Abstract
It is well accepted that for transcriptional silencing in budding yeast, the evolutionarily conserved lysine deacetylase Sir2, in concert with its partner proteins Sir3 and Sir4, establishes a chromatin structure that prevents RNA polymerase II (Pol II) transcription. However, the mechanism of repression remains controversial. Here, we show that the recruitment of Pol II, as well as that of the general initiation factors TBP and TFIIH, occurs unimpeded to the silent HMRa1 and HMLα1/HMLα2 mating promoters. This, together with the fact that Pol II is Ser5 phosphorylated, implies that SIR-mediated silencing is permissive to both preinitiation complex (PIC) assembly and transcription initiation. In contrast, the occupancy of factors critical to both mRNA capping and Pol II elongation, including Cet1, Abd1, Spt5, Paf1C, and TFIIS, is virtually abolished. In agreement with this, efficiency of silencing correlates not with a restriction in Pol II promoter occupancy but with a restriction in capping enzyme recruitment. These observations pinpoint the transition between polymerase initiation and elongation as the step targeted by Sir2 and indicate that transcriptional silencing is achieved through the differential accessibility of initiation and capping/elongation factors to chromatin. We compare Sir2-mediated transcriptional silencing to a second repression mechanism, mediated by Tup1. In contrast to Sir2, Tup1 prevents TBP, Pol II, and TFIIH recruitment to the HMLα1 promoter, thereby abrogating PIC formation.
In eukaryotes, transcription occurs in the context of chromatin. A traditional view is that nucleosomes exert their regulatory role by impeding the access of proteins, both gene-specific regulators and general transcription factors (GTFs), to the DNA (33). In support of this idea, when the TATA box is assembled into a nucleosome in vitro, its accessibility for TATA binding protein (TBP) is reduced by at least four orders of magnitude (27), while the accessibility of a high-affinity heat shock element for its cognate HSF activator is reduced by more than three orders of magnitude (66). However, it is now appreciated that in vivo, chromatin modification and remodeling complexes, in combination with histone variants and the intrinsically low affinity of many gene promoters for histones, collaborate in making the euchromatic template accessible to these and other regulatory factors (reviewed in references 36 and 71).
Heterochromatin, the cytologically condensed compartment of the eukaryotic nucleus, likewise is a substrate of chromatin-remodeling complexes and other regulatory factors (reviewed in reference 16), yet genes residing in heterochromatin generally are transcriptionally silent. A key feature of heterochromatin is its ability to repress gene expression in a position-dependent but sequence-independent fashion. Thus, the position of a gene on the chromosome, rather than its associated enhancer, upstream activation sequence (UAS), and promoter elements, can dictate its expression state. The budding yeast Saccharomyces cerevisiae does not contain condensed chromatin at the cytological level; however, it does contain domains of silent chromatin that resemble, in both their molecular and epigenetic characteristics, the repressed heterochromatic domains of higher eukaryotes (46).
In S. cerevisiae, silent chromatin is found at the telomeres, the ribosomal DNA repeats, and the two cryptic mating-type loci, HMR and HML, located near the right and left telomeres of chromosome III, respectively (14, 51). The silent mating loci bear genes (a1 and a2 at HMR and α1 and α2 at HML) that encode transcriptional regulators. Their activation in a wild-type cell requires their transposition to a centromere-proximal euchromatic site, MAT, located on the same chromosome. This transposition, which occurs only in homothallic haploid cells, is initiated by the HO double-stranded DNA endonuclease that cuts a specific site within the MAT locus. The double-stranded break subsequently is repaired by nonreciprocal homologous recombination between the mating-type genes located in MAT and those of the opposite mating type found at either HMR or HML, which act as the donors of mating information. The directionality of mating-type interconversion is determined by a recombinational enhancer located proximal to HML and that, when active (as is the case in a cells), increases the probability that HML will serve as the donor of mating-type information. When the enhancer is repressed, as is the case in α cells, HMR serves as the donor (reviewed in references 64 and 72).
Silencing at the HM loci is controlled by cis-acting elements termed silencers. These contain binding sites for sequence-specific factors (ORC, Rap1, and Abf1) that trigger the formation of a specialized chromatin structure through the concerted recruitment of the Sir2, Sir3, and Sir4 silencing proteins (reviewed in references 18 and 43). The silencing complex horizontally propagates along the chromatin fiber through iterative cycles of H4 K16 deacetylation catalyzed by Sir2, an evolutionarily conserved NAD+-dependent lysine deacetylase (26, 63). The deacetylation of H4 K16 and the resultant production of O-acetyl-ADP-ribose are necessary for the formation of a trimeric complex between Sir2/Sir4 and Sir3 (25, 38). The resultant chromatin structure consists of positioned, hypoacetylated, and hypomethylated nucleosomes (7, 49, 52, 70).
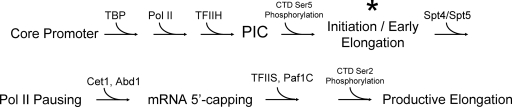
Transcription is a multistep process, and each step is highly regulated. Initially, sequence-specific activators bind to UAS elements (enhancers); these, in turn, recruit polymerase II (Pol II) and GTFs to the core promoter, leading to the formation of the preinitiation complex (PIC) (schematically summarized in Fig. 1). Transcription initiation requires the general factor TFIIH, the ATPase subunit of which unwinds the DNA, leading to the formation of an open complex. Also, the kinase subunit of TFIIH phosphorylates Ser5 residues of the carboxy-terminal domain (CTD) of Rpb1, the large Pol II subunit (53). Early elongation often is accompanied by a pause, during which the pre-mRNA is capped at its 5′ end. Following this step, which generally takes place when the nascent mRNA chain is 25 to 35 nucleotides long (48), Pol II engages in productive elongation concomitant with the phosphorylation of Ser2 residues with the CTD and the recruitment of elongation factors, including TFIIS, DSIF (Spt4/Spt5), and the Paf1 complex (Paf1C) (53). (As indicated in Fig. 1, Spt4/Spt5 may play an additional role in instigating the Pol II pause following early elongation [53].)
FIG. 1.
Order of recruitment of initiation, capping, and elongation factors at a typical Pol II gene. Factors and activities depicted are principally those evaluated in this study and are not meant to be comprehensive. This work demonstrates that SIR-dependent silent chromatin is permissive to steps in the transcriptional cascade upstream of the initiation-elongation transition (asterisk) while being restrictive to steps downstream of it.
How silent chromatin represses gene transcription remains poorly understood, but theoretically it could act at any one of the above steps. Early observations suggested that silent chromatin is resistant to the activity of endogenous and exogenous nucleases, as well as DNA repair and modification enzymes (reviewed in references 15 and 51). This contributed to the idea that silent chromatin represses transcription through its ability to sterically hinder the access of sequence-specific proteins. This model is intuitively appealing, because it is consistent with the in vitro reconstitution studies discussed above that demonstrate the reduced accessibility of DNA sequences when assembled into stable nucleosomes. However, such a model cannot account for how the silent HM loci remain fully permissive to the binding of other sequence-specific factors, including enzymes that mediate homologous recombination, site-specific recombination, and retrotransposition (9, 24, 29, 75).
Investigations of an ectopically silenced heat shock transgene cast additional doubt on the steric hindrance mechanism. These studies showed that despite efficient, SIR-dependent silencing, the hsp82 promoter remained accessible, as measured by nuclease hypersensitivity (35). Consistently with this, UAS and TATA genomic footprints were retained (57), and essentially normal levels of the activator HSF, the initiation factor TBP, and Pol II itself were present (56). An analysis of the naturally silenced HMRa1 promoter supported these conclusions, as both TBP and Pol II were detected in the SIR-repressed state (56). These findings gave rise to the notion that SIR acts at a point downstream of both activator binding and PIC recruitment to silence transcription. More recently, a third model has been proposed: PIC interference. This model posits that SIR is permissive to activator binding, yet transcription is abolished because of a failure to recruit RNA polymerase. In support of this, at several SIR-silenced URA3 transgenes as well as at both the HML and HMR mating loci, Pol II, along with the general initiation factors TFIIB and TFIIE, could not be detected (8).
Here, we use chromatin immunoprecipitation (ChIP) to quantitatively measure the abundance of initiation, capping, and elongation factors at the naturally silenced HMRa1 and HMLα1/HMLα2 promoters. We employ two genetic backgrounds and rigorous controls for both nonspecific immunoprecipitation (IP) and spurious PCR amplification. We find, consistent with predictions of the downstream inhibition model, that three components of the PIC, namely, TBP, Pol II, and TFIIH, are present within the silent HMR and HML promoters. Furthermore, Pol II is efficiently phosphorylated at Ser5 within its CTD, indicating that polymerase is not only present but also has initiated transcription. In striking contrast, the occupancy of 5′-capping enzymes and elongation factors is virtually eliminated, and the recruitment of Mediator is restricted. Our results pinpoint the transition between Pol II initiation and elongation as the step targeted by SIR and provide important insight into how silent chromatin can abrogate gene expression.
MATERIALS AND METHODS
Yeast strains.
Strains used in this study are derived from the S288C and SLY101 genetic backgrounds (Table 1). SLY101 is congenic to W303 (35). MATα strains of the SLY101 background used here bear hsp82 alleles flanked by HMR-E silencers (57). sir2Δ strains were generated using one-step transplacement of the SIR2 open reading frame (ORF) with a PCR-amplified DNA fragment bearing the KANMX marker and gene-specific flanking sequences (21) and were confirmed by genomic PCR in conjunction with mating-type assays (cells bearing sir2Δ lose the ability to mate with cells of the opposite identity). To excise the KANMX marker, cells were transformed with the plasmid pSH47 that bears a URA3+ marker and a Cre recombinase regulated under a GAL1 promoter (21) and then were induced in 2% galactose for 2.5 h, followed by screening for kanamycin-sensitive colonies that then were cured of the plasmid on medium containing 5-fluoorotic acid. Strains with an HM locus deletion were obtained from the parental sir2Δ or sir4Δ strain by replacing the corresponding mating-type gene with the KANMX marker and were further confirmed by genomic PCR as well as mating type assays (cells bearing the deletion of either SIR2 or SIR4 combined with a single HM locus deletion regain the ability to mate with cells of the opposite identity). Tandem affinity purification (TAP)-tagged strains were obtained from Open Biosystems, and each tagged allele was confirmed by genomic PCR. To C-terminally tag the chromosomal KIN28 gene with the 9-Myc epitope, we performed a one-step gene transplacement of strains EAS2001, EAS2011, and LG1101 (Table 1). The transforming DNA was PCR amplified using the plasmid pWZV87 as the template (30). The proper targeting of the KIN28-9Myc-KlTRP1 fragment was confirmed by genomic PCR.
