anti-MED1 immunoblotting.
In order to specifically address the functional role of MED1 in TR-dependent transcription, we immunodepleted MED1 from HeLa cell nuclear extracts using anti-MED1 antibodies (40). A mock-depleted nuclear extract using preimmune antibodies served as a control. The antibodies against MED1 quantitatively removed greater than 80% of the MED1, as determined by immunoblotting, whereas protein levels of TATA binding protein, TFIIB, TFIIF (RAP30), and other components of the basal transcription apparatus remained unchanged (Fig. 8A and data not shown). Consistent with a recent study reporting that MED1 exists in only a subpopulation of the total cellular Mediator complexes (41), significant levels of Mediator subunits MED17, MED24, and CycC were still present in the MED1-depleted nuclear extract (Fig. 8A). Nonetheless, we did observe that some Mediator subunits (e.g., MED6 and MED7) appeared to be semidepleted by various degrees, along with MED1. Pertinent to its role in NR coactivation, immunodepletion of MED1 from the HeLa nuclear extract resulted in a 7.5-fold reduction in TR-dependent transcription from the TRE3Δ53 promoter template (Fig. 8B, lanes 2 and 3).
We next examined whether addition of purified recombinant MED1 could restore TR-dependent transcription in vitro. Importantly, addition of wild-type bv-FL-MED1 to the MED1-depleted extract restored TR-dependent transcription to levels approximately 80% of that observed in the mock-depleted extract (Fig. 8B, lanes 2 and 6). To define the regions of MED1 required for TR coactivation, we next tested whether the MED1 deletion mutants CΔ454 and CΔ918 could restore TR-dependent transcription in the MED1-depleted extract. Although addition of CΔ454 had no significant effect, addition of CΔ918 restored transcription to nearly 70% of the level observed in the mock-depleted extract (Fig. 8B, lanes 2, 4, and 5). These findings most likely reflect the presence of the two LXXLL motifs, located between amino acids 604 and 645 of CΔ918 (Fig. 6B) but deleted in CΔ454, that are essential for targeting MED1 to RXR/TR heterodimers in the presence of T3 (29).
To investigate whether MED1 phosphorylation affects its coactivator activity in vitro, FL-MED1 or the MED1-ERK mutant was preincubated in kinase buffer containing or lacking ERK and then added to MED1-depleted nuclear extract and assayed for restoration of TR-dependent transcription (Fig. 8C). Transcription reaction mixtures containing prephosphorylated MED1 but lacking the template-specific activator RXR/TR served as controls for nonspecific affects of ERK (Fig. 8C, lanes 1, 2, 5, and 6). Intriguingly, we found that when FL-MED1 was prephosphorylated with ERK in vitro, TR-dependent transcription was stimulated ∼2.5-fold over levels conferred by FL-MED1 preincubated in mock kinase buffer alone (Fig. 8C, lanes 3 and 4). By contrast, preincubation of the MED1-ERK mutant with ERK in vitro had no stimulatory affect on TR-dependent transcription (Fig. 8C, lanes 7 and 8). Immunoblot assays confirmed that MED1 protein levels in each transcription reaction mixture were relatively equal (Fig. 8D). Given that ERK phosphorylation of MED1 did not significantly affect binding to TR (Fig. 4C), the findings here lend support to the idea that MED1 phosphorylation promotes its association with the Mediator complex, likely via mechanisms involving, but not limited to, stabilized interactions with MED7.
DISCUSSION
Mediator plays an essential role in eukaryotic transcription by acting as a functional adaptor between DNA-bound regulatory factors and the RNA Pol II-associated basal apparatus (15, 22). In humans, MED1 is a pivotal component of the complex that serves as a direct binding target for NRs and other types of gene-specific activators. Paradoxically, MED1 is only variably associated with Mediator, and the mechanisms regulating its entry into the core complex are unclear. Furthermore, surprisingly little is known about which Mediator subunits directly tether MED1 to the core complex. In light of our recent discovery that human MED1 is a substrate for MAPK-ERK, we hypothesized that MED1 phosphorylation might influence its interaction with other proteins, possibly potentiating its functional activity.
