Abstract
Twofold reductions in telomerase RNA levels cause telomere shortening in both humans and the yeast Saccharomyces cerevisiae. To test whether multiple genes that affect telomere length act by modulating telomerase RNA abundance, we used real-time reverse transcription-PCR to screen S. cerevisiae deletion strains reported to maintain shorter or longer telomeres to determine the levels of their telomerase RNA (TLC1) abundance. Of 290 strains screened, 5 had increased TLC1 levels; 4 of these maintained longer telomeres. Twenty strains had decreased TLC1 levels; 18 of these are known to maintain shorter telomeres. Four strains with decreased TLC1 RNA levels contained deletions of subunits of Paf1C (polymerase II-associated factor complex). While Paf1C had been implicated in the transcription of both polyadenylated and nonpolyadenylated RNAs, Paf1C had not been associated previously with the noncoding telomerase RNA. In Paf1C mutant strains, TLC1 overexpression partially rescues telomere length and cell growth defects, suggesting that telomerase RNA is a critical direct or indirect Paf1C target. Other factors newly identified as affecting TLC1 RNA levels include cyclin-dependent kinase, the mediator complex, protein phosphatase 2A, and ribosomal proteins L13B and S16A. This report establishes that a subset of telomere length genes act by modulating telomerase RNA abundance.
Telomere length is a quantitative trait affected by more than 273 genes in Saccharomyces cerevisiae (1, 9, 12). To put this number into perspective, genes representing nearly 5% of the ∼6,000-gene yeast genome (15) influence telomeres. Some of these genes directly impact telomere homeostasis, either by contributing to the activity of the telomerase enzyme or by affecting the physical state of telomere ends. Most telomere length genes, however, lack obvious connections to telomere biology. The functions of these genes include vesicular transport, chromatin modification, ribosome assembly, and transcription. We reasoned that some of the telomere length genes with unexplained functions may affect telomerase activity by modulating the abundance of a core telomerase component, the noncoding telomerase RNA.
The fact that telomerase RNA levels can regulate telomere length is well established. In humans, dyskeratosis congenita (28), the bone marrow failure disease, is characterized by short telomeres resulting from telomerase RNA haploinsufficiency (11, 26, 27). In yeast studies, we have recently demonstrated a causal relationship between a twofold reduction in TLC1 (telomerase component 1) RNA, resulting from tlc1Δ heterozygosity, and telomere shortening: ectopic expression of TLC1 in TLC1/tlc1Δ cells rescues the telomere length defect (30). The mechanism of telomere length regulation revealed by telomerase RNA haploinsufficiency might be more general. That is, genes responsible for the transcription, processing, and stability of the telomerase RNA might influence telomere length.
Two published examples support this hypothesis. Reductions in TLC1 abundance, caused either by inability of the RNA to associate with the Sm heteroheptamer or by the mtr10Δ mutation, have been correlated with shortened telomeres (10, 40). Sm proteins are best known for binding near the 3′ ends and mediating the nuclear localization of small nuclear RNPs involved in mRNA splicing, so the idea that they influence telomerase, which is also a noncoding nuclear RNP, seemed reasonable. Mtr10p was first identified as an importin required for poly(A) RNA export through its nuclear importation of Npl3p, an mRNA-binding protein (34).
Given that yeast cells maintain telomerase RNA at a low, limiting level (30), it seemed likely that regulatory networks beyond Sm binding and MTR10 exist to modulate the transcription, 3′ end formation, and stability of TLC1. To search for genes in the telomerase RNA biogenesis pathway, we measured TLC1 RNA abundance in 290 candidate strains. In order to measure TLC1 RNA with the requisite sensitivity, we used real-time reverse transcription-PCR (RT-PCR) assays developed in our laboratory (30). While TLC1-specific oligonucleotides have been included on DNA microarrays designed for high-throughput screening for factors involved in noncoding RNA biogenesis (35), the steady-state levels of TLC1 were too low to yield signal above background. In the present study, our real-time RT-PCR approach was sensitive enough to identify 25 gene deletions (including mtr10Δ) that caused a 1.5-fold or greater change in telomerase RNA levels. The effect of these genes could be direct or could be mediated through as-yet-unidentified intermediates.
This screening uncovered the importance of an RNA polymerase II (Pol II)-associated transcriptional complex, called the Paf1 complex, in TLC1 biogenesis. Approximately 2% of cellular RNA Pol II is associated with Pol II-associated factor complex (Paf1C) (31) in complexes that are distinct from Srb-mediator Pol II holoenzymes (42). Paf1C has been shown to affect 3′ end formation of both polyadenylated and nonpolyadenylated Pol II transcripts (38), including SDA1 and MAK21 mRNAs (important for ribosome biogenesis) (36) and SNR47, a noncoding snoRNA (41). TLC1 RNA is transcribed by RNA Pol II, and a fraction of the RNA is polyadenylated (6). In this report, we show that cis-acting elements in both the promoter and the transcribed region of the TLC1 gene respond to Paf1C to enhance TLC1 RNA accumulation in S. cerevisiae.
Thus, this study identified a set of yeast genes that affect the steady-state level of TLC1 and are therefore candidates for being directly involved in TLC1 biogenesis. More generally, this report contributes to the field of telomere biology by identifying a previously unappreciated pathway through which a subset of telomere length genes act: the modulation of telomerase RNA abundance.
MATERIALS AND METHODS
Real-time RT-PCR screening for altered telomerase RNA abundance.
We selected 290 candidate gene deletion strains from the S. cerevisiae haploid gene deletion collection (14, 49). The MATa collection was purchased from Open Biosystems. Candidate strains bud16Δ, ctr9Δ, paf1Δ, set1Δ, sla2Δ, and mtr10Δ were not included in the MATa collection and were purchased individually or were purchased as heterozygous diploids and then sporulated. The isogenic wild-type control strain BY4733 (3) was purchased from ATCC (American Type Culture Collection; catalog no. 200895).
Deletion strains were grown in yeast extract-peptone-dextrose medium (2% dextrose) containing 200 μg/ml G418. Yeast total RNA was prepared from ∼108 cells harvested from cultures grown to cell densities between 9 × 106 and 2 × 107 cells/ml (as determined by counting with a hemocytometer). The cells were frozen in liquid nitrogen, stored at −80°C, resuspended in RLT lysis buffer (Qiagen), and subjected to four rounds of 30 s of glass-bead lysis in a FastPrep machine (QBiogene). The lysates were processed according to the RNeasy Mini kit protocol (Qiagen), using an on-column DNase treatment (Qiagen). Total RNA concentrations were quantified using a NanoDrop-1000 spectrophotometer (NanoDrop) and were serially diluted to 1.25 ng/μl into water containing 10 ng/μl MS2 carrier RNA (Roche). In vitro-transcribed standard RNAs TLC1(1261A55), ACT1, and U2 were taken from stocks described previously (30).
The total RNAs and standard RNAs were reverse transcribed as described previously (30), using the random nonamer/oligo(dT) primer mix provided in a QuantiTect RT kit (Qiagen). To quantify cDNAs corresponding to total TLC1 [the major poly(A)-negative species and the minor 3′-extended poly(A)-positive species], TLC1 containing the 3′ extension, U2, and ACT1, we used our previously published (30) real-time PCR scorpion assays with primer pairs p38 and p39, p90 and p91, p34 and p35, and p32 and p33, respectively. Real-time PCRs were set up essentially as described previously (30), with the following modifications. The p38-p39 (TLC1), p34-p35 (U2), and p32-p33 (ACT1) 20-μl assays were run simultaneously for each sample in 96-well plates in a LightCycler 480 instrument (Roche). LightCycler 480 probe master mix (Roche) was used for these real-time PCRs in accordance with Roche's protocol. The LightCycler 480 cycling parameters were 95°C denaturation for 10 min followed by 45 cycles of 95°C for 10 s, 57°C for 30 s (with a single fluorescence acquisition), and 72°C for 10 s followed by a 50°C cooling period for 10 s. Each 96-well plate contained samples to create five-point standard curves for TLC1, U2, and ACT1, a no-template control (water), and duplicate RT reaction mixtures from the wild-type strain and 12 candidate strains. The p90-p91 (3′ extended TLC1) assays were run in a LightCycler 2.0 instrument as described previously (30).
Real-time RT-PCR data analysis.