TABLE 1.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| SLY101 | MATα ade−can1-100 cyh2rhis3-11,15 leu2-3,112 trp1-1 ura3 | 35 |
| EAS2011 | SLY101; hsp82-2001 | 57 |
| EAS2001 | EAS2011; sir4Δ2::HIS3 | 57 |
| LG1001 | EAS2001; hmraΔ::KANMX | This study |
| LG1101 | SLY101; MATa | This study |
| LG1102 | LG1101; sir2Δ::KANMX | This study |
| LG1103 | LG1101; sir2Δ; hmlαΔ::KANMX | This study |
| JHD10 | EAS2011; KIN28-9Myc | This study |
| JHD11 | EAS2001; KIN28-9Myc | This study |
| SBK701 | LG1101, KIN28-9Myc | This study |
| LG11 | SBK701; sir2Δ::KANMX | This study |
| S288Ca | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| S288Cα | MATα his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Research Genetics |
| LG2881 | S288Cα; sir2Δ::KANMX | This study |
| LG2882 | S288Cα; sir2Δ; hmraΔ::KANMX | This study |
| LG2883 | S288Ca; sir2Δ::KANMX | This study |
| LG2884 | S288Ca; sir2Δ; hmlαΔ::KANMX | This study |
| TAP-SPT5 | S288Ca; SPT5-TAP | Open Biosystems |
| LG101 | TAP-SPT5; sir2Δ::KANMX | This study |
| TAP-DST1 | S288Ca; DST1-TAP | Open Biosystems |
| LG102 | TAP-DST1; sir2Δ::KANMX | This study |
| TAP-RTF1 | S288Ca; RTF1-TAP | Open Biosystems |
| LG103 | TAP-RFT1; sir2Δ::KANMX | This study |
| TAP-GAL11 | S288Ca; GAL11-TAP | Open Biosystems |
| LG104 | TAP-GAL11; sir2Δ::KANMX | This study |
| TAP-SRB4 | S288Ca; SRB4-TAP | Open Biosystems |
| LG105 | TAP-SRB4; sir2Δ::KANMX | This study |
| TAP-TFB1 | S288Ca; TFB1-TAP | Open Biosystems |
| LG106 | TAP-TFB1; sir2Δ::KANMX | This study |
Cultivation.
S. cerevisiae strains were cultivated at 30°C to early log phase (1 × 107 to 2 × 107 cells/ml) in rich yeast extract-peptone-dextrose broth supplemented with 0.03 mg/ml adenine.
ChIP.
ChIP was performed essentially as described previously (56). Fifty-milliliter cultures were cross-linked with 1% formaldehyde and then converted to spheroplasts with lyticase (4 mg/ml; ICN Biomedicals or Sigma) and lysed using 1 volume of 0.5-mm glass beads for 30 min at 4°C on an Eppendorf 5432 mixer. Chromatin was sheared to a mean size of 0.5 to 0.7 kb with a Branson 250 sonifier equipped with a microtip that used three 25-s pulses at constant power and an output setting of 22 W. The clarified supernatant (final volume, 3.0 ml) was used in IPs as described below. The sources of antibodies were the following: Cet1 and Abd1, Steve Buratowski (Harvard Medical School); Sir3, Rohinton Kamakaka (University of California—Santa Cruz); yTBP, Michael Green (University of Massachusetts Medical Center); Ser5-phosphorylated CTD (monoclonal antibody H14; Covance); Myc (monoclonal antibody 9E10; Santa Cruz Biotech); and mouse Pol II CTD (raised in rabbits immunized with glutathione S-transferase-CTD [expression vector obtained from David Bentley, University of Colorado Health Sciences Center]).
IPs typically were achieved by adding 5 μl antiserum to 300 μl of chromatin lysate, followed by mixing the solution on a nutator at 4°C overnight. Pansorbin cells (40 μl; Calbiochem) then were added, and the incubation was continued for an additional 3 h. For the Ser5-P-CTD ChIPs, chromatin was isolated in the presence of a phosphatase inhibitor cocktail (10 mM each of NaF, NaN3, pNPP, NaPPi, and β-glycerophosphate). Chromatin lysate (150 μl) was preincubated with 2.5 μl of a 50% slurry of anti-mouse immunoglobulin M (IgM)-agarose beads (Sigma) preblocked with 1 mg/ml bovine serum albumin and 0.3 mg/ml of salmon sperm DNA for 3 h at 4°C. H14 antibody (2.5 μl) then was added to the clarified supernatant and permitted to incubate overnight at 4°C. A total of 2.5 μl of fresh preblocked anti-mouse IgM-agarose beads then was added, and the mixture was incubated at 4°C for 3 h. Beads then were washed as previously described (56). All TAP ChIPs were performed as described previously (28) using IgG-agarose beads (Sigma) and no antibody.
Following washing and the reversal of formaldehyde-induced cross-links, DNA was ethanol precipitated and dissolved in 30 μl Tris-EDTA (TE). For input samples, 200 μl of soluble chromatin was ethanol precipitated and dissolved in TE, and cross-links were reversed. The chromatin was reprecipitated, and DNA was purified and dissolved in 50 μl TE. Generally, DNA representing 0.02 to 0.4% of the total chromatin sample (input) or 6 to 12% of the IP was amplified. In addition to template DNA, the 50-μl reaction mixtures contained 2.5 mM MgCl2; 400 μM each of dCTP, dGTP, dTTP, and dATP; and 1 μCi of [α-32P]dATP (6,000 Ci/mmol). After 2 min of denaturation at 93°C and the addition of 1.25 U Taq DNA polymerase, the temperature was lowered to 60°C for 1 min, followed by 32 s at 72°C. Samples then were subjected to a program of 25 cycles, each consisting of 1 min at 93°C, 1 min at 60°C, and 32 s at 72°C. PCR products were precipitated, electrophoresed on 8% Tris-borate-EDTA polyacrylamide gels, dried, exposed to a Phosphor screen, and quantified on a Storm 860 PhosphorImager (Molecular Dynamics) using ImageQuant 5.2 software.
The following gene-specific primers were employed: HMLα1/HMLα2 promoter (251-bp PCR product), GCCCACTTCTAAGCTGATTTCAATCTCTCC and GGCTTCGAAGTAAACATATTGTGAATGTCG; HMLα1 3′ untranslated region (3′-UTR) (240 bp), CCATTTAGTTTTTAGTACGATTGC and CCAAACTTACGATCTTTGGACC; HMRa1 promoter (139 bp), GTTCTTTCGGGGAAACTGTATAAAACTTCC and GTTAAACAGAGTTCTGTTTATGTTTTCCGCC; HMRa1 3′-UTR (170 bp), CCAACATTTTCGTATATGGCG and CTTGTGCAAATTCCAACTAAAGG; HMR-E (156 bp), CGAACGATCCCCGTCCAAGTTATGAGC and CAGGAGTACCTGCGCTTATTCTCAAAC; ARS504 (73 bp), GTCAGACCTGTTCCTTTAAGAGG and CATACCCTCGGGTCAAACAC; PMA1 promoter (322 bp), GGTACCGCTTATGCTCCCCTCC and GATTTTCTTTAACTAGCTGGGG; HSP82 promoter (396 bp), CACCCCCCCTCTCTCAACACAGTAATCC and GGACTCTATTTTCTATCAGGTATGATTTCTTCAACTC; HSP82 ORF (198 bp), GTTCTACTCGGCTTTCTCCAAAAATATC and CAGCCTTTAGAGATTCACCAGTGATGTAG; and HSP82 3′-UTR (275 bp), GAGTTGACGAAGGTGGTGCTCAAGACAAG and CCTATTCAAGGCCATGATGTTCTACCTAATC. These primer pairs were used at the following concentrations: HMLα1/HMLα2 promoter (50 pmol), HMLα1 3′-UTR (50 pmol), HMRa1 promoter (25 pmol), HMRa1 3′-UTR (25 pmol), HMR-E (25 pmol), ARS504 (15 pmol), PMA1 promoter (50 pmol), HSP82 promoter (40 pmol), HSP82 ORF (12.5 pmol), and HSP82 3′-UTR (12.5 pmol).
Quantification of the data was done essentially as described before (74). To calculate the abundance of a given gene sequence (Qgene) present in an IP, we used the following formula: Qgene = IPgene/inputgene. In this calculation, input is used solely for the purpose of normalizing the amplification efficiency of each genomic locus in the multiplex PCR; it is not used to normalize sample-to-sample variation in recovery. Instead, as described below, the coamplified ARS504 locus is used for this purpose. To eliminate any contribution of nonspecific IP, we subtracted the signal arising from a mock IP (Pansorbin cells or agarose beads only) in the TBP, Cet1, Abd1, and H14 ChIPs; the signal from chromatin immunoprecipitated from a nontagged strain for the Myc- and TAP-tagged ChIPs; or the signal obtained from preimmune serum for the CTD ChIPs prior to calculating Qgene. The gel background value alone was subtracted from the ARS504 value, a nontranscribed locus that served as an internal recovery (nonspecific IP) control to which all Qgene values were normalized. The Qgene/QARS504 quotient in the derepressed state was set at 1.0, to which all other values were normalized. For Sir3 ChIPs, the abundance of the gene sequence was quantified relative to that of HMRa1, with the QHMRa1/QARS504 quotient for the SIR+ sample normalized to 1.0. To derive the P values listed in Table 2, a two-tailed t test was conducted using the TTEST function on Excel 2003, using two-sample equal variance as the parameter.
TABLE 2.