Here, we report that MED1 phosphorylation by ERK is required for its NR transcriptional-coactivator activity, as determined by transient-transcription assays (Fig. 3). Remarkably, both thyroid and steroid hormones were found to stimulate MED1 phosphorylation in vivo via extranuclear activation of ERK (Fig. 1 and 2). Pertinent to the molecular mechanism, we found that ERK phosphorylation of MED1 enhanced its association with Mediator (Fig. 5), but not with NRs (Fig. 4). Significantly, we found that MED1 directly binds to the MED7 subunit and that MED1 phosphorylation enhanced this interaction (Fig. 6 and 7). Finally, we found that ERK phosphorylation of MED1 enhanced TR-dependent transcription in vitro (Fig. 8). Our findings thus suggest that ERK phosphorylation of MED1 may be a regulatory mechanism that promotes its association with Mediator, at least in part, via interactions with MED7.
Human Mediator is comprised of at least 30 subunits arranged into four subcomplexes termed the head, tail, middle, and Cdk8 modules (2). Our findings show that human MED1 directly interacts with MED7 of the middle module, which is consistent with electron microscopy studies showing MED1 peripherally associated with the middle region of the human Mediator complex (2, 33, 34). Previous yeast studies also revealed a MED1-MED7 interaction in vitro (14), as did a pairwise yeast two-hybrid study (12), which, similar to our data, mapped the MED1 interaction domain of yeast MED7 to the C terminus. In the study here, we further mapped the minimal MED7 interaction domain of MED1 to amino acids 454 and 690. Given that ERK phosphorylates MED1 at Thr-1032 and Thr-1457 (26), phosphorylation is not an absolute requirement for MED7 binding. Nevertheless, we consistently found that ERK phosphorylation of FL-MED1 markedly enhanced its association with MED7 both in vitro and in vivo (Fig. 6 and 7). Our data thus suggest that phosphorylation at Thr-1032 and Thr-1457 may generate additional binding surfaces for MED7 and/or stabilize primary MED1-MED7 interactions, possibly via phosphorylation-dependent conformational changes. Our work is reminiscent of an earlier study in which phosphorylation of the NR coactivator SRC-3/AIB-1 enhanced its interaction with a number of cofactors despite the fact that the phosphorylation sites did not necessarily overlap with the interaction domains (38). The structural determinants underlying the phosphorylation-dependent MED1-MED7 interaction are currently under investigation.
Although the interaction between MED1 and MED7 appears to be conserved from yeast to humans, we do not rule out additional MED1 interactions with other Mediator subunits. Indeed, yeast MED1 can interact with as many as four subunits in the middle module (MED4, MED5, MED7, and MED9) and possibly one subunit in the tail module (MED14) (12, 14). Similarly, mutagenesis of human MED1 revealed two N-terminal domains (amino acids 108 to 212 and 215 to 390) that appear to be required for association with core Mediator (20). Furthermore, since association with RNA Pol II or various DNA-binding activators (including NRs) dramatically rearranges the overall conformation of the Mediator complex (25, 32, 33), particularly the middle and head modules (6), its conceivable that MED1 might exhibit differential interactions with several distinct Mediator subunits, depending on the transcriptional state of the core complex and/or the phosphorylation state of MED1. In this regard, the findings reported here are important in that they begin to better define how MED1 is tethered to the Mediator complex within the context of activated cellular signal transduction pathways.
A novel finding of this study is that thyroid and gonadal steroids are potent activators of MED1 phosphorylation. In view of the classic action of nuclear hormones, in which ligand directly binds cognate receptor and affects gene expression, the question arises as to the functional significance of this extraneous event. One scenario is that MED1 phosphorylation facilitates a feed-forward action of thyroid and steroid hormones in which specific NR coactivators, as well as NRs, become activated. Accordingly, nuclear hormones entering target cells rapidly activate ERK, leading to, among other events, phosphorylation of MED1, thus promoting its association with the core Mediator complex. Concomitantly, nuclear hormones bind to their cognate NRs, triggering allosteric changes that promote coactivator binding. Inherent in this model is ERK-directed phosphorylation of NRs themselves, which has been shown to promote nuclear localization, interaction with cofactors, increased transcriptional activity, and receptor turnover (16, 17). Moreover, the model would encompass phosphorylation and activation of other coactivators, including SRC-3 (38) and PGC-1 (28), that function in concert with Mediator. Importantly, rapid phosphorylation of both NRs and their coactivators likely potentiates subsequent NR-coactivator complex formation so that a maximal transcriptional response is rapidly achieved and sustained. Future studies should reveal whether other cellular signal transduction pathways target MED1 or other components of the Mediator complex for regulatory phosphorylation and affect their functional roles in NR-dependent gene expression.
