Abstract
The SNAIL transcription factor contains C-terminal tandem zinc finger motifs and an N-terminal SNAG repression domain. The members of the SNAIL family have recently emerged as major contributors to the processes of development and metastasis via the regulation of epithelial-mesenchymal transition events during embryonic development and tumor progression. However, the mechanisms by which SNAIL represses gene expression are largely undefined. Previously we demonstrated that the AJUBA family of LIM proteins function as corepressors for SNAIL and, as such, may serve as a platform for the assembly of chromatin-modifying factors. Here, we describe the identification of the protein arginine methyltransferase 5 (PRMT5) as an effector recruited to SNAIL through an interaction with AJUBA that functions to repress the SNAIL target gene, E-cadherin. PRMT5 binds to the non-LIM region of AJUBA and is translocated into the nucleus in a SNAIL- and AJUBA-dependent manner. The depletion of PRMT5 in p19 cells stimulates E-cadherin expression, and the SNAIL, AJUBA, and PRMT5 ternary complex can be found at the proximal promoter region of the E-cadherin gene, concomitant with increased arginine methylation of histones at the locus. Together, these data suggest that PRMT5 is an effector of SNAIL-dependent gene repression.
The SNAG family of zinc finger transcription factors in vertebrates include GFI-1A, GFI-1B, the insulinoma-associated protein IA-1, the homeobox protein GSH-1, and the SNAIL/SLUG family. These proteins play important roles in the regulation of development, stem cell self-renewal, and tumor progression (5, 22, 49). They share a common set of functional domains: a C-terminal DNA binding domain composed of five to seven Cys2-His2 zinc fingers and a highly conserved N-terminal repression domain designated SNAG. The SNAG motif was first identified from the GFI-1 protein and comprises the first 21 amino acid residues in the N terminus. The SNAG domain is a potent and transferable repression motif (22, 49). However, unlike other repression domains which are associated with zinc finger proteins, such as the KRAB domain and the BTB-POZ domain, whose mechanisms of repression are well established, little is known about the mechanisms of the SNAG domain-mediated repression (9, 15).
The SNAIL protein has emerged as a potent regulator of the processes of embryonic development and tumor progression through the regulation of the epithelial-mesenchymal transition (EMT) (5, 36). In mammalian cells, SNAIL induces EMT at least partially through repression of the E-cadherin gene, thereby altering cell adhesion (6). The SNAIL protein has been found in multiprotein complexes containing histone deacetylases (HDACs), mSIN3A, and LOXL2/3 (39, 40). However, the biological significance of these interactions and how SNAIL mediates functional protein complex assembly at specific promoters in the context of chromatin remain undefined.
We previously identified novel corepressors that directly bind to the SNAG domains of GFI-1 and SNAIL by using yeast two-hybrid assays (3). The AJUBA family of LIM pro teins were identified as prospective candidates which bind to the minimal SNAG domain (3). AJUBA is a multiple LIM domain-containing protein and belongs to the AJUBA/zyxin family of LIM proteins (19). This family includes the AJUBA subfamily AJUBA, LIMD1, and WTIP and the zyxin subfamily zyxin, LPP, and TRIP6. Only the AJUBA subfamily, and not the zyxin subfamily, associated with SNAG domain-containing proteins (31). The AJUBA/zyxin family is characterized by three tandem C-terminal LIM domains and unique N-terminal regions designated the PreLIM regions (19, 26). The AJUBA protein is predominantly cytoplasmic, yet is recruited to E-cadherin-adhesive complexes during epithelium formation and can shuttle between the nucleus and cytoplasm (27). The AJUBA protein may function as a scaffold protein to assemble multiple cytoplasmic protein complexes involved in the processes of cell adhesion, migration, mitosis, and cell differentiation (14, 19, 23). However, its role in the nucleus as a regulator of gene expression is poorly defined.
The in vitro and in vivo studies of the interaction between AJUBA and SNAIL demonstrated that AJUBA functions as a SNAIL corepressor to repress the E-cadherin gene and is recruited to the endogenous E-cadherin promoter in a SNAIL-dependent manner (31). The expression of AJUBA orthologs during the development of Xenopus parallels that of SNAIL, and AJUBA orthologs cooperate with SNAIL and SLUG during the development of the neural crest in Xenopus (31). Since AJUBA itself does not contain an apparent enzymatic activity, we postulated that AJUBA may recruit other effectors to the SNAG domain of SNAIL to modify chromatin structure.
In this study, we purified AJUBA-interacting proteins and we describe the protein arginine methyltransferase 5 (PRMT5) as a candidate in this role. PRMT5 is a type II protein arginine methyltransferase and plays important roles in the regulation of gene transcription (29). Our studies provide strong evidence that PRMT5 is a key component of the SNAIL-silencing complex through binding to AJUBA.
MATERIALS AND METHODS
Plasmids.
