Abstract
PUF family proteins are among the best-characterized regulatory RNA-binding proteins in nonmammalian species, but relatively little is known about mRNA targets or functions of mammalian PUF proteins. In this study, we used ribonomic analysis to identify and analyze mRNAs associated with ribonucleoproteins containing an endogenous human PUF protein, Pum1. Pum1-associated mRNAs were highly enriched for genes encoding proteins that function in transcriptional regulation and cell cycle/proliferation, results consistent with the posttranscriptional RNA regulon model and the proposed ancestral functions of PUF proteins in stem cell biology. Analysis of 3′ untranslated region sequences of Pum1-associated mRNAs revealed a core Pum1 consensus sequence, UGUAHAUA. Pum1 knockdown demonstrated that Pum1 enhances decay of associated mRNAs, and relocalization of Pum1 to stress granules suggested that Pum1 functions in repression of translation. This study is the first in vivo genome-wide mRNA target identification of a mammalian PUF protein and provides direct evidence that human PUF proteins regulate stability of associated mRNAs. Comparison of Pum1-associated mRNAs to mRNA targets of PUF proteins from Saccharomyces cerevisiae and Drosophila melanogaster demonstrates how a well-conserved RNA-binding domain and cognate binding sequence have been evolutionarily rewired to regulate the collective expression of different sets of functionally related genes.
PUF family RNA-binding proteins (RBPs) are among the best-characterized regulators of posttranscriptional gene expression in nonmammalian eukaryotes. Named for the founding members Pumilio (32) and FBF (61), PUF proteins are represented throughout the eukaryotic lineage (47). The common feature of PUF proteins is the PUF homology domain (PUF-HD), an RNA-binding domain typically consisting of eight imperfect repeats of a 32-amino-acid sequence (60). The overall sequences of PUF proteins from different species are not highly similar outside of the PUF-HD, although the PUF-HD is incredibly well-conserved (46). This extreme conservation of the PUF-HD suggests that posttranscriptional regulation of gene expression by PUF proteins has remained important throughout evolution. Although there is a growing body of knowledge concerning PUF proteins in nonmammalian model organisms, relatively little is known about the functions or mRNA targets of PUF proteins in mammals.
Genetic analyses have revealed diverse functions of PUF proteins, such as embryo patterning in Drosophila melanogaster (32), germ line establishment and maintenance in Drosophila (14, 33, 42) and Caenorhabditis elegans (4, 10, 30, 53, 61), and mitochondrial function in Saccharomyces cerevisiae (16). PUF proteins have been found to function as repressors of gene expression through both repression of translation and enhancement of decay of target mRNAs (13, 19, 20, 41, 55). Crystal structure analysis of the PUF-HD from Drosophila Pumilio revealed that it forms a crescent shape (12), with protein-RNA interactions occurring on the inner concave surface and protein-protein interactions on the outer surface.
Genome-wide target identification of five S. cerevisiae PUF proteins, Puf1 to -5 (17), and the single Drosophila PUF protein, Pumilio (18), demonstrated that each protein binds to a specific group of functionally related mRNAs distinct from those mRNAs bound by any other PUF protein (except for Puf1 and Puf2, which bind overlapping sets of mRNAs). Puf3 was found to bind almost exclusively to mRNAs of nuclear-encoded mitochondrial proteins, and this binding was later demonstrated to have functional consequences when it was shown that Puf3 regulates stability of target messages in a condition-specific manner (13) and regulates mitochondrial biogenesis and motility in S. cerevisiae (16). These experiments represent a compelling example of the posttranscriptional RNA operon/regulon model (26, 28), demonstrating a mechanism through which expression of functionally related genes can be coordinately regulated at the level of the mRNA. Targets of Drosophila Pumilio also contained potential RNA regulons, most notably the vacuolar-type ATPase and the embryo-patterning cascade, which Pumilio mutants are known to disrupt (18). These studies also demonstrated for the first time that while the cis-trans interactions between PUF proteins and target mRNAs are similar, target messages of PUF proteins are not conserved through evolution, at least from S. cerevisiae to Drosophila (18).
Mammalian genomes contain two PUF genes, Pum1 and Pum2 (46), both of which have been studied to a limited extent. The Pum2 gene has been knocked out in the mouse, but the only obvious phenotype was smaller testis size with no effect on fertility (58). Several potential human Pum2 mRNA targets have been discovered by various groups (15, 31, 48, 56); however, the in vivo genome-wide targets of the protein have not been identified. Expression of reporter constructs containing Pum2 target 3′ untranslated regions (UTRs) was shown to be repressed by Pum2 overexpression, but the mechanism of repression was not determined (31). Rat Pum2 was found to localize to stress granules in hippocampal neurons, with Pum2 knockdown interfering with stress granule formation and Pum2 overexpression inducing aggregates that costained with stress granule markers (52). Even less is known about Pum1. The human Pum1 RNA-binding domain can bind to the Nanos response element, an mRNA sequence bound by Drosophila Pumilio, and has been found by crystal structure analysis to interact with RNA in a very modular fashion, with each of the eight PUF repeats directly contacting a single RNA base (54). Recombinant Pum1 was shown in vitro to interact with the CNOT8 protein, a member of the CCR4-NOT deadenylase complex (20), suggesting that enhancement of target mRNA deadenylation and decay may be a conserved mechanism of PUF protein function (57).
Due to the lack of knowledge regarding both targeting and function of mammalian PUF proteins, this study was undertaken to identify genome-wide mRNAs associated with Pum1 protein, along with mechanisms by which Pum1 regulates these mRNAs. By determining the genome-wide targets of a PUF protein from a third species, humans, we gained insight as to how the use of a highly conserved RNA-binding domain and cognate binding sequence has changed throughout evolution by regulating different sets of functionally related mRNAs.
MATERIALS AND METHODS
Cell culture.
HeLa S3 cells, used for all experiments except immunofluorescence (IF) assays, were grown in Ham's F-12 supplemented with 10% fetal bovine serum. HeLa CCL2 cells were used for IF and were grown in Dulbecco's modified Eagle's medium supplemented with nonessential amino acids and 10% fetal bovine serum. All cells were grown in humidified incubators at 37°C and 5% CO2.
Immunoprecipitations.
Immunoprecipitation (IP) reactions were performed as described previously (27, 50). Briefly, 10 μg of anti-Pum1 antibody (goat polyclonal; Bethyl Labs) was incubated overnight with protein G-agarose beads. The beads were washed, then buffer and cell lysate were added, and the reaction mixtures were tumbled for 4 hours at 4°C. After this incubation, the beads were thoroughly washed again and then either boiled in 2× Laemli buffer for IP-Western experiments or 1 ml of Trizol was added and RNA extracted for microarray and reverse transcription-PCR (RT-PCR) experiments. Identical IPs performed with beads precoated with preimmune goat serum were used as a negative control.
Western blot assays.
Protein samples were loaded onto 4 to 20% Tris-HCl-polyacrylamide gel electrophoresis gels. After electrophoresis, proteins were transferred to nitrocellulose membranes, and then these membranes were blocked and probed with anti-Pum1 antibody. Visualization was performed with horseradish peroxidase-linked secondary antibody and enhanced chemiluminescence detection.