ADDENDUM IN PROOF
Since the submission of this paper, it has been demonstrated by Ge et al. (K. Ge, Y.-W. Cho, H. Guo, T. B. Hong, M. Guermah, M. Ito, H. Yu, M. Kalkum, and R. G. Roeder, Mol. Cell. Biol. 28:1081-1091, 2008) that METD1 interacts with the MED7 subunit of the Mediator complex.
Acknowledgments
We thank H. Yoshikawa, R. Young, L. Myers, and J. W. Conaway for plasmids and N. Selvamurugan and B. Lewis for critically reading the manuscript and for helpful discussions.
This work was supported by NIH grant DK054030 awarded to J.D.F.
Footnotes
Published ahead of print on 7 April 2008.
REFERENCES
- 1.Belakavadi, M., and J. D. Fondell. 2006. Role of the mediator complex in nuclear hormone receptor signaling. Rev. Physiol. Biochem. Pharmacol. 15623-43. [DOI] [PubMed] [Google Scholar]
- 2.Chadick, J. Z., and F. J. Asturias. 2005. Structure of eukaryotic Mediator complexes. Trends Biochem. Sci. 30264-271. [DOI] [PubMed] [Google Scholar]
- 3.Cheskis, B. J. 2004. Regulation of cell signalling cascades by steroid hormones. J. Cell Biochem. 9320-27. [DOI] [PubMed] [Google Scholar]
- 4.Conaway, J. W., L. Florens, S. Sato, C. Tomomori-Sato, T. J. Parmely, T. Yao, S. K. Swanson, C. A. Banks, M. P. Washburn, and R. C. Conaway. 2005. The mammalian Mediator complex. FEBS Lett. 579904-908. [DOI] [PubMed] [Google Scholar]
- 5.Crawford, S. E., C. Qi, P. Misra, V. Stellmach, M. S. Rao, J. D. Engel, Y. Zhu, and J. K. Reddy. 2002. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J. Biol. Chem. 2773585-3592. [DOI] [PubMed] [Google Scholar]
- 6.Davis, J. A., Y. Takagi, R. D. Kornberg, and F. A. Asturias. 2002. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell 10409-415. [DOI] [PubMed] [Google Scholar]
- 7.Davis, P. J., F. B. Davis, and V. Cody. 2005. Membrane receptors mediating thyroid hormone action. Trends Endocrinol. Metab. 16429-435. [DOI] [PubMed] [Google Scholar]
- 8.Edwards, D. P. 2005. Regulation of signal transduction pathways by estrogen and progesterone. Annu. Rev. Physiol. 67335-376. [DOI] [PubMed] [Google Scholar]
- 9.Fondell, J. D. 2002. Gene activation by thyroid hormone receptor in vitro and purification of the TRAP coactivator complex. Methods Mol. Biol. 202195-214. [DOI] [PubMed] [Google Scholar]
- 10.Frade, R., M. Balbo, and M. Barel. 2000. RB18A, whose gene is localized on chromosome 17q12-q21.1, regulates in vivo p53 transactivating activity. Cancer Res. 606585-6589. [PubMed] [Google Scholar]
- 11.Gordon, D. F., E. A. Tucker, K. Tundwal, H. Hall, W. M. Wood, and E. C. Ridgway. 2006. MED220/thyroid receptor-associated protein 220 functions as a transcriptional coactivator with Pit-1 and GATA-2 on the thyrotropin-beta promoter in thyrotropes. Mol. Endocrinol. 201073-1089. [DOI] [PubMed] [Google Scholar]
- 12.Guglielmi, B., N. L. van Berkum, B. Klapholz, T. Bijma, M. Boube, C. Boschiero, H. M. Bourbon, F. C. Holstege, and M. Werner. 2004. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 325379-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Halachmi, S., E. Marden, G. Martin, H. MacKay, C. Abbondanza, and M. Brown. 1994. Estrogen receptor-associated proteins: possible mediators of hormone-induced transcription. Science 2641455-1458. [DOI] [PubMed] [Google Scholar]
