Abstract
Krüppel-like factors (KLFs) recognize CACCC and GC-rich sequences in gene regulatory elements. Here, we describe the disruption of the murine basic Krüppel-like factor gene (Bklf or Klf3). Klf3 knockout mice have less white adipose tissue, and their fat pads contain smaller and fewer cells. Adipocyte differentiation is altered in murine embryonic fibroblasts from Klf3 knockouts. Klf3 expression was studied in the 3T3-L1 cellular system. Adipocyte differentiation is accompanied by a decline in Klf3 expression, and forced overexpression of Klf3 blocks 3T3-L1 differentiation. Klf3 represses transcription by recruiting C-terminal binding protein (CtBP) corepressors. CtBPs bind NADH and may function as metabolic sensors. A Klf3 mutant that does not bind CtBP cannot block adipogenesis. Other KLFs, Klf2, Klf5, and Klf15, also regulate adipogenesis, and functional CACCC elements occur in key adipogenic genes, including in the C/ebpα promoter. We find that C/ebpα is derepressed in Klf3 and Ctbp knockout fibroblasts and adipocytes from Klf3 knockout mice. Chromatin immunoprecipitations confirm that Klf3 binds the C/ebpα promoter in vivo. These results implicate Klf3 and CtBP in controlling adipogenesis.
Mammalian Krüppel-like factor (KLF) proteins contain three characteristic classical zinc fingers near their C termini (29). These domains recognize CACCC and related GC-rich elements in promoters and enhancers. Although the DNA-binding domains of the KLF family proteins share similar functions, their N-terminal domains are highly variable and display different molecular functions, with some operating primarily in gene activation and others in the repression of transcription.
Several members of the family are abundant in many (and possibly all) tissues, while other KLFs are restricted in their expression. In addition to detailed analysis of expression profiles, gene knockout experiments in mice have been useful in defining the major biological roles of different family members. For instance, Klf1 (also known as erythroid Krüppel-like factor gene [Eklf]) is highly expressed in erythroid tissues, and knockout experiments have demonstrated that it is required for definitive erythropoiesis (4, 20, 25). Klf1 knockout embryos die in utero around embryonic day 14.5 (E14.5) from failure to fully activate the adult β-globin gene. Chromatin immunoprecipitation experiments and rescue experiments in Klf1 knockout cell lines have confirmed that Klf1 binds to a CACCC box in the β-globin gene promoter and drives adult globin expression (8).
The biological roles of many of the other KLF proteins have similarly been defined. KLFs are important in numerous cellular processes, including the control of proliferation, differentiation, and apoptosis, and different family members contribute to the development of different tissues (5, 13, 29). Recently, a combination of cellular and knockout experiments has revealed that several family members contribute to adipogenesis. Klf2 is expressed early in adipogenesis, and later its expression decreases (2, 35). Accordingly, using the well-characterized 3T3-L1 cell model system, Klf2 has been shown to be an inhibitor of adipogenesis. It has been proposed that it inhibits the activation of the peroxisome proliferator-activated receptor γ (PPARγ) gene (PPARγ); PPARγ is a transcription factor known to drive adipogenesis (2). In contrast, Klf5 appears to drive adipogenesis. It is expressed early during differentiation, with maximal levels observed at 3 h after the onset of 3T3-L1 differentiation (21). Homozygous Klf5 knockout embryos die early in gestation, but heterozygous animals survive and show a deficiency in white adipose tissue (21). Furthermore, Klf5+/− murine embryonic fibroblasts (MEFs) are impaired in their ability to differentiate into adipocytes (21). Overexpression of Klf5 in 3T3-L1 cells enhances adipogenesis, whereas a dominant-negative form blocks differentiation (21). It has been proposed that Klf5 activates expression of the PPARγ gene (21). Klf15 is another positive regulator of adipogenesis. Klf15 expression increases dramatically during 3T3-L1 differentiation (19), and overexpression in 3T3-L1 cells results in enhanced adipocyte formation (2). Ectopic expression of Klf15 in NIH 3T3 or C2C12 cells can direct these nonadipocyte cell lines down the adipocyte lineage (19). This effect is mediated by PPARγ indicating that Klf15 acts upstream of PPARγ in adipocyte differentiation (19). Other KLFs, such as Klf6, Klf7, and possibly Klf11, have also been implicated in adipogenesis and metabolic control (6, 14, 17). The demonstration that various KLFs are instrumental in regulating adipocyte differentiation is consistent with the fact that functional CACCC binding sites are found in the control regions of key adipogenic genes, such as C/ebpα (12, 30) and Pparγ (2, 19, 21).
We have been investigating another member of the KLF family, basic Krüppel-like factor (Klf3). Klf3 is primarily a repressor of transcription. It contains an N-terminal repression domain that includes a Pro-X-Asp-Leu-Ser motif that recruits the well-characterized corepressor C-terminal binding protein (CtBP) (32). CtBP binds and is activated by NADH. Accordingly, it is viewed as a “metabolic sensor” that responds to the redox state of the cell (36). The Klf3 gene is broadly expressed (10) but its physiological roles have not been defined. In order to identify biological processes that are dependent on Klf3, we have disrupted the Klf3 gene by homologous recombination in murine embryonic stem cells and generated Klf3-deficient mice.
Klf3 knockout mice are smaller than their wild-type littermates, but while most organs are proportionally smaller, there is a marked reduction in white adipose tissue. Analysis of the fat pads of the knockout animals shows that they contain smaller and fewer cells. In order to determine whether there is a defect in adipogenesis in these mice, MEFs from the knockout embryos were studied. These cells showed an increased propensity to differentiate into lipid-forming cells in culture. The molecular role of Klf3 was investigated using the 3T3-L1 system. It was found that Klf3 mRNA and protein levels were high initially but declined as adipogenesis ensued. Forced expression of Klf3, but not of a Klf3 mutant unable to bind the corepressor CtBP, blocked adipogenesis. This result implicates CtBP in adipogenesis.
Microarray analyses have previously suggested that CtBP may be involved in repressing C/ebpα (11). We confirmed the derepression of C/ebpα in CtBP-deficient murine embryonic fibroblasts and examined C/ebpα levels in Klf3 knockout adipocytes and in Klf3-deficient murine embryonic fibroblasts. In all cases, C/ebpα was found to be derepressed. Finally, chromatin immunoprecipitation experiments on undifferentiated 3T3-L1 cells (where Klf3 is abundant) show that Klf3 is associated with the C/ebpα promoter in vivo. We conclude that Klf3 and CtBP play a role in normal adipogenesis, at least in part through the repression of key target genes, such as C/ebpα.
MATERIALS AND METHODS
Generation of knockout mice.
Klf3−/− mice were generated by standard methods. Details of constructs are available on request. Experiments were performed on mice maintained on the FVB/NJ background. All mice were housed at a temperature of 24 to 28°C with a 12-h light-dark cycle and free access to water and standard chow. Male and female mice were caged separately unless they were used specifically for breeding purposes. Mice were weaned at 3 weeks of age, and tail snips were taken for genomic DNA isolation. Ethical approval for the use of animals used in this project was obtained from the Animal Care and Ethics Committee (Sydney University; approval no. L02/1-2005/3/4048).
Genotyping.
Genomic DNA was isolated from tail biopsies of 3-week-old mice anesthetized with isoflurane (VCA I.S.O.; Veterinary Companies of Australia Pty Ltd., Kings Park, NSW, Australia). For embryos, a small portion of the head was used. Each tissue sample was digested overnight at 55°C in 350 μl of DNA lysis solution (50 mM Tris [pH 8.0], 100 mM EDTA, 100 mM NaCl, and 1% [wt/vol] sodium dodecyl sulfate) plus 35 μl of proteinase K (10 mg/ml). Following protein digestion, 2 μl of RNase A (5 mg/ml) was added, and samples were incubated at 37°C for 30 min. DNA was extracted by adding 250 μl of phenol-chloroform-isoamyl alcohol (Progen Industries, Darra, QLD, Australia), with vigorous shaking for 2 min at room temperature. The phases were separated by centrifugation at 13,000 × g for 2 min, and the aqueous phase was extracted with 250 μl of chloroform, with vigorous shaking for 2 min at room temperature. Phases were separated as above, and the DNA was precipitated from the aqueous phase by adding 250 μl of isopropanol. Samples were vortexed and centrifuged at 13,000 × g for 15 min at 4°C. DNA pellets were air dried for 30 min at room temperature and then resuspended in 250 μl of embryo transfer water (Sigma Chemical Company, St. Louis, MO) at 4°C overnight.