RNA extraction.
Trizol (Invitrogen) was used for all RNA extractions according to the manufacturer's protocol.
RT-PCR.
Reverse transcription was performed with the iScript cDNA synthesis kit from Bio-Rad according to the manufacturer's protocols, using a combination oligo(dT) and random hexamers for priming. End-point PCR was carried out in the linear range (30 cycles), and products were resolved on 1% agarose gels and stained with ethidium bromide. Quantitative PCR (qPCR) was performed using a Roche Lightcycler with Sybr green detection (Invitrogen) and the ΔΔCT analysis method, using either β2-microglobulin or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) for normalization. All real-time PCR primer sets were designed so the products would span multiple exons, and amplification of a single product of the correct size was confirmed by melting curve analysis and agarose gel electrophoresis.
Microarray analysis.
Arrays were printed at the Duke Microarray Facility using the Genomics Solutions OmniGrid 300 arrayer. The arrays contained the Human Operon v3.0.2 oligonucleotide set (Oligo Source), which consists of 34,602 unique optimized 70-mers. RNA quality was ascertained using an Agilent 2100 bioanalyzer (Agilent Technologies). All arrays were subject to Loess normalization within each array and scale normalization across all arrays using the Array Magic package in R (7). Replicate probes were collapsed to one probe corresponding to the median value of all the replicates. Gene set enrichment analysis (GSEA) was used to calculate t scores in comparing the Pum1 IP to the negative IP results (38, 49).
Gaussian mixture modeling was performed multiple times on the t score distribution to estimate the mean, standard deviation, and weight of each mixture using the Mixtools package in R (43, 59). Each iteration of mixture modeling was initialized at different values in the distribution, and the parameters from the model with the highest likelihood were used to create log of odds (LOD) scores of Pum1 ribonucleoprotein (RNP) association by comparing the weighted probability density functions of the Pum1-associated versus background distributions. LOD scores greater than 0 have a higher probability of being in the Pum1-RNP-associated distribution compared to background distribution; therefore, a LOD score of 0 was used as a cutoff for determining Pum1-associated probes.
Functional enrichment.
Pum1-associated and total expressed genes were loaded into either WebGestalt (62) or Panther (37) using appropriate gene identifiers. Pum1 target genes were compared to the total expressed genes to determine functional enrichments. To calculate significance of enrichment, WebGestalt uses a hypergeometric test and Panther uses a modified Fisher test with a Bonferroni correction for multiple testing.
Motif finding.
3′UTRs of Pum1 target and total expressed genes were obtained from a local pipeline which uses information from the PolyA_DB database (63) combined with other data to define 3′UTRs by genomic coordinates (35), and sequences were obtained from the latest human genome build based on these coordinates. MEME analysis (5) was run locally using 3′UTRs of the top 100 Pum1-associated mRNAs as a training set.
Pum1 knockdown.
Pum1 knockdown was performed using a mixture of two Ambion Silencer small interfering RNAs (siRNAs) targeting the Pum1 mRNA and siPort NeoFx reagent (Ambion) for transfection, following the manufacturer's instructions. Protein knockdown of approximately 70 to 95% was confirmed by Western blotting for all experiments. Assays were performed 40 to 48 h after transfection of siRNAs. A nontargeting siRNA (Ambion) was used as a negative control.
mRNA decay rates.
mRNA levels of target messages were determined by RT-qPCR at hourly time points after addition of 5 μg/ml actinomycin D. The nontarget message GAPDH was used for normalization. Exponential decay curves were fit to points representing the means of three biological replicates, and half-lives were calculated based on the equations describing these best-fit curves.
Immunofluorescence.
IF was performed essentially as described elsewhere (25). Briefly, cells were grown on glass coverslips precoated with 0.1% gelatin and then fixed in 4% paraformaldehyde followed by 100% methanol. Coverslips were then blocked, incubated with appropriate antibodies, and mounted to slides. Pum1 and Ddx6 antibodies and Pum1 blocking peptide were from Bethyl Labs, and HuR antibody was supernatant from hybridoma 3A2 cells. Images were captured using a Zeiss Axioplan 2 microscope with a 63× objective and Axiovision 4.6.3 SP2 software. Images were then combined and minimally processed using Adobe Photoshop, with all images processed identically. For cell stress, sodium arsenite was added to growth medium at a concentration of 0.5 mM, and cells were fixed as described after 45 min of stress.
Orthologues of Puf3 and Pumilio target genes were determined using the online database mining tool Biomart (www.biomart.org).
Microarray accession numbers.
All microarray data were submitted to the GEO database and can be found under the accession number GSE 11301.
RESULTS
Identification of Pum1-associated mRNAs.
In this study, we used the RNA-binding protein immunoprecipitation-chip (RIP-chip) method to identify genome-wide mRNAs associated with RNPs containing Pum1 (21, 27, 50). All experiments were performed in human HeLa cells, and all Pum1 IPs were performed with a goat polyclonal antibody raised against a small peptide region specific to the Pum1 protein. Negative control IPs were performed with preimmune goat serum. Before performing RIP-chip, we first confirmed specific IP of Pum1 protein from cytoplasmic lysate by IP-Western blotting, using identical conditions except that in the final step protein was extracted from the beads and used for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting. We were able to efficiently recover Pum1 protein in our IP reactions and correspondingly deplete a substantial amount from the original starting material (Fig. 1A). After confirmation of efficient Pum1 protein recovery, we next sought to determine whether specific mRNAs associated with Pum1 were enriched in the Pum1 IP pellet. We chose cyclin B1 as a potential target due to its conservation as a PUF target in other species (3, 40, 45, 47). RT-PCR analysis confirmed specific enrichment of cyclin B1 mRNA in the Pum1 IP compared to the negative control (Fig. 1B). After confirming that specific mRNAs associated with the Pum1 protein could be identified using these procedures, we next performed RIP-chip in biological triplicates using custom spotted cDNA microarrays. Each biological replicate consisted of a Pum1 IP sample, a negative control IP sample, and a total RNA sample, which were each hybridized to a separate microarray along with a common reference sample. Total RNA microarrays were used to identify the transcriptome of the cells from which IPs were performed, providing an accurate background for subsequent analyses. A probe was considered present in the transcriptome if the signal from the spot was at least twofold greater than local background in all three total RNA or Pum1 IP microarrays. t scores, based on the t statistic, for Pum1 IP versus the negative control IP were calculated for all probes on the microarrays (49). A visual inspection of the t score values of the probes (Fig. 2, histogram) suggested two distributions: a background distribution and a Pum1-associated distribution.
FIG. 1.
A. IP-Western blot of Pum1, probed with anti-Pum1-specific antibody. In, input; Sup, supernatant. (Only ∼20% of the IP sample was loaded.) B. RT-PCR analysis of RNA recovered from Pum1 IP. Pum1, Pum1 IP; Neg, negative IP; Tot, total RNA.
FIG. 2.
A. Distribution of Pum1 IP versus negative IP t scores for three biological replicates of RIP-chip. Pum1-associated (solid black curve), background (dashed black curve), and the sum of Pum1-associated and background (gray curve) probability distributions are shown, as defined by Gaussian mixture modeling.