The TLC1/U2, TLC1/ACT1, and U2/ACT1 ratios listed in Table S1 in the supplemental material were obtained through a multistep process. First, the absolute quantification module of the LightCycler 480 basic software interpolated the number of TLC1, U2, and ACT1 molecules present in each PCR based on the internal standard curves, as described previously (30). These raw values were entered into a database; all the data were subsequently treated as a single pool rather than as representing discrete 96-well plates.
Second, the raw TLC1/U2 and TLC1/ACT1 ratios for each sample were calculated to correct for differences in the input amounts of total RNA in the RT reaction mixtures. (The U2/ACT1 ratio was also calculated as a way to flag samples whose U2 or ACT1 abundance noticeably differed from that of the wild type.) More than one real-time RT-PCR was performed for each candidate. The ratios are listed in Table S1 in the supplemental material.
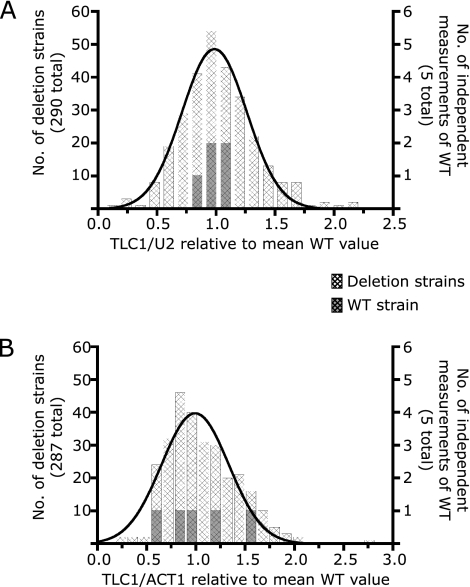
Third, the ratios for each candidate were normalized against the relevant wild-type ratios. These final numbers (shown in the Fig. 1 histograms) facilitate quick assessment of how different the telomerase RNA levels in each candidate deletion strain are from those of the wild type.
FIG. 1.
Distribution of TLC1 levels in 290 candidate deletion strains. For each strain listed in Table S1 in the supplemental material, the mean ratios of TLC1/U2 (panel A) and TLC1/ACT1 (panel B), normalized to the relevant wild-type (WT) mean ratio, were grouped into bins and plotted as a histogram. U2 is an snRNA (small nuclear RNA involved in mRNA splicing), and ACT1 is the actin mRNA. For comparison, measurements of the wild-type ratios from five independent RNA preparations are shown (dark cross-hatched bars). The sensitivity of ACT1 mRNA levels to cell density (30) may contribute to the spread in the TLC1/ACT1 ratios. A Gaussian distribution (black curve) was fitted to each histogram. The parameters of the Gaussian fits are mean = 0.986, standard deviation = 0.274, and R2 = 0.979 (A) and mean = 0.992, standard deviation = 0.342, and R2 = 0.909 (B).
Yeast strains and plasmids.
Strains denoted BY were S288C descendants (3), and strains denoted JJ were derived from D273-10b (46). Full genotypes of the following JJ strains were published previously (2): JJ662 (wild type), JJ665 (cdc73Δ), JJ1328 (ctr9Δ), JJ576 (paf1Δ), JJ1336 (leo1Δ), JJ1303 (rtf1Δ), JJ1202 (paf1Δ ctr9Δ), JJ1361 (paf1Δ leo1Δ), and JJ1326 (paf1Δ rtf1Δ). The pTLC1 plasmid (see Fig. 3) used in this study was pRS426-TLC1.
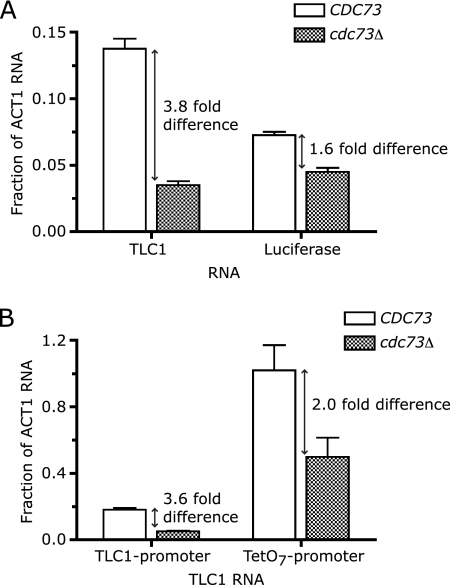
FIG. 3.
Ectopic expression of telomerase RNA partially rescues the telomere length and growth defects of Paf1C mutant cells. (A) Telomeres in cells containing either an empty high-copy-number (2μm) plasmid (lanes with no plasmid indicated) or a 2μm plasmid expressing TLC1 (pTLC1) were analyzed by Southern blotting. The changes in telomere length caused by increasing TLC1 RNA levels are indicated, as are TLC1 levels relative to ACT1 mRNA levels. Chr IV, chromosome IV. (B) Telomeres in strains (BY background) lacking telomere length maintenance genes that affect telomerase-independent pathway(s) were analyzed by Southern blotting. These strains contained either an empty 2μm plasmid or a 2μm plasmid expressing TLC1 (pTLC1). The changes in telomere length relative to wild type (WT) are indicated (diagonal numbers), as are the changes in telomere length between cells expressing an empty 2μm plasmid and pTLC1 (horizontal numbers). (C) Overexpressing TLC1 partially rescues the temperature sensitivity of Paf1C mutants in the JJ strain background. The same strains from which DNA was harvested for the Southern blot shown in panel A were grown to late log phase and suspended at a concentration of 108 cells/ml. Serial dilutions (100-fold; 108, 106, and 104 cells/ml) were then spotted on plates and incubated at either 30°C or 37°C. p-empty, empty high-copy-number (2μm) plasmid.
The luciferase reporter strains (see Fig. 4) were generated in two steps. First, the promoter for vector pJJ1358+TLC1 was made by cloning the TLC1 promoter into the KpnI site of the luciferase reporter integrating vector pJJ1358 (36). We assumed the TLC1 promoter sequences were contained within the 252-bp intergenic region between the closest upstream gene (SNR161) and TLC1. This promoter region was amplified with AMp166 (5′-GGGGTACCTCTAGAACTTGTGTTAGTTTATAAATAAATTTTATATCACTATATGTGTGG) and AMp167 (5′-GGGGTACCTAGTTTTATTCTCAAACCAGAAAAATCACACTAAAAGCTAC). The full sequence of the promoter for pJJ1358+TLC1 is available upon request. Second, the integrating vector was cleaved at the unique NcoI site within the URA3 marker and transformed into JJ662 (wild type) and JJ665 (cdc73Δ). Integrants were selected on synthetic media lacking uracil. End-point PCR was used to verify that the TLC1-promoter-luciferase-reporter cassette had integrated at the URA3 locus (the primers used were AMp180 [5′-CACATGCAGCTCCCGGAGACGGTCACAGCTTGTC] and AMp185 [5′-GTTACTTGGTTCTGGCGAGGTATTGGATAGTTCC]). Note that we generated luciferase reporter strains only in the JJ background; the BY strain background is ura3Δ0 and completely lacks a URA3 sequence into which the cassette could integrate.
FIG. 4.
The Paf1 complex affects both promoter-dependent and promoter-independent aspects of TLC1 RNA accumulation. (A) Loss of Cdc73p affects luciferase mRNA production from the TLC1 promoter. Total RNA was extracted from CDC73 and cdc73Δ strains expressing the TLC1 promoter-luciferase reporter and was reverse transcribed. TLC1 RNA and luciferase mRNA levels were then quantified by real-time RT-PCR and normalized to the same internal ACT1 mRNA measurement. Each bar represents the mean of the results obtained with four independent RNA preparations; the error bars represent standard errors of the means. (B) Loss of Cdc73p affects TLC1 expression, even when driven from a heterologous promoter. Total RNA was extracted from CDC73 and cdc73Δ strains expressing TLC1 either from its endogenous TLC1 promoter or from the heterologous TetO7 promoter and was reverse transcribed. TLC1 RNA was then quantified by real-time RT-PCR and normalized to ACT1 mRNA levels. (The same results were obtained when TLC1 was normalized to U2 snRNA levels; data not shown.) Each bar represents the mean of the results obtained with at least four independent RNA preparations; the error bars represent standard errors of the means.