Mice were genotyped using a single PCR to simultaneously identify neo and Klf3 sequences. The sequences of forward and reverse primers used are as follows: neo, 5′-TGATGCAATGCGGCGGCTGCATAC-3′ (forward) and 5′-CAGAAGAACTCGTCAAGAAGGCGA-3′ (reverse); Klf3, 5′-AAATGCACCTGGGAAGGCTGCAC-3′ (forward) and 5′-CAGACTAGCATGTGGCGTTTCCTG-3′ (reverse).
The PCR was carried out in a 25-μl solution containing 1 μl of genomic DNA, 2.5 μl of 10× REDTaq buffer, 1 μl of 10 mM deoxynucleoside triphosphate mixture (2.5 mM each), 0.2 μl of each 100 μM primer, and 1.25 units of REDTaq DNA polymerase (Sigma). The thermal parameters of the PCR were 94°C for 2 min and 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 60 s, followed by one final cycle of 72°C for 5 min.
Organ weight analysis.
Twelve-week-old mice were weighed and then sacrificed with a lethal injection of Euthanasia Fort Solution (Apex Laboratories Pty. Ltd., Somersby, NSW, Australia). Various organs were immediately collected and weighed.
Histological analysis of tissues.
Four-day-old mice or fresh epididymal pads from 12-week-old male mice were collected and fixed in 4% neutral buffered formaldehyde solution. Hematoxylin-eosin (H-E) staining was performed at the Histopathology Laboratory (Department of Pathology, Blackburn Building, University of Sydney).
Adipocyte size determination.
Isolated adipocytes were obtained from excised epididymal fat pads of 4-month-old mice by collagenase as previously described (27). Cells were then fixed with 2% OsO4 in phosphate-buffered saline overnight at 37°C. The adipocyte diameter of 800 to 1,500 cells was determined using Photoshop software (Adobe Systems Inc.) from images of adipocyte suspensions obtained by light microscopy. Since adipocytes have 95% lipid content and are spherical in shape, cell volumes were estimated from the diameter.
Total DNA content from adipose tissue.
Epididymal white adipose tissue was dissected from 12-week-old mice and immediately frozen in liquid nitrogen. Approximately 100 mg of tissue was homogenized using a PRO 200 homogenizer (PROScientific Inc., Oxford, CT), and genomic DNA was extracted using TRI reagent (Sigma) as described by the manufacturer. DNA per mg of tissue was measured by UV spectrophotometry, and the total DNA content from fat pads was determined by multiplying the obtained DNA concentration for each sample by the average epididymal white adipose tissue weight for each genotype.
Cell culture, induction of adipocyte differentiation, and Oil Red O staining.
3T3-L1 cells were cultured at 37°C and 5% CO2 in high-glucose Dulbecco's modified Eagle's medium (HG-DMEM) supplemented with 10% (vol/vol) heat inactivated fetal calf serum (FCS) and 1% (vol/vol) penicillin, streptomycin, and glutamine (PSG). Two days after the cells reached confluence, adipocyte differentiation was induced (designated day 0). The medium was replaced with differentiation medium (HG-DMEM, 10% FCS, 1% PSG, 2 μg/ml insulin, 1 μM dexamethasone, 0.5 mM 3-isobutyl-1-methylxanthine). On day 3 postinduction the medium was changed to postdifferentiation medium (HG-DMEM, 10% FCS, 1% PSG, 2 μg/ml insulin). On day 6 and thereafter, the cells were maintained in standard medium, which was replenished every 3 to 4 days.
MEFs were prepared from E14.5 Klf3−/− embryos or their wild-type littermates. The outgrowing primary cell population was passaged every 2 to 3 days upon confluence and continuously cultured in HG-DMEM supplemented with 10% FCS and 1% PSG. MEFs were induced to differentiate as described above for 3T3-L1 cells.
We are grateful to Jeffrey Hildebrand (University of Pittsburgh, Pittsburgh, PA) for the gift of the Ctbp−/− and Ctbp+/− MEFs. These were cultured in the same medium as described above for 3T3-L1 cells.
EcoPack 2-293 packaging cells (Clontech, Mountain View, CA) were grown in the same medium as described above for 3T3-L1 cells.
Oil Red O staining of differentiated cells was performed to visualize intracellular lipid droplets. Cells were fixed in 4% neutral buffered formaldehyde for 30 min at room temperature. Cells were then rinsed quickly in water, followed by 60% isopropanol for 5 min. Oil Red O (Sigma) (0.3% in isopropanol) was diluted with water (3:2), filtered, and then used to stain the cells for 5 min at room temperature. Cells were washed with water. Where indicated, cells were counterstained with hematoxylin for 1 min and washed for a final time with water. The stained cells were visualized by light microscopy.
Real-time RT-PCR.
RNA was isolated from 3T3-L1 cells at the indicated times (see Fig. 5A), white adipose tissue of 12-week-old mice, and Klf3 and Ctbp MEFs. RNA extraction, cDNA synthesis and real-time reverse transcription-PCR (RT-PCR) were performed as previously described (10). The sequences of forward and reverse primers used are as follows: Klf3, 5′-TACAGGAGAAAAGCCGTACAAATG-3′ and 5′-TCATCAGACCGAGCGAACTTC-3′; C/ebpα, 5′-GAGCCGAGATAAAGCCAAACA-3′ and 5′-CGGTCATTGTCACTGGTCAACT-3′; C/ebpβ, 5′-CAAGCTGAGCGACGAGTACA-3′ and 5′-CAGCTGCTCCACCTTCTTCT-3′; C/ebpδ, 5′-CGCAGACAGTGGTGAGCTTG-3′ and 5′-CTTGCGCACAGCGATGTTGTT-3′; Pparγ, 5′-CTTGCTGTGGGGATGTCTCACAA-3′ and 5′-GATCTCCGCCAACAGCTTCTCCT-3′; Klf15, 5′-TACACCAAGAGCAGCCACCT-3′ and 5′-AACTCATCTGAGCGGGAAAA-3′; Klf2, 5′-ACCAAGAGCTCGCACCTAAA-3′ and 5′-TCCTTCCCAGTTGCAATGAT-3′; Klf5, 5′-CCGGAGACGATCTGAAACAC-3′ and 5′-GGAGCTGAGGGGTCAGATACTT-3′; 18S, 5′-CACGGCCGGTACAGTGAAAC-3′ and 5′-AGAGGAGCGAGCGACCAA-3′; Pgm2, 5′-GGAGCAGGGATTTCTCGAATGAA-3′ and 5′-GGTCACTGAACTGCTTTTCCAGGT-3′; Tbc1d1, 5′-AATGTGGTTCAGCCCACAGACAT-3′ and 5′-CCAGTGCAATCTTTTTGGTGTCA-3′; Tlr1, 5′-TTCAAGTGTGCAGCTGATTGCTC-3′ and 5′-CAGTGCTAACGTGCCGAAGAGAT-3′; Tlr6, 5′-TAAGGAATTTGGCAACCTGACGA-3′ and 5′-CAAGTGAGCAACTGGGAGCAGAT-3′.