The Myc epitope-tagged pMEX-Myc-Ajuba plasmids have been described previously (19). All AJUBA mutants and truncations were made by using QuikChange site-directed mutagenesis procedures following the manufacturer's protocol (Stratagene, La Jolla, CA), and all mutants were confirmed by DNA sequencing. The AJUBA cDNAs were subcloned from pMEX-Myc-Ajuba via digestion with BamHI and XhoI and inserted into the pcDNA3.1-N-Flag vector to create a Flag epitope-tagged AJUBA fusion protein in the N terminus. The pcDNA-RFP-Ajuba and pGL2-E-cadherin luciferase reporter plasmids have been previously described (42, 45). The pcDNA-Flag-Prmt5 plasmid was provided by G. Dreyfuss (16). The Sport6-CMV-Snail (murine) plasmid was purchased from Open Biosystems (Huntsville, AL).
The glutathione S-transferase (GST)-AJUBA protein GST-AJUBA (aa 244-350) was generated by PCR amplification of the DNA fragment encoding amino acid residues 244 to 350 of murine AJUBA. The GST-AJUBA mutant (GST-AJUBA LLL-AAA) containing leucine to alanine mutations was generated by using a site-directed mutagenesis kit (Invitrogen). The PCR products were cloned into the BamHI and EcoRI sites of pGEX-4T-1. The truncated Prmt5 plasmids containing PRMT5 (aa 1-170), PRMT5 (aa 169-422), and PRMT5 (aa 421-637) were generated by amplifying the DNA fragment encoding the indicated residues of human PRMT5 and cloning the resulting fragments into the pET-28a vector.
Cell culture, transfections, and luciferase reporter assays.
HEK293 cells, U2OS cells, and p19 cells were maintained in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum, 2 mM l-glutamine, and penicillin (50 U/ml)-streptomycin (50 μg/ml) at 37°C under 5% CO2 in a humidified chamber.
For transfection, HEK293 cells were seeded at 5 × 104 cells per well in 24-well plates. The β-galactosidase plasmid (50 ng) and pGL2-E-cad-Luc reporter (200 ng), along with SNAIL- and/or AJUBA-encoding plasmids, were transiently transfected into the cells with Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA). Twenty-four hours posttransfection, cells were harvested and lysed. The luciferase and β-galactosidase activities were measured with a luciferase reporter gene assay kit (Promega, Madison, WI) and a β-galactosidase kit (Clontech, Mountain View, CA), respectively. The transfection efficiency among plates was normalized to the β-galactosidase activity level, and all transfections were repeated three times in duplicate.
Affinity purification of a native AJUBA complex and size fractionation.
To purify AJUBA-associated proteins, a Flag-tagged, full-length AJUBA cDNA in the pcDNA3.1 vector was stably expressed in HEK293 cells. Single-cell clones were selected with G418 and screened by Western blotting using anti-Flag antibody. A cell clone expressing the Flag-AJUBA protein at a level comparable to that of the endogenous AJUBA was chosen for the purification. A total of 5 × 109 cells were lysed in buffer A containing 20 mM Tris-HCl (pH 8.0), 150 mM NaCl, 2.5 mM EDTA, 0.5% NP-40, 0.2 mM phenylmethylsulfonyl fluoride (PMSF), and 0.5 mM dithiothreitol. Cell lysates were precleared with the protein A-agarose beads for 2 h and then incubated with the anti-Flag agarose M2 beads (Sigma, St. Louis, MO) at 0.5 ml of beads per 100 mg of cell lysate for 2 h to overnight with rotation. The M2 beads were washed four times with buffer BC500 containing 20 mM Tris-HCl (pH 7.8), 500 mM KCl, 0.2 mM EDTA, 10% glycerol, 10 mM β-mercaptoethanol, 0.2% NP-40, 0.2 mM PMSF, and 1 μg/ml of aprotinin, leupeptin, and pepstatin. The protein complex was eluted with the Flag peptides (Sigma) at 100 μg/ml in buffer BC100 containing 20 mM Tris-HCl (pH 7.8), 50 mM KCl, 0.2 mM EDTA, 10% glycerol, 10 mM β-mercaptoethanol, 0.2 mM PMSF, and 1 μg/ml of aprotinin, leupeptin, and pepstatin. The eluted proteins were resolved on 4 to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels for Western blotting and silver and colloidal staining analyses. The proteins were excised from the gels and identified by standard mass spectrometry at the Wistar Institute Cancer Center Proteomics Core Facility.