In vivo occupancy of initiation, capping, and elongation factors at silent and derepressed HM locia
| Factor/strain background | Occupancy level at HML
|
Occupancy level at HMR
|
||||
|---|---|---|---|---|---|---|
| SIR+ | sir2Δ | P value | SIR+ | sir2Δ/sir4Δ | P value | |
| Rpb1/S288C | 1.51 ± 0.19 | 1 ± 0.05 | 0.038 | 1.07 ± 0.09 | 1 ± 0.04 | 0.220 |
| TBP/S288C | 1.53 ± 0.14 | 1 ± 0.12 | 0.031 | 1.19 ± 0.09 | 1 ± 0.09 | 0.110 |
| Kin28/SLY101 | 1.95 ± 0.14 | 1 ± 0.37 | 0.014 | 1.39 ± 0.34 | 1 ± 0.12 | 0.340 |
| Tfb1/S288C | 1.51 ± 0.16 | 1 ± 0.32 | 0.039 | |||
| Ser5-P-Rpb1/S288 | 1.28 ± 0.07 | 1 ± 0.09 | 0.045 | 1.29 ± 0.06 | 1 ± 0.11 | 0.060 |
| Cet1/S288C | 0.04 ± 0.02 | 1 ± 0.11 | 0.001 | 0.06 ± 0.04 | 1 ± 0.09 | 0.002 |
| Abd1/S288C | 0.11 ± 0.06 | 1 ± 0.15 | 0.005 | 0.11 ± 0.07 | 1 ± 0.19 | 0.009 |
| Spt5/S288C | −0.09 ± 0.02 | 1 ± 0.38 | 0.003 | |||
| Dst1/S288C | 0.08 ± 0.18 | 1 ± 0.47 | 0.012 | |||
| Rtf1/S288C | 0.17 ± 0.05 | 1 ± 0.24 | 0.027 | |||
Occupancy levels were quantified as described in Materials and Methods. Values represent means ± standard errors of the means (n = 3 or 4). P values were determined using a two-tailed t test. Statistically significant differences in factor occupancy (P < 0.05) are highlighted in boldface.
Northern analysis.
For Northern analyses, total cellular RNA was isolated from 10-ml aliquots of each strain examined for ChIP assays (aliquots were removed prior to the addition of formaldehyde) and purified, electrophoresed, and blotted to a Gene Screen as described previously (57). Blots were hybridized overnight to a1-, α1-, or α2-specific probes at 55, 65, and 52.5°C, respectively, to visualize mating-type transcripts or the HSP82 probe at 45°C to detect the HSP82 transcript, washed, exposed to a PhosphorImager, and then rehybridized (without stripping) to the ACT1 probe at 55°C. We note that the α2 probe shares sequence similarity with the a2 gene and therefore cross-hybridizes to the a2 mRNA; nonetheless, the two transcripts can be distinguished by size and individually quantified. Gel-purified templates were generated by PCR from yeast genomic DNA. All hybridization probes were synthesized by 25 cycles of linear PCR in the presence of 5 mM MgCl2; 300 μM each of dCTP, dGTP, dTTP; 3 μM of dATP; 100 μCi of [α-32P]dATP; and 1.25 U of Taq DNA polymerase. The following probes were used (coordinates relative to ATG): a1, +156 to +326; a2, +356 to +562, α1, +101 to +496; α2, +63 to +322; HSP82, +2167 to +2228; and ACT1, +606 to +1000.
RESULTS
Pol II and TBP are efficiently recruited to hyperrepressed HM promoters.
To investigate the mechanism by which Sir proteins silence transcription, we used ChIP to measure factor abundance at the HM mating-type loci, the relevant features of which are schematically illustrated in Fig. 2A. We analyzed SIR+ parental strains and isogenic sir2Δ or sir4Δ deletion strains in parallel, allowing a direct comparison between heterochromatic and euchromatic gene states. In SIR+ strains, Sir3 is recruited to both HM loci, yet it appears to be more abundant at HML than at HMR (Fig. 2B). This might reflect the presence of two functionally autonomous silencers at this locus (HML-E and HML-I) as opposed to HMR, which has only one, HMR-E (6, 50, 58). The efficiency of Sir2/3/4 recruitment has been shown to correlate with both the dosage and arrangement of silencers (56) (L. Gao and D. S. Gross, unpublished data). As expected, in either a sir2Δ or sir4Δ mutant, Sir3 recruitment is abolished (Fig. 2B). Concomitantly, the HM loci are transcriptionally derepressed (Fig. 2C, lanes 2 and 5). The exception to this is the α1 gene, which fails to express in either sir2Δ strain, as it is the target of Tup1 repression in sir mutants (22). The role played by Tup1 is further considered below.
FIG. 2.
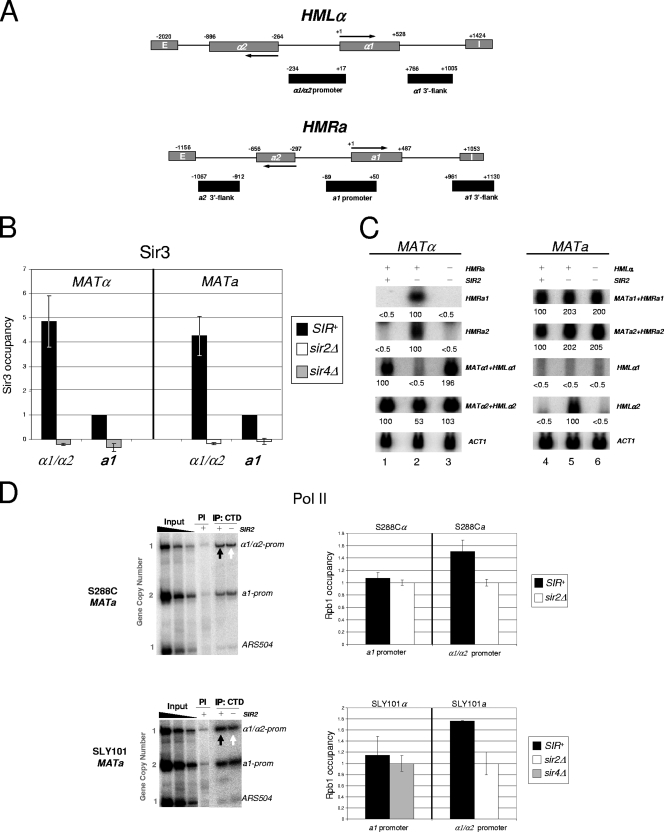
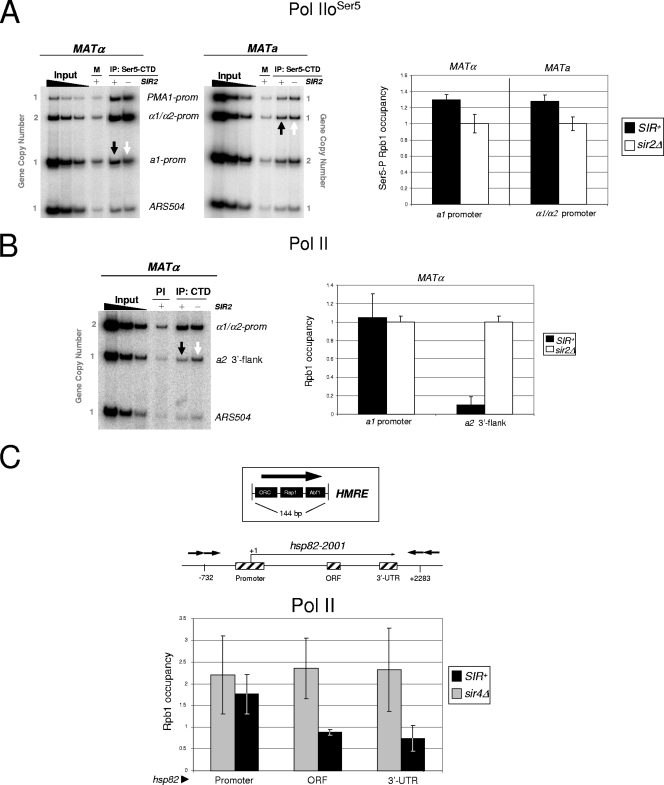
SIR extinguishes transcription at the HM loci yet is fully permissive to Pol II recruitment. (A) Physical maps of HMLα and HMRa. These loci are located near the left and right ends of chromosome III, respectively. Locations of mating-type genes, flanking silencer elements (termed E and I), and PCR amplicons (black rectangles) are shown. Coordinates are provided relative to the ATG codons (+1) of the HMLα1 and HMRa1 ORFs. Note that the HMRa2 promoter cannot be specifically amplified due to extensive sequence identity with both the promoter and ORF sequences of the HMLα2 gene. To validate the specificity of the hybridizations and PCRs, chromosomal deletions of HMLα (−1781 to +950) and HMRa (−1038 to +952) were engineered into selected strains (Table 1 and Fig. 2C and 5A). (B) Deletion of either SIR2 or SIR4 abolishes Sir3 association with the silent mating-type promoters. A summary of Sir3 ChIP assays in SIR+, sir2Δ, and sir4Δ strains (SLY101 background) is shown. Sir3 abundance at each promoter is normalized relative to its abundance at HMRa1 in a SIR+ strain, which was arbitrarily set at 1. Depicted are the means ± standard deviations of two independent experiments. (C) Northern analysis of isogenic MATα and MATa strains (S288C background). Transcript abundance was quantified using a PhosphorImager and was normalized to that for ACT1. For each probe and strain combination, the signal of the derepressed HM gene was arbitrarily assigned a value of 100; the others are expressed relative to it. In the case of HMLα1, which is not derepressed in a sir2Δ mutant (see the text), the signal above the background was negligible in all three MATa strains. Values represent the means of two independent experiments. A plus sign signifies the presence of a gene or locus; a minus sign signifies its deletion. (D) In vivo cross-linking analysis of the large Pol II subunit (Rpb1) at the α1/α2 and a1 promoters and a nontranscribed (ORF-less) locus, ARS504, in both S288C and SLY101 genetic backgrounds. On the left are representative gels containing multiplex PCRs of input and immunoprecipitated DNAs isolated from formaldehyde-cross-linked, sonicated chromatin (sheared to a mean size of 0.5 to 0.7 kb) isolated from MATa strains. Chromatin was immunoprecipitated with either preimmune serum (PI) or anti-CTD antiserum (IP), cross-links were reversed, and purified DNA was subjected to multiplex PCR in the presence of [α-32P]dATP using primers specific for the regions indicated, followed by electrophoresis and detection by a PhosphorImager. Input samples, derived from the SIR+ parent strain, represent threefold serial dilutions (0.2, 0.067, and 0.022%) of soluble chromatin used in each IP. Black arrows indicate signals arising from silent HM loci; white arrows indicate signals arising from their derepressed counterparts (sir2Δ background). As these MATa strains bear two copies of the a1 promoter (one at MAT and the other at HMR), as indicated, unambiguous measurement of factor occupancy is possible only for HMLα1/HMLα2. The lower image is a two-part composite derived from the same gel. On the right are summaries of Rpb1 occupancy at the indicated promoters in SIR+, sir2Δ, and sir4Δ cells based on ChIP assays such as those illustrated. In each pairwise comparison, Rpb1 occupancy of the sir mutant is set at 1.0, with its occupancy in the SIR+ strain normalized to that. Depicted are summaries of four (S288C) or two (SLY101) biological replicates (means ± standard errors of the means or ± standard deviations, respectively).