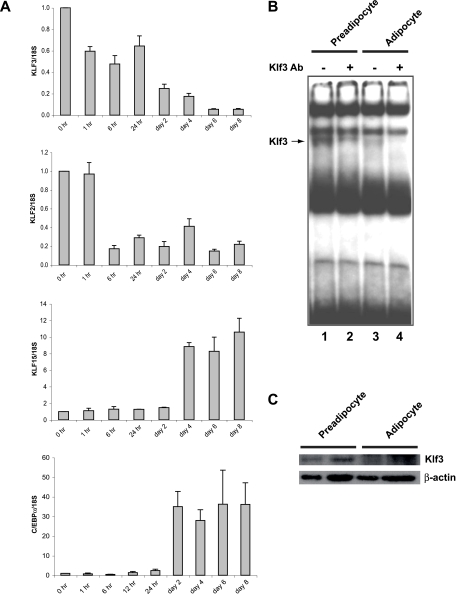
FIG. 5.
Klf3 expression during 3T3-L1 adipocyte differentiation. (A) Total RNA was extracted from 3T3-L1 cells at the indicated times before and after induction of differentiation. The expression levels of Klf3, Klf2, Klf15, and C/ebpα were determined by quantitative real-time RT-PCR and normalized to 18S rRNA levels. These levels were then further normalized with respect to the level in 3T3-L1 cells at day 0, set at 1. Results are expressed as means plus SEM (n = 3). (B and C) Nuclear extracts were made from cells at day 0 (preadipocytes) and day 10 (adipocytes) of 3T3-L1 differentiation. An EMSA using a double-stranded oligonucleotide probe containing a CACCC sequence shows that Klf3 is present in preadipocytes and reduced in adipocytes (B). Anti-Klf3 antibody supershift reactions confirm that these species are Klf3 (lanes 2 and 4). Note that supershift of the Klf3 band reveals the presence of another CACCC box binding protein that runs similarly to Klf3. All data are representative of at least three independent experiments. Klf3 protein was detected by Western blot analysis using an anti-Klf3 antibody (C). β-Actin was used as a loading control. Duplicates are shown for each condition. All data are representative of at least three independent experiments.
Nuclear extract preparation and EMSAs.
Nuclear extracts were prepared from 3T3-L1 preadipocytes and adipocytes, and electrophoretic mobility shift assays (EMSAs) were carried out as described previously (9). Generation of the anti-Klf3 antibody has been described previously (9). The oligonucleotides used in the synthesis of radiolabeled probes were from the mouse β-major globin promoter: 5′-TAGAGCCACACCCTGGTAAG-3′ and 5′-CTTACCAGGGTGTGGCTCTA-3′.
Western blot analysis.
The Western blot procedure was carried out as previously described (24). Briefly, nuclear extracts from 3T3-L1 preadipocytes and adipocytes were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis, electrotransferred to nitrocellulose membrane, and analyzed with antibodies against Klf3 as previously described (9) and β-actin (Sigma). Detection was carried out using Western Lightning Chemiluminescence Reagent Plus (Perkin Elmer Life Sciences, Boston, MA) and X-ray film (Eastman Kodak Company, Rochester, NY).
Retroviral vectors.
Full-length Klf3 and Klf3ΔDL (in which the DL residues in the PXDLS motif are mutated [see below]) sequences were excised from pMT2-Klf3 and pcDNA3-Klf3ΔDL and inserted into the BamHI and EcoRI sites of a pMXs vector containing a puromycin resistance gene (generously supplied by Toshio Kitamura, University of Tokyo, Tokyo, Japan) to generate the retroviral vectors pMXs-Klf3 and pMXs-Klf3ΔDL.
Retroviral infection.
To generate retrovirus, EcoPack 2-293 packaging cells were transfected with 5 μg of pMXs-Klf3, pMXs-Klf3ΔDL, or empty pMXs with the use of FuGENE 6 (Roche Applied Science, Indianapolis, IN). After 8 h, the cells were incubated in fresh medium. At 48 h after transfection, the supernatant containing viral particles was harvested and passed through a filter (pore size, 0.45 μm). 3T3-L1 cells were plated at a cell density of 1.5 × 105 cells/60-mm plate 12 h before infection. These cells were then incubated with the viral supernatants in the presence of 8 μg/ml Polybrene (Sigma) for 24 h. Cells successfully transduced were selected on the basis of their resistance to puromycin (2.5 μg/ml). Differentiation of the stable transformants was then performed as described above, and cells were stained with Oil Red O at day 5 of differentiation.
ChIP assays.
Approximately 5 × 106 3T3-L1 cells were used for each chromatin immunoprecipitation (ChIP). Cross-linking was performed by treatment of undifferentiated cells with ethylene glycol bissuccinimidylsuccinate (Sigma) (to a final concentration of 1.5 mM) for 30 min at room temperature, followed by formaldehyde (Sigma) (final concentration, 1%) for 10 min. Cross-linking was terminated with glycine (final concentration, 20 mM). After preparation of nuclear extracts, chromatin was sonicated for 20 min at 4°C, using a 30-s on and 30-s off protocol. Chromatin was precleared with protein A agarose beads (blocked previously with rabbit serum) at 4°C overnight. Precleared chromatin samples were immunoprecipitated with a rabbit polyclonal anti-Klf3 antibody (previously described in Crossley et al. [9]) or rabbit serum as a negative control. Immunoprecipitated chromatin samples were reverse cross-linked, purified, and subjected to real-time RT-PCR analysis. DNA prepared from chromatin samples served as a positive control (input). The real-time RT-PCR primers were designed to amplify regions around the mouse C/ebpα promoter, spaced approximately 1 kb apart: −5000, 5′-CCCAGGCAGAACAAAACATAGG-3′ and 5′-GGGCAGGCCATTGTTTTGTA-3′; −4000, 5′-TGCTTTACTTGGTGCCTCTTTG-3′ and 5′-TTGGTGTCGACTCCTTCATGAA-3′; −3000, 5′-CGGGAGCCTGGCATCTT-3′ and 5′-TTCCAGGAGCGGCTACAGA-3′; −2000, 5′-AGTGTGTTCCAGCTTCCTTCAAC-3′ and 5′-GAGGCTGACATCCTCGCTTATG-3′; −1000, 5′-GCTCCACGCTGGGTAGCA-3′ and 5′-CCAGGCCAGAGCGATAGGA-3′; +1, 5′-AGGAGAAGGCGGGCTCTAAG-3′ and 5′-ATCGAAGGCGCCAGTAGGA-3′; +1000, 5′-CGGTGCGGGCAAAGC-3′ and 5′-CGTACCCGGTACTCGTTGCT-3′.
Statistics.
All data are presented as means plus standard errors of the means (SEM). Differences between groups were analyzed using analysis of variance (Statview Statistical Analysis Software, version 4.57). Values of P of <0.05 were considered significant.
RESULTS
Klf3−/− mice possess less white adipose tissue.
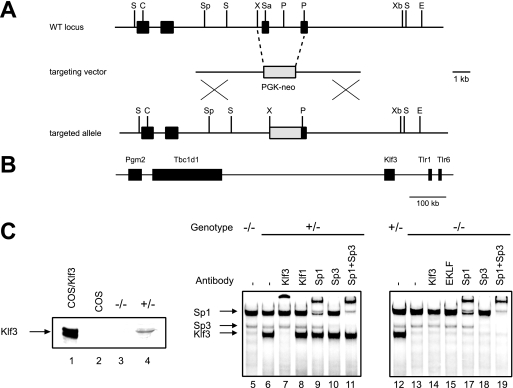
In order to assess the biological role of Klf3, we carried out the targeted disruption of the Klf3 gene as shown in Fig. 1A. The targeting vector is designed to delete a portion of the zinc finger region and ablate DNA-binding and Klf3 activity. Targeting was achieved in 129 embryonic stem cells and chimeric 129/C57BL/6 mice were generated. These were bred to C57BL/6 animals, and germ line transmission of the targeted allele was achieved. Heterozygotes were bred together to generate homozygous animals, and tissues from these animals were analyzed to verify that no functional Klf3 protein was detectable. No Klf3 protein could be detected by Western blotting, and the more sensitive EMSAs confirmed that no functional Klf3 was present in knockout tissues (Fig. 1C). The targeting construct used to disrupt the Klf3 gene contained the neo cassette, which was retained in the knockout animals. To confirm that there were no changes in expression of genes surrounding the neo cassette (Fig. 1B), real-time RT-PCR was performed on RNA-extracted Klf3 null and wild-type MEFs. No significant alterations in expression were observed in the four genes directly surrounding the site of neo insertion (data not shown).