In order to objectively define Pum1 target mRNAs based on a distribution of t scores, we employed Gaussian mixture modeling (GMM). GMM uses probabilistic modeling to identify single Gaussian distributions in a population consisting of a mixture of multiple Gaussian distributions (43). GMM uses expectation maximization modeling, which is prone to converging on a local optimum; therefore, several iterations of mixture modeling were performed which initialized at different values in the distribution, and the model with the greatest probability was then used to define Pum1-associated mRNAs. Both Pum1 and background distributions were defined as an equation relating t scores to probability (Fig. 2, curves), and using these equations we calculated the LOD for each probe appearing in the Pum1 distribution versus the background distribution. Those probes with a greater likelihood of being in the Pum1 distribution (LOD > 0) were considered Pum1-associated mRNAs (targets). While one might expect that this cutoff would result in a high false-positive rate, downstream analysis proved that a LOD cutoff of >0 was appropriate. A summary of probes, t scores, and LOD scores is available in Table S1 of the supplemental material. Use of GMM and creation of LOD scores allowed us to objectively define probabilities of probes being in the Pum1 distribution versus the background distribution and thus allowed for a less arbitrary determination of probes that could be considered Pum1 associated. Probes were collapsed into unique genes, and of the 6,539 unique genes represented in total, 726 (11.1%) were considered Pum1 targets on this basis.
We used RT-qPCR to confirm select targets by measuring their enrichment in the Pum1 IP versus either the negative IP or total RNA, using the nontarget message β2-microglobulin for normalization and GAPDH mRNA as a negative control. qPCR analysis confirmed levels of enrichment up to 240-fold for target messages in the Pum1 IP, with similar levels of enrichment versus either the negative IP or total RNA (Fig. 3). Target messages confirmed by RT-qPCR represented a range of LOD scores greater than zero, thus confirming these targets serve as partial validation of the LOD cutoff of >0 when defining Pum1-associated mRNAs, as noted above.
FIG. 3.
RT-qPCR confirmation of Pum1-associated mRNAs. Log2 enrichments versus either total RNA (A) or negative IP (B), normalized to β2-microglobulin, are shown. Select target messages are represented by black bars and nontargets by gray bars. Error bars represent the standard errors of the means of three biological replicates.
Pum1 target mRNA analysis.
After defining mRNAs associated with the Pum1 RNP, we proceeded to determine if the proteins encoded by Pum1 target mRNAs were functionally related, as is predicted by the RNA regulon model (26, 28). We analyzed Pum1 target genes using two Web-based programs, Panther (37) and WebGestalt (62), which search gene lists for significant enrichment in gene ontology (GO) categories and other functional groupings. We also analyzed Pum1 targets using GSEA, which compares a rank-ordered gene list of interest to other gene sets in the GSEA Molecular Signatures database to discover significant correlations between sets of genes (49). Results from WebGestalt and Panther, as seen in Table 1, were very similar. The Panther program searches for both positively and negatively enriched categories and applies a strict Bonferroni correction for multiple testing when calculating significance (37); thus, fewer categories were found to be enriched in the Panther analysis, and the statistical significance of that enrichment was lower. WebGestalt makes no correction for multiple testing; thus, the results obtained by this method, while having lower P values, may be less biologically significant. However, results obtained using either WebGestalt or Panther were in agreement. One of the more striking results of both the WebGestalt and Panther analyses was the large number of target genes and extreme significance of enrichment of GO categories involved in transcription and regulation of transcription. Another noteworthy result was the enrichment of mRNAs representing genes involved in regulation of the cell cycle and cell proliferation and differentiation, a result consistent with proposed ancestral functions of PUF proteins in stem cell biology (57). GSEA of the Pum1 IP rank-ordered gene list created from LOD scores revealed a high degree of correlation with various gene sets, including gene sets whose mRNA levels were found to decrease after UVC and UVB exposure, whose mRNAs increased after cytomegalovirus (CMV) infections, a gene set consisting of HOX genes, and gene sets related to the cell cycle (Table 1). This result provides further support for a role for the Pum1 protein in stem cell function, as well as a role for Pum1 in response to stress (see below).
TABLE 1.
Genes represented by Pum1-associated mRNA in various GO categories
| Analysis method and GO category (positively or negatively enriched) | No. of genes | P value |
|---|---|---|
| Panther | ||
| Positively enriched | ||
| mRNA transcription | 100 | 9.66E-04 |
| mRNA transcription regulation | 71 | 9.40E-03 |
| Transcription factor | 103 | 3.02E-05 |
| Other transcription factor | 32 | 4.42E-02 |
| Nucleoside, nucleotide, and nucleic acid metabolism | 173 | 2.57E-02 |
| Cell proliferation and differentiation | 51 | 1.76E-02 |
| Negatively enriched | ||
| Oxidoreductase | 5 | 9.90E-07 |
| Reductase | 0 | 1.15E-02 |
| Dehydrogenase | 2 | 2.85E-02 |
| Electron transport | 2 | 1.32E-02 |
| WebGestalt | ||
| Positively enriched | ||
| Regulation of biological processes | 220 | 1.17E-09 |
| Regulation of transcription | 138 | 5.48E-10 |
| Transcription factor activity | 67 | 1.10E-09 |
| Regulation of cell cycle | 42 | 2.71E-03 |
| Ubiquitin cycle | 30 | 5.80E-03 |
| Wnt receptor signaling pathway | 11 | 1.95E-03 |
| Small GTPase-mediated signal transduction | 32 | 3.75E-04 |
| Cell communication | 128 | 2.13E-04 |
| GSEAa | ||
| Positively enriched | ||
| UVC high all DN | 117 | <0.001 |
| UVB NHEK1 DN | 66 | <0.001 |
| CMV/HCMV time course all up | 76 | <0.001 |
| HOX genes | 10 | <0.001 |
| Cell cycle | 28 | <0.001 |
For GSEA results, the numbers of genes reported represent core enrichment. The GO category names for GSEA are those used in the analysis software.
Within the set of Pum1 target genes are several specific functional relationships that represent putative RNA operons/regulons, such as that between cyclin B1, Cdc2, p21, and Wee1. The cyclin B1-Cdc2 complex is a key regulator of the G2/M transition of the mitotic cell cycle, with p21 and Wee1 acting as negative regulators of Cdc2 (www.biocarta.com). Although it may seem counterintuitive that Pum1 would regulate expression of both one protein and a second protein that negatively regulates the first protein, this situation has been seen in C. elegans, where a PUF protein represses a mitogen-activated protein (MAP) kinase and a gene that inactivates the same MAP kinase, thereby ensuring continued repression of the MAP kinase gene after PUF-mediated repression of both proteins is relieved (31). PCNA, GSK3β, p21, and p27 form another cell cycle-related functional group, as all of these proteins function as inhibitors of cyclin D (www.biocarta.com). One of the most striking potential Pum1 regulons is that of the E2F transcription factors: four of the five E2Fs that were represented in the total mRNA population were found to be Pum1 targets (E2F3 to E2F6 are targets, and E2F1 is not a target), showing an overlap of two highly enriched categories in Pum1 targets: cell cycle/proliferation and transcription (11). A large number of RNA processing and RNA-binding protein genes were also found to be Pum1 targets, among them the histone stem-loop-binding protein, DICER, Pum1 itself, and the other human PUF protein, Pum2.