We first investigated the occupancy of Pol II. As expected, polymerase is present at the derepressed HMLα1/HMLα2 promoters (sir2Δ background). This is most clearly seen in MATa strains that bear α1 and α2 genes only at HML (Fig. 2D). Strikingly, Pol II occupancy remains high in a SIR+ context (black arrows) when HMLα1 and HMLα2 transcription is extinguished (Fig. 2C, lane 4), Sir3 occupancy is high (Fig. 2B), and local nucleosomes are both hypoacetylated (7) and H3 K4 hypomethylated (52). Importantly, Pol II occupancy at the silent α1/α2 promoters is well above that seen at a nontranscribed euchromatic region (ARS504) (Fig. 2D) and is observed in two distinct genetic backgrounds (S288C and SLY101). A virtually identical pattern of Pol II occupancy is seen at HMRa1 in MATα cells, and these data, along with those for HMLα1/HMLα2, are quantified in Fig. 2D. To confirm that the PCR amplification was specific, we constructed strains individually deleted for either HML or HMR and subjected these to ChIP analysis as described above. As expected, neither deletion strain evinced a significant PCR product (see Fig. 5A, lane 5 of each gel). Taken together, these findings argue that SIR is permissive to the recruitment of Pol II at stably silenced target genes.
FIG. 5.
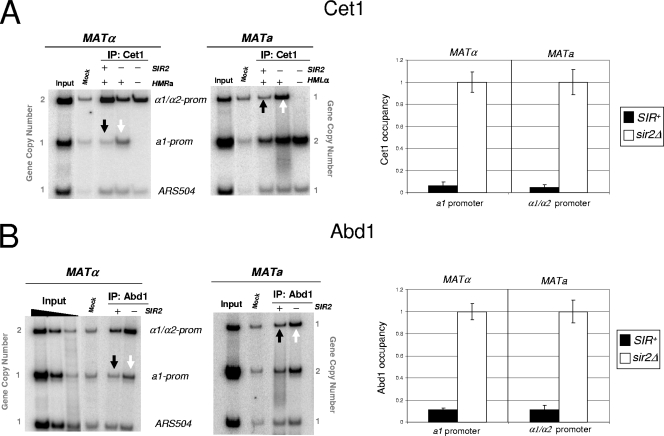
SIR restricts the occupancy of capping enzymes at both HMR and HML. (A) On the left is an in vivo cross-linking analysis of Cet1 at the a1 and α1/α2 promoters (prom) in isogenic S288C strains. “Input” represents 0.067% of soluble chromatin, derived from the S288C parental strain, used in each IP. Mock IP, sheared chromatin (derived from the S288C parental strain) precipitated with Pansorbin cells only. Note that lane 5 of each gel represents an analysis of strains deleted for both SIR2 and the indicated HM locus (see the legend to Fig. 2A). A summary of four independent experiments (means ± standard errors of the means) for each strain is provided on the right. (B) In vivo cross-linking analysis of Abd1 at the a1 and α1/α2 promoters, conducted as described for panel A.
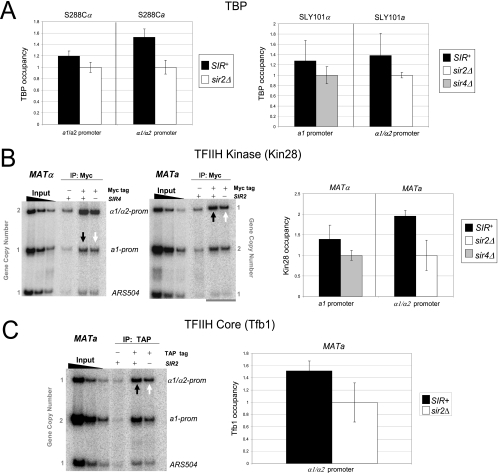
We next investigated whether TBP was present at the silent mating promoters. While TBP is typically the first PIC component recruited to the core promoter (Fig. 1), exceptions exist (2), and the absence of TBP at the HM loci could explain silencing. However, TBP can be readily detected, irrespective of the strain background (Fig. 3A), at a level that is at least equivalent to its occupancy in the euchromatic state. The occupancy of Pol II and TBP at the hyperrepressed a1 and α1/α2 promoters is consistent with the downstream inhibition model of transcriptional silencing while simultaneously arguing against both steric hindrance and PIC interference models.
FIG. 3.
Silent chromatin is permissive to the recruitment of both TBP and TFIIH. (A) TBP ChIP analysis of the a1 and α1/α2 promoters in isogenic S288C and SLY101 strains (α or a mating type, as indicated). Assays were conducted as described in the legend to Fig. 2D. Shown are summaries of either four (S288C strains) or two (SLY101 strains) independent experiments (means ± standard errors of the means [SEM] or ± standard deviations, respectively). (B) In vivo cross-linking analysis of Kin28 (Myc tagged). On the left are representative ChIP gels, and on the right are summaries of three independent experiments (means ± SEM) of MATα and MATa strains (SLY101 background). KIN28-Myc is present at its native chromosomal locus and is the only copy of KIN28 in these strains; thus, the epitope-tagged version is functional. Black and white arrows indicate silent and derepressed states, respectively. Input samples are as described in the legend to Fig. 2D. (C) In vivo cross-linking analysis of the core TFIIH subunit, Tfb1 (TAP tagged). TFB1-TAP is present at its native chromosomal locus (S288C background). A representative gel is illustrated on the left (black and white arrows indicate silent and derepressed states, respectively), and the means ± SEM of three independent experiments are provided on the right.
Silent chromatin is permissive to TFIIH recruitment.
TFIIH is typically the last component assembled within the PIC during the activation of Pol II genes (45). Although its presence is not required for the stable association of TBP (34), Pol II transcription is critically dependent on TFIIH. Therefore, given that both TBP and Pol II are present at the hyperrepressed HM loci, transcriptional silencing might arise from the impaired recruitment of this nine-subunit, 438-kDa complex. However, as shown in the gel analysis of Fig. 3B, the essential TFIIH subunit, Kin28 (TFIIH kinase), is efficiently recruited to the heterochromatic HMRa1 and HMLα1/HMLα2 promoters, and its abundance at each locus equals or exceeds that seen in the sir2Δ or sir4Δ euchromatic state. To rule out the possibility that Kin28, as part of the TFIIK kinase subcomplex, is recruited independently of core TFIIH, we also examined the occupancy of Tfb1, an essential core subunit of TFIIH. As shown in Fig. 3C, the Tfb1 occupancy of the HMLα1/HMLα2 promoter is significant in the SIR-induced heterochromatic state and actually exceeds its occupancy in the sir2Δ-induced euchromatic state. We conclude that TFIIH is present, and abundant, within the SIR-silenced HM promoters. The paradoxical enhanced occupancy of TFIIH at α1/α2 in the SIR+ strain parallels findings for Pol II and TBP (Fig. 2D, 3A) and is further considered below.
Silent chromatin is permissive to Ser5 phosphorylation of the Pol II CTD, yet Pol II arrests at or near the promoter.
A critical function of the TFIIH kinase is to phosphorylate the Pol II large subunit at Ser5 within the CTD heptad repeat (23). The Ser5-phosphorylated isoform of Pol II is characteristic of the polymerase that has initiated transcription (53). Therefore, a potential way that SIR could act is by inhibiting the phosphorylation of Ser5 residues within the Pol II CTD, thereby aborting transcriptional initiation. However, the Ser5-phosphorylated isoform of Pol II is present at the silent HM promoters, and its abundance is comparable to that seen in the sir2Δ euchromatic state (Fig. 4A). This result provides additional evidence for the presence of Pol II at the silent a1 and α1/α2 genes and further suggests that Pol II has initiated transcription.
FIG. 4.
Silent chromatin is permissive to the Ser5 phosphorylation of the CTD, yet Pol II fails to elongate through the gene coding region. (A) Pol IIoSer5 ChIP analysis of the a1, α1/α2, and PMA1 promoters (prom) of the indicated S288C strains detected using the monoclonal antibody H14. M, mock IP (sheared chromatin precipitated with preblocked anti-IgM-agarose beads only). Black and white arrows indicate signals arising from silenced and derepressed states, respectively. In this experiment, the PMA1 promoter serves as a positive control for Ser5-phosphorylated Rpb1 occupancy; ARS504 serves as a negative control. On the right is a summary of four independent experiments (means ± standard errors of the means [SEM]). (B) In vivo cross-linking analysis of Pol II within the 3′ flank of the HMRa2 gene in strain S288Cα (left). Also evaluated are the α1/α2 promoter and ARS504, which serve as positive and negative ChIP controls, respectively. PI, immunoprecipitation with preimmune serum. On the right is a comparison of Pol II occupancy at the HMRa1 promoter region and the HMRa2 3′ flank, obtained from independent PCRs of the same chromatin DNA templates (n = 3; results are given as means ± SEM). The occupancy for each locus is normalized to that seen in the sir2Δ strain, which is set at 1.0. (C) Pol II stalls at the SIR-silenced hsp82 promoter. The upper graphic is a schematic of the HMR-E silencer element, illustrating binding sites for ORC, Rap1, and Abf1, and the location of integrated silencers flanking the hsp82-2001 gene (silencers are symbolized by arrows) (57). Also illustrated are PCR amplicons (cross-hatched rectangles) centered at coordinates −170, +1400, and +2100 relative to the ATG (+1) of hsp82-2001. The lower graphic is a summary of Pol II occupancy at hsp82-2001 in SIR+ and sir4Δ strains (means ± SEM; n = 3). Note that although there is virtually no difference in Pol II promoter occupancy in SIR+ versus sir4Δ cells, there are significant differences in Pol II occupancy within the ORF and 3′-UTR (P < 0.05; two-tailed t test).
We next investigated whether phosphorylated Pol II is capable of productive elongation by testing its presence at the 3′ end of HMRa2 in a SIR+ strain. We focused on a2, since the small sizes of the a1, α1, and α2 ORFs (378, 525, and 630 bp, respectively), along with the presence of closely abutting (and similarly regulated) genes 3′ of both HMRa1 and HMLα1 (see Fig. 8), prevent a definitive analysis of Pol II localization at these genes. Figure 4B reveals that Pol II is virtually undetectable within the 3′-flanking region of HMRa2, a site located ∼1 kb downstream of the a2 promoter. This contrasts with Pol II abundance at the HMRa1 promoter in SIR+ cells as well as its abundance within the 3′ flank of HMRa2 in sir2Δ cells and is consistent with the stalling of Ser5-phosphorylated Pol II at the 5′ end of HMRa2. We extended this analysis to a previously characterized hsp82 transgene, ectopically silenced by integrated, flanking HMR-E silencer elements (57). Consistent with the above results, Pol II occupancy at the hsp82-2001 promoter is undiminished by SIR, in contrast to its significantly reduced occupancy within the gene's ORF and 3′-UTR (Fig. 4C).