- 14.Kang, J. S., S. H. Kim, M. S. Hwang, S. J. Han, Y. C. Lee, and Y. J. Kim. 2001. The structural and functional organization of the yeast mediator complex. J. Biol. Chem. 27642003-42010. [DOI] [PubMed] [Google Scholar]
- 15.Kornberg, R. D. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30235-239. [DOI] [PubMed] [Google Scholar]
- 16.Lange, C. A. 2004. Making sense of cross-talk between steroid hormone receptors and intracellular signaling pathways: who will have the last word? Mol. Endocrinol. 18269-278. [DOI] [PubMed] [Google Scholar]
- 17.Levin, E. R. 2005. Integration of the extranuclear and nuclear actions of estrogen. Mol. Endocrinol. 191951-1959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis, B. A., and D. Reinberg. 2003. The mediator coactivator complex: functional and physical roles in transcriptional regulation. J. Cell Sci. 1163667-3675. [DOI] [PubMed] [Google Scholar]
- 19.Lin, H. Y., F. B. Davis, J. K. Gordinier, L. J. Martino, and P. J. Davis. 1999. Thyroid hormone induces activation of mitogen-activated protein kinase in cultured cells. Am. J. Physiol. 2761014-1024. [DOI] [PubMed] [Google Scholar]
- 20.Malik, S., M. Guermah, C. X. Yuan, W. Wu, S. Yamamura, and R. G. Roeder. 2004. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol. Cell. Biol. 248244-8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malik, S., and R. G. Roeder. 2005. Dynamic regulation of pol II transcription by the mammalian Mediator complex. Trends Biochem. Sci. 30256-263. [DOI] [PubMed] [Google Scholar]
- 22.Malik, S., and R. G. Roeder. 2003. Isolation and functional characterization of the TRAP/mediator complex. Methods Enzymol. 364257-284. [DOI] [PubMed] [Google Scholar]
- 23.Migliaccio, A., G. Castoria, M. Di Domenico, A. De Falco, A. Bilancio, and F. Auricchio. 2002. Src is an initial target of sex steroid hormone action. Ann. N. Y. Acad. Sci. 963185-190. [DOI] [PubMed] [Google Scholar]
- 24.Misra, P., E. D. Owuor, W. Li, S. Yu, C. Qi, K. Meyer, Y. J. Zhu, M. S. Rao, A. N. Kong, and J. K. Reddy. 2002. phosphorylation of transcriptional coactivator peroxisome proliferator-activated receptor (PPAR)-binding protein (PBP). Stimulation of transcriptional regulation by mitogen-activated protein kinase. J. Biol. Chem. 27748745-48754. [DOI] [PubMed] [Google Scholar]
- 25.Naar, A. M., D. J. Taatjes, W. Zhai, E. Nogales, and R. Tjian. 2002. Human CRSP interacts with RNA polymerase II CTD and adopts a specific CTD-bound conformation. Genes Dev. 161339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandey, P. K., T. S. Udayakumar, X. Lin, D. Sharma, P. S. Shapiro, and J. D. Fondell. 2005. Activation of TRAP/Mediator subunit TRAP220/Med1 is regulated by mitogen-activated protein kinase-dependent phosphorylation. Mol. Cell. Biol. 2510695-10710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pearson, G., F. Robinson, T. Beers Gibson, B. E. Xu, M. Karandikar, K. Berman, and M. H. Cobb. 2001. Mitogen-activated protein (MAP) kinase pathways: regulation and physiological functions. Endocr. Rev. 22153-183. [DOI] [PubMed] [Google Scholar]
- 28.Puigserver, P., J. Rhee, J. Lin, Z. Wu, J. C. Yoon, C. Y. Zhang, S. Krauss, V. K. Mootha, B. B. Lowell, and B. M. Spiegelman. 2001. Cytokine stimulation of energy expenditure through p38 MAP kinase activation of PPARγ coactivator-1. Mol. Cell 8971-982. [DOI] [PubMed] [Google Scholar]