Identification of the Pum1 USER.
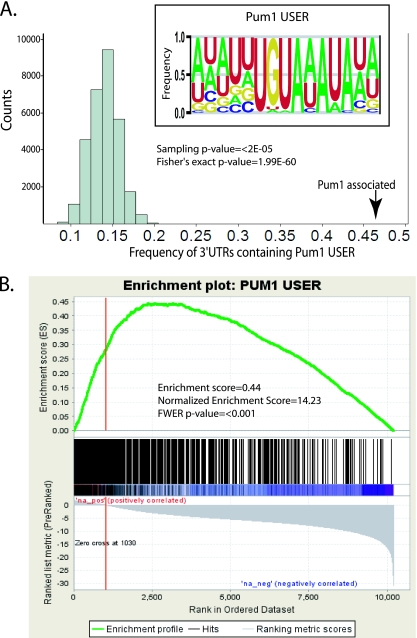
Previous analyses of PUF protein target mRNAs revealed a consensus sequence present in the 3′UTRs of target messages (17, 18). We used the motif finding program MEME (5) to search for a consensus sequence in the 3′UTRs of Pum1 targets. The 3′UTR sequences of the top 100 Pum1-associated mRNAs, by LOD score, were used as a training set for MEME analysis, resulting in discovery of the consensus sequence shown in the inset in Fig. 4. Contained in the Pum1 target consensus sequence is the 8-nucleotide core sequence UGUAHAUA, which has been shown by X-ray crystallographic analysis to be directly bound by the Pum1 PUF-HD (54). We searched for the occurrence of this core sequence in the 3′UTRs of the Pum1 targets not used in the MEME training set and found it occurred at least once in 46.5% of Pum1 target 3′UTRs but only in 13.5% of total mRNA 3′UTRs. To determine the likelihood of this enrichment occurring by chance, we created 50,000 sets of random 3′UTRs and determined the frequency of 3′UTRs in each set that contained the core consensus sequence at least once. The random sets of 3′UTRs were chosen from the mRNAs expressed in HeLa cells, and each set contained the same number of 3′UTRs as the Pum1-associated set. The occurrence of the Pum1 core consensus sequence was determined for each of these sets, resulting in a distribution with a mean of 13.5% and a maximum of 20.1% (Fig. 4). This represents an extremely significant enrichment of the core Pum1 consensus sequence in Pum1 targets (P < 2 × 10−5) even after excluding those 3′UTRs used as the training set. We also used Fisher's exact test to determine the significance of enrichment of this sequence in Pum1 targets and calculated P to be 1.99E-60. As elements of this 8-nucleotide core sequence have been shown to be important for target mRNA regulation by PUF proteins (6, 9, 19, 24, 31, 39), we will henceforth refer to this sequence as the Pum1 USER (untranslated sequence element for regulation) (28).
FIG. 4.
A. Calculated frequency of 3′UTRs containing the Pum1 USER, UGUAHAUA, among Pum1-associated mRNAs compared with 50,000 randomly chosen sets of mRNAs. P values derived from this sampling and Fisher's exact test are shown. Random sets of mRNAs were derived from mRNAs present in the HeLa cell transcriptome, and each set contained the same number of mRNAs as the Pum1-associated set. Inset shows the consensus Pum1 target sequence, including the 8-nucleotide Pum1 USER, as determined by MEME analysis. B. GSEA of enrichment of the Pum1 USER based on LOD scores. Enrichment scores and the family-wise error rate (FWER) P value are shown. The vertical red line in the graphs represents LOD of 0.
Although it may be unexpected to find that only about half of Pum1 target messages contained the Pum1 USER, there are a number of likely explanations for this outcome. The RIP-chip method is optimized to isolate entire RNPs (27, 50), and thus some of those messages associated with Pum1 RNPs are not expected be directly bound by Pum1. The search for the Pum1 USER was also based on a simple string (UGUAHAUA) rather than a more descriptive and flexible position-specific weight matrix. Because the position-specific weight matrix more accurately describes flexibility in the consensus sequence, it could also identify sequences in 3′UTRs to which Pum1 binds with almost as high an affinity as the consensus sequence which were not recognized by a simple string search. Finally, only the 3′UTRs of mRNAs were searched for the Pum1 USER; thus, any USERs present in the 5′UTRs or coding sequences would not be identified.
In order to more thoroughly explore enrichment of the Pum1 USER in Pum1-associated messages, we also used GSEA (49). This analysis used all genes with the Pum1 USER in their 3′UTR as a gene set and calculated the running enrichment of genes containing the Pum1 USER in the Pum1 RIP-chip data ordered by LOD score. As can be seen in Fig. 4B, the peak of the running enrichment score occurs after LOD of 0, demonstrating that our earlier LOD cutoff of >0 is valid for determining targets and is likely conservative. The normalized enrichment score of 14.23 and family-wise error rate P value of <0.001 for this analysis again demonstrate the extreme enrichment of this sequence in the 3′UTRs of Pum1-associated mRNAs.
Role of Pum1 in decay of target messages.
We next sought to determine whether Pum1 enhances decay of associated mRNAs, as has been shown for other PUF proteins (19, 41). Thus, we knocked down Pum1 protein using siRNA and then assayed decay rates of target messages. Extent of Pum1 protein depletion was determined to be approximately 70 to 95% by Western blotting for all experiments. To determine mRNA decay rates, actinomycin D was used to inhibit transcription and qPCR was performed to determine the percentages of transcripts remaining at multiple time points after treatment, as normalized to GAPDH and averaged across three biological replicates. An exponential decay curve was fit to the mean of these measurements, and half-lives of messages were determined based on the equation describing this curve (Fig. 5). Control experiments were performed with an siRNA not known to target any mRNAs. All but one of the Pum1 target mRNAs assayed showed increased stability during Pum1 knockdown, although there was a range in the degree of increased stability. As Pum1 protein has been shown to interact with CNOT8 protein (20), a member of the CCR4-NOT deadenylase complex, it will be interesting to determine in future studies whether Pum1's effect on stability of associated mRNAs is at least partially mediated by deadenylation via this complex.
FIG. 5.
Decay rates of Pum1 target mRNAs, normalized to GAPDH, as determined by RT-qPCR after Pum1 knockdown followed by treatment with actinomycin D (ActD) to inhibit transcription. Black boxes represent Pum1 knockdown, and gray diamonds represent the control. Error bars represent standard errors of the means of three biological replicates.
Subcellular localization of Pum1 protein following oxidative stress.