FIG. 1.
Targeted disruption of Klf3 in mice. (A) Generation of Klf3 knockout mice. Schematic representation of the wild-type (WT) Klf3 locus (with a partial restriction map), the targeting vector, and the targeted allele. Black boxes represent exons. The zinc finger domain is contained in the last two exons. The gray box represents the phosphoglycerate kinase (PGK)-neo cassette. Abbreviations: S, StuI; C, ClaI; Sp, SpeI; X, XhoI; Sa, SalI; P, PmlI; Xb, XbaI. (B) Schematic showing genomic location of murine Klf3 and neighboring genes on chromosome 5. Pgm2, glucose phosphomutase 2; Tbc1d1, TBC1 domain family, member 1; Tlr1, Toll-like receptor 1; and Tlr6, Toll-like receptor 6. (C) Western blotting (left panel) using anti-Klf3 serum, showing full-length Klf3 ectopically expressed in COS cells (lane 1), COS cells alone (lane 2), and spleen samples from Klf3 knockout and heterozygous littermates (lanes 3 and 4, respectively). The right panels (lanes 5 to 18) show EMSAs using the β-major globin gene promoter −90 CACCC box. The bands generated by Klf3, Sp1, and Sp3 are indicated by arrows and are supershifted by appropriate antisera.
To avoid variations that might arise from the mixed genetic background, mice were back-crossed to the FVB/NJ strain for 10 generations. The population was monitored for viability. Examination of the genotypes at 3 weeks of age revealed that the proportion of knockouts is less than expected from Mendelian ratios. Klf3−/− mice from heterozygous crosses make up only 17% (rather than 25%) of the total offspring at weaning (n = 353; P < 0.005) (data not shown). When genotype ratios were measured in utero at E14.5, the fraction of Klf3−/− mice was within Mendelian expectations (n = 49) (data not shown). To date there are no obvious defects to explain the reduction in viability of the knockout after E14.5.
At birth, the Klf3−/− mice appear virtually indistinguishable from their wild-type and heterozygous littermates. However, by the time of weaning, the Klf3−/− mice are visibly smaller than their wild-type counterparts (see Fig. S1A in the supplemental material). To further define this phenotype, body weight was measured weekly from 3 to 12 weeks of age (see Fig. S1B in the supplemental material). At each time point the Klf3−/− mice were significantly smaller than both their wild-type and heterozygous counterparts (P < 0.0001). The size reductions were observed in both male and female Klf3 knockout mice.
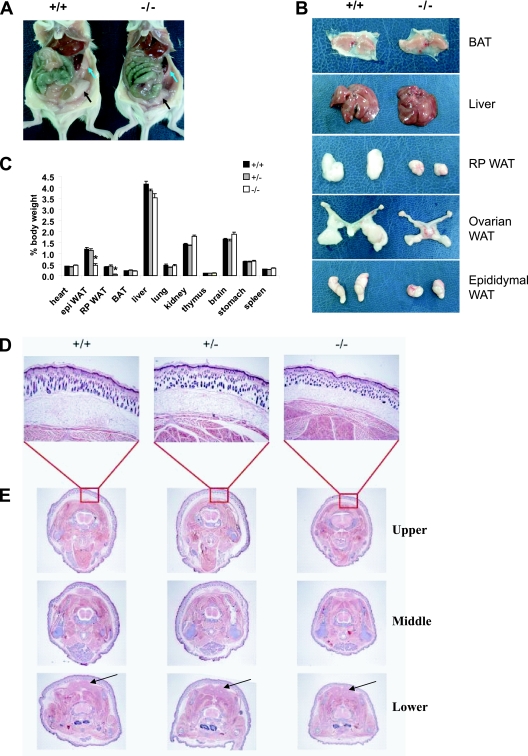
Klf3−/− mice also appeared leaner than their wild-type and heterozygous littermates (see Fig. S1A in the supplemental material). We hypothesized that perhaps the reduced size of Klf3−/− mice could be partially attributed to a deficiency in white adipose tissue, reminiscent of the impaired adipogenesis in Klf5+/− mice (21). To investigate this, the sizes of gonadal (ovarian in females and epididymal in males) and retroperitoneal white adipose tissue depots were compared between wild-type, heterozygous, and Klf3−/− mice at 12 weeks of age. Macroscopic analyses demonstrated that, indeed, the Klf3−/− mice had dramatic reductions in the size of their fat pads compared to wild-type mice (Fig. 2A and B). These decreases were observed in both males and females. In contrast to white adipose tissue depots, other tissues such as brown adipose tissue and liver exhibited no gross abnormalities in morphology or size relative to other organs (Fig. 2B). Multiple organs from wild-type, heterozygous, and Klf3−/− mice were weighed and normalized for body weight (Fig. 2C). Compared with tissues from wild-type mice, the gonadal and retroperitoneal fat pads were reduced to 38% and 15% of normal size, respectively, in Klf3−/− males (P < 0.001). Similar reductions were found in Klf3−/− females (P < 0.05) (data not shown). Other tissues including heart, lung, thymus, stomach, and spleen were equivalent in mice of different genotypes although there was a slight but detectable decrease in the size of the liver, and small increases in kidney and brain size were observed in knockouts for both sexes (P < 0.05).
FIG. 2.
Klf3−/− mice have reduced fat mass compared to wild-type mice. (A) Peritoneal cavity of wild-type and Klf3−/− mice showing ovarian (black arrows) and retroperitoneal (blue arrows) white adipose tissue (WAT). (B) Organs were dissected from 12-week-old mice. A reduction in size was seen in Klf3−/− white adipose tissue samples, while the gross morphology of the brown adipose tissue (BAT) and liver appeared similar between genotypes. (C) Relative weights of various organs. Values are plotted as a percentage of body weight at 12 weeks of age. The results are expressed as means plus SEM from nine wild-type, five Klf3+/−, and eight Klf3−/− male mice. *, P < 0.001 for Klf3−/− versus wild-type/Klf3 +/− mice. epi, epididymal; RP, retroperitoneal. (D and E) Reduced subcutaneous white adipose tissue in Klf3−/− neonates. Transverse sections were taken from the neck and scapula of wild-type, heterozygous, and Klf3−/− mice at 4 days of age (n = 3 for each genotype). Images in panel D are at a magnification of ×40. Panel E shows sequential sections taken at the upper, middle, and lower torso at a magnification of ×8 (n = 3 for each genotype). The unstained rings underlying the skin represent areas of subcutaneous white adipose tissue. Brown adipose tissue is indicated by arrows in the lower panel. Sections were stained with H-E.
To further pinpoint the stage of development at which the reduction in white adipose tissue is first evident, neonates were studied. Transverse sections of 4-day-old wild-type, heterozygous, and Klf3−/− neonates were taken and stained with (H-E). Sections were taken at three different levels through the mouse body at the levels of the neck and scapula (Fig. 2E). Klf3−/− neonates had reduced subcutaneous white adipose tissue (Fig. 2D and E, the unstained band underlying the skin) compared to wild-type and heterozygous neonates. Again, it appears that the amount of brown adipose tissue is similar among all genotypes (Fig. 2E, arrows in the lower panel), suggesting that there is a particular deficiency in white adipose tissue, in addition to the general reduction in size that is detectable even at this early stage of development.
Fat pads in Klf3−/− mice have smaller and fewer cells.