FIG. 8.
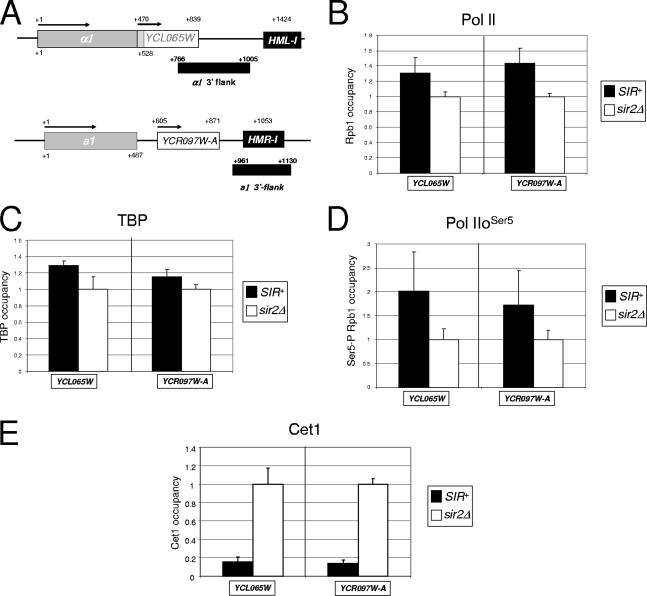
The SIR-regulated YCL065W and YCR097W-A genes are fully permissive to Pol II and TBP recruitment and to Ser5 CTD phosphorylation, but they are restrictive to the recruitment of capping enzyme. (A) Physical maps of HMLα1/YCL065W and HMRa1/YCR097W-A showing the location and orientation of ORFs and the location of the HML-I and HMR-I silencers. Also shown are PCR amplicons used for this analysis (black rectangles). Coordinates are numbered relative to the ATG codons of the α1 and a1 ORFs. (B) Summary of Pol II CTD ChIP assays (S288C background) for YCL065W and YCR097W-A. ChIP analysis and the quantification of factor occupancy were performed as described in the legend to Fig. 2D. Bars represent means ± standard errors of the means (SEM) for four independent biological replicates. (C to E) TBP, Pol IIoSer5, and Cet1 abundance at YCL065W and YCR097W-A. ChIP analysis and quantification were performed as described above (depicted are means ± SEM; n = 4).
SIR restricts the recruitment of both 5′-capping enzymes and elongation factors.
The inability of Pol IIoSer5 to recover from a stalled state at the 5′ end of silent genes could reflect the presence of a number of obstacles. For example, SIR-stabilized nucleosomes could physically block the transit of RNA polymerase despite the normal recruitment of factors critical to productive elongation. Alternatively, SIR could impair Pol II processivity by preventing the access of these factors. To distinguish between these possibilities, we initially tested for the presence of capping enzyme, given that capping takes place early following initiation (Fig. 1). Indeed, Cet1/Ceg1's recruitment to the transcription elongation complex (TEC) is triggered by the Ser5 phosphorylation of the CTD (32, 54). Strikingly, Cet1 is present only at background levels within the silent a1 and α1/α2 gene promoters (Fig. 5A), although it is abundant under derepressing conditions. Quantification of four biological replicates indicates that Cet1 occupancy is reduced 16- to 20-fold by SIR repression (Fig. 5A). This impairment in Cet1 recruitment stands in stark contrast to the essentially complete accessibility of TBP, Pol II, and TFIIH to silent chromatin.
We next addressed whether the occupancy of the 5′ mRNA cap methylase, Abd1, also is influenced by SIR. Abd1 binds the CTD independently of Cet1/Ceg1, and its interaction requires both Ser5 phosphorylation and the TFIIH kinase (54). Given this, it was possible that despite the absence of Cet1 at the silent a1 and α1/α2 genes, Abd1 recruitment occurred unimpaired. However, as shown in Fig. 5B, the recruitment of Abd1, like that of Cet1, is highly restricted (∼10-fold). This restriction has important mechanistic implications, given that Abd1 has been functionally linked to subsequent elongation in both S. cerevisiae and the fission yeast Saccharomyces pombe (20, 55). Taken together, the data argue that silent chromatin is highly restrictive to the recruitment of capping machinery.
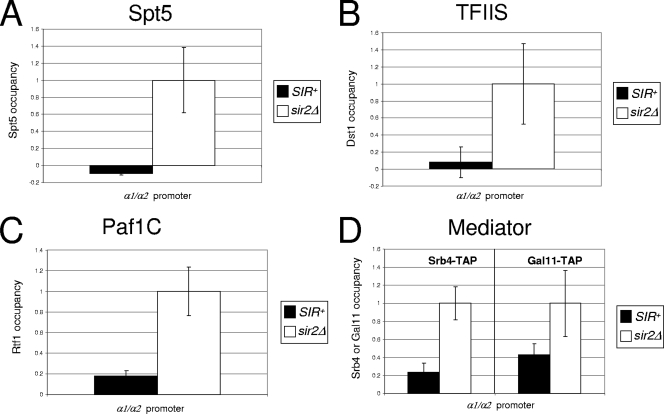
The absence of capping enzymes raises the possibility that SIR additionally restricts other factors whose association with the TEC takes place either concomitantly with or subsequent to that of capping enzymes. We focused on three elongation factors: Spt5, TFIIS, and the Paf1 complex (Paf1C). The essential elongation factor Spt5, as part of the Spt4/Spt5 complex, has been implicated in the control of early transcription (Fig. 1) and physically and functionally interacts with both Ceg1/Cet1 and Abd1 (37, 41). As shown in Fig. 6A, SIR reduces the recruitment of Spt5 to the silent α1/α2 promoter to an undetectable level. The sir2Δ mutation alleviates this block, and Spt5 is efficiently recruited concomitantly with transcriptional activation.
FIG. 6.
SIR strongly reduces the occupancy of elongation factors Spt5, TFIIS, and Paf1C and partially restricts the recruitment of Mediator. (A to D) In vivo cross-linking analysis of the indicated TAP-tagged factors at the HMLα/HMLα promoter in SIR+ and sir2Δ S288Ca derivatives, conducted as described for Fig. 3C (in all cases, means ± standard errors of the means are shown [n = 3]).
TFIIS, an elongation factor that reactivates stalled Pol II, is localized principally at the 5′ end of genes (53). Given the role of TFIIS in releasing inappropriately paused Pol II, its absence from silent coding regions might further underlie the inability of Pol II to elongate within silent chromatin. Indeed, as shown in Fig. 6B, the occupancy of TFIIS (Dst1) at HMLα1/HMLα2 is reduced 12-fold by SIR.
To investigate the effect of SIR silencing on Paf1C recruitment, we examined the occupancy of its Rtf1 subunit. Paf1C associates with Ser5-phosphorylated Pol II and genetically and physically interacts with the Spt4/Spt5 complex (61), as well as with Spt6, FACT, and Chd1. Moreover, Paf1C plays a critical role in regulating transcription-associated histone modifications, including H2B ubiquitylation and H3 K4 methylation, and does this through mediating the interaction between Pol II and the enzymes responsible for these modifications. As shown in Fig. 6C, SIR strongly impedes the binding of Rtf1 and, by extension, of Paf1C to the silent HMLα promoters, thereby providing a basis for the absence of activating histone modifications within silent chromatin (reviewed in references 15 and 46) as well as further evidence accounting for the failure of Pol II to escape from the gene's 5′ end.
Efficiency of silencing inversely correlates with the presence of capping enzyme.
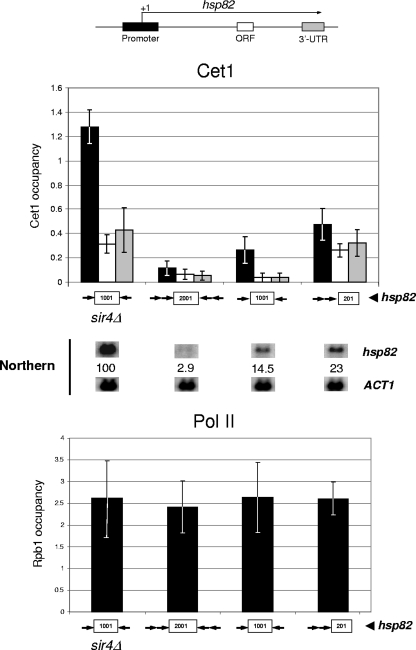
To investigate the functional link between capping enzyme recruitment and transcriptional silencing in more detail, we employed silencer-flanked hsp82 transgenes. As discussed above, these consist of the native HSP82 heat shock gene flanked by chromosomally integrated HMR-E silencers. The efficiency of transcriptional silencing at these alleles correlates with Sir2 recruitment: hsp82-2001, flanked by four silencers, exhibits ∼95% of the level of Sir2 observed at HMR and is repressed >30-fold; hsp82-1001, flanked by two silencers and containing ∼80% of the level of Sir2, is repressed 5- to 10-fold; and hsp82-201, bearing tandem upstream silencers and containing ∼30% of Sir2, is silenced 2- to 4-fold (56, 57). If the reduction in recruitment of capping enzymes is linked to silencing, then the degree to which these factors are prevented from accessing the hsp82 transgenes should correlate with the degree to which they are silenced. This is in fact what is seen: despite Pol II being present at normal levels at the 5′ ends of all three silenced hsp82 alleles, Cet1 recruitment is reduced by almost 10-fold at hsp82-2001, 5-fold at hsp82-1001, and 3-fold at hsp82-201 (Fig. 7). Therefore, restriction in Cet1 occupancy, unlike that of Pol II, directly correlates with SIR-mediated silencing.
FIG. 7.
Efficiency of SIR-dependent silencing strongly correlates with a restriction in recruitment of the Cet1 capping enzyme. Illustrated is a summary of Cet1 and Pol II (Rpb1) occupancy of SIR-silenced hsp82 transgenes (top and bottom histograms). Factor occupancy at the euchromatic (derepressed) hsp82-1001 allele also is depicted (sir4Δ background). These transgenes differ in the dosage and arrangement of integrated silencers that flank the hsp82 promoter and coding region (symbolized by arrows; see the legend to Fig. 4C). Cet1 abundance is quantified for the promoter, ORF, and 3′-UTR (the location of amplicons and shading key are indicated at the top); Pol II abundance is quantified for the promoter only. For both analyses, bars represent means ± standard errors of the means of three biological replicates. A representative Northern analysis of each strain also is displayed, with hsp82 transcript levels, normalized to ACT1, indicated (values represent the means of two independent experiments).