The tetO7 promoter-TLC1 strain was constructed in two steps. First, we generated a PCR cassette to replace the 50 nucleotides upstream of the TLC1 coding sequence with the Kanr-tetO7-TATACYC1 heterologous promoter by use of AMp101 (5′-CATTGACATTTTCATAGGGTACCTATCTTCCTCTCTAGTTTTATTggatcccccgaattgatc), AMp102 (5′-AAAACTTCCTCTTTAGCAATGGTGACATATAGATCTCAAGGTTCTagcggataacaatttcacacagga), and the plasmid containing RP188, the Kanr-tetO7-TATACYC1 template (21). (Note that the uppercase primer sequences correspond to TLC1 sequence and the lowercase to the promoter cassette amplified from RP188.) Second, this PCR cassette was integrated into the TLC1 locus in strain R1158 (21). (R1158 contains the tTA transactivator integrated into the same BY MATa strain used in the genome deletion collection.) Colony-purified G418-resistant colonies were confirmed by PCR. TLC1 expression from the tetO7 promoter was effectively shut off in the presence of 10 μg/ml doxycycline, as verified by Northern blot analysis and real-time RT-PCR.
To generate the tetO7 promoter-TLC1 cdc73Δ strain, the tetO7 promoter-TLC1 strain was crossed to the BY cdc73Δ strain and the resulting diploid was sporulated. The spores were genotyped both by PCR, for detection of the presence of the tetO7 and cdc73Δ alleles, and by growth on plates lacking uracil, to select for strains containing the tTA transactivator, which is essential for tetO7 promoter activity.
Luciferase mRNA quantification.
Because TLC1 RNA is present in low abundance in cells (30), we reasoned that the TLC1 promoter may be too weak to drive sufficient luciferase expression for luminescence measurements. Instead, we monitored luciferase mRNA expression directly by real-time RT-PCR. The luciferase reporter strains (CDC73 TLC1 promoter-luciferase and cdc73Δ TLC1 promoter-luciferase) were grown to log phase and harvested. Total RNA was extracted, diluted, and reverse-transcribed, as described previously (30). The cDNA was amplified using the following luciferase-specific scorpion primer/probe set (designed by DxS Limited and synthesized by Biosearch Technologies): AMp164 (5′-F-CCGCGCTATGAAACGATATGGGCGCGG-Q-B-GCATACGACGATTCTGTGATTTGTA, where F is 6-carboxyfluorescein, Q is Black Hole Quencher 1, and B is hexaethylene glycol), and AMp165 (5′-ACGTACGCGGAATACTTCGAA). This primer set produced a 90-bp PCR product, which was measured in the LightCycler 480 instrument as described above. To determine the efficiency of the real-time PCR luciferase assay, we generated a five-point standard curve by use of 10-fold serial dilutions of luciferase control RNA purchased from Promega (catalog no. L4561); this Coleoptera luciferase mRNA corresponds to the sequence of Promega's pGEM-luc vector. Both the reaction efficiency (1.94) and the error (0.007) indicated a robust assay.
Initial measurements of 12 independent isolates from luciferase reporter strains revealed various luciferase mRNA levels, in multiples of the lowest observed number, suggesting that some of the strains contained more than one luciferase reporter cassette. To test this, we generated a standard curve using linearized pJJ1358+TLC1 promoter plasmid and measured luciferase copy numbers in the genomic DNA prepared from the 12 strains. Five showed multiple (two to four) integrations, which correlated with the observed mRNA expression results. To verify that these strains had multiple integrations, we designed an end-point PCR assay, using AMp180 (5′-CACATGCAGCTCCCGGAGACGGTCACAGCTTGTC) and AMp181 (5′-GGTTCACGTAGTGGGCCATCGCCCTGATAGACGG), that amplified a product only when head-to-tail copies of the luciferase cassette were present. A PCR product was generated for all the suspected multiple-integrant strains and none for the single-integrant strains. We used the single-integrant strains for the analyses (see Fig. 4B).
Northern and Southern blot analyses.
Northern and Southern blot analyses were performed as described previously (53). Relative changes in telomere length were measured using ImageQuant TL.
RESULTS
Selection of gene deletion strains.
The 290 candidate strains were selected by the following criteria: (i) 273 strains had been found to have altered telomere lengths in large-scale telomere length screenings (1, 12); (ii) 11 had been reported to have a clumpy cell morphology (49) similar to that of the observed microscopic phenotype of tlc1Δ cells (A. D. Mozdy and T. R. Cech, unpublished data); (iii) 5 had been found to show deletions of genes that interact with the Paf1 complex and were added during the study, after we identified the importance of the Paf1 complex; and (iv) 1 (mtr10Δ) had been previously shown to have short telomeres because of reduced TLC1 levels (10) and therefore served as a positive control.
A subset of telomere length genes affects telomerase RNA abundance.
We measured telomerase RNA (TLC1) levels in 290 nonessential gene deletion strains by using real-time RT-PCR assays we had described previously (30). Five independent RNA preparations from wild-type cells showed similar TLC1/U2 RNA ratios (Fig. 1A) but more variability in their TLC1/ACT1 RNA ratios (Fig. 1B), suggesting that some of the spread observed for the deletion strains was due to experimental variability. We therefore required that a strain have both TLC1/U2 and TLC1/ACT1 ratios that differed from those of the wild type by at least 1.5-fold in order to be scored as having a significantly different TLC1 RNA level; the 25 strains that met this standard are listed in Table 1. This conservative criterion excludes candidates whose TLC1 count appears altered relative to the presence of only one normalizer (either U2 or ACT1). While such candidates may indeed impact TLC1, we erred on the stringent side to limit false positives arising from genes that instead affect U2 or ACT1 RNAs. For example, the TLC1/U2 ratio in the lea1Δ strain is twofold higher than in wild-type cells (the TLC1/ACT1 ratio indicates a lesser, 1.3-fold effect). Lea1p is a component of the U2 snRNP and is necessary for normal accumulation of U2 (44). The inflated TLC1/U2 ratio in lea1Δ cells is therefore at least partially due to depressed U2 levels. The TLC1/U2, TLC1/ACT1, and U2/ACT1 ratios for all 290 strains can be found in Table S1 in the supplemental material.
TABLE 1.
Telomere length genes that affect telomerase RNA levels by at least 1.5-fold
| Gene | Open reading frame | Gene ontology annotation | Telomere phenotype (reduction)a | Telomere phenotype reference(s) | Ratio normalized against WTb
|
No. of RNA preparations | TLC1 transcripts WT in sizec | ||
|---|---|---|---|---|---|---|---|---|---|
| TLC1/U2 | TLC1/ACT1 | 3′-extended TLC1/total TLC1 | |||||||
| MTR10 | YOR160W | hnRNP import into nucleus | Short | 10 | 0.15 | 0.23 | 2.33 | 1 | NDd |
| CDC73 | YLR418C | Transcription, Paf1 complex | Short (160 bp) | This study (1, 12) | 0.20 | 0.25 | 2.14 | 4 | Yes |
| CTR9 | YOL145C | Transcription, Paf1 complex | Short (170 bp) | This study | 0.25 | 0.35 | 1.57 | 3 | Yes |
| PAF1 | YBR279W | Transcription, Paf1 complex | Short (110 bp) | This study (12) | 0.25 | 0.38 | 1.91 | 4 | Yes |
| LEO1 | YOR123C | Transcription, Paf1 complex | Short (110 bp) | This study (12) | 0.40 | 0.43 | 2.12 | 4 | Yes |
| NUT3 | YBL093C | Transcription, mediator complex | Short (140 bp) | This study | 0.43 | 0.59 | 1.16 | 3 | Yes |
| ARF1 | YDL192W | Vesicular transport | Slightly short | 12 | 0.48 | 0.54 | 1.65 | 2 | ND |
| YKU70 | YMR284W | Telomerase RNP | Very short | 1, 12 | 0.53 | 0.67 | 1.61 | 2 | Yes |
| TPD3 | YAL016W | Protein phosphatase type 2A regulator | Short | 1 | 0.54 | 0.60 | 0.72 | 2 | ND |
| RPL13B | YMR142C | Ribosome biogenesis/assembly | Slightly short | 1, 12 | 0.55 | 0.64 | 0.88 | 2 | Yes |
| HUR1 | YGL168W | DNA replication | Short (75 bp) | This study (1) | 0.56 | 0.59 | 0.64 | 3 | Yes |
| VPS34 | YLR240W | Vesicular transport | Short | 1, 12 | 0.56 | 0.65 | 0.70 | 2 | ND |
| AGP2 | YBR132C | Amino acid transport | Short | 1 | 0.57 | 0.55 | 1.03 | 2 | Yes |
| ARV1 | YLR242C | Vesicular transport and sterol metabolism | Short | 1 | 0.59 | 0.63 | 0.90 | 2 | ND |
| UGO1 | YDR470C | Mitochondrion | Slightly long | 1 | 0.59 | 0.57 | 0.75 | 2 | ND |
| SMI1 | YGR229C | Cell wall organization/biogenesis | Short | 1 | 0.59 | 0.59 | 1.05 | 2 | ND |
| HDA2 | YDR295C | Transcription, histone deacetylation | Short | 1 | 0.61 | 0.63 | 1.26 | 2 | ND |
| HFI1 | YPL254W | Transcription, SAGA complex adaptor | Short (120 bp) | This study (12) | 0.61 | 0.63 | 1.38 | 3 | ND |
| CWP2 | YKL096W-A | Cell wall organization/biogenesis | ND | 0.62 | 0.58 | 1.85 | 2 | Yes | |
| PHO85 | YPL031C | Signal transduction kinase | Short | 1 | 0.65 | 0.64 | 1.16 | 2 | ND |
| RPS16A | YMR143W | Ribosome biogenesis/assembly | Slightly long | 1, 12 | 1.54 | 1.66 | 0.84 | 2 | Yese |
| SRB2 | YHR041C | Transcription, mediator complex | Slightly short | 1, 12 | 1.65 | 1.78 | 0.35 | 1 | ND |
| ARD1 | YHR013C | Protein acetylation | Very long | 12 | 1.90 | 1.58 | ND | 1 | ND |
| PPE1 | YHR075C | Protein phosphatase 2A methylesterase | Long | 1 | 1.93 | 1.55 | ND | 1 | ND |
| REF2f | YDR195W | RNA 3′-end processing | Slightly long | 12 | 1.44 | 2.75 | 1.77 | 3 | Yes |
Telomere length classifications follow those in previous reports (1, 12): slightly short (<50 bp shorter than WT), short (50 to 200 bp shorter than WT), very short (>150 bp shorter than WT), slightly long (<50 bp longer than WT), long (50 to 300 bp longer than WT), and very long (>150 bp longer than WT). Note that some categories overlap because the reported telomere length categories differ (1, 12). For telomeres examined in this study, specific telomere length reductions are indicated in parentheses.