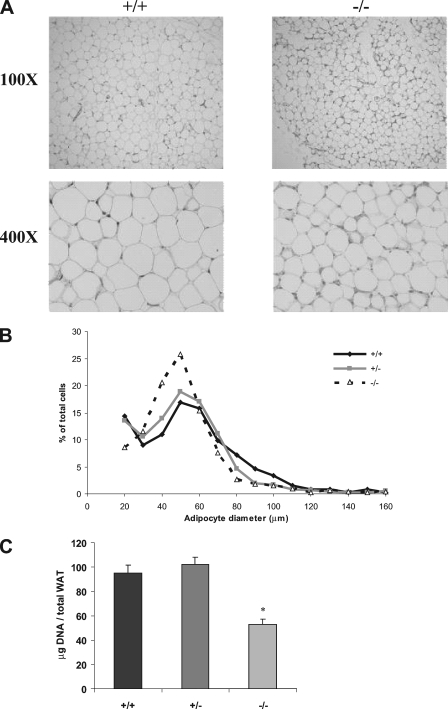
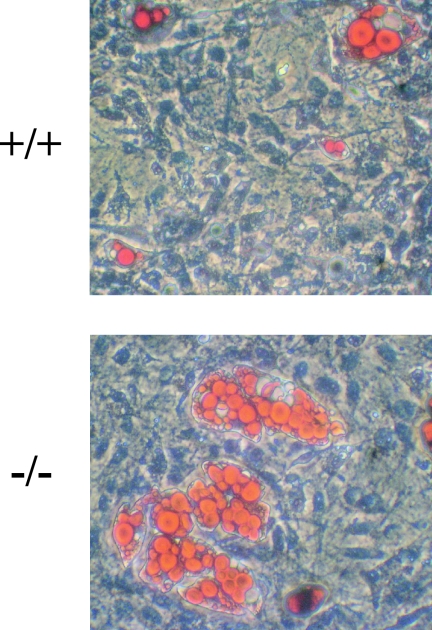
To characterize the apparent white adipose tissue deficiency at the cellular level, we investigated whether Klf3−/− mice had less fat because the size of the individual cells was reduced and/or because there were fewer cells present in fat pads. Histological sections of white adipose tissue stained with H-E suggest that the cells from Klf3−/− mice are smaller in diameter than wild-type cells (Fig. 3A). To confirm this result, cells were isolated by collagenase digestion, and their diameters were measured. It was confirmed that in Klf3−/− animals there was a distinct shift toward cells of a smaller diameter (Fig. 3B).
FIG. 3.
White adipose tissue from Klf3−/− mice has smaller and fewer adipocytes. (A) Histological sections of epididymal white adipose tissue were stained with H-E. (B) Size distribution of adipocytes isolated from the epididymal white adipose tissue of wild-type, Klf3+/−, and Klf3−/− mice. Adipocytes were isolated by collagenase digestion and fixed with osmium tetroxide (n = 3 for each genotype). Adipocyte diameter was determined for 885 wild-type, 1,037 Klf3 +/−, and 1,506 Klf3−/− adipocytes using Photoshop software. (C) DNA content was used as a measure of cell number. Results are expressed as means plus SEM of five animals per genotype. *, P < 0.001 for Klf3−/− versus wild-type/Klf3+/− mice. WAT, white adipose tissue.
To determine how much smaller the Klf3−/− cells were compared to wild type, the measured diameter of each cell was used to estimate cell volume (assuming cells are spherical). On average, the Klf3−/− cells were 61% of the size of wild-type cells (see Fig. S2 in the supplemental material). This reduction in size is not sufficient to explain the total deficit of white adipose tissue, as the mice were observed to have 38% (rather than 61%) of the amount of gonadal white adipose tissue observed in wild-type littermates (Fig. 2C) (note that the size measurements were carried out only in male mice). We therefore sought to investigate if the fat pads also contained fewer cells.
We used the DNA content of epididymal fat pads to provide an estimate of the number of cells in the fat pads. Compared to wild-type samples, epididymal pads from Klf3−/− mice have only 56% of the amount of total DNA content (P < 0.001) (Fig. 3C), suggesting that there are approximately 56% of the number of cells in the tissue. This number is obviously an estimate that relies on the assumption that the proportion of diploid cells is equivalent in the two samples. But, importantly, when this reduction (0.56) is combined with the smaller size of the cells (0.61), the result (0.56 × 0.61 = 0.34) approximates the total amount of gonadal white adipose tissue observed, i.e., 38% (Fig. 2C). This suggests that the reduction in the size and number of cells in the fat of Klf3 null mice can account for the observed deficit in white adipose tissue.
MEFs from Klf3−/− animals display enhanced adipocyte differentiation.
In vivo many processes directly or indirectly influence fat accumulation. Thus, the reduction in white adipose tissue observed in the Klf3 null neonates and in the 12-week-old animals could be due to a combination of factors. One possibility is that the reduction in white adipose tissue is due to alterations in cellular proliferation rates in Klf3 null animals. To address this hypothesis, proliferation rates of Klf3 null and wild-type MEFs were assayed by both fluorescence-activated cell sorting cell cycle analysis and the CellTiter 96 assay (Promega). No differences in cell cycle or proliferation were observed (data not shown). To investigate whether the reduction in adipose tissue could be due to a specific alteration in adipocyte differentiation, in vitro cell culture experiments were carried out. MEFs were derived from E14.5 Klf3 null and wild-type embryos and exposed to inducers of adipocyte differentiation. The cells were then stained and counterstained with Oil Red O and hematoxylin, respectively, at day 8 after hormonal induction. A proportion of these fibroblasts acquired characteristics of adipocytes, as assessed by the cytoplasmic accumulation of lipid droplets. In three independent experiments, Klf3 null MEFs displayed an enhanced ability to accumulate lipid compared to wild-type cells in culture (Fig. 4).
FIG. 4.
Enhanced adipocyte differentiation in Klf3-deficient cells. Fibroblasts derived from E14.5 wild-type (+/+) and Klf3 knockout (−/−) mouse embryos were induced to undergo adipocyte differentiation. Cells were stained with Oil Red O and counterstained with hematoxylin on day 8. Images are representative of at least three independent experiments.
Klf3 levels decrease upon adipocyte differentiation of 3T3-L1 cells in culture.
To further define the function of Klf3 in adipogenesis, we switched to the 3T3-L1 cell system. 3T3-L1 cells can be induced to differentiate in culture. Several gene regulatory proteins of the KLF family, such as Klf2, Klf5, and Klf15, which have been shown to play important roles in the differentiation of adipocytes, are dynamically regulated during 3T3-L1 differentiation (2, 19, 21) (Fig. 5A). We monitored Klf3 expression during the differentiation of 3T3-L1 cells. Total RNA was collected at various stages throughout 3T3-L1 differentiation and analyzed by real-time RT-PCR. Klf3 mRNA was expressed most highly at day 0 in 3T3-L1 preadipocytes and diminished upon adipocyte differentiation (Fig. 5A). As expected, other factors such as C/ebpα, C/ebpβ, C/ebpγ, Pparγ, and aP2 were induced with 3T3-L1 differentiation, whereas Klf2 declined (Fig. 5A and data not shown). To determine if the decline in Klf3 mRNA was also reflected in protein levels and DNA-binding activity, Western blotting and EMSAs were carried out. These experiments confirmed that Klf3 mRNA, protein, and DNA-binding activity all decline during the differentiation of 3T3-L1 cells (Fig. 5B, C).
Overexpression of Klf3 inhibits adipocytic differentiation of 3T3-L1 cells in vitro.
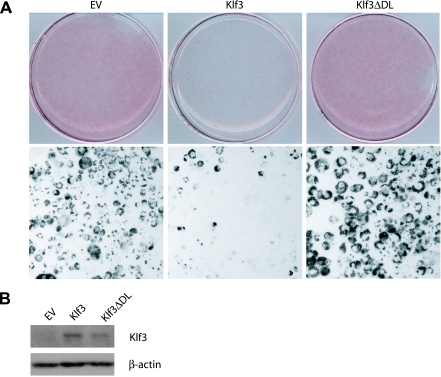
The dynamic regulation of Klf3 suggested that it may be a direct regulator of adipocyte differentiation. To test whether Klf3 shutdown was critical to proper adipocyte differentiation, we used a retroviral system to enforce the ectopic overexpression of Klf3 and monitored the effect on adipocyte differentiation. As judged by Oil Red O staining, forced expression of Klf3 significantly inhibited lipid accumulation (Fig. 6A). The forced expression of Klf3 had no appreciable effect on cell viability or number (data not shown).