The fractionation of the cell extracts (Superose 6) was carried out according to the manufacturer's instructions and has been described previously (20, 21). Briefly, the column was equilibrated in buffer BC500 prior to the loading of the cell extracts. HEK293 cells were harvested at 48 h posttransfection. Whole-cell extract (1 ml) was prepared in buffer A and was loaded onto the preequilibrated column, which was run at 0.35 ml/min in the cold room and was collected at 0.5 ml per fraction. The protein complex from each fraction was precipitated by using trichloroacetic acid and was resuspended in 100 μl of 0.1 N NaOH solution. To prepare the protein samples for Western blotting, 100 μl of 5× Laemmli buffer was added into each fraction and heated for 5 min, and 50 μl of the protein sample was resolved on a NuPAGE gel (Invitrogen). The proteins were visualized by Western blotting with anti-Myc and anti-Flag monoclonal antibodies.
Coimmunoprecipitation, Western blotting, immunofluorescence, and antibodies.
Myc-Ajuba, Flag-Prmt5, and/or Flag-Snail plasmids were transiently transfected into HEK293 cells, and at 24 h posttransfection, the cells were lysed in buffer A. Coimmunoprecipitations were performed with either anti-Myc or anti-Flag antibodies. The Western blotting and immunofluorescence analyses were previously described (24, 46). Mouse monoclonal anti-Myc (Invitrogen) and anti-Flag (Sigma) and rabbit polyclonal anti-SNAIL (Santa Cruz Biotechnology), anti-H4R3 (UPSTATE, Charlottesville, VA), and anti-E-cadherin (Cell Signaling, Danvers, MA) antibodies were purchased. The rabbit polyclonal anti-AJUBA antibody was raised by immunizing rabbits with a bacterially expressed six-His fusion protein of murine AJUBA (amino acids 1 to 216) as the antigen.
siRNA knockdown, methyltransferase inhibitor MTA treatment, and reverse transcriptase PCR.
Smart-pool small interfering RNAs (siRNAs) targeting murine Ajuba and Prmt5 (Dharmacon, Lafayette, CO) were transfected into the cells with the Lipofectamine 2000 reagent (Invitrogen). 5′-Deoxy-5′-methyl-thioadenosine (MTA; Sigma) was dissolved in dimethyl sulfoxide. p19 cells were seeded at 1.5 × 105 cells/well in 6-well cell culture plates on day 0. MTA was added into the medium on day 1 at concentrations of 100 μM and 200 μM for 48 h.
Total RNA from p19 cells was isolated with an RNeasy kit (Qiagen, Valencia, CA). The RNA was treated with RQ DNase I to remove any genomic DNA contamination. Two micrograms of the treated total RNA was used for cDNA synthesis in a 20-μl reaction mixture with Superscript II reverse transcriptase (Invitrogen). The primer pairs used for E-cadherin and GAPDH amplification were sense, 5′-GAGAACGGTGGTCAAAGAGC-3′, and antisense, 5′-CATCTCCCATGGTGCCACAC-3′, and sense, 5′-ACCACAGTCCATGCCATCAC-3′, and antisense, 5′-TCCACCCCCTGTTGCTGTA-3′, respectively. The PCR amplification was carried out by using Taq DNA polymerase (Promega) at 94°C for 15 s, 60°C for 15 s, and 72°C for 60 s.
ChIP.
Chromatin immunoprecipitation (ChIP) experiments were carried out in HEK293 cells stably expressing the Flag-SNAIL cDNA and in p19 cells. HEK293-Flag-SNAIL and HEK293-vector cell lines were established by the transfection of pcDNA3.1 Flag-SNAIL and parental vectors into HEK293 cells and selected with zeocin at 400 ng/ml. The expression of the Flag-SNAIL protein was confirmed by Western blotting. To prepare cells for ChIP, HEK293-Flag-SNAIL and HEK293-vector cells were grown in 150-mm plates to 70 to 90% confluence and fixed by the addition of 574 μl of 37% formaldehyde directly into 20 ml of growth medium to a final concentration of 1% for 20 min in the cell culture incubator. The cross-linking reaction was stopped by the addition of 1.25 ml of 2 M glycine in phosphate-buffered saline buffer at room temperature for 5 min. Cells were harvested, and the ChIP assays were performed according to the protocol supplied with the EZ-CHIP kit (Upstate; catalog no. 17-371). The immunoprecipitated DNAs were amplified with primer set 1: 5′-AATCAGAACCGTGCAGGTCC-3′ and 5′-ACAGGTGCTTTGCAGTTCCG-3′. This 250-bp amplicon flanks the three E-boxes located in the proximal promoter region of the E-cadherin gene. Primer set 2, 5′-GGCTCAAGCTATCCTTGCAC-3′ and 5′-GTGCAGTGGCTCATGTCTGT-3′, was used to amplify a 197-bp fragment carried by exon 16 of the E-cadherin gene. The PCR fragments were cloned, and their identities were confirmed by DNA sequencing. For quantitation, the PCR products were resolved on 2% agarose gels and visualized with ethidium bromide.