The mean ratio for each gene deletion strain was normalized against the relevant mean wild-type (WT) ratio (the ratio for TLC1/U2 is 0.0406, for TLC1/ACT1 is 0.155, and for TLC1-3′ ext/total TLC1 is 0.160).
TLC1 transcripts were examined by Northern blot analysis. Two major TLC1 transcripts are visible in total RNA isolated from wild-type cells: a 1.157-kb mature form and a larger, polyadenylated, 3′ extended form.
ND, not determined.
The mature, 1.157-kb TLC1 transcript is present in cells lacking Rps16Ap; however, the 3′ extended form is not discernible by Northern analysis.
The ref2Δ strain does not fit the strict criterion that both the TLC1/U2 and TLC1/ACT1 ratios must differ from that of the WT by at least 1.5-fold. It is included in this table because of all the strains examined, telomerase RNA levels in this strain are the most dramatically increased. The normalized U2/ACT1 ratio (1.9) indicates that U2 snRNA levels in ref2Δ cells are also elevated, which could account for the observed TLC1/U2 ratio.
As is consistent with observations that telomerase abundance influences telomere length (8, 30), 18 of the 20 deletion strains that contained less TLC1 than wild type are known to have short telomeres, while 4 of the 5 deletion strains that contained more TLC1 than wild type had long telomeres (Table 1). Of the 290 candidate strains, deletion of MTR10 had the largest impact (an approximately sevenfold decrease) on telomerase RNA abundance. Because Mtr10p is an importin that is already known to contribute to TLC1 RNA levels (10, 30), its appearance in Table 1 provides some validation of our screening.
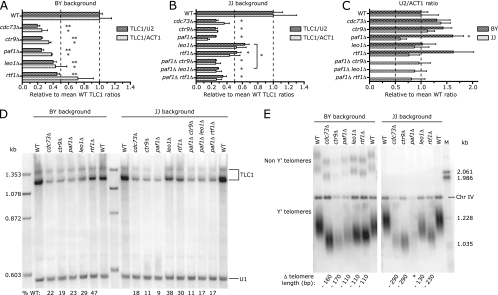
The next largest effects (greater than twofold decreases) were observed with cdc73Δ, ctr9Δ, paf1Δ, and leo1Δ cells. The products of these four genes physically and functionally interact within Paf1C (31). In addition to exhibiting reduced overall levels of TLC1, the Paf1C mutant strains contain altered levels of the 3′-extended form of TLC1 relative to the mature form (Table 1). Because the screening unambiguously revealed the importance of the Paf1 complex in TLC1 biogenesis, we focused the remainder of this study on Paf1C. The other hits are addressed in the Discussion.
The Paf1 complex is required for telomerase RNA to accumulate to wild-type levels.
The Paf1C is comprised of Cdc73p, Ctr9p, Paf1p, Leo1p, and Rtf1p (31). Four of the five subunits were identified in the real-time RT-PCR screening as altering the TLC1/U2 and TLC1/ACT1 ratios by at least 1.5-fold; the rtf1Δ strain missed the cutoff for inclusion in Table 1 because its TLC1/ACT1 ratio was 1.4-fold lower than that of the wild type. To further explore the role of the Paf1 complex in TLC1 transcription/stability, we examined TLC1 RNA from strains lacking each of the five subunits in two genetic backgrounds (BY strains and JJ strains). Both real-time RT-PCR and Northern blot analyses indicated that TLC1 is reduced approximately fourfold in strains lacking Cdc73p, Paf1p, or Ctr9p, and approximately twofold in strains lacking Leo1p or Rtf1p, in both backgrounds (Fig. 2A, B, and D). This two- to fourfold drop in TLC1 RNA levels was consistent with the pattern reported for other Paf1C transcriptional targets in paf1Δ cells (5). The TLC1 transcripts appeared to be similar to the wild type in size, although reduced in abundance (Fig. 2D). The U2/ACT1 ratios were not depressed in the Paf1C mutants (Fig. 2C), demonstrating that the Paf1 complex does not affect all noncoding RNAs transcribed by Pol II.
FIG. 2.
The Paf1 complex is necessary for wild-type telomerase RNA accumulation and telomere maintenance. (A, B, and C) TLC1, U2, and ACT1 RNA levels were measured by real-time RT-PCR and normalized as indicated. At least three independent RNA preparations were analyzed for each strain shown, except for the JJ paf1Δ strain, where n = 2. Error bars represent the standard errors of the means. TLC1 levels in each deletion strain were compared to TLC1 levels in the isogenic wild-type (WT) strain by use of a Student's two-tailed t test (*, P < 0.05; **, P < 0.001). The bracket in panel B indicates a significant difference between leo1Δ cells and paf1Δ leo1Δ cells in TLC1 abundance levels. (D) The same RNA samples were analyzed by Northern blotting; both the 3′ extended and mature forms of TLC1 are indicated by the bracket. (E) To assess telomere length in the Paf1C mutant strains, genomic DNA was prepared and analyzed via Southern blotting. A change in telomere length relative to wild-type results is indicated. Hybridization to a chromosome IV (Chr IV) restriction fragment serves as an internal length standard and a loading control. Note that the XhoI digest patterns differ between the non-Y′ telomeres of the BY and the JJ strain backgrounds. *, insufficient genomic DNA to measure telomere fragment length due to poor viability of the JJ paf1Δ strain.
We measured TLC1 in paf1Δ leo1Δ and paf1Δ rtf1Δ double mutants (Fig. 2B), because leo1Δ and rtf1Δ mutations have been reported to suppress paf1Δ phenotypes such as slow growth, temperature sensitivity, and decreased expression of several Paf1C transcriptional targets (31). For example, the level of mRNA of CLN1, the G1 cyclin, is reduced threefold in paf1Δ cells but is restored to wild-type levels in paf1Δ rtf1Δ double mutants (31). In contrast, neither the loss of Leo1 nor the loss of Rtf1 suppressed the low TLC1 levels observed in paf1Δ cells (Fig. 2B; see results for paf1Δ leo1Δ and paf1Δ rtf1Δ double mutants). Instead, we found that paf1Δ is epistatic to leo1Δ with respect to TLC1 abundance. The difference between leo1Δ cells and paf1Δ leo1Δ cells in TLC1 levels is statistically significant (P < 0.05 using a Student's t test). (The difference between rtf1Δ and paf1Δ rtf1Δ cells in TLC1 levels is not statistically significant [P = 0.07].)