To further study the function of Pum1, we used immunofluorescence analysis to determine its subcellular localization during cell growth and induced stress. Growing cells displayed a diffuse, granular cytoplasmic Pum1 staining pattern, and upon arsenite-induced oxidative stress Pum1 protein relocalized to large cytoplasmic granules (Fig. 6, left panels). A blocking peptide was used to confirm specificity of the Pum1 antibody (not shown). Stress granules and processing bodies (p-bodies) are both candidate large cytoplasmic granules known to be induced by oxidative stress (2); thus, to determine if these granules were stress granules, p-bodies, or both, we costained with a stress granule marker, HuR (Fig. 6A, middle) or a p-body marker, Ddx6 (Fig. 6B, middle panels). Costaining confirmed that Pum1 protein was present in the same aggregates as HuR after arsenite treatment (Fig. 6A, middle panel and zoom image), indicating that Pum1 protein is a component of stress granules. Pum1 also costained with exogenously transfected constructs expressing the recombinant fluorescent proteins G3BP-green fluorescent protein, TIA1-yellow fluorescent protein (YFP), and FAST-YFP (not shown), all of which are known components of stress granules (25). Ddx6-containing p-bodies were sometimes juxtaposed but did not significantly overlap with Pum1 granules after induction of stress (Fig. 6B, middle panel and zoom image), indicating that Pum1 is not a component of p-bodies and providing further evidence that Pum1 is a component of stress granules. Cells that had been transfected with a plasmid expressing Dcp1a-YFP protein, another p-body marker (25), were also stained for Pum1, and again Pum1 granules appeared juxtaposed but distinct from Dcp1a-containing p-bodies (not shown). Cells were also stained for Pum1 and HuR or Ddx6 after heat shock, with results again indicating that Pum1 is a component of stress granules but not p-bodies (data not shown). The observation that Pum1 is a stress granule component suggests that it may have a role in regulating translation of target messages. Unlike the related protein Pum2 (52), Pum1 knockdown did not interfere with stress granule formation after arsenite treatment, indicating that Pum1 is not necessary to form these structures in HeLa cells (data not shown).
FIG. 6.
Coimmunofluorescence of Pum1 (red) and HuR (A) or Ddx6 (B) (in green in corresponding middle panel) in unstressed and stressed HeLa cells. DNA was stained with 4′,6′-diamidino-2-phenylindole (DAPI) and is shown in blue. Magnified regions show a stress granule containing both Pum1 and HuR (A) and a Pum1-containing stress granule juxtaposed with Ddx6-containing p-bodies (B).
Conservation of PUF protein target mRNAs and potential RNA regulons.
One of the main goals of this study was to compare Pum1 target genes to target genes of PUF proteins in other species in order to observe how the posttranscriptional regulatory adaptors of the PUF-HD and cognate binding sequence have been rewired throughout evolution. The four amino acids that directly contact RNA in each of the eight repeats of the PUF-HD (54) are completely conserved between S. cerevisiae Puf3, Drosophila Pumilio, and human Pum1, as is the 8-nucleotide sequence to which each PUF-HD likely binds (17, 18). Neither the RNA-contacting amino acids nor consensus binding sequence of any other yeast Puf protein show this level of conservation; thus, only Puf3 was considered in this analysis. mRNA targets of Puf 3 and Pumilio have been previously determined using RIP-chip, and it was found that there was little conservation of target genes between them (17, 18). In order to determine if there is conservation of targets between Pum1 and Puf3 or Pumilio, we first determined how many Puf3 or Pumilio target genes have human orthologues that are expressed in HeLa cells and then determined how many of those genes with human orthologues are also Pum1 targets (Fig. 7A). Puf3 target genes have 89 human orthologues, and only 7 of these are also Pum1 target genes, representing no significant enrichment for Pum1 targets among Puf3 targets. Pumilio targets in the adult Drosophila ovary have 502 human orthologues, with 73 also being Pum1 target genes. This represents a slightly significant enrichment of Pum1 targets among Pumilio targets, with a sampling P value of 0.0183 and a Fisher's exact P value of 0.036. Statistical significance via sampling was determined by creating 10,000 sets of random genes containing the same number of genes as the Puf3 or Pumilio target genes with orthologues and then determining the proportion of those genes that were Pum1 targets. A possible caveat to this analysis is that each RIP-chip experiment was performed in a different cell type and thus each PUF protein had the potential to associate with different mRNAs in vivo. To address this issue we limited the universe of this analysis to only genes that were expressed in HeLa cells as defined by our study.
FIG. 7.
A. The number of S. cerevisiae Puf3 and Drosophila Pumilio target genes that have human orthologues and the number of these that are also Pum1 targets in HeLa cells. The statistical significance of this conservation of targets is indicated. B. The motif UGUAHAUA is predominant in yeast Puf3, fly Pumilio, and human Pum1 associated mRNAs, while the proteins encoded by each PUF protein's target mRNAs are not conserved, consistent with the notion of evolutionary rewiring of RNA regulon networks. Data on Puf3 and Pumilio targets were derived from references 17 and 18.
In addition to identities of genes encoded by target mRNAs, we also considered whether functional relationships among target genes, or potential RNA regulons (26, 28), were conserved (Fig. 7A). Target genes of Puf3 are almost entirely nuclear-encoded mitochondrial proteins (17), while those of Pumilio and Pum1 are not enriched for mitochondrial functions (18). The potential RNA regulon of cell cycle regulation is not conserved between Pumilio and Pum1, although there are many cyclin mRNAs among the Pumilio targets (18). Conversely, Pum1 targets are not enriched for components of the V-type ATPase or the embryo patterning cascade, functional groupings found to be enriched among Pumilio targets (8, 18). Pum1 targets and Pumilio targets are both enriched for the GO categories of transcription factor and membrane-bound organelle (18), showing that while most of the individual genes regulated by the proteins have changed, the types of genes regulated are similar. Combined with data regarding conservation of individual targets, this observation indicates that neither targets nor functions of PUF proteins have been conserved between S. cerevisiae and humans, yet some targets and functions have been conserved between Drosophila and humans. Thus, by keeping the PUF-HD and cognate binding sequence as static modules but changing the sets of mRNAs that are regulated, evolution has been able to rewire posttranscriptional regulation of gene expression networks, as previously suggested through experimental and computational analysis of evolutionary conservation of potential posttranscriptional regulatory elements (8, 18, 21, 26, 36).
DISCUSSION
While various genetic and biochemical studies have revealed mRNA targets and functions of PUF proteins in nonmammalian model organisms, little is currently known about the interactions or functions of PUF proteins in mammals. This study represents both the first in vivo genome-wide mRNA target identification of a mammalian PUF protein and the first direct demonstration of a function of a mammalian PUF protein in regulating its target mRNAs. By comparing targets of PUF proteins in three organisms, we were also able to observe how mRNA targets of these proteins have changed through evolution, despite conserved cis-trans interactions between the PUF-HD and cognate binding sequence.
Advantages of the RIP-chip method.
The RIP-chip method used in this study is a biochemical convention that allows the recovery of endogenous RNP complexes, which are more physiologically relevant than a single RBP bound to a single RNA. Isolation of entire RNPs also allows the identification of other components of these RNPs, including microRNAs (27). In addition, the RIP-chip method allows the study of RNPs during dynamic situations where RNP constituents may be changing, possibly resulting in different functional outcomes. RIP-chip is a relatively simple method with optimized conditions that provide an environment where all of the components necessary for an RBP's binding and function are present.
Although the RIP-chip procedure used in this study was similar to that used in previous studies, some differences are noteworthy. After determining t scores of Pum1 IP versus negative IP for all of the probes on the microarrays, we used Gaussian mixture modeling to objectively determine the two distributions representing the Pum1-associated and background messages. GMM allowed us to make a more sophisticated, probabilistic determination of which probes were Pum1 associated and which represented background. By calculating probabilities of association, this method also allows for more advanced downstream analysis of target mRNAs, as probabilities of association can be used rather than a simple discrete definition of which mRNAs are targets. For example, GSEA would not have been possible by simply defining which mRNAs were targets, because it depended on the continuous metric of a rank-ordered list of mRNAs created from LOD scores.