p19-siAjuba and p19-siLuc (small interfering luciferase) cells were established as previously described (31). Retroviral vectors containing short hairpin RNAs targeting murine Ajuba (GGAGAGCCGTCACTCGTAC) and the luciferase gene were introduced into p19 cells and selected with puromycin. The resulting pooled p19-siAjuba and p19-siLuc cells were verified by Western blotting and were used for ChIP assays. The ChIP assays were performed essentially as described above for HEK293 cells. The primer sets used to amplify the murine E-cadherin promoter were set 1 forward, 5′-AGACAGGGGTGGAGGAAGTT-3′, and reverse, 5′-GGGCAGGAGTCTAGCAGAAG-3′, and set 2 forward, 5′-AGGTATCTTGGTGTGGGTGCAACT-3′, and reverse, 5′-ACGCCAAGAAACTTAAGTGGTGCC-3′. The resulting DNA samples were analyzed with real-time PCR as described previously (32).
RESULTS
PRMT5 was identified as one of the AJUBA-interacting proteins.
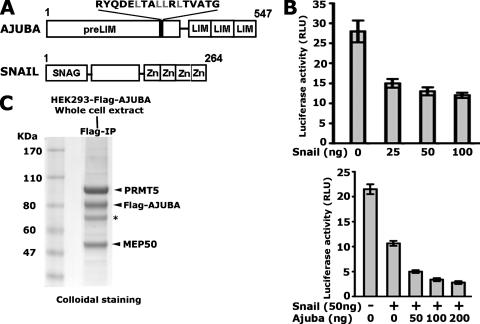
To isolate potential AJUBA-interacting proteins which may repress the SNAIL target genes, such as E-cadherin, we chose a cell system which supports SNAIL-mediated represssion. Since HEK293 cells have been widely used for the purification of protein complexes, we first tested whether SNAIL can repress E-cadherin expression in this cell line. The E-cadherin-Luc reporter contains three SNAIL binding sites, which are located in the proximal promoter region of the E-cadherin gene; the promoter is responsive to SNAIL in a variety of cell types (6, 39, 44). HEK293 cells were transiently transfected with SNAIL- and AJUBA-encoding plasmids alone or in combination with the E-cadherin-Luc reporter. SNAIL by itself was able to repress the transcription of the E-cadherin promoter-driven luciferase reporter in a dosage-dependent manner (Fig. 1B, top panel). When AJUBA and SNAIL were coexpressed in HEK293 cells, we observed increased repression over that with SNAIL alone (Fig. 1B, bottom panel). These results demonstrate that HEK293 cells contain the factors required for SNAIL and AJUBA to repress E-cadherin. To purify these factors, we selected a cell clone stably expressing the Flag-AJUBA protein at levels comparable to the endogenous levels and performed affinity purification (Fig. 1C and data not shown). As expected, the major protein eluted in the affinity purification that migrated at a molecular mass of 80 kDa was the Flag-AJUBA protein. Other proteins, migrating at 95 kDa, 70 kDa, and 50 kDa, were observed in multiple independent purification experiments. The major protein at 95 kDa was identified as PRMT5 and was pursued as a candidate AJUBA-interacting protein. The MEP50 (50 kDa) protein, a known cofactor for PRMT5, was also observed in the purification (17, 18).
FIG. 1.
Affinity purification of AJUBA-interacting proteins. (A) Diagram showing the architectures of the SNAIL and AJUBA proteins. The shaded bar in the PreLIM region contains a potential nuclear box motif. (B) AJUBA can augment SNAIL-mediated repression of the promoter-luciferase reporter activities of E-cadherin in HEK293 cells. Error bars show standard deviations. RLU, relative light units; +, present; −, absent. (C) Colloidal staining shows the potential AJUBA-interacting proteins purified from HEK293 cells. The asterisk shows vimentin. The prestained molecular mass marker (Benchmarker; Invitrogen) migrates 15 to 20 kDa faster than the predicted molecular size. Molecular sizes are shown on the left. IP, immunoprecipitation.
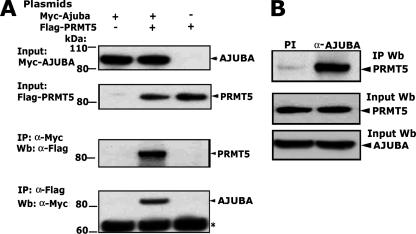
To confirm the interaction between AJUBA and PRMT5, we first transiently cotransfected full-length Myc-AJUBA and Flag-PRMT5 cDNAs into HEK293 cells. Reciprocal coimmunoprecipitations showed that PRMT5 and AJUBA can interact with each other in HEK293 cells (Fig. 2A). To further confirm that an AJUBA-PRMT5 interaction occurs with the endogenous proteins, whole-cell lysates from p19 cells were used for immunoprecipitation. Indeed, the endogenous AJUBA was able to immunoprecipitate the endogenous PRMT5 (Fig. 2B). Collectively, these data suggest that PRMT5 is a novel AJUBA-interacting protein.
FIG. 2.