FIG. 6.
Reduced adipocyte differentiation in 3T3-L1 cells stably expressing Klf3. (A) 3T3-L1 preadipocytes were retrovirally infected with empty vector (EV), Klf3, or Klf3ΔDL and subjected to Oil Red O staining to assess for lipid accumulation at day 5 of differentiation. Macroscopic (upper panel) and microscopic (magnification, ×100; lower panel) views are shown. Images are representative of at least three independent experiments. (B) 3T3-L1 cells were retrovirally infected with EV, Klf3, or Klf3ΔDL and differentiated. Nuclear extracts were prepared and subjected to Western blot analysis using the indicated antibodies. All data are representative of at least three independent experiments.
Klf3 represses transcription by recruiting the corepressor protein CtBP through a PXDLS motif in its repression domain (32). In order to determine whether this interaction is required for inhibiting adipocyte differentiation, a Klf3 mutant that is unable to bind CtBP was tested. This mutant (Klf3ΔDL) carries a point mutation in the PXDLS motif of Klf3 but is otherwise intact (32). Forced overexpression of the mutant Klf3ΔDL had no effect on differentiation (Fig. 6A). Figure 6B confirms that both the Klf3 and Klf3ΔDL proteins were detectably overexpressed. Together, these data indicate that Klf3 plays a role in adipocyte differentiation and that this role requires the recruitment of CtBP.
Derepression of C/ebpα in the absence of Klf3.
We hypothesized that Klf3 and its corepressor CtBP may be exerting an effect on adipogenesis by repressing a gene that is important in adipocyte differentiation. Several observations suggested C/ebpα as a possible candidate. First, C/ebpα has been reported as a gene repressed by CtBP (11). Second, defects in C/ebpα generate a phenotype reminiscent of the Klf3 knockout phenotype, that is, reduction in white adipose tissue but normal brown adipose tissue (18). It has been shown that the C/ebpα gene promoter is bound and regulated by the KLF-related protein Sp1 (30), suggesting that the C/ebpα gene promoter contains functional CACCC boxes through which Klf3 may also operate. Finally, the C/ebpα expression pattern during 3T3-L1 differentiation is consistent with repression by Klf3 (Fig. 5A); i.e., C/ebpα is low when Klf3 levels are high and vice versa. We therefore investigated whether Klf3 represses C/ebpα.
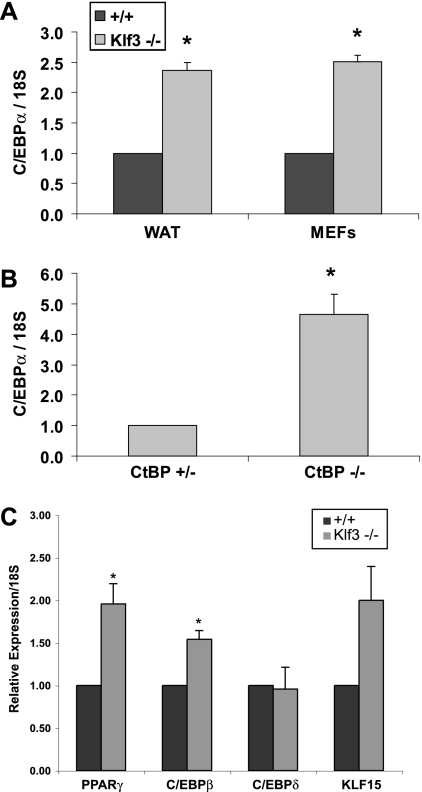
We first compared C/ebpα mRNA levels in normal and Klf3 knockout undifferentiated MEFs. As shown in Fig. 7A, C/ebpα is derepressed in Klf3 knockout cells. We also examined CtBP-deficient MEFs to confirm the previous microarray experiments that indicated that C/ebpα levels are elevated in these cells (11). Real-time RT-PCR analysis confirmed that C/ebpα levels were approximately fourfold higher in the absence of CtBP (Fig. 7B). Finally, we examined RNA from the white adipose tissue of Klf3−/− mice and their wild-type counterparts. We again observed two to three times the amount of C/ebpα in Klf3 null white adipose tissue compared to wild-type white adipose tissue (Fig. 7A). Taken together, these results indicate that C/ebpα is consistently found to be derepressed in the absence of Klf3 or its corepressor CtBP. We also examined the expression of other adipogenic genes in white adipose tissue. Real-time RT-PCR analysis indicated an approximately twofold increase in levels of Pparγ, C/ebpβ, and Klf15 in Klf3 null white adipose tissue (Fig. 7C). We did not observe any significant alteration in the expression of C/ebpδ, Klf2, or Klf5. These findings suggest that Klf3 acts as a regulator of adipocyte differentiation at least in part through repressing C/ebpα expression.
FIG. 7.
C/ebpα is derepressed in the absence of Klf3 and CtBP. Quantitative real-time RT-PCR analysis of wild-type (+/+) and Klf3 −/− white adipose tissue and MEFs (A), CtBP+/− and CtBP−/− MEFs (B), and wild-type and Klf3−/− white adipose tissue (C). Expression levels were normalized to the level of 18S rRNA. These levels were then further normalized with respect to the level in wild-type sample or CtBP+/− MEFs, set at 1. Error bars indicate SEM. P values are defined as follows, and significant differences are indicated by asterisks: <0.05 for Klf3−/− versus wild-type (A; n = 4 for each genotype), <0.05 for CtBP−/− versus CtBP+/− (B; n = 3 for each genotype), and <0.05 for Klf3−/− versus wild-type (C; n = 3 for each genotype).
Klf3 binds the endogenous C/ebpα promoter in 3T3-L1 cells.
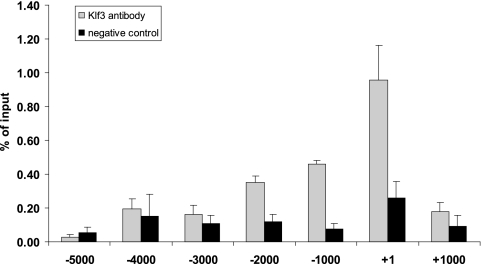
To determine whether Klf3 directly binds the C/ebpα promoter in vivo to mediate repression, we performed ChIP assays. ChIP material was obtained from 3T3-L1 preadipocytes, in which Klf3 is abundant, and was immunoprecipitated with an anti-Klf3 antibody. A primer walk across the C/ebpα gene was conducted to compare the amounts of Klf3 at the C/ebpα promoter and at regions up- and downstream. Klf3 occupancy was seen most strongly at the region around the transcriptional start site (Fig. 8). This region contains multiple CACCC boxes matching the Klf3 binding site consensus (data not shown). There was minimal Klf3 binding detected in regions other than the C/ebpα promoter. In addition to the negative control regions of DNA upstream and downstream of the promoter, immunoprecipitation with a nonimmune serum control showed no significant enrichment at the C/ebpα promoter. Taken together, these results suggest that Klf3 directly associates with the C/ebpα promoter and contributes to its repression in vivo.
FIG. 8.
Klf3 binds at to the C/ebpα promoter. 3T3-L1 preadipocytes were fixed and chromatin material was subjected to ChIP analysis. Chromatin was immunoprecipitated using an anti-Klf3 antibody or rabbit serum as a negative control. Primers used for real-time RT-PCR were targeted to various regions spaced 1 kb apart along the C/ebpα promoter.
DISCUSSION
The molecular pathways that regulate adipogenesis have been extensively studied. PPAR and C/EBP family transcription factors have been shown to play a dominant role, particularly in driving the terminal differentiation and maturation of adipocytes (22, 28). More recently, KLF proteins have been shown to regulate both the early (2, 21) and, in the case of Klf15 (19), also the later stages of adipogenesis. The KLFs have been shown to act, at least in part, by regulating the expression of PPARγ. Klf2 inhibits expression of the PPARγ gene (2), while Klf5 and Klf15 drive it (19, 21). Similarly, the KLF-related proteins Sp1 and Sp3 have been implicated in regulating C/ebpα expression (12, 30).