- 29.Ren, Y., E. Behre, Z. Ren, J. Zhang, Q. Wang, and J. D. Fondell. 2000. Specific structural motifs determine TRAP220 interactions with nuclear hormone receptors. Mol. Cell. Biol. 205433-5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stumpf, M., C. Waskow, M. Krotschel, D. van Essen, P. Rodriguez, X. Zhang, B. Guyot, R. G. Roeder, and T. Borggrefe. 2006. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc. Natl. Acad. Sci. USA 10318504-18509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taatjes, D. J., M. T. Marr, and R. Tjian. 2004. Regulatory diversity among metazoan co-activator complexes. Nat. Rev. Mol. Cell Biol. 5403-410. [DOI] [PubMed] [Google Scholar]
- 32.Taatjes, D. J., A. M. Naar, F. Andel III, E. Nogales, and R. Tjian. 2002. Structure, function, and activator-induced conformations of the CRSP coactivator. Science 2951058-1062. [DOI] [PubMed] [Google Scholar]
- 33.Taatjes, D. J., T. Schneider-Poetsch, and R. Tjian. 2004. Distinct conformational states of nuclear receptor-bound CRSP-Med. complexes. Nat. Struct. Mol. Biol. 11664-671. [DOI] [PubMed] [Google Scholar]
- 34.Taatjes, D. J., and R. Tjian. 2004. Structure and function of CRSP/Med2: a promoter-selective transcriptional coactivator complex. Mol. Cell 14675-683. [DOI] [PubMed] [Google Scholar]
- 35.Udayakumar, T. S., M. Belakavadi, K. H. Choi, P. K. Pandey, and J. D. Fondell. 2006. Regulation of Aurora-A kinase gene expression via GABP recruitment of TRAP220/MED1. J. Biol. Chem. 28114691-14699. [DOI] [PubMed] [Google Scholar]
- 36.Wada, O., H. Oishi, I. Takada, J. Yanagisawa, T. Yano, and S. Kato. 2004. BRCA1 function mediates a TRAP/DRIP complex through direct interaction with TRAP220. Oncogene 236000-6005. [DOI] [PubMed] [Google Scholar]
- 37.Wang, Q., D. Sharma, Y. Ren, and J. D. Fondell. 2002. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J. Biol. Chem. 27742852-42858. [DOI] [PubMed] [Google Scholar]
- 38.Wu, R. C., J. Qin, P. Yi, J. Wong, S. Y. Tsai, M. J. Tsai, and B. W. O'Malley. 2004. Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic reponses to multiple cellular signaling pathways. Mol. Cell 15937-949. [DOI] [PubMed] [Google Scholar]
- 39.Yuan, C. X., M. Ito, J. D. Fondell, Z. Y. Fu, and R. G. Roeder. 1998. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc. Natl. Acad. Sci. USA 957939-7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang, J., and J. D. Fondell. 1999. Identification of mouse TRAP100: a transcriptional coregulatory factor for thyroid hormone and vitamin D receptors. Mol. Endocrinol. 131130-1140. [DOI] [PubMed] [Google Scholar]
- 41.Zhang, X., A. Krutchinsky, A. Fukuda, W. Chen, S. Yamamura, B. T. Chait, and R. G. Roeder. 2005. MED1/TRAP220 exists predominantly in a TRAP/Mediator subpopulation enriched in RNA polymerase II and is required for ER-mediated transcription. Mol. Cell 1989-100. [DOI] [PubMed] [Google Scholar]
- 42.Zhu, Y., C. Qi, S. Jain, M. M. Le Beau, R. Espinosa III, G. B. Atkins, M. A. Lazar, A. V. Yeldandi, M. S. Rao, and J. K. Reddy. 1999. Amplification and overexpression of peroxisome proliferator-activated receptor binding protein (PBP/PPARBP) gene in breast cancer. Proc. Natl. Acad. Sci. USA 9610848-10853. [DOI] [PMC free article] [PubMed] [Google Scholar]