Genes abutting HMLα1 and HMRa 1 are silenced by SIR but permissive to Pol II recruitment.
To extend the generality of our observations, we tested whether Pol II, TBP, or Cet1 is recruited to two additional silenced genes, YCL065W and YCR097W-A. These cryptic genes are located immediately downstream of HMLα1 and HMRa1, respectively (Fig. 8A), and they are expressed in a sir2Δ mutant but not in the SIR+ parental strain (data not shown). As illustrated in Fig. 8B and C, Pol II and TBP are in fact present and abundant at the YCL065W and YCR097W-A promoters in the SIR+ strain. We then asked whether either Pol IIoSer5 or Cet1 is present, given our findings with HMLα1/HMLα2 and HMRa1. Strikingly, Pol IIoSer5 is present at these genes (Fig. 8D), while Cet1 is not (Fig. 8E). These results recapitulate findings for the HMLα1/HMLα2 and HMRa1 promoters and, intriguingly, suggest the presence of two additional SIR-regulated genes within the HM loci.
SIR partially restricts recruitment of Mediator.
Finally, we examined the recruitment of Mediator, a transcriptional coregulator thought to bridge sequence-specific activators with RNA Pol II. Consistent with a general role in transcription, Mediator has been detected at the 5′ ends of many, if not most, genes (3). In light of this, it has been proposed that Mediator should be considered a general transcription factor that is equivalent to components of the PIC (65). An alternative view is that Mediator primarily acts as a gene-specific coactivator (13). Mediator has been detected in holoenzyme preparations of Pol II (31) but is not corecruited with polymerase at certain genes (5, 10). As shown in Fig. 6D, the occupancy at HMLα1/HMLα2 of both head (Srb4) and tail (Gal11) Mediator subunits is reduced 50 to 75% by SIR. This observation argues that Mediator is recruited to HML independently of Pol II and that its access, unlike that of Pol II, is restricted.
DISCUSSION
SIR is permissive to GTF recruitment and PIC assembly.
Here, we have investigated the mechanism by which SIR regulates transcription in Saccharomyces cerevisiae. We demonstrate that at SIR-repressed genes, steps in the transcription cascade that occur upstream of PIC recruitment are fully permitted, while those occurring downstream are essentially abolished. This conclusion arises from a quantitative assessment of factor occupancy at two naturally silenced targets of SIR, HMRa1 and HMLα1/HMLα2, and in two distinct genetic backgrounds. At these genes transcription is extinguished, yet three GTFs—TBP, Pol II, and TFIIH—are present and abundant within their promoters. The presence of TFIIH, the last factor typically recruited to the PIC, implies that the initiation complex is fully assembled. Additionally, the presence of Ser5-phosphorylated Pol II suggests that Kin28 is functional and that Pol II has initiated transcription. Nonetheless, the recruitment of downstream factors involved in mRNA capping and transcript elongation is virtually abolished (considered more fully below). Importantly, PIC assembly in silent chromatin is not a peculiarity of the mating-type promoters. We observed that two other genes, YCL065W and YCR097W-A, are similarly regulated: Pol II and TBP are present at normal levels, and Pol II is Ser5 phosphorylated, yet there is no detectable transcription in a SIR+ background. Both, however, are expressed in a sir2Δ mutant.
Our detection of Pol II at silent HMRa1 and HMLα1/HMLα2 is at odds with a recent, quantitative ChIP analysis that gave rise to the PIC interference model (8). That study employed strains of the S288C background, as done here, and investigated the presence of Pol II at the silent HM promoters using both a CTD-specific antibody (8WG16) and a phospho-Ser5-CTD-specific antibody (H14, as employed here [Fig. 4A]). However, the authors of that study failed to detect the presence of Pol II using either antibody; likewise, they were unable to detect either TFIIB or TFIIE (8). While the reason for this is unclear, three lines of evidence argue against the PIC interference model. First, as discussed above, we detected TBP and TFIIH, in addition to Pol II, at the hyperrepressed promoters. Normal levels of Pol II are consistent with the unimpaired function of Kin28, given that Kin28 thermal inactivation strongly reduces Pol II promoter occupancy (42, 54). Second, the ChIP methodology we employed is sufficiently sensitive to detect differences in factor occupancy at these genes, as demonstrated by the fact that we observed large differences in the occupancy of downstream factors at euchromatic versus heterochromatic promoters. Third, a recent high-resolution genome-wide analysis of Pol II density supports the notion that Pol II is present at SIR-silenced promoters (62). In this ChIP-chip analysis, the Rpb3 subunit of Pol II was detected at genome-average levels within the HM loci as well as within the promoters of telomere-linked genes (62). In telling contrast, Pol II density was significantly reduced within the coding regions of telomeric genes, paralleling our observations at both native (HMRa2) and transgenic (hsp82-2001) targets, and as expected if SIR permits Pol II recruitment but prevents its productive elongation.
PIC recruitment, assembly, and normal function within the context of silent chromatin are all the more striking given that H3 K4 trimethylation, a mark that stabilizes TBP binding to nucleosomes in mammalian cells (69) and SAGA binding to nucleosomes in yeast (47), is greatly reduced at HMR and HML (44, 52), as are two other covalent marks of active chromatin, H3 K36 trimethylation and H2B K123 ubiquitylation (12, 67). Thus, silent chromatin is impoverished in all three marks that correlate with transcriptional activation in S. cerevisiae, methylation, ubiquitylation, and, as discussed above, acetylation, yet it is permissive to the binding, at core promoters, of critical components of the transcriptional machinery.
SIR imposes a 5′ arrest on Pol II by restricting recruitment of capping and elongation factors.
In contrast to the unhindered access of Pol II and GTFs to silent promoters, the occupancy of the capping enzymes Cet1/Ceg1 and Abd1 and elongation factors Spt5, TFIIS, and Paf1C is highly restricted. Of particular significance is the tight inverse correlation between silencing efficiency and Cet1 occupancy (Fig. 7). This suggests a mechanistic link between a block in capping enzyme recruitment and silencing. Abd1 harbors a transcription elongation activity independent of cap methylation, and the inactivation of this function reduces Pol II occupancy at the 5′ end and/or Pol II processivity (55). Therefore, SIR, by impeding capping enzyme recruitment, may contribute to Pol II stalling. Such stalling takes place at or near the gene's 5′ end, given that Pol II abundance is considerably reduced (relative to that of the sir2Δ-derepressed state) at points downstream (Fig. 4B, C). We have tested for the presence of short 5′ transcripts associated with the silenced hsp82-2001 gene. None could be found with primer extension assays (56). Therefore, the silencing of stably repressed loci such as HML, HMR, and hsp82-2001 is unlikely to involve posttranscriptional processing events, as recently shown for Sir2-regulated telomeric reporter genes and the nontranscribed spacer regions within the ribosomal DNA array (68).
Regarding the actual step targeted, our data are compatible with at least two possibilities. First, SIR may prevent the transition between initiation and promoter escape, a rate-limiting step for Pol II in vitro. Alternatively, SIR may permit early elongation yet increase the inherent tendency of the early elongation complex to pause. While arrested elongation complexes can be rescued by TFIIS (1), SIR prevents TFIIS recruitment. An interesting aspect of SIR-induced promoter-proximal pausing is that it must occur without the participation of the Spt4/Spt5 complex. We speculate that the Sir2/3/4 complex plays that role through its deacetylation and stable positioning of nucleosomes.
What underlies the differential accessibility of initiation and capping/elongation factors to silent chromatin?
An important implication of our findings is that SIR-mediated silent chromatin is differentially accessible to initiation and capping/elongation factors. Although our work does not address how this might be achieved, there are several possibilities. One way is via molecular sieving. This idea is appealing, given the longstanding view that chromatin can repress gene expression by excluding factors from accessing their target DNA sequences (see Introduction). However, the proteins with the most severely restricted access, Cet1/Ceg1, Abd1, TFIIS, and Spt4/5, tend to be relatively small, with molecular masses of 115, 50, 35, and 127 kDa, respectively. In contrast, the accessible factors Pol II and TFIIH have respective masses of 550 and 438 kDa. As TBP occupancy may signify the presence of TFIID, with a mass of ∼1.2 MDa, it is unlikely that SIR-silenced chromatin acts by molecular sieving.
Alternatively, SIR may prevent polymerase elongation as a consequence of the structural features of silent chromatin itself. Hypoacetylated nucleosomes are very stable, with adjacent nucleosomes possessing the potential to interact with each other through ionic bonding (39) and arrays of H4 K16 hypoacetylated nucleosomes capable of forming 30-nm-like fibers (59). Thus, Pol II, although able to gain access to silent promoters, may be unable to elongate through hypoacetylated nucleosomes complexed with Sir2/3/4. The inability of polymerase to elongate through stabilized silent nucleosomes may prevent the stable association of capping and elongation factors with the TEC. Additionally, it is possible that Sir2 inhibits the recruitment of one or more capping/elongation factors by virtue of its intrinsic lysine deacetylase activity. This could occur by direct deacetylation of an elongation factor or via the deacetylation of H4 AcK16, which may serve as a binding site for one or more downstream factors. Although our data indicate that at least five factors are excluded from silent chromatin, it is conceivable that a subset of them, or an as-yet-unidentified factor, actually is targeted, abrogating subsequent steps in the transcriptional cascade.
Importantly, our data appear to rule out a third mechanism, elongational arrest via the proteolysis of stalled Pol II complexes. Inappropriately stalled Pol II is targeted for proteasome-mediated degradation by Rsp5-dependent ubiquitylation (4). However, Ser5 phosphorylation, a signature of SIR repression, strongly inhibits Pol II ubiquitylation (60). A schematic summarizing our findings at SIR-repressed HMLα1/HMLα2 is illustrated in Fig. 9A.