Our results indicate that a repressive KLF, Klf3, is also involved in the control of adipogenesis. Its abundance in uninduced 3T3-L1 cells and its shutdown as adipogenesis proceeds suggest that it may play an early role in inhibiting adipogenesis. As Klf3 expression diminishes, C/ebpα expression concomitantly increases. Forced overexpression of Klf3 in the 3T3-L1 system blocks adipogenesis, but a mutant form of Klf3 that cannot bind the corepressor CtBP is ineffective. This result implicates CtBP in the control of adipogenesis.
There is now significant evidence linking CtBP with metabolic control and adipogenesis. CtBP responds to NADH levels, and one proposal is that it dimerizes (or multimerizes) in the presence of NADH and that its conformation is altered so that it is able to bind to Pro-X-Asp-Leu-Ser motifs found in its partner transcriptional repressors (1, 15, 31, 37). Thus, CtBP is viewed as a metabolic sensor that can modulate gene expression in response to the redox state of the cell. Studies in CtBP null MEFs have indicated that it represses several genes involved in metabolism and the regulation of lipids, including lactate dehydrogenase 2 (B chain), glyceraldehyde-3-phosphate dehydrogenase, glucose phosphate isomerase, lipoprotein lipase 1, and the elongation of long chain fatty acids genes (Elo2 and Elo3) (11). Furthermore CtBP has recently been found to regulate lipid droplet accumulation in fibroblasts (3). Further evidence linking CtBP to adipogenesis comes from the observation that CtBP binds RIP140 (34), a nuclear hormone receptor that regulates fat accumulation (7, 16). Interestingly, Sir2, another corepressor protein that responds to changes in NAD+/NADH ratios, has also been implicated in adipogenesis. In contrast to CtBP, Sir2 is active when NAD+ (rather than NADH) levels are high. Upon food withdrawal, Sir2 represses PPARγ expression (26). Thus, it appears that both Sir2 and CtBP may regulate adipogenesis in response to NAD+/NADH ratios.
Our results indicate that Klf3 and CtBP repress the C/ebpα promoter. This is consistent with the fact that functional CACCC boxes have been identified in the C/ebpα promoter (30) and that C/ebpα has previously been listed as a gene that is derepressed in Ctbp knockout cells (11). The phenotype of the Klf3 knockout is also consistent with a defect in C/ebpα expression. That is, C/ebpα has been shown to be important for white but not brown adipose tissue (18), and we observed a deficiency in white but not brown adipose tissue in the Klf3 knockout. We also sought to use ChIP assays to test for the presence of CtBP at the C/ebpα promoter. Although we observed some enrichment around the start of transcription, high background levels with existing CtBP antiserum hindered interpretation of these experiments (data not shown), so we have not yet directly shown that CtBP is present. Nevertheless, the observations that the Klf3 mutant unable to bind CtBP is unable to block adipogenesis in 3T3-L1 cells and that C/ebpα is derepressed in CtBP null MEFs support for a role for CtBP in the process.
On the other hand, while our results suggest that derepression of C/ebpα interferes with normal adipogenesis and contributes to the Klf3 phenotype, this derepression alone may not fully explain the mouse phenotype and cellular results. For instance, it is unexpected that the Klf3 knockout mouse has less fat, whereas in culture Klf3 appears to have an inhibitory role and Klf3 knockout fibroblasts differentiate into adipocytes more readily than normal fibroblasts. We propose two possible explanations. C/EBPα is known to be antimitotic (33); thus, it is possible that in culture in vitro its derepression and premature accumulation result in an increase in lipid-accumulating cells, but in mice in vivo the premature expression of this antimitotic factor prevents expansion of adipocyte progenitors and results in a smaller fat pad. It is also likely that indirect processes influence fat pad size in vivo. Klf3 is broadly expressed, and there may be subtle defects in many tissues that affect physiology and/or behavior. Results from a preliminary feeding study suggest that the knockouts may feed less (data not shown), so this could well partly explain the phenotype in vivo. Although there are likely to be multiple factors influencing the ultimate in vivo phenotype, the results from 3T3-L1 cells, taken together with the altered adipogenesis in Klf3 knockout fibroblasts in culture, suggest that Klf3 does play a direct role in adipogenesis and that derepression of C/ebpα is one contribution to the phenotype observed in the mice.
Precisely how Klf3 fits within the hierarchy of KLFs that control adipogenesis remains to be defined. The Klf3 gene is driven by Klf1 in erythroid cells (10) and by Klf4 in skin cells (23). During 3T3-L1 differentiation the decline in Klf3 levels coincides with the decline in Klf2 (Lklf), suggesting the possibility that Klf3 is driven by Klf2 during adipogenesis. Klf1, Klf2, and Klf4 form a closely related cluster of activating KLFs within the KLF family, and it is possible that they regulate similar sets of genes, including Klf3, in different tissues. Future experiments will explore the interplay between different KLFs in different tissues during development.
Supplementary Material
Acknowledgments
This work was supported by NIH grant NHLBI HL073443 and grants from the Australian ARC and NHMRC (M.C.). N.S., B.J., S.E., and A.F. acknowledge the support of the Australian Postgraduate Award.
We are grateful to David James for help with adipocyte biology.
Footnotes
Published ahead of print on 7 April 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Balasubramanian, P., L.-J. Zhao, and G. Chinnadurai. 2003. Nicotinamide adenine dinucleotide stimulates oligomerization, interaction with adenovirus E1A and an intrinsic dehydrogenase activity of CtBP. FEBS Lett. 537157-160. [DOI] [PubMed] [Google Scholar]
- 2.Banerjee, S. S., M. W. Feinberg, M. Watanabe, S. Gray, R. L. Haspel, D. J. Denkinger, R. Kawahara, H. Hauner, and M. K. Jain. 2003. The Kruppel-like factor KLF2 inhibits peroxisome proliferator-activated receptor-gamma expression and adipogenesis. J. Biol. Chem. 2782581-2584. [DOI] [PubMed] [Google Scholar]
- 3.Bartz, R., J. Seemann, J. K. Zehmer, G. Serrero, K. D. Chapman, R. G. Anderson, and P. Liu. 2007. Evidence that mono-ADP-ribosylation of CtBP1/BARS regulates lipid storage. Mol. Biol. Cell 183015-3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieker, J. J. 1996. Isolation, genomic structure, and expression of human erythroid Kruppel-like factor (EKLF). DNA Cell Biol. 15347-352. [DOI] [PubMed] [Google Scholar]
- 5.Bieker, J. J. 2001. Kruppel-like factors: three fingers in many pies. J. Biol. Chem. 27634355-34358. [DOI] [PubMed] [Google Scholar]
- 6.Cao, S., M. E. Fernandez-Zapico, D. Jin, V. Puri, T. A. Cook, L. O. Lerman, X.-Y. Zhu, R. Urrutia, and V. Shah. 2005. KLF11-mediated repression antagonizes Sp1/sterol-responsive element-binding protein-induced transcriptional activation of caveolin-1 in response to cholesterol signaling. J. Biol. Chem. 2801901-1910. doi: 10.1074/jbc.M407941200. [DOI] [PubMed] [Google Scholar]