PRMT5 is a novel AJUBA-interacting protein. (A) The exogenously expressed PRMT5 and AJUBA proteins interact in HEK293 cells. Molecular sizes are shown on the left. The asterisk shows nonspecific bands. (B) The endogenous AJUBA and PRMT5 proteins interact in p19 cells. Immunoprecipitation was performed with anti-AJUBA polyclonal antibody, and preimmune rabbit immunoglobulin G was used as control. The immunoprecipitated proteins were resolved on sodium dodecyl sulfate-polyacrylamide gel electrophoresis gels, and Western blotting was performed with monoclonal anti-PRMT5 antibody. +, present; −, absent; α, anti; IP, immunoprecipitation; Wb, Western blot; PI, preimmune serum.
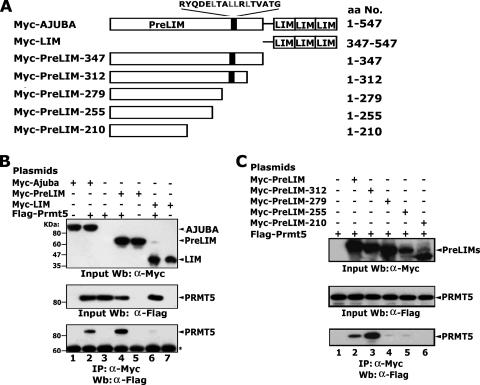
The PreLIM region of AJUBA binds to PRMT5.
The amino acid sequence of the AJUBA protein predicts two structurally distinct regions: the PreLIM region in the N terminus and the LIM domains in the C terminus (Fig. 3A). To determine which regions in AJUBA are responsible for the PRMT5 binding, plasmids encoding Myc-tagged, full-length AJUBA and the two truncated forms, PreLIM and LIM, were cotransfected with plasmid encoding Flag-PRMT5 into HEK293 cells. The results of the immunoprecipitation experiments showed that the PreLIM region bound PRMT5 while the LIM region did not (Fig. 3B, lanes 2, 4, and 6). To further identify which specific motifs or residues in the PreLIM region are critical for the interaction between AJUBA and PRMT5, we made progressive C-terminal deletions in the PreLIM region (Fig. 3A). The results of these experiments confirmed that PreLIM bound to PRMT5. Interestingly, the PreLIM protein comprising amino acids 1 to 312 showed stronger binding than the complete PreLIM region, suggesting that the region encompassing amino acids 312 to 347 may contain inhibitory motifs that limit PRMT5 binding (Fig. 3C, lanes 2 and 3). The proteins comprising amino acids 1 to 279, 1 to 255, and 1 to 210 of PreLIM failed to bind PRMT5. These data indicate that the region between residues 279 and 312 of AJUBA is essential for PRMT5 binding.
FIG. 3.
A single domain in the PreLIM region is essential for PRMT5 binding. (A) Diagram showing the progressive deletions of AJUBA. (B) The PreLIM region of AJUBA binds to PRMT5. Molecular sizes are shown on the left. The asterisk shows nonspecific bands. (C) The region between amino acid residues 279 and 312 of AJUBA binds to PRMT5. Plasmids were transfected into HEK293 cells. Immunoprecipitations (IP) were carried out with anti-Myc antibody, and Western blotting (Wb) was performed with anti-Flag antibody. +, present; −, absent; α, anti.
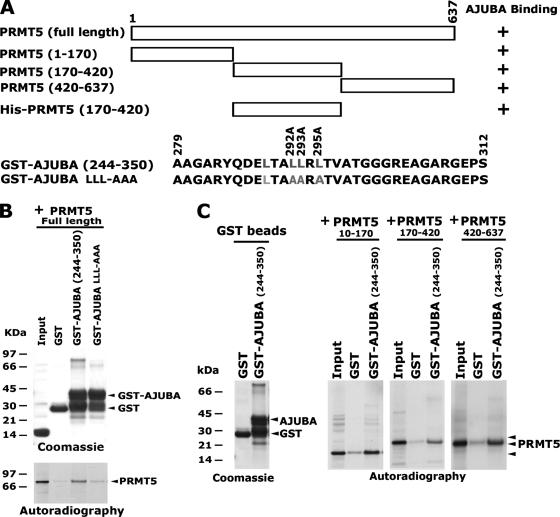
To identify the regions in the PRMT5 protein that can interact with AJUBA, we used the full-length PRMT5 and three truncated mutants (Fig. 4A). These fragments were translated in vitro and used for GST-AJUBA (aa 244-350) binding assays. The full-length PRMT5 and the three truncated mutants bound to wild-type (WT) GST-AJUBA, but not to the mutant GST-AJUBA LLL-AAA (Fig. 4B and C). These data suggest that multiple domains in the PRMT5 protein can interact with AJUBA. To further determine whether AJUBA can directly bind PRMT5, we used bacterially expressed GST-AJUBA and His-PRMT5 (aa 170-420) proteins for in vitro binding assays. The results demonstrated that GST-AJUBA (aa 244-350) directly bound His-PRMT5 (aa 170-420), while the AJUBA mutant showed no binding (data not shown).