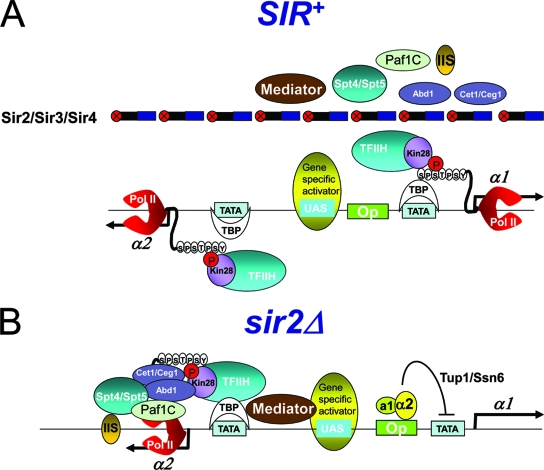
FIG. 9.
Schematic summary of factor occupancy at SIR-repressed, Tup1/Ssn6-repressed, and transcriptionally derepressed HMLα promoters. (A) SIR permits the access of TBP, Pol II, and TFIIH to the silent HMLα1/HMLα2 promoters (this study) and, by analogy with other SIR-regulated loci, a gene-specific activator(s) (8, 56, 57). Ser5 phosphorylation of the Pol II CTD also is permitted, yet Pol II is stalled at the genes' 5′ ends. In contrast to GTFs, access of the Cet1/Ceg1 and Abd1 capping enzymes and the Spt5, TFIIS, and Paf1C elongation factors is strongly inhibited. The access of Mediator also is restricted, although not as severely. (B) Tup1/Ssn6, recruited to the HMLα1 promoter by the a1/α2 repressor in sir2Δ mutants, aborts the assembly of the PIC, as suggested by occupancy data presented here (summarized in Table 2) and by analogy with other Tup1-regulated promoters (34, 73). Mediator participates in Tup1 repression (17); thus, although it was not specifically tested here, it could be present. In the same cell, the HMLα2 promoter is transcriptionally derepressed; consequently, GTFs, Mediator, and downstream factors are abundant.
Silent yeast promoters and quiescent mammalian promoters are both characterized by the presence of stalled polymerase.
Of relevance to our findings is a recent genome-wide analysis of human cells that demonstrated the presence of Pol II at the vast majority of promoters of protein-encoding genes, including >50% of inactive promoters (19). Interestingly, just as we have seen for silenced yeast promoters, Pol II association with inactive human promoters likely is due to transcription initiation without transcript accumulation. This phenomenon was seen in three different cell types (embryonic stem cells, hepatocytes, and B cells), so it likely represents the prevalent mechanism by which gene transcription is controlled in humans. While it is intriguing that this basic attribute, PIC assembly and Pol II initiation at an otherwise inactive gene promoter, is evolutionarily conserved, the mechanism by which these human genes are kept in a quiescent state is unknown. Our findings that SIR-silenced, heterochromatic genes are repressed via the differential recruitment of initiation and capping/elongation factors provide a potential model by which this large and important class of genes is transcriptionally regulated.
Tup1/Ssn6 represses the α1 promoter by interfering with PIC recruitment.
Our results indicate that TBP, Pol II, Kin28, and Pol IIoSer5 occupancy at the HMLα1/HMLα2 promoters paradoxically is more abundant in the context of SIR silencing than in the euchromatic (sir2Δ or sir4Δ), derepressed state. This contrasts with the essentially equivalent occupancy of the same factors at the HMRa1 promoter and provides a stark contrast to the null occupancy of capping/elongation factors at either HML or HMR (summarized in Table 2). A probable explanation is that the α1 promoter is subject to a second form of negative regulation, conferred by the a1/α2 haploid gene-specific repressor (22), present in sir haploids and wild-type diploids. This repressor negatively regulates the transcription of linked promoters by the recruitment of the Tup1/Ssn6 corepressor, which in turn recruits type I and type II histone deacetylases (reviewed in references 11 and 40). Previous studies have demonstrated that Tup1 prevents the recruitment of TBP and Pol II to the promoters it regulates (34, 73). Therefore, our data, in combination with these previous observations, suggest that at the HMLα1 promoter, Tup1 and SIR use distinct mechanisms to repress transcription. Tup1 blocks the recruitment of TBP, Pol II, and TFIIH, thereby aborting PIC assembly (illustrated in Fig. 9B), while the Sir2/3/4 complex permits the recruitment of these factors but abrogates transcription by restricting the access of capping and elongation factors, resulting in the irreversible stalling of Pol II at the gene's 5′ end.
Acknowledgments
We thank Kelly Tatchell for yeast strains and helpful discussions; Sebastian Chavez for sharing unpublished data; Steve Buratowski, Michael Green, and Rohinton Kamakaka for generous gifts of antibodies; David Bentley for the glutathione S-transferase-CTD construct; and members of our laboratory for their critical comments on an earlier version of the manuscript.
This work was supported by grants awarded to D.S.G. from the National Science Foundation (MCB-0450419; MCB-0747227).
Footnotes
Published ahead of print on 7 April 2008.
REFERENCES
- 1.Adelman, K., M. T. Marr, J. Werner, A. Saunders, Z. Ni, E. D. Andrulis, and J. T. Lis. 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol. Cell 17103-112. [DOI] [PubMed] [Google Scholar]
- 2.Agalioti, T., S. Lomvardas, B. Parekh, J. Yie, T. Maniatis, and D. Thanos. 2000. Ordered recruitment of chromatin modifying and general transcription factors to the IFN-β promoter. Cell 103667-678. [DOI] [PubMed] [Google Scholar]
- 3.Andrau, J. C., L. van de Pasch, P. Lijnzaad, T. Bijma, M. G. Koerkamp, J. van de Peppel, M. Werner, and F. C. Holstege. 2006. Genome-wide location of the coactivator Mediator: binding without activation and transient Cdk8 interaction on DNA. Mol. Cell 22179-192. [DOI] [PubMed] [Google Scholar]
- 4.Beaudenon, S. L., M. R. Huacani, G. Wang, D. P. McDonnell, and J. M. Huibregtse. 1999. Rsp5 ubiquitin-protein ligase mediates DNA damage-induced degradation of the large subunit of RNA polymerase II in Saccharomyces cerevisiae. Mol. Cell. Biol. 196972-6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bhoite, L. T., Y. Yu, and D. J. Stillman. 2001. The Swi5 activator recruits the Mediator complex to the HO promoter without RNA polymerase II. Genes Dev. 152457-2469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brand, A. H., L. Breeden, J. Abraham, R. Sternglanz, and K. Nasmyth. 1985. Characterization of a “silencer” in yeast: a DNA sequence with properties opposite to those of a transcriptional enhancer. Cell 4141-48. [DOI] [PubMed] [Google Scholar]
- 7.Braunstein, M., A. B. Rose, S. G. Holmes, C. D. Allis, and J. R. Broach. 1993. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 7592-604. [DOI] [PubMed] [Google Scholar]
- 8.Chen, L., and J. Widom. 2005. Mechanism of transcriptional silencing in yeast. Cell 12037-48. [DOI] [PubMed] [Google Scholar]
- 9.Cheng, T.-H., and M. R. Gartenberg. 2000. Yeast heterochromatin is a dynamic structure that requires silencers continuously. Genes Dev. 14452-463. [PMC free article] [PubMed] [Google Scholar]
- 10.Cosma, M. P., S. Panizza, and K. Nasmyth. 2001. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 71213-1220. [DOI] [PubMed] [Google Scholar]
- 11.Courey, A. J., and S. Jia. 2001. Transcriptional repression: the long and the short of it. Genes Dev. 152786-2796. [DOI] [PubMed] [Google Scholar]
- 12.Emre, N. C., K. Ingvarsdottir, A. Wyce, A. Wood, N. J. Krogan, K. W. Henry, K. Li, R. Marmorstein, J. F. Greenblatt, A. Shilatifard, and S. L. Berger. 2005. Maintenance of low histone ubiquitylation by Ubp10 correlates with telomere-proximal Sir2 association and gene silencing. Mol. Cell 17585-594. [DOI] [PubMed] [Google Scholar]
- 13.Fan, X., D. M. Chou, and K. Struhl. 2006. Activator-specific recruitment of Mediator in vivo. Nat. Struct. Mol. Biol. 13117-120. [DOI] [PubMed] [Google Scholar]
- 14.Fox, C. A., and K. H. McConnell. 2005. Toward biochemical understanding of a transcriptionally silenced chromosomal domain in Saccharomyces cerevisiae. J. Biol. Chem. 2808629-8632. [DOI] [PubMed] [Google Scholar]
- 15.Gao, L., and D. S. Gross. 2006. Using genomics and proteomics to investigate mechanisms of transcriptional silencing in Saccharomyces cerevisiae. Brief. Funct. Genomics Proteomics 5280-288. [DOI] [PubMed] [Google Scholar]
- 16.Grewal, S. I., and S. Jia. 2007. Heterochromatin revisited. Nat. Rev. Genet. 835-46. [DOI] [PubMed] [Google Scholar]
- 17.Gromöller, A., and N. Lehming. 2000. Srb7p is a physical and physiological target of Tup1p. EMBO J. 196845-6852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross, D. S. 2001. Sir proteins as transcriptional repressors. Trends Biochem. Sci. 26685-686. [DOI] [PubMed] [Google Scholar]
- 19.Guenther, M. G., S. S. Levine, L. A. Boyer, R. Jaenisch, and R. A. Young. 2007. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 13077-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guiguen, A., J. Soutourina, M. Dewez, L. Tafforeau, M. Dieu, M. Raes, J. Vandenhaute, M. Werner, and D. Hermand. 2007. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J. 261552-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Güldener, U., S. Heck, T. Fiedler, J. Beinhauer, and J. H. Hegemann. 1996. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 242519-2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harashima, S., A. M. Miller, K. Tanaka, K. Kusumoto, Y. Mukai, K. Nasmyth, and Y. Oshima. 1989. Mating-type control in Saccharomyces cerevisiae: isolation and characterization of mutants defective in repression by a1-α2. Mol. Cell. Biol. 94523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hengartner, C. J., V. E. Myer, S.-M. Liao, C. J. Wilson, S. S. Koh, and R. A. Young. 1998. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 243-53. [DOI] [PubMed] [Google Scholar]
- 24.Holmes, S. G., and J. R. Broach. 1996. Silencers are required for inheritance of the repressed state in yeast. Genes Dev. 101021-1032. [DOI] [PubMed] [Google Scholar]
- 25.Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie, S. P. Gygi, and D. Moazed. 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 224167-4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Imai, S.-I., C. M. Armstrong, M. Kaeberlein, and L. Guarente. 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403795-800. [DOI] [PubMed] [Google Scholar]
- 27.Imbalzano, A. N., H. Kwon, M. R. Green, and R. E. Kingston. 1994. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature 370481-485. [DOI] [PubMed] [Google Scholar]