- 7.Christian, M., E. Kiskinis, D. Debevec, G. Leonardsson, R. White, and M. G. Parker. 2005. RIP140-targeted repression of gene expression in adipocytes. Mol. Cell. Biol. 259383-9391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coghill, E., S. Eccleston, V. Fox, L. Cerruti, C. Brown, J. Cunningham, S. Jane, and A. Perkins. 2001. Erythroid Kruppel-like factor (EKLF) coordinates erythroid cell proliferation and hemoglobinization in cell lines derived from EKLF null mice. Blood 971861-1868. [DOI] [PubMed] [Google Scholar]
- 9.Crossley, M., E. Whitelaw, A. Perkins, G. Williams, Y. Fujiwara, and S. H. Orkin. 1996. Isolation and characterization of the cDNA encoding BKLF/TEF-2, a major CACCC-box-binding protein in erythroid cells and selected other cells. Mol. Cell. Biol. 161695-1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Funnell, A. P., C. A. Maloney, L. J. Thompson, J. Keys, M. Tallack, A. C. Perkins, and M. Crossley. 2007. Erythroid Kruppel-like factor directly activates the basic Kruppel-like factor gene in erythroid cells. Mol. Cell. Biol. 272777-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grooteclaes, M., Q. Deveraux, J. Hildebrand, Q. Zhang, R. H. Goodman, and S. M. Frisch. 2003. C-terminal-binding protein corepresses epithelial and proapoptotic gene expression programs. Proc. Natl. Acad. Sci. USA 1004568-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jiang, M. S., and M. D. Lane. 2000. Sequential repression and activation of the CCAAT enhancer-binding protein-alpha (C/EBPalpha) gene during adipogenesis. Proc. Natl. Acad. Sci. USA 9712519-12523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaczynski, J., T. Cook, and R. Urrutia. 2003. Sp1- and Kruppel-like transcription factors. Genome Biol. 4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kanazawa, A., Y. Kawamura, A. Sekine, A. Iida, T. Tsunoda, A. Kashiwagi, Y. Tanaka, T. Babazono, M. Matsuda, K. Kawai, T. Iiizumi, T. Fujioka, M. Imanishi, K. Kaku, Y. Iwamoto, R. Kawamori, R. Kikkawa, Y. Nakamura, and S. Maeda. 2005. Single nucleotide polymorphisms in the gene encoding Kruppel-like factor 7 are associated with type 2 diabetes. Diabetologia 481315-1322. [DOI] [PubMed] [Google Scholar]
- 15.Kumar, V., J. E. Carlson, K. A. Ohgi, T. A. Edwards, D. W. Rose, C. R. Escalante, M. G. Rosenfeld, and A. K. Aggarwal. 2002. Transcription corepressor CtBP is an NAD+-regulated dehydrogenase. Mol. Cell 10857-869. [DOI] [PubMed] [Google Scholar]
- 16.Leonardsson, G., J. H. Steel, M. Christian, V. Pocock, S. Milligan, J. Bell, P. W. So, G. Medina-Gomez, A. Vidal-Puig, R. White, and M. G. Parker. 2004. Nuclear receptor corepressor RIP140 regulates fat accumulation. Proc. Natl. Acad. Sci. USA 1018437-8442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li, D., S. Yea, S. Li, Z. Chen, G. Narla, M. Banck, J. Laborda, S. Tan, J. M. Friedman, S. L. Friedman, and M. J. Walsh. 2005. Kruppel-like factor-6 promotes preadipocyte differentiation through histone deacetylase 3-dependent repression of DLK1. J. Biol. Chem. 28026941-26952. [DOI] [PubMed] [Google Scholar]
- 18.Linhart, H. G., K. Ishimura-Oka, F. DeMayo, T. Kibe, D. Repka, B. Poindexter, R. J. Bick, and G. J. Darlington. 2001. C/EBPα is required for differentiation of white, but not brown, adipose tissue. Proc. Natl. Acad. Sci. USA 9812532-12537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mori, T., H. Sakaue, H. Iguchi, H. Gomi, Y. Okada, Y. Takashima, K. Nakamura, T. Nakamura, T. Yamauchi, N. Kubota, T. Kadowaki, Y. Matsuki, W. Ogawa, R. Hiramatsu, and M. Kasuga. 2005. Role of Kruppel-like factor 15 (KLF15) in transcriptional regulation of adipogenesis. J. Biol. Chem. 28012867-12875. [DOI] [PubMed] [Google Scholar]
- 20.Nuez, B., D. Michalovich, A. Bygrave, R. Ploemacher, and F. Grosveld. 1995. Defective haematopoiesis in fetal liver resulting from inactivation of the EKLF gene. Nature 375316-318. [DOI] [PubMed] [Google Scholar]
- 21.Oishi, Y., I. Manabe, K. Tobe, K. Tsushima, T. Shindo, K. Fujiu, G. Nishimura, K. Maemura, T. Yamauchi, N. Kubota, R. Suzuki, T. Kitamura, S. Akira, T. Kadowaki, and R. Nagai. 2005. Kruppel-like transcription factor KLF5 is a key regulator of adipocyte differentiation. Cell Metab. 127-39. [DOI] [PubMed] [Google Scholar]
- 22.Otto, T. C., and M. D. Lane. 2005. Adipose development: from stem cell to adipocyte. Crit. Rev. Biochem. Mol. Biol. 40229-242. [DOI] [PubMed] [Google Scholar]
- 23.Patel, S., Z. F. Xi, E. Y. Seo, D. McGaughey, and J. A. Segre. 2006. Klf4 and corticosteroids activate an overlapping set of transcriptional targets to accelerate in utero epidermal barrier acquisition. Proc. Natl. Acad. Sci. U S A 10318668-18673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perdomo, J., A. Verger, J. Turner, and M. Crossley. 2005. Role for SUMO modification in facilitating transcriptional repression by BKLF. Mol. Cell. Biol. 251549-1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Perkins, A. C., A. H. Sharpe, and S. H. Orkin. 1995. Lethal beta-thalassaemia in mice lacking the erythroid CACCC-transcription factor EKLF. Nature 375318-322. [DOI] [PubMed] [Google Scholar]
- 26.Picard, F., M. Kurtev, N. Chung, A. Topark-Ngarm, T. Senawong, R. Machado De Oliveira, M. Leid, M. W. McBurney, and L. Guarente. 2004. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 429771-776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodbell, M. 1964. Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. J. Biol. Chem. 239375-380. [PubMed] [Google Scholar]
- 28.Rosen, E. D., and O. A. MacDougald. 2006. Adipocyte differentiation from the inside out. Nat. Rev. Mol. Cell Biol. 7885-896. [DOI] [PubMed] [Google Scholar]
- 29.Suske, G., E. Bruford, and S. Philipsen. 2005. Mammalian SP/KLF transcription factors: bring in the family. Genomics 85551-556. [DOI] [PubMed] [Google Scholar]
- 30.Tang, Q.-Q., M.-S. Jiang, and M. D. Lane. 1999. Repressive effect of Sp1 on the C/EBPα gene promoter: role in adipocyte differentiation. Mol. Cell. Biol. 194855-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thio, S. S. C., J. V. Bonventre, and S. I.-H. Hsu. 2004. The CtBP2 co-repressor is regulated by NADH-dependent dimerization and possesses a novel N-terminal repression domain 10.1093/nar/gkh344. Nucleic Acids Res. 321836-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner, J., and M. Crossley. 1998. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 175129-5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Umek, R. M., A. D. Friedman, and S. L. McKnight. 1991. CCAAT-enhancer binding protein: a component of a differentiation switch. Science 251288-292. [DOI] [PubMed] [Google Scholar]
- 34.Vo, N., C. Fjeld, and R. H. Goodman. 2001. Acetylation of nuclear hormone receptor-interacting protein RIP140 regulates binding of the transcriptional corepressor CtBP. Mol. Cell. Biol. 216181-6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu, J., S. V. Srinivasan, J. C. Neumann, and J. B. Lingrel. 2005. The KLF2 transcription factor does not affect the formation of preadipocytes but inhibits their differentiation into adipocytes. Biochemistry 4411098-11105. [DOI] [PubMed] [Google Scholar]
- 36.Zhang, Q., C. C. Fjeld, A. C. Nottke, and R. H. Goodman. 2006. CtBP as a redox sensor in transcriptional repression, p. 68-76. In G. Chinnadurai (ed.), CtBP family proteins. Landes Bioscience, Austin, TX.
- 37.Zhang, Q., D. W. Piston, and R. H. Goodman. 2002. Regulation of corepressor function by nuclear NADH. Science 2951895-1897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.