FIG. 4.
Multiple domains in PRMT5 interact with AJUBA. (A) Diagram showing the full-length PRMT5, its truncations, and the GST-AJUBA constructs. +, positive. (B) Full-length PRMT5 binds WT GST-AJUBA, but not the mutant. Molecular sizes are shown on the left. +, present. (C) All three PRMT5 truncations can bind the WT GST-AJUBA. Bacterially expressed and purified GST-AJUBA (aa 244-350) and its mutant with mutations of leucines to alanines were incubated with in vitro-translated PRMT5 proteins. Molecular sizes are shown on the left. +, present.
SNAIL, AJUBA, and PRMT5 are found in the same complex.
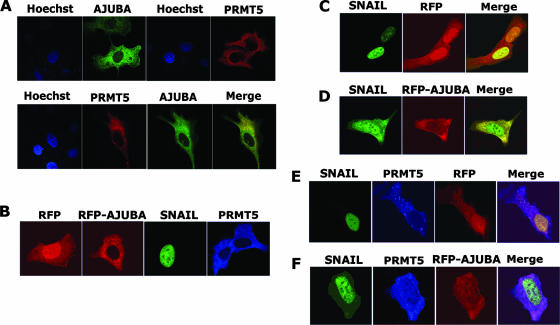
To determine if the AJUBA protein functions as an adaptor to bridge SNAIL and PRMT5 proteins, we first performed immunofluorescence assays to detect the subcellular localization of AJUBA and PRMT5. Plasmids encoding Myc-AJUBA and Flag-PRMT5 were transiently transfected into U2OS cells. When expressed alone, both the AJUBA and PRMT5 proteins were predominantly cytoplasmic, with similar distribution patterns (Fig. 5A, top panel). When the proteins were coexpressed, the distribution of each protein was not significantly changed (Fig. 5A, bottom panel). To further visualize their subcellular localization in the presence of SNAIL, we transfected plasmids encoding SNAIL, Flag-PRMT5, red fluorescent protein (RFP)-AJUBA, and/or pcDNA-RFP into U2OS cells. Consistent with previous observations, when expressed alone the SNAIL protein was localized in the nucleus, while the AJUBA and PRMT5 proteins were predominantly cytoplasmic. The signal from RFP alone was found in both the cytoplasm and the nucleus. The RFP-AJUBA fusion protein showed localization identical to that of the WT AJUBA (Fig. 5B). However, the coexpression of AJUBA and SNAIL affected the localization of both: the SNAIL protein was retained in the cytoplasm, and concomitantly, a significant amount of the AJUBA protein was localized in the nucleus (Fig. 5D). In contrast, the coexpression of PRMT5 and SNAIL revealed no apparent effect on either localization (Fig. 5E). Strikingly, when AJUBA, SNAIL, and PRMT5 were coexpressed, a significant amount of the PRMT5 protein was relocated to the nucleus and colocalized with nuclear SNAIL and AJUBA (Fig. 5F). These observations suggest that an interaction exists between SNAIL and PRMT5 in the presence of AJUBA.
FIG. 5.
PRMT5 is translocated into the nucleus through interaction with AJUBA and SNAIL. (A) Subcellular localization of AJUBA and PRMT5 in U2OS cells. The plasmids encoding Myc-AJUBA and Flag-PRMT5 were transiently transfected into U2OS cells, and immunofluorescent images were taken with confocal microscopy. AJUBA is predominately cytoplasmic when expressed alone, and PRMT5 is cytoplasmic with a distribution pattern similar to that of AJUBA (top panel). The coexpression of AJUBA and PRMT5 is shown in the bottom panel. (B) Individual expression of PRMT5, SNAIL, RFP-AJUBA, and RFP in U2OS cells. When expressed alone, the SNAIL protein is localized in the nucleus, while the RFP-AJUBA and PRMT5 proteins are in the cytoplasm; RFP alone is found in both cytoplasm and nucleus. (C) The coexpression of RFP and SNAIL does not affect their localization. (D) The coexpression of AJUBA and SNAIL affects both of their localizations: the SNAIL protein is retained in the cytoplasm, and concomitantly, a significant amount of the AJUBA protein is localized in the nucleus. (E) The coexpression of PRMT5, RFP, and SNAIL reveals no apparent effect on either localization. (F) The coexpression of RFP-AJUBA, SNAIL, and PRMT5 results in a significant amount of the PRMT5 protein being relocated to the nucleus and colocalized with nuclear SNAIL and AJUBA.