- 28.Kim, M., S. H. Ahn, N. J. Krogan, J. F. Greenblatt, and S. Buratowski. 2004. Transitions in RNA polymerase II elongation complexes at the 3′ ends of genes. EMBO J. 23354-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klar, A. J., J. B. Hicks, and J. N. Strathern. 1982. Directionality of yeast mating-type interconversion. Cell 28551-561. [DOI] [PubMed] [Google Scholar]
- 30.Knop, M., K. Siegers, G. Pereira, W. Zachariae, B. Windsor, K. Nasmyth, and E. Schiebel. 1999. Epitope tagging of yeast genes using a PCR-based strategy: more tags and improved practical routines. Yeast 15963-972. [DOI] [PubMed] [Google Scholar]
- 31.Koleske, A. J., and R. A. Young. 1994. An RNA polymerase II holoenzyme responsive to activators. Nature 368466-469. [DOI] [PubMed] [Google Scholar]
- 32.Komarnitsky, P., E.-J. Cho, and S. Buratowski. 2000. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 142452-2460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kornberg, R. D., and Y. Lorch. 1991. Irresistible force meets immovable object: transcription and the nucleosome. Cell 67833-836. [DOI] [PubMed] [Google Scholar]
- 34.Kuras, L., and K. Struhl. 1999. Binding of TBP to promoters in vivo is stimulated by activators and requires pol II holoenzyme. Nature 399609-613. [DOI] [PubMed] [Google Scholar]
- 35.Lee, S., and D. S. Gross. 1993. Conditional silencing: the HMRE mating-type silencer exerts a rapidly reversible position effect on the yeast HSP82 heat shock gene. Mol. Cell. Biol. 13727-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, B., M. Carey, and J. L. Workman. 2007. The role of chromatin during transcription. Cell 128707-719. [DOI] [PubMed] [Google Scholar]
- 37.Lindstrom, D. L., S. L. Squazzo, N. Muster, T. A. Burckin, K. C. Wachter, C. A. Emigh, J. A. McCleery, J. R. Yates III, and G. A. Hartzog. 2003. Dual roles for Spt5 in pre-mRNA processing and transcription elongation revealed by identification of Spt5-associated proteins. Mol. Cell. Biol. 231368-1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liou, G. G., J. C. Tanny, R. G. Kruger, T. Walz, and D. Moazed. 2005. Assembly of the SIR complex and its regulation by O-acetyl-ADP-ribose, a product of NAD-dependent histone deacetylation. Cell 121515-527. [DOI] [PubMed] [Google Scholar]
- 39.Luger, K., A. W. Mader, R. K. Richmond, D. F. Sargent, and T. J. Richmond. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389251-260. [DOI] [PubMed] [Google Scholar]
- 40.Malavé, T. M., and S. Y. Dent. 2006. Transcriptional repression by Tup1-Ssn6. Biochem. Cell Biol. 84437-443. [DOI] [PubMed] [Google Scholar]
- 41.Mandal, S. S., C. Chu, T. Wada, H. Handa, A. J. Shatkin, and D. Reinberg. 2004. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc. Natl. Acad. Sci. USA 1017572-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mason, P. B., and K. Struhl. 2003. The FACT complex travels with elongating RNA polymerase II and is important for the fidelity of transcriptional initiation in vivo. Mol. Cell. Biol. 238323-8333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moazed, D. 2001. Common themes in the mechanisms of gene silencing. Mol. Cell 8489-498. [DOI] [PubMed] [Google Scholar]
- 44.Oki, M., and R. T. Kamakaka. 2005. Barrier function at HMR. Mol. Cell 19707-716. [DOI] [PubMed] [Google Scholar]
- 45.Orphanides, G., T. Lagrange, and D. Reinberg. 1996. The general transcription factors of RNA polymerase II. Genes Dev. 102657-2683. [DOI] [PubMed] [Google Scholar]
- 46.Pirrotta, V., and D. S. Gross. 2005. Epigenetic silencing mechanisms in budding yeast and fruit fly: different paths, same destinations. Mol. Cell 18395-398. [DOI] [PubMed] [Google Scholar]
- 47.Pray-Grant, M. G., J. A. Daniel, D. Schieltz, J. R. Yates, and P. A. Grant. 2005. Chd1 chromodomain links histone H3 methylation with SAGA- and SLIK-dependent acetylation. Nature 433434-438. [DOI] [PubMed] [Google Scholar]
- 48.Rasmussen, E. B., and J. T. Lis. 1993. In vivo transcriptional pausing and cap formation on three Drosophila heat shock genes. Proc. Natl. Acad. Sci. USA 907923-7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ravindra, A., K. Weiss, and R. T. Simpson. 1999. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol. Cell. Biol. 197944-7950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rivier, D. H., J. L. Ekena, and J. Rine. 1999. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics 151521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rusche, L. N., A. L. Kirchmaier, and J. Rine. 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72481-516. [DOI] [PubMed] [Google Scholar]
- 52.Santos-Rosa, H., A. J. Bannister, P. M. Dehe, V. Geli, and T. Kouzarides. 2004. Methylation of H3 lysine 4 at euchromatin promotes Sir3p association with heterochromatin. J. Biol. Chem. 27947506-47512. [DOI] [PubMed] [Google Scholar]
- 53.Saunders, A., L. J. Core, and J. T. Lis. 2006. Breaking barriers to transcription elongation. Nat. Rev. Mol. Cell Biol. 7557-567. [DOI] [PubMed] [Google Scholar]
- 54.Schroeder, S. C., B. Schwer, S. Shuman, and D. Bentley. 2000. Dynamic association of capping enzymes with transcribing RNA polymerase II. Genes Dev. 142435-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schroeder, S. C., D. A. Zorio, B. Schwer, S. Shuman, and D. Bentley. 2004. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol. Cell 13377-387. [DOI] [PubMed] [Google Scholar]
- 56.Sekinger, E. A., and D. S. Gross. 2001. Silenced chromatin is permissive to activator binding and PIC recruitment. Cell 105403-414. [DOI] [PubMed] [Google Scholar]
- 57.Sekinger, E. A., and D. S. Gross. 1999. SIR repression of a yeast heat shock gene: UAS and TATA footprints persist within heterochromatin. EMBO J. 187041-7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shei, G.-J., and J. R. Broach. 1995. Yeast silencers can act as orientation-dependent gene inactivation centers that respond to environmental signals. Mol. Cell. Biol. 153496-3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shogren-Knaak, M., H. Ishii, J. M. Sun, M. J. Pazin, J. R. Davie, and C. L. Peterson. 2006. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science 311844-847. [DOI] [PubMed] [Google Scholar]
- 60.Somesh, B. P., J. Reid, W. F. Liu, T. M. Sogaard, H. Erdjument-Bromage, P. Tempst, and J. Q. Svejstrup. 2005. Multiple mechanisms confining RNA polymerase II ubiquitylation to polymerases undergoing transcriptional arrest. Cell 121913-923. [DOI] [PubMed] [Google Scholar]
- 61.Squazzo, S. L., P. J. Costa, D. L. Lindstrom, K. E. Kumer, R. Simic, J. L. Jennings, A. J. Link, K. M. Arndt, and G. A. Hartzog. 2002. The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 211764-1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Steinmetz, E. J., C. L. Warren, J. N. Kuehner, B. Panbehi, A. Z. Ansari, and D. A. Brow. 2006. Genome-wide distribution of yeast RNA polymerase II and its control by Sen1 helicase. Mol. Cell 24735-746. [DOI] [PubMed] [Google Scholar]
- 63.Suka, N., K. Luo, and M. Grunstein. 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Cell 32378-383. [DOI] [PubMed] [Google Scholar]
- 64.Szeto, L., M. K. Fafalios, H. Zhong, A. K. Vershon, and J. R. Broach. 1997. Alpha2p controls donor preference during mating type interconversion in yeast by inactivating a recombinational enhancer of chromosome III. Genes Dev. 111899-1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Takagi, Y., and R. D. Kornberg. 2006. Mediator as a general transcription factor. J. Biol. Chem. 28180-89. [DOI] [PubMed] [Google Scholar]
- 66.Taylor, I. C., J. L. Workman, T. J. Schuetz, and R. E. Kingston. 1991. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 51285-1298. [DOI] [PubMed] [Google Scholar]
- 67.Tompa, R., and H. D. Madhani. 2007. Histone H3 lysine 36 methylation antagonizes silencing in Saccharomyces cerevisiae independently of the Rpd3S histone deacetylase complex. Genetics 175585-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Vasiljeva, L., M. Kim, N. Terzi, L. M. Soares, and S. Buratowski. 2008. Transcription termination and RNA degradation contribute to silencing of RNA polymerase II transcription within heterochromatin. Mol. Cell 29313-323. [DOI] [PubMed] [Google Scholar]
- 69.Vermeulen, M., K. W. Mulder, S. Denissov, W. W. Pijnappel, F. M. van Schaik, R. A. Varier, M. P. Baltissen, H. G. Stunnenberg, M. Mann, and H. T. Timmers. 2007. Selective anchoring of TFIID to nucleosomes by trimethylation of histone H3 lysine 4. Cell 13158-69. [DOI] [PubMed] [Google Scholar]
- 70.Weiss, K., and R. T. Simpson. 1998. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating type locus HMLa. Mol. Cell. Biol. 185392-5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams, S. K., and J. K. Tyler. 2007. Transcriptional regulation by chromatin disassembly and reassembly. Curr. Opin. Genet. Dev. 1788-93. [DOI] [PubMed] [Google Scholar]
- 72.Wu, X., and J. E. Haber. 1996. A 700 bp cis-acting region controls mating-type dependent recombination along the entire left arm of yeast chromosome III. Cell 87277-285. [DOI] [PubMed] [Google Scholar]
- 73.Zaman, Z., A. Z. Ansari, S. S. Koh, R. Young, and M. Ptashne. 2001. Interaction of a transcriptional repressor with the RNA polymerase II holoenzyme plays a crucial role in repression. Proc. Natl. Acad. Sci. USA 982550-2554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao, J., J. Herrera-Diaz, and D. S. Gross. 2005. Domain-wide displacement of histones by activated heat shock factor occurs independently of Swi/Snf and is not correlated with RNA polymerase II density. Mol. Cell. Biol. 258985-8999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zou, S., N. Ke, J. M. Kim, and D. F. Voytas. 1996. The Saccharomyces retrotransposon Ty5 integrates preferentially into regions of silent chromatin at the telomeres and mating loci. Genes Dev. 10634-645. [DOI] [PubMed] [Google Scholar]