As is consistent with the most dramatic reductions in TLC1 RNA, telomeres were shortest in cdc73Δ, paf1Δ, and ctr9Δ cells in both the BY and JJ backgrounds (Fig. 2E). A subtle difference between BY and JJ backgrounds for the leo1Δ and rtf1Δ strains was observed: TLC1 levels were more depressed in leo1Δ than in rtf1Δ BY cells, while the opposite was observed for JJ cells.
More telomerase RNA partially rescues the telomere length and growth defects in Paf1C mutant cells.
Cells lacking Paf1C components have shortened telomeres and reduced TLC1 RNA levels. To test whether insufficient telomerase RNA is responsible for the telomere length defect, we expressed TLC1 from a high-copy plasmid in Paf1C mutant strains. Because TLC1 RNA expressed from its endogenous promoter fails to accumulate to wild-type levels in Paf1C mutants, we measured TLC1 RNA by real-time RT-PCR to see whether high-copy-number plasmid expression could boost steady-state TLC1 levels. As shown at the bottom of Fig. 3A (TLC1/ACT1 ratios), Paf1C mutant strains expressing the high-copy TLC1 plasmid contained at least 10-fold more TLC1 RNA than the Paf1C strains carrying an empty plasmid and at least 3-fold more TLC1 RNA than wild-type cells. This excess TLC1 RNA was processed normally; the 3′ extended/total TLC1 ratios were indistinguishable between cdc73Δ cells expressing endogenous and high-copy plasmid levels of TLC1 (data not shown). After fewer than 50 generations posttransformation with the high-copy-number TLC1 plasmid, telomeres were markedly lengthened in cdc73Δ and ctr9Δ cells and slightly lengthened in paf1Δ cells (Fig. 3A). Telomeres were also slightly lengthened in leo1Δ cells from the BY background but slightly shortened in those from the JJ background.
It seemed possible that TLC1 overexpression might cause general telomere lengthening, even in short-telomere strains that do not have reduced telomerase RNA levels. We therefore tested three strains with normal TLC1 levels (see Table S1 in the supplemental material): dcc1Δ, vps28Δ, and upf3Δ. In addition to the data reported here, previous double-mutant analyses of dcc1Δ tlc1Δ, vps28Δ tlc1Δ, and upf3Δ tlc1Δ indicated that the effects of these genes on telomere length were not exclusively dependent on the presence of telomerase (12). While TLC1 overexpression had subtle effects on telomere lengths in dcc1Δ, vps28Δ, and upf3Δ cells (Fig. 3B), the lack of a clear gain in telomere length as observed in cdc73Δ and ctr9Δ cells after the same generation time suggested that excess telomerase RNA does not generally impact short telomeres in fewer than 50 generations. The rescue of short telomeres in cdc73Δ and ctr9Δ cells, then, is likely due to reversing the cause of the short telomeres, i.e., to insufficient TLC1 RNA levels.
The observed partial, rather than complete, restoration of telomeres to wild-type length may be explained by the short generation time. If we were to passage these cells through more generations, telomeres might fully recover. To avoid ambiguities in interpretation, given the observation that TLC1 overexpression for 250 generations causes telomere lengthening in wild-type cells (30), presumably via an adaptive cellular process that responds to increased steady-state TLC1 levels over time, we did not track telomere length beyond 50 generations. Instead, we looked for quick and dramatic telomere lengthening in a restricted time frame during which the telomeres of wild-type cells are unaffected by increased telomerase RNA.
We noticed that Paf1C mutant strains containing the high-copy-number TLC1 plasmid grew better in liquid culture than the same strains containing an empty plasmid. To examine this growth phenotype more closely, we spotted serially diluted cells onto plates and grew them at 30°C and 37°C (Fig. 3C). As previously reported, the cdc73Δ, ctr9Δ, paf1Δ, and rtf1Δ mutations caused temperature sensitivity in the JJ background (2). This temperature sensitivity phenotype was partially rescued by TLC1 overexpression (Fig. 3C, top right panel). Paf1C mutants in the BY background were not as sensitive to high temperature (Fig. 3C, bottom panels). This observation raises the possibility that temperature sensitivity is related to telomere length, because telomeres are shorter in the JJ Paf1C mutants than in the BY Paf1C mutants (Fig. 2E). Another possibility is that increased telomerase RNA affects the levels or functions of associated proteins that impact temperature sensitivity. A practical outcome of the observation that excess telomerase RNA can partially rescue telomere length and growth defects of Paf1C mutants is that growing difficult Paf1C mutant strains can be augmented by transforming these cells with a high-copy-number TLC1 plasmid.
Paf1C has both promoter-dependent and promoter-independent effects on telomerase RNA.
Although the Paf1 complex accompanies Pol II for all transcriptionally active yeast genes (32), its loss affects only a small subset of transcripts (36). At least some of these targets rely on Paf1C for posttranscriptional processing (36, 41) but not for initiation, raising the following question: is Paf1C required at the promoter (e.g., for initiation of TLC1 transcription) or for downstream events, which could include transcript elongation and 3′ end formation? To address this question, we performed two sets of experiments. First, we tested the sensitivity of the TLC1 promoter to the cdc73Δ mutation by use of a luciferase reporter. Second, we tested the sensitivity of the TLC1 transcribed region to cdc73Δ by use of a heterologous promoter driving TLC1 expression.
We measured accumulation of telomerase RNA and luciferase mRNA, both driven by the TLC1 promoter, in CDC73 and cdc73Δ strains. As expected, telomerase RNA levels were reduced nearly fourfold in cdc73Δ cells compared to CDC73 cell results (Fig. 4A, left bars). There was also less luciferase mRNA in cdc73Δ cells than in CDC73 cells, although the effect was smaller (a 1.6-fold reduction) (Fig. 4A, right bars). A Student's t test revealed this 1.6-fold difference to be significant (P < 0.001). This depression in luciferase mRNA production was not due to an effect of cdc73Δ on the luciferase transcript itself; when a luciferase construct lacking the TLC1 promoter was integrated into the URA3 locus so that luciferase transcription was driven from the nearest promoter element, luciferase mRNA accumulated to the same low level in CDC73 and cdc73Δ strains (data not shown). This reporter analysis indicated that Paf1C is required for normal transcription from the TLC1 promoter but that this promoter effect alone is too small to account for the observed reduction of TLC1 RNA levels in cdc73Δ cells.
To examine the effect of cdc73Δ on the transcribed TLC1 region, we replaced the endogenous TLC1 promoter with a heterologous promoter, tetO7 (29). We then measured TLC1 RNA levels in the following four isogenic strains: CDC73 TLC1 promoter-TLC1, cdc73Δ TLC1 promoter-TLC1, CDC73 TetO7 promoter-TLC1, and cdc73Δ TetO7promoter-TLC1. TLC1 RNA accumulated to approximately sixfold-higher levels when expressed from the tetO7 promoter compared to the results seen with its endogenous promoter (Fig. 4B). The important comparison (between CDC73 tetO7 promoter-TLC1 cells and cdc73Δ tetO7 promoter-TLC1 cells) revealed a twofold decrease in TLC1 RNA levels upon loss of Cdc73p (Fig. 4B, right bars). These data indicated that the Paf1 complex also plays a promoter-independent role in TLC1 biogenesis. The decrease in TLC1 levels observed in cdc73Δ cells, then, is multiplicative: an approximately twofold promoter-dependent role and a twofold promoter-independent role for Paf1C account for the approximately fourfold total effect on TLC1 RNA accumulation.
DISCUSSION
Overview of the telomerase RNA abundance screening.
Our real-time RT-PCR screening identified 24 genes not known previously to affect TLC1 steady-state abundance and replicated the finding (10) that MTR10 affects TLC1 biogenesis. These genes affect diverse biological functions (Table 1). The most-represented function is transcription; we found four members of the Pol II-associated complex Paf1C (CDC73, CTR9, PAF1, and LEO1), two members of the Pol II-associated mediator complex (NUT3 and SRB2), and two genes that modulate the histone acetylation state (HDA2 and HFI1). These gene products could be directly involved in TLC1 RNA transcription or they could affect the expression of an intermediary protein that contributes to TLC1 RNA accumulation. The functional breakdown of the remaining genes is as follows: (i) vesicular transport (ARF1, VPS34, and ARV1); (ii) RNA processing (MTR10 and REF2); (iii) protein phosphatase 2A activity (TPD3 and PPE1); (iv) telomerase RNP production (YKU70); (v) signal transduction (PHO85); (vi) ribosome biogenesis (RPL13B and RPS16A); and (vii) genes whose broad biological functions are unclear (HUR1, AGP2, UGO1, SMI1, CWP2, and ARD1).