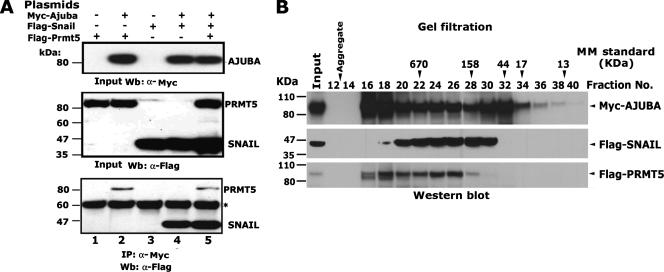
To verify such an interaction, we performed coimmunoprecipitation to determine whether the SNAIL, AJUBA, and PRMT5 proteins can form a ternary complex. We transfected the Myc-AJUBA-encoding plasmid, together with the Flag-SNAIL- and Flag-PRMT5-encoding plasmids, into HEK293 cells. We observed that AJUBA could simultaneously interact with PRMT5 and SNAIL (Fig. 6A, lane 5). To further confirm this observation, we performed size exclusion fractionation of whole-cell extracts prepared from HEK293 cells expressing Myc-AJUBA, Flag-SNAIL, and Flag-PRMT5. The AJUBA protein eluted in three well-defined peaks (Fig. 6B, fractions 16 to 18, 24 to 26, and 32), whereas SNAIL eluted in a single peak (fractions 24 to 26), and PRMT5 eluted in two peaks (fractions 18 and 24 to 26). These observations suggest that SNAIL, AJUBA, and PRMT5 can form a ternary protein complex in vivo and can be coeluted in fractions 24 to 26. Additionally, AJUBA and PRMT5 may form additional complexes of higher complexity, as evidenced by their coelution in fraction 18.
FIG. 6.
The AJUBA protein can simultaneously immunoprecipitate SNAIL and PRMT5. (A) Coimmunoprecipitation of the SNAIL, AJUBA, and PRMT5 proteins. The plasmids encoding Myc-AJUBA, Flag-SNAIL, and Flag-PRMT5 were transiently transfected into HEK293 cells, and immunoprecipitation (IP) was done with anti-Myc antibody and Western blotting (Wb) with anti-Flag antibody. The asterisk shows nonspecific bands. +, present; −, absent; α, anti. (B) Western blots show the eluted SNAIL, AJUBA, and PRMT5 proteins from the Superose 6 sizing column. Whole-cell extract (8 mg) was prepared from HEK293 cells transiently expressing Myc-AJUBA, Flag-SNAIL, and Flag-PRMT5 and was loaded onto a Superose 6 gel filtration column. Molecular sizes are shown on the left. MM, molecular mass.
SNAIL, AJUBA, and PRMT5 bind to the endogenous promoter of E-cadherin.
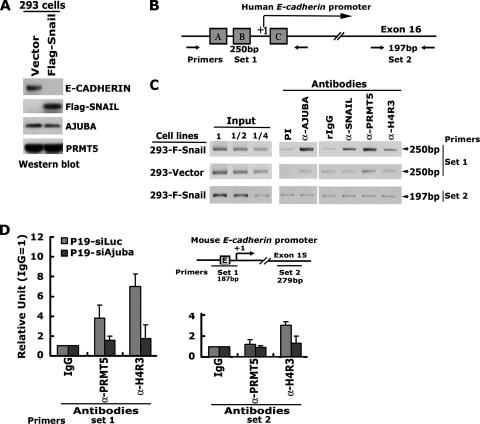
To examine whether SNAIL, AJUBA, and PRMT5 form a functional multiprotein complex to repress the SNAIL target gene in living cells, we employed ChIP assays to examine their association with the endogenous promoter of the E-cadherin gene. Clonal HEK293-Flag-SNAIL cells were established by stable transfection, and protein expression was confirmed by Western blotting (Fig. 7A). The expression of exogenous Flag-SNAIL induced morphological changes in HEK293 cells, as has been observed in other cell types (data not shown) (1, 8, 25, 48). Downregulation of the E-cadherin gene expression was observed in Flag-SNAIL-transfected HEK293 cells (Fig. 7A). The immunoprecipitated DNA fragments were examined by PCR amplification using primer sets 1 and 2 (Fig. 7B). We observed that the proximal promoter of the E-cadherin gene flanking the three SNAIL binding sites was highly enriched by antibodies to SNAIL, AJUBA, and PRMT5 in HEK293-Flag-SNAIL cells. However, in the HEK293-vector cells this enrichment was not observed (Fig. 7C). Using an antibody which detects methylated histone H4 at arginine 3 (H4R3), we detected increased H4 methylation at the promoter of the E-cadherin gene (Fig. 7C). The fragment residing in exon 16 was not enriched by any of these antibodies. Together, these data suggest that the association of SNAIL, AJUBA, and PRMT5 with the E-cadherin gene occurs at the proximal promoter of the E-cadherin gene.
FIG. 7.