Pum1 and the RNA regulon model.
The observation that many of the mRNAs associated with Pum1 belong to a relatively small number of functional groupings is consistent with the RNA operon/regulon model (22, 26, 28). This model describes how multiple genes with related functions can be coordinately regulated at the level of the mRNA. Indeed, many aspects of the RNA regulon model are reflected in the results from this study. For example, one key aspect is that posttranscriptional regulation at the level of the mRNA accommodates the multifunctionality of eukaryotic proteins by allowing a single gene to participate in multiple regulons (cis-combinatorial). Thus, although Pum1 targets are enriched for regulators of progression through the cell cycle yet we observed no function for Pum1 in cell cycle progression (data not shown), it is likely that these genes also function in processes that are not related to the normal progression of the cell cycle, and these alternate processes may in fact be affected by Pum1 perturbation. It will be interesting to determine whether Pum1 functions in the meiotic cell cycle, especially since PUF proteins are known to function in the germ line of various organisms, including mouse (33, 58, 61). The cell proliferation- and differentiation-related targets of Pum1 may reflect a role for Pum1 in stem cell biology, a proposed ancestral function of PUF proteins (57). Another key aspect of the RNA regulon model is that regulation of mRNAs can be trans-combinatorial, with multiple trans factors specifying the fate of a single mRNA. There is precedent for this combinatorial trans-regulation in yeast, where several mRNAs have been shown to be regulated by multiple PUF proteins (23, 51). This combinatorial regulation provides an explanation for the range of effects of Pum1 knockdown on target message stability, as each target message is likely regulated by various other RBPs and microRNAs. Although the level of Pum2 protein in HeLa cells is relatively low compared to some other commonly used cell lines (not shown), it is possible that Pum2 may act redundantly on some or all of the Pum1 target mRNAs, thus dampening the effect on stability seen during Pum1 knockdown. It is also likely that not every copy of a Pum1 target mRNA will be bound by a Pum1 protein, especially since Pum1 associates with mRNAs representing over 10% of expressed genes. Thus, only a portion of each Pum1 target will be stabilized by Pum1 knockdown, since not every copy would normally have been regulated by Pum1. The RNA regulon model also states that RNPs may be dynamic with changing cellular conditions, and thus it will be of interest to determine how stability and translation of Pum1 target messages change following cell stress, especially since we have observed a change in the localization of Pum1 protein upon both oxidative stress and heat shock. There is precedent for this condition-specific regulation in yeast, where Puf3 differentially regulates stability of target messages when the yeast are grown on different carbon sources (13). It will also be important to determine how the targets of human Pum1 and Pum2 change during development and stem cell maturation, processes known to involve PUF proteins in Drosophila (32, 33) and C. elegans (29).
Evolutionary rewiring of PUF-HD and PUF-USER.
Previous studies of RBPs that bind AU-rich elements were unable to draw strong conclusions regarding conservation of RNA-protein interactions among species for two main reasons. First, the RBPs in question bind RNA through the widely represented RRM motif, and thus true orthologues between species were difficult to discern (1). Second, the sequences to which AU-rich element RBPs bind are not generally unique and involve elements that use both sequence and structure (34). Neither of these problems presents itself when considering PUF proteins. The PUF-HD is extremely well-conserved across species, yet each species has few PUF genes; mammalian genomes contain only two (47). The sequence to which the PUF-HD binds is also very well-conserved, with a UGUR followed by an AU-rich sequence identified in most PUF-binding sites and the 8-nucleotide core motif, UGUAHAUA, almost identical between Puf3, Pumilio, and Pum1 (17, 18, 54). Comparison of target genes of these three proteins revealed that they likely regulate different processes; Puf3 binds messages of genes with mitochondrial function (17), but there is no enrichment for genes with mitochondrial functions among targets of Pumilio (18) or Pum1. Targets of Pumilio and Pum1 do not share specific functional relationships, and although there is some conservation of target genes, most of the Pum1 targets and Pumilio targets are different (Fig. 7). These observations show that the modules of the PUF-HD and cognate binding sequence have remained fixed through evolution, while the identities of target messages have changed phylogenetically. This study provides experimental evidence of a rewiring process that was predicted through analysis of conservation of potential posttranscriptional regulatory elements, showing how a conserved cis-trans interaction can be evolutionarily rewired to coordinate the expression of different subsets of genes in different species (8, 18, 21, 26, 36).
Pum1 as a regulator of regulators.
Many of the genes whose mRNAs are bound by Pum1 are known regulators of gene expression and cellular processes, and thus Pum1 could be described as a regulator of regulators (26, 36, 44). The GO category containing the most Pum1 targets is “regulation of biological processes,” and target genes of Pum1 contain genes that regulate gene expression at the transcriptional, posttranscriptional, and posttranslational levels. GO analysis revealed that transcription factor genes and ubiquitin cycle genes are enriched in Pum1 targets, and many of the target genes involved in nucleic acid metabolism encode proteins that bind and process RNA. Pum1 targets are also enriched for GTPase-mediated signal transduction genes and other genes involved in signaling pathways that themselves could be described as regulatory, as their activation or repression typically results in changes in gene expression. Thus, Pum1 could be described as a regulator of regulators, because it associates with genes that regulate multiple levels of gene expression, as well as genes encoding members of signaling pathways that trigger changes in gene expression.
Pum1's function as a regulator of regulators is also evident when observing genes that are not represented in Pum1 targets. The GO categories of electron transport and oxidoreductase were significantly depleted among Pum1 targets, indicating that Pum1 does not regulate mRNAs involved in processes that are not regulatory. Pum1 targets did not show significant enrichment of any GO categories involving metabolism (other than nucleic acid metabolism, which is related to transcription and RNA processing), again demonstrating the lack of a role for Pum1 in nonregulatory processes and supporting its role as a regulator of regulators.
Importance of Pum1 target mRNA identification for study of gene expression.
Posttranscriptional regulation of gene expression has been increasingly studied in recent years, although global aspects have not been widely explored. Study of PUF family RNA-binding proteins has shown how these proteins can regulate a variety of subsets of mRNAs in a variety of organisms, with results typically supporting the proposed ancestral role of PUF proteins in stem cell biology (57). Due to the likely functions of PUF proteins in human stem cells, it is of interest to determine the role of human PUF proteins in regulating gene expression. This work serves to vastly expand the body of knowledge concerning PUF protein mRNA targeting and function in mammals, which will aid future studies in elucidating how PUF proteins act to regulate gene expression at the posttranscriptional level, particularly in stem cell biology, growth, and differentiation.
Supplementary Material
Acknowledgments
We thank Nancy Kedersha for generous contribution of constructs encoding stress granule and p-body markers and intellectual input, Blanche Capel and her group for assistance with immunofluorescence, Uwe Ohler and William Majoros for access to the 3′UTR database, Sayan Mukherjee for valuable advice on statistical analyses, and Shelton Bradrick and Keene lab members for helpful comments regarding the manuscript.