Because the screening identified multiple members of the Paf1 complex, we used the Osprey network visualization tool (4) to look for other interactions between the genes listed in Table 1. The following pairs interact genetically: genes SMI1 and PHO85 (synthetic lethality) (25, 45); genes PHO85 and HFI1 (synthetic lethality) (20, 24); genes HFI1 and NUT3, genes SRB2 and ARD1, and genes ARD1 and HUR1 (phenotypic suppression) (7); genes HUR1 and YKU70, genes YKU70 and CDC73, and genes ARD1 and CDC73 (phenotypic enhancement) (7); and genes PPE1 and TPD3 (synthetic rescue) (19). This final pair is interesting, because tpd3Δ cells contain decreased telomerase RNA and short telomeres whereas ppe1Δ cells contain increased telomerase RNA and long telomeres. The opposing effects make sense in light of the opposing functions of Ppe1p and Tpd3p, both of which regulate protein phosphatase 2A activity. Protein phosphatase 2A activity is decreased in tpd3Δ cells and increased in ppe1Δ cells (19); this observation, in conjunction with our analysis, indicates that protein phosphatase 2A activity leads to increased transcription or stability of telomerase RNA. It will be interesting to test whether other protein phosphatase 2A holoenzyme components (RRD2, PPH21, CDC55, and PPM1) affect telomerase RNA levels and/or telomere length.
NUT3 and SRB2 interact physically (13) within the mediator complex. Yeast mediator is a 25-protein complex that regulates Pol II transcriptional initiation (22). Of the 15 nonessential mediator components, 12 (NUT3, SRB5, SOH1, NUT1, GAL11, MED1, PGD1, SRB2, SRB8, SRB9, SRB10, and SRB11) are reported to affect telomere length (1, 12). These 12 define components of the head, middle, tail, and cyclin-dependent kinase (CDK) mediator modules (16). The CDK module is a negative regulatory module (17) that represses transcription of ∼3% of the yeast genome (18). Deletion of any of the four CDK components (SRB8 to SRB11) results in long telomeres and elevated levels of telomerase RNA (see Table S1 in the supplemental material). The subtle (<1.5-fold) increases in TLC1 levels observed by real-time PCR were confirmed by Northern analysis (data not shown), suggesting that TLC1 is a novel target of CDK repression. On the other hand, loss of NUT3, the head component, leads to a twofold drop in TLC1 levels (Table 1). Altogether, our data suggest that mediator is important for TLC1 transcription but that its effect is negatively regulated by the CDK module.
Three of the genes listed in Table 1, CTR9, NUT3, and CWP2, were not previously known to affect telomerase and telomeres. This is not surprising, given that the published genome-wide screenings for telomere length defects were estimated to have missed ∼40% of genes that affect telomere length (9). We analyzed ctr9Δ cells because the Paf1C is clearly important for TLC1 accumulation. We analyzed nut3Δ and cwp2Δ cells solely because their clumpy cell morphology is reminiscent of the tlc1Δ phenotype we observed (Mozdy and Cech, unpublished). We do not know whether or how telomerase RNA levels are linked to cellular morphology; however, clumpiness was either a reasonable or a fortuitous criterion for inclusion in our screening because it led to the identification of two additional genes that affect telomerase RNA levels.
The Paf1 complex affects promoter-dependent and promoter-independent steps of TLC1 production.
This study revealed that the Paf1 complex is important for TLC1 biogenesis, with quantitatively similar effects on the transcribed region of the gene (observed with a heterologous promoter) and on the DNA sequences preceding the start site for transcription—the putative promoter. We were unable to test whether these effects are direct or indirect using either of two obvious approaches: use of directed mutations of Paf1C-responsive cis elements within TLC1 or use of a PAF1 conditional allele. First, no significant consensus sequence larger than 4 bp has been identified in the promoters of the Paf1C primary target genes (36), so it is unclear what element(s) in the TLC1 promoter the Paf1C might recognize. In addition, no common elements in the 3′ untranslated regions of Paf1 targets been identified (36). Second, the long half-life of TLC1 precludes distinguishing direct from indirect effects in a time course following shutoff of a conditional PAF1 allele. TLC1's half-life has been reported to exceed 1 h (23) and to be at least 2 h (6). To conclude that a transcript is a direct Paf1C target, a previous study required that the transcript undergo a twofold change in expression over a 2-h time period following shutoff of a tetracycline-regulated PAF1 (36). By the time (∼2 h) a twofold change could be detected in TLC1 abundance, other shorter-lived transcripts would have been affected, obfuscating interpretation.
For the remainder of this discussion, we assume that the TLC1 gene is the direct target of Paf1C, although we do not discount the possibility that Paf1C regulates production of other factors that in turn interact with the promoter region and the transcribed region of TLC1. Even with this assumption, the mechanism of the Paf1C effect is complicated to address, because the Paf1 complex has been implicated in all stages of Pol II transcription, from initiation to elongation to 3′ end formation. Cdc73p forms a stoichiometric complex with Pol II (42), and Paf1C components are found at promoters and throughout the coding regions of genes (37, 43). The Paf1C interacts with elongation complexes that modify chromatin via histone methylation and ubiquitination (33, 48, 50, 51) and with 3′ end formation complexes, including the THO component of the transcription export complex (5) and Nrd1p and Nab3p, the snoRNA terminators (41). Although many of these Paf1C interaction partners (BRE1, BRE2, CCR4, CDH1, CTK1, HPR1, MFT1, RAD6, SET1, SPT4, and THP2) were included in this screening, none appeared to impact TLC1 RNA levels as much as Paf1C did. While it is intriguing that the THO complex members HPR1, MFT1, and THP2 have subtle but clear effects on TLC1 levels (see Table S1 in the supplemental material), this observation is insufficient to suggest that RNA export is the critical step in the Paf1C-guided journey to TLC1 RNA maturation.
For many of the known Paf1C transcriptional targets, the critical Paf1C role was shown to be promoter-independent, posttranscriptional processing (36, 41). Specifically, Paf1C is important for the proper utilization of a subset of poly(A) sites; loss of the Paf1C leads to reduced utilization of proximal poly(A) sites and increased utilization of distal poly(A) sites for primary target genes SDA1 and MAK21 (36). TLC1 RNA is unusual in having both poly(A)+ and poly(A)− forms and in having an A-U-rich 3′ region with multiple candidate polyadenylation sites (6). Decreased utilization of a major TLC1 poly(A) site in Paf1C mutants could partially account for reduced TLC1 RNA. The published U1221G mutation in a TLC1 polyadenylation efficiency element (nucleotides 1221 to 1226) (6) leads to a ∼1.4-fold reduction in the predominant, mature form of TLC1 (∼1.157 kb long), a greater than threefold reduction in the less-abundant, 3′-extended, polyadenylated form (∼1.35 kb long), and the appearance of a larger transcript that is visible on a Northern blot but is too faint to be quantified (Mozdy and Cech, unpublished). Although no discrete elongated TLC1 species was discernible by Northern blot analysis in total RNA extracted from paf1C mutant strains, there was a clear increase in the ratio of 3′-extended TLC1 to total TLC1 in the absence of Paf1C (Table 1). The observation that a TLC1 cis element predicted to guide polyadenylation enhances both TLC1 accumulation and proper 3′ end formation gives plausibility to a model in which the Paf1C enhances utilization of TLC1 polyadenylation signals that generate a stable RNA.
However, there are also reasons to believe that the role of Paf1C in TLC1 transcription may be more complex. First, no aberrant 3′ ends were observed for TLC1 transcripts isolated from Paf1C mutant cells (Fig. 2D), in contrast to previous results for other Paf1C targets (36, 41). Second, TLC1 RNA analysis results deviate from the short half-life pattern of other primary targets that exhibit decreased abundance upon loss of Paf1 (36). The average yeast mRNA half-life is ∼21 min (18, 47), while the average mRNA half-life of a primary Paf1C target whose level is depressed upon loss of Paf1 is 12 min (36). The half-life of TLC1 is over an hour (23). Third, unlike other primary Paf1C targets which are relatively unaffected by the cdc73Δ mutation (5), reduction of TLC1 levels was more extensive in cdc73Δ cells than in any other Paf1C mutant. Cdc73p anchors the rest of the Paf1C to Pol II and chromatin (32), so the fact that its loss has a large effect on TLC1 levels suggests that Paf1C may function in TLC1 promoter-dependent initiation.