SNAIL, AJUBA, and PRMT5 are associated with the E-cadherin gene at the proximal promoter. (A) Western blot analysis of the expression of the Flag-SNAIL, AJUBA, PRMT5, and E-cadherin proteins in HEK293 cells indicates that the E-cadherin gene is downregulated by the overexpression of Flag-SNAIL. (B) Diagram illustrating the human E-cadherin promoter and the PCR primers used for ChIP. A, B, and C in the boxes indicate the three SNAIL-binding sites in the promoter. (C) PCR analysis of the immunoprecipitated DNA fragments. Molecular sizes are shown on the right. PI, preimmune serum. Numbers above the input lanes indicate dilutions of the input DNAs in water (1, no dilution). (D) Analysis of the ChIP assay results using real-time PCR in p19-siAjuba and p19-siLuc cells, and diagram illustrating the murine E-cadherin promoter and the PCR primers used for ChIP. E in the box in the diagram indicates the SNAIL binding site. Error bars show standard deviations. IgG, immunoglobulin G; α, anti.
We have shown that short hairpin RNA-mediated knockdown of AJUBA in p19 cells resulted in the upregulation of E-cadherin (31). Thus, we asked whether endogenous SNAIL and AJUBA can be found at the promoter region of the E-cadherin gene in p19 cells. We performed ChIP assays of p19-siAjuba and control p19-siLuc cells. Similar to the results seen for HEK293 cells, both SNAIL and AJUBA can bind to the proximal region of the E-cadherin promoter in p19-siLuc cells. However, in p19-siAjuba cells SNAIL binding to the E-cadherin promoter decreased, and no AJUBA was found to bind to this region (31). The PRMT5 protein was also found to bind to the proximal region of the E-cadherin promoter, and there was increased H4 methylation at the same locus in p19-siLuc cells, while knockdown of AJUBA abolished PRMT5 binding and decreased H4 methylation at the E-cadherin promoter in p19-siAjuba cells (Fig. 7D). Taken together, these data suggest that endogenous SNAIL, AJUBA, and PRMT5 proteins can bind to the same locus in the E-cadherin promoter and that AJUBA is required for PRMT5 binding to this region.
Modulation of AJUBA and PRMT5 in p19 cells results in upregulation of E-cadherin expression.
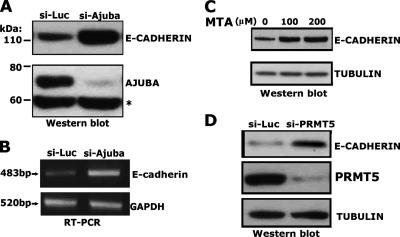
Since the nucleus-localized SNAIL-AJUBA-PRMT5 complex can be found at the E-cadherin promoter, we sought to determine whether this well-established SNAIL target gene was targeted by this ternary complex. It has been previously found that the induction of SNAIL protein in p19 cells repressed E-cadherin gene expression, while the depletion of SNAIL protein resulted in the upregulation of the SNAIL target genes (10, 35). To test the roles of AJUBA and PRMT5 in the regulation of the expression of endogenous E-cadherin, an siRNA targeting murine Ajuba was transfected into the p19 cells. The level of the AJUBA protein was significantly decreased by the siRNA (Fig. 8A). Moreover, the expression of E-cadherin at both the mRNA and protein levels was significantly increased (Fig. 8A and B). These results suggest that AJUBA is involved in the repression of E-cadherin gene expression.
FIG. 8.
Modulation of AJUBA and PRMT5 in p19 cells results in upregulation of E-cadherin expression. (A) Western blotting results show that siRNA targeting AJUBA can stimulate E-cadherin gene expression in p19 cells. Molecular sizes are shown on the left. The asterisk shows nonspecific bands. (B) Reverse transcriptase PCR (RT-PCR) analysis of the E-cadherin mRNA level in siAjuba p19 cells. Molecular sizes are shown on the left. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (C and D) The methyltransferase inhibitor MTA stimulates E-cadherin expression in p19 cells (C), and similarly, siRNA knockdown of PRMT5 in p19 cells results in the upregulation of E-cadherin gene expression (D), shown by the results of Western blotting.
To determine the role of PRMT5 in the regulation of E-cadherin gene expression, we treated p19 cells with the methyltransferase inhibitor MTA, which has been shown to block PRMT5 function (43). Treatment of the p19 cells with MTA at doses of 100 μM and 200 μM stimulated E-cadherin expression compared to the results for the dimethyl sulfoxide control (Fig. 8C). As a more-specific approach, we employed siRNA knockdown of Prmt5. An siRNA targeting Prmt5 was transfected into p19 cells, and the level of PRMT5 protein was significantly decreased (Fig. 8D). Moreover, E-cadherin expression was increased in PRMT5 knockdown cells, as shown by the results of Western blot analyses. Collectively, these data strongly suggest that both AJUBA and PRMT5 mediate the SNAIL-dependent repression of E-cadherin and that