Footnotes
Published ahead of print on 14 April 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anantharaman, V., E. V. Koonin, and L. Aravind. 2002. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 301427-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, P., and N. Kedersha. 2006. RNA granules. J. Cell Biol. 172803-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Asaoka-Taguchi, M., M. Yamada, A. Nakamura, K. Hanyu, and S. Kobayashi. 1999. Maternal Pumilio acts together with Nanos in germline development in Drosophila embryos. Nat. Cell Biol. 1431-437. [DOI] [PubMed] [Google Scholar]
- 4.Bachorik, J. L., and J. Kimble. 2005. Redundant control of the Caenorhabditis elegans sperm/oocyte switch by PUF-8 and FBF-1, two distinct PUF RNA-binding proteins. Proc. Natl. Acad. Sci. USA 10210893-10897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bailey, T. L., N. Williams, C. Misleh, and W. W. Li. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34W369-W373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernstein, D., B. Hook, A. Hajarnavis, L. Opperman, and M. Wickens. 2005. Binding specificity and mRNA targets of a C. elegans PUF protein, FBF-1. RNA 11447-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buness, A., W. Huber, K. Steiner, H. Sultmann, and A. Poustka. 2005. arrayMagic: two-colour cDNA microarray quality control and preprocessing. Bioinformatics 21554-556. [DOI] [PubMed] [Google Scholar]
- 8.Chan, C. S., O. Elemento, and S. Tavazoie. 2005. Revealing posttranscriptional regulatory elements through network-level conservation. PLoS Comput. Biol. 1e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Crittenden, S. L., D. S. Bernstein, J. L. Bachorik, B. E. Thompson, M. Gallegos, A. G. Petcherski, G. Moulder, R. Barstead, M. Wickens, and J. Kimble. 2002. A conserved RNA-binding protein controls germline stem cells in Caenorhabditis elegans. Nature 417660-663. [DOI] [PubMed] [Google Scholar]
- 10.Crittenden, S. L., C. R. Eckmann, L. Wang, D. S. Bernstein, M. Wickens, and J. Kimble. 2003. Regulation of the mitosis/meiosis decision in the Caenorhabditis elegans germline. Philos. Trans. R. Soc. Lond. B 3581359-1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeGregori, J., and D. G. Johnson. 2006. Distinct and overlapping roles for E2F family members in transcription, proliferation and apoptosis. Curr. Mol. Med. 6739-748. [DOI] [PubMed] [Google Scholar]
- 12.Edwards, T. A., S. E. Pyle, R. P. Wharton, and A. K. Aggarwal. 2001. Structure of Pumilio reveals similarity between RNA and peptide binding motifs. Cell 105281-289. [DOI] [PubMed] [Google Scholar]
- 13.Foat, B. C., S. S. Houshmandi, W. M. Olivas, and H. J. Bussemaker. 2005. Profiling condition-specific, genome-wide regulation of mRNA stability in yeast. Proc. Natl. Acad. Sci. USA 10217675-17680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forbes, A., and R. Lehmann. 1998. Nanos and Pumilio have critical roles in the development and function of Drosophila germline stem cells. Development 125679-690. [DOI] [PubMed] [Google Scholar]
- 15.Fox, M., J. Urano, and R. A. Reijo Pera. 2005. Identification and characterization of RNA sequences to which human PUMILIO-2 (PUM2) and deleted in Azoospermia-like (DAZL) bind. Genomics 8592-105. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Rodriguez, L. J., A. C. Gay, and L. A. Pon. 2007. Puf3p, a Pumilio family RNA binding protein, localizes to mitochondria and regulates mitochondrial biogenesis and motility in budding yeast. J. Cell Biol. 176197-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerber, A. P., D. Herschlag, and P. O. Brown. 2004. Extensive association of functionally and cytotopically related mRNAs with Puf family RNA-binding proteins in yeast. PLoS Biol. 2E79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerber, A. P., S. Luschnig, M. A. Krasnow, P. O. Brown, and D. Herschlag. 2006. Genome-wide identification of mRNAs associated with the translational regulator PUMILIO in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1034487-4492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstrohm, A. C., B. A. Hook, D. J. Seay, and M. Wickens. 2006. PUF proteins bind Pop2p to regulate messenger RNAs. Nat. Struct. Mol. Biol. 13533-539. [DOI] [PubMed] [Google Scholar]
- 20.Goldstrohm, A. C., D. J. Seay, B. A. Hook, and M. Wickens. 2007. PUF protein-mediated deadenylation is catalyzed by Ccr4p. J. Biol. Chem. 282109-114. [DOI] [PubMed] [Google Scholar]
- 21.Halbeisen, R. E., A. Galgano, T. Scherrer, and A. P. Gerber. 2008. Post-transcriptional gene regulation: from genome-wide studies to principles. Cell Mol. Life Sci. 65798-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hieronymus, H., and P. A. Silver. 2004. A systems view of mRNP biology. Genes Dev. 182845-2860. [DOI] [PubMed] [Google Scholar]
- 23.Hook, B. A., A. C. Goldstrohm, D. J. Seay, and M. Wickens. 2007. Two yeast PUF proteins negatively regulate a single mRNA. J. Biol. Chem. 28215430-15438. [DOI] [PubMed] [Google Scholar]
- 24.Jackson, J. S., Jr., S. S. Houshmandi, F. Lopez Leban, and W. M. Olivas. 2004. Recruitment of the Puf3 protein to its mRNA target for regulation of mRNA decay in yeast. RNA 101625-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kedersha, N., and P. Anderson. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 43161-81. [DOI] [PubMed] [Google Scholar]
- 26.Keene, J. D. 2007. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 8533-543. [DOI] [PubMed] [Google Scholar]
- 27.Keene, J. D., J. M. Komisarow, and M. B. Friedersdorf. 2006. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protocols 1302-307. [DOI] [PubMed] [Google Scholar]
- 28.Keene, J. D., and S. A. Tenenbaum. 2002. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 91161-1167. [DOI] [PubMed] [Google Scholar]
- 29.Kimble, J., and S. L. Crittenden. 2007. Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans. Annu. Rev. Cell Dev. Biol. 23405-433. [DOI] [PubMed] [Google Scholar]
- 30.Kraemer, B., S. Crittenden, M. Gallegos, G. Moulder, R. Barstead, J. Kimble, and M. Wickens. 1999. NANOS-3 and FBF proteins physically interact to control the sperm-oocyte switch in Caenorhabditis elegans. Curr. Biol. 91009-1018. [DOI] [PubMed] [Google Scholar]
- 31.Lee, M. H., B. Hook, G. Pan, A. M. Kershner, C. Merritt, G. Seydoux, J. A. Thomson, M. Wickens, and J. Kimble. 2007. Conserved regulation of MAP kinase expression by PUF RNA-binding proteins. PLoS Genet. 3e233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehmann, R., and C. Nusslein-Volhard. 1987. Involvement of the Pumilio gene in the transport of an abdominal signal in the Drosophila embryo. Nature 329167-170. [Google Scholar]