The human homolog of CDC73, HRPT2 (hyperparathyroidism-jaw tumor syndrome 2), has been implicated in human cancers (39, 52, 54). It will be interesting to determine whether telomerase RNA levels are affected in tumors harboring mutant HRPT2.
Supplementary Material
Acknowledgments
We are grateful to Joan Betz and Judith Jaehning for the JJ Paf1C mutant strains and the pJJ1358 plasmid and to Timothy Hughes for the R1158 strain and the RP188 plasmid. David Whitcombe of DxS Limited generously designed the luciferase scorpion primer set. We also thank David Zappulla and Karen Goodrich for technical assistance and fruitful discussions.
A.D.M. was a Research Associate of the Howard Hughes Medical Institute.
Footnotes
Published ahead of print on 14 April 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Askree, S. H., T. Yehuda, S. Smolikov, R. Gurevich, J. Hawk, C. Coker, A. Krauskopf, M. Kupiec, and M. J. McEachern. 2004. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc. Natl. Acad. Sci. USA 1018658-8663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Betz, J. L., M. Chang, T. M. Washburn, S. E. Porter, C. L. Mueller, and J. A. Jaehning. 2002. Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268272-285. [DOI] [PubMed] [Google Scholar]
- 3.Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li, P. Hieter, and J. D. Boeke. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14115-132. [DOI] [PubMed] [Google Scholar]
- 4.Breitkreutz, B. J., C. Stark, and M. Tyers. 2003. The GRID: the general repository for interaction datasets. Genome Biol. 4R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang, M., D. French-Cornay, H. Y. Fan, H. Klein, C. L. Denis, and J. A. Jaehning. 1999. A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol. Cell. Biol. 191056-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapon, C., T. R. Cech, and A. J. Zaug. 1997. Polyadenylation of telomerase RNA in budding yeast. RNA 31337-1351. [PMC free article] [PubMed] [Google Scholar]
- 7.Collins, S. R., K. M. Miller, N. L. Maas, A. Roguev, J. Fillingham, C. S. Chu, M. Schuldiner, M. Gebbia, J. Recht, M. Shales, H. Ding, H. Xu, J. Han, K. Ingvarsdottir, B. Cheng, B. Andrews, C. Boone, S. L. Berger, P. Hieter, Z. Zhang, G. W. Brown, C. J. Ingles, A. Emili, C. D. Allis, D. P. Toczyski, J. S. Weissman, J. F. Greenblatt, and N. J. Krogan. 2007. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 446806-810. [DOI] [PubMed] [Google Scholar]
- 8.Cristofari, G., and J. Lingner. 2006. Telomere length homeostasis requires that telomerase levels are limiting. EMBO J. 25565-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edmonds, D., B. J. Breitkreutz, and L. Harrington. 2004. A genome-wide telomere screen in yeast: the long and short of it all. Proc. Natl. Acad. Sci. USA 1019515-9516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ferrezuelo, F., B. Steiner, M. Aldea, and B. Futcher. 2002. Biogenesis of yeast telomerase depends on the importin mtr10. Mol. Cell. Biol. 226046-6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu, D., and K. Collins. 2003. Distinct biogenesis pathways for human telomerase RNA and H/ACA small nucleolar RNAs. Mol. Cell 111361-1372. [DOI] [PubMed] [Google Scholar]
- 12.Gatbonton, T., M. Imbesi, M. Nelson, J. M. Akey, D. M. Ruderfer, L. Kruglyak, J. A. Simon, and A. Bedalov. 2006. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gavin, A. C., M. Bosche, R. Krause, P. Grandi, M. Marzioch, A. Bauer, J. Schultz, J. M. Rick, A. M. Michon, C. M. Cruciat, M. Remor, C. Hofert, M. Schelder, M. Brajenovic, H. Ruffner, A. Merino, K. Klein, M. Hudak, D. Dickson, T. Rudi, V. Gnau, A. Bauch, S. Bastuck, B. Huhse, C. Leutwein, M. A. Heurtier, R. R. Copley, A. Edelmann, E. Querfurth, V. Rybin, G. Drewes, M. Raida, T. Bouwmeester, P. Bork, B. Seraphin, B. Kuster, G. Neubauer, and G. Superti-Furga. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415141-147. [DOI] [PubMed] [Google Scholar]
- 14.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418387-391. [DOI] [PubMed] [Google Scholar]
- 15.Goffeau, A., B. G. Barrell, H. Bussey, R. W. Davis, B. Dujon, H. Feldmann, F. Galibert, J. D. Hoheisel, C. Jacq, M. Johnston, E. J. Louis, H. W. Mewes, Y. Murakami, P. Philippsen, H. Tettelin, and S. G. Oliver. 1996. Life with 6000 genes. Science 274546, 563-567. [DOI] [PubMed] [Google Scholar]
- 16.Guglielmi, B., N. L. van Berkum, B. Klapholz, T. Bijma, M. Boube, C. Boschiero, H. M. Bourbon, F. C. Holstege, and M. Werner. 2004. A high resolution protein interaction map of the yeast Mediator complex. Nucleic Acids Res. 325379-5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hengartner, C. J., V. E. Myer, S. M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 243-53. [DOI] [PubMed] [Google Scholar]
- 18.Holstege, F. C., E. G. Jennings, J. J. Wyrick, T. I. Lee, C. J. Hengartner, M. R. Green, T. R. Golub, E. S. Lander, and R. A. Young. 1998. Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95717-728. [DOI] [PubMed] [Google Scholar]
- 19.Hombauer, H., D. Weismann, I. Mudrak, C. Stanzel, T. Fellner, D. H. Lackner, and E. Ogris. 2007. Generation of active protein phosphatase 2A is coupled to holoenzyme assembly. PLoS Biol. 5e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huang, D., J. Moffat, and B. Andrews. 2002. Dissection of a complex phenotype by functional genomics reveals roles for the yeast cyclin-dependent protein kinase Pho85 in stress adaptation and cell integrity. Mol. Cell. Biol. 225076-5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes, T. R., M. J. Marton, A. R. Jones, C. J. Roberts, R. Stoughton, C. D. Armour, H. A. Bennett, E. Coffey, H. Dai, Y. D. He, M. J. Kidd, A. M. King, M. R. Meyer, D. Slade, P. Y. Lum, S. B. Stepaniants, D. D. Shoemaker, D. Gachotte, K. Chakraburtty, J. Simon, M. Bard, and S. H. Friend. 2000. Functional discovery via a compendium of expression profiles. Cell 102109-126. [DOI] [PubMed] [Google Scholar]
- 22.Kornberg, R. D. 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30235-239. [DOI] [PubMed] [Google Scholar]
- 23.Larose, S., N. Laterreur, G. Ghazal, J. Gagnon, R. J. Wellinger, and S. A. Elela. 2007. RNase III-dependent regulation of yeast telomerase. J. Biol. Chem. 2824373-4381. [DOI] [PubMed] [Google Scholar]
- 24.Lenburg, M. E., and E. K. O'Shea. 2001. Genetic evidence for a morphogenetic function of the Saccharomyces cerevisiae Pho85 cyclin-dependent kinase. Genetics 15739-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lesage, G., A. M. Sdicu, P. Menard, J. Shapiro, S. Hussein, and H. Bussey. 2004. Analysis of beta-1,3-glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics 16735-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ly, H., M. Schertzer, W. Jastaniah, J. Davis, S. L. Yong, Q. Ouyang, E. H. Blackburn, T. G. Parslow, and P. M. Lansdorp. 2005. Identification and functional characterization of 2 variant alleles of the telomerase RNA template gene (TERC) in a patient with dyskeratosis congenita. Blood 1061246-1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marrone, A., D. Stevens, T. Vulliamy, I. Dokal, and P. J. Mason. 2004. Heterozygous telomerase RNA mutations found in dyskeratosis congenita and aplastic anemia reduce telomerase activity via haploinsufficiency. Blood 1043936-3942. [DOI] [PubMed] [Google Scholar]
- 28.Marrone, A., A. Walne, and I. Dokal. 2005. Dyskeratosis congenita: telomerase, telomeres and anticipation. Curr. Opin. Genet. Dev. 15249-257. [DOI] [PubMed] [Google Scholar]
- 29.Mnaimneh, S., A. P. Davierwala, J. Haynes, J. Moffat, W. T. Peng, W. Zhang, X. Yang, J. Pootoolal, G. Chua, A. Lopez, M. Trochesset, D. Morse, N. J. Krogan, S. L. Hiley, Z. Li, Q. Morris, J. Grigull, N. Mitsakakis, C. J. Roberts, J. F. Greenblatt, C. Boone, C. A. Kaiser, B. J. Andrews, and T. R. Hughes. 2004. Exploration of essential gene functions via titratable promoter alleles. Cell 11831-44. [DOI] [PubMed] [Google Scholar]
- 30.Mozdy, A. D., and T. R. Cech. 2006. Low abundance of telomerase in yeast: implications for telomerase haploinsufficiency. RNA 121721-1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mueller, C. L., and J. A. Jaehning. 2002. Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 221971-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mueller, C. L., S. E. Porter, M. G. Hoffman, and J. A. Jaehning. 2004. The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14447-456. [DOI] [PubMed] [Google Scholar]
- 33.Ng, H. H., S. Dole, and K. Struhl. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 27833625-33628. [DOI] [PubMed] [Google Scholar]
- 34.Pemberton, L. F., J. S. Rosenblum, and G. Blobel. 1997. A distinct and parallel pathway for the nuclear import of an mRNA-binding protein. J. Cell Biol. 1391645-1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng, W. T., M. D. Robinson, S. Mnaimneh, N. J. Krogan, G. Cagney, Q. Morris, A. P. Davierwala, J. Grigull, X. Yang, W. Zhang, N. Mitsakakis, O. W. Ryan, N. Datta, V. Jojic, C. Pal, V. Canadien, D. Richards, B. Beattie, L. F. Wu, S. J. Altschuler, S. Roweis, B. J. Frey, A. Emili, J. F. Greenblatt, and T. R. Hughes. 2003. A panoramic view of yeast noncoding RNA processing. Cell 113919-933. [DOI] [PubMed] [Google Scholar]
- 36.Penheiter, K. L., T. M. Washburn, S. E. Porter, M. G. Hoffman, and J. A. Jaehning. 2005. A posttranscriptional role for the yeast Paf1-RNA polymerase II complex is revealed by identification of primary targets. Mol. Cell 20213-223. [DOI] [PubMed] [Google Scholar]
- 37.Pokholok, D. K., N. M. Hannett, and R. A. Young. 2002. Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol. Cell 9799-809. [DOI] [PubMed] [Google Scholar]
- 38.Rosonina, E., and J. L. Manley. 2005. From transcription to mRNA: PAF provides a new path. Mol. Cell 20167-168. [DOI] [PubMed] [Google Scholar]
- 39.Rozenblatt-Rosen, O., C. M. Hughes, S. J. Nannepaga, K. S. Shanmugam, T. D. Copeland, T. Guszczynski, J. H. Resau, and M. Meyerson. 2005. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol. Cell. Biol. 25612-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seto, A. G., A. J. Zaug, S. G. Sobel, S. L. Wolin, and T. R. Cech. 1999. Saccharomyces cerevisiae telomerase is an Sm small nuclear ribonucleoprotein particle. Nature 401177-180. [DOI] [PubMed] [Google Scholar]
- 41.Sheldon, K. E., D. M. Mauger, and K. M. Arndt. 2005. A requirement for the Saccharomyces cerevisiae Paf1 complex in snoRNA 3′ end formation. Mol. Cell 20225-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shi, X., M. Chang, A. J. Wolf, C. H. Chang, A. A. Frazer-Abel, P. A. Wade, Z. F. Burton, and J. A. Jaehning. 1997. Cdc73p and Paf1p are found in a novel RNA polymerase II-containing complex distinct from the Srbp-containing holoenzyme. Mol. Cell. Biol. 171160-1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simic, R., D. L. Lindstrom, H. G. Tran, K. L. Roinick, P. J. Costa, A. D. Johnson, G. A. Hartzog, and K. M. Arndt. 2003. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 221846-1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stevens, S. W., D. E. Ryan, H. Y. Ge, R. E. Moore, M. K. Young, T. D. Lee, and J. Abelson. 2002. Composition and functional characterization of the yeast spliceosomal penta-snRNP. Mol. Cell 931-44. [DOI] [PubMed] [Google Scholar]
- 45.Tong, A. H., G. Lesage, G. D. Bader, H. Ding, H. Xu, X. Xin, J. Young, G. F. Berriz, R. L. Brost, M. Chang, Y. Chen, X. Cheng, G. Chua, H. Friesen, D. S. Goldberg, J. Haynes, C. Humphries, G. He, S. Hussein, L. Ke, N. Krogan, Z. Li, J. N. Levinson, H. Lu, P. Menard, C. Munyana, A. B. Parsons, O. Ryan, R. Tonikian, T. Roberts, A. M. Sdicu, J. Shapiro, B. Sheikh, B. Suter, S. L. Wong, L. V. Zhang, H. Zhu, C. G. Burd, S. Munro, C. Sander, J. Rine, J. Greenblatt, M. Peter, A. Bretscher, G. Bell, F. P. Roth, G. W. Brown, B. Andrews, H. Bussey, and C. Boone. 2004. Global mapping of the yeast genetic interaction network. Science 303808-813. [DOI] [PubMed] [Google Scholar]
- 46.Ulery, T. L., D. A. Mangus, and J. A. Jaehning. 1991. The yeast IMP1 gene is allelic to GAL2. Mol. Gen. Genet. 230129-135. [DOI] [PubMed] [Google Scholar]
- 47.Wang, Y., C. L. Liu, J. D. Storey, R. J. Tibshirani, D. Herschlag, and P. O. Brown. 2002. Precision and functional specificity in mRNA decay. Proc. Natl. Acad. Sci. USA 995860-5865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Warner, M. H., K. L. Roinick, and K. M. Arndt. 2007. Rtf1 is a multifunctional component of the paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol. Cell. Biol. 276103-6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson, B. Andre, R. Bangham, R. Benito, J. D. Boeke, H. Bussey, A. M. Chu, C. Connelly, K. Davis, F. Dietrich, S. W. Dow, M. El Bakkoury, F. Foury, S. H. Friend, E. Gentalen, G. Giaever, J. H. Hegemann, T. Jones, M. Laub, H. Liao, N. Liebundguth, D. J. Lockhart, A. Lucau-Danila, M. Lussier, N. M'Rabet, P. Menard, M. Mittmann, C. Pai, C. Rebischung, J. L. Revuelta, L. Riles, C. J. Roberts, P. Ross-MacDonald, B. Scherens, M. Snyder, S. Sookhai-Mahadeo, R. K. Storms, S. Veronneau, M. Voet, G. Volckaert, T. R. Ward, R. Wysocki, G. S. Yen, K. Yu, K. Zimmermann, P. Philippsen, M. Johnston, and R. W. Davis. 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285901-906. [DOI] [PubMed] [Google Scholar]
- 50.Wood, A., J. Schneider, J. Dover, M. Johnston, and A. Shilatifard. 2005. The Bur1/Bur2 complex is required for histone H2B monoubiquitination by Rad6/Bre1 and histone methylation by COMPASS. Mol. Cell 20589-599. [DOI] [PubMed] [Google Scholar]
- 51.Xiao, T., C. F. Kao, N. J. Krogan, Z. W. Sun, J. F. Greenblatt, M. A. Osley, and B. D. Strahl. 2005. Histone H2B ubiquitylation is associated with elongating RNA polymerase II. Mol. Cell. Biol. 25637-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yart, A., M. Gstaiger, C. Wirbelauer, M. Pecnik, D. Anastasiou, D. Hess, and W. Krek. 2005. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol. Cell. Biol. 255052-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zappulla, D. C., K. Goodrich, and T. R. Cech. 2005. A miniature yeast telomerase RNA functions in vivo and reconstitutes activity in vitro. Nat. Struct. Mol. Biol. 121072-1077. [DOI] [PubMed] [Google Scholar]
- 54.Zhu, B., S. S. Mandal, A. D. Pham, Y. Zheng, H. Erdjument-Bromage, S. K. Batra, P. Tempst, and D. Reinberg. 2005. The human PAF complex coordinates transcription with events downstream of RNA synthesis. Genes Dev. 191668-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.