- 33.Lin, H., and A. C. Spradling. 1997. A novel group of Pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 1242463-2476. [DOI] [PubMed] [Google Scholar]
- 34.Lopez de Silanes, I., M. Zhan, A. Lal, X. Yang, and M. Gorospe. 2004. Identification of a target RNA motif for RNA-binding protein HuR. Proc. Natl. Acad. Sci. USA 1012987-2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majoros, W. H., and U. Ohler. 2007. Spatial preferences of microRNA targets in 3′ untranslated regions. BMC Genomics 8152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mesarovic, M. D., S. N. Sreenath, and J. D. Keene. 2004. Search for organising principles: understanding in systems biology. Syst. Biol (Stevenage) 119-27. [DOI] [PubMed] [Google Scholar]
- 37.Mi, H., N. Guo, A. Kejariwal, and P. D. Thomas. 2007. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 35D247-D252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mootha, V. K., C. M. Lindgren, K.-F. Eriksson, A. Subramanian, S. Sihag, J. Lehar, P. Puigserver, E. Carlsson, M. Ridderstrale, E. Laurila, N. Houstis, M. J. Daly, N. Patterson, J. P. Mesirov, T. R. Golub, P. Tamayo, B. Spiegelman, E. S. Lander, J. N. Hirschhorn, D. Altshuler, and L. C. Groop. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34267-273. [DOI] [PubMed] [Google Scholar]
- 39.Murata, Y., and R. P. Wharton. 1995. Binding of pumilio to maternal hunchback mRNA is required for posterior patterning in Drosophila embryos. Cell 80747-756. [DOI] [PubMed] [Google Scholar]
- 40.Nakahata, S., Y. Katsu, K. Mita, K. Inoue, Y. Nagahama, and M. Yamashita. 2001. Biochemical identification of Xenopus Pumilio as a sequence-specific cyclin B1 mRNA-binding protein that physically interacts with a Nanos homolog, Xcat-2, and a cytoplasmic polyadenylation element-binding protein. J. Biol. Chem. 27620945-20953. [DOI] [PubMed] [Google Scholar]
- 41.Olivas, W., and R. Parker. 2000. The Puf3 protein is a transcript-specific regulator of mRNA degradation in yeast. EMBO J. 196602-6611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Parisi, M., and H. Lin. 1999. The Drosophila Pumilio gene encodes two functional protein isoforms that play multiple roles in germline development, gonadogenesis, oogenesis and embryogenesis. Genetics 153235-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson, K. 1894. Contributions to the mathematical theory of evolution. Philos. Trans. R. Soc. Lond. A 18571-110. [Google Scholar]
- 44.Pullmann, R., Jr., H. H. Kim, K. Abdelmohsen, A. Lal, J. L. Martindale, X. Yang, and M. Gorospe. 2007. Analysis of turnover and translation regulatory RNA-binding protein expression through binding to cognate mRNAs. Mol. Cell. Biol. 276265-6278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sonoda, J., and R. P. Wharton. 2001. Drosophila Brain tumor is a translational repressor. Genes Dev. 15762-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spassov, D. S., and R. Jurecic. 2002. Cloning and comparative sequence analysis of PUM1 and PUM2 genes, human members of the Pumilio family of RNA-binding proteins. Gene 299195-204. [DOI] [PubMed] [Google Scholar]
- 47.Spassov, D. S., and R. Jurecic. 2003. The PUF family of RNA-binding proteins: does evolutionarily conserved structure equal conserved function? IUBMB Life 55359-366. [DOI] [PubMed] [Google Scholar]
- 48.Spik, A., S. Oczkowski, A. Olszak, P. Formanowicz, J. Blazewicz, and J. Jaruzelska. 2006. Human fertility protein PUMILIO2 interacts in vitro with testis mRNA encoding Cdc42 effector 3 (CEP3). Reprod. Biol. 6103-113. [PubMed] [Google Scholar]
- 49.Subramanian, A., P. Tamayo, V. K. Mootha, S. Mukherjee, B. L. Ebert, M. A. Gillette, A. Paulovich, S. L. Pomeroy, T. R. Golub, E. S. Lander, and J. P. Mesirov. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 10215545-15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tenenbaum, S. A., C. C. Carson, P. J. Lager, and J. D. Keene. 2000. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc. Natl. Acad. Sci. USA 9714085-14090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ulbricht, R. J., and W. M. Olivas. 2007. Puf1p acts in combination with other yeast Puf proteins to control mRNA stability. RNA 14246-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vessey, J. P., A. Vaccani, Y. Xie, R. Dahm, D. Karra, M. A. Kiebler, and P. Macchi. 2006. Dendritic localization of the translational repressor Pumilio 2 and its contribution to dendritic stress granules. J. Neurosci. 266496-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Walser, C. B., G. Battu, E. F. Hoier, and A. Hajnal. 2006. Distinct roles of the Pumilio and FBF translational repressors during C. elegans vulval development. Development 1333461-3471. [DOI] [PubMed] [Google Scholar]
- 54.Wang, X., J. McLachlan, P. D. Zamore, and T. M. Hall. 2002. Modular recognition of RNA by a human Pumilio-homology domain. Cell 110501-512. [DOI] [PubMed] [Google Scholar]
- 55.Wharton, R. P., J. Sonoda, T. Lee, M. Patterson, and Y. Murata. 1998. The Pumilio RNA-binding domain is also a translational regulator. Mol. Cell 1863-872. [DOI] [PubMed] [Google Scholar]
- 56.White, E. K., T. Moore-Jarrett, and H. E. Ruley. 2001. PUM2, a novel murine Puf protein, and its consensus RNA-binding site. RNA 71855-1866. [PMC free article] [PubMed] [Google Scholar]
- 57.Wickens, M., D. S. Bernstein, J. Kimble, and R. Parker. 2002. A PUF family portrait: 3′UTR regulation as a way of life. Trends Genet. 18150-157. [DOI] [PubMed] [Google Scholar]
- 58.Xu, E. Y., R. Chang, N. A. Salmon, and R. A. Reijo Pera. 2007. A gene trap mutation of a murine homolog of the Drosophila stem cell factor Pumilio results in smaller testes but does not affect litter size or fertility. Mol. Reprod. Dev. 74912-921. [DOI] [PubMed] [Google Scholar]
- 59.Young, D. S., D. R. Hunter, R. T. Elmore, F. Xuan, T. P. Hettmansperger, and H. Thomas. 2007. The Mixtools package: tools for mixture models. R Package version 0.2.0. http://www.r-project.org.
- 60.Zamore, P. D., J. R. Williamson, and R. Lehmann. 1997. The Pumilio protein binds RNA through a conserved domain that defines a new class of RNA-binding proteins. RNA 31421-1433. [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang, B., M. Gallegos, A. Puoti, E. Durkin, S. Fields, J. Kimble, and M. P. Wickens. 1997. A conserved RNA-binding protein that regulates sexual fates in the C. elegans hermaphrodite germ line. Nature 390477-484. [DOI] [PubMed] [Google Scholar]
- 62.Zhang, B., S. Kirov, and J. Snoddy. 2005. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 33W741-W748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhang, H., J. Hu, M. Recce, and B. Tian. 2005. PolyA_DB: a database for mammalian mRNA polyadenylation. Nucleic Acids Res. 33D116-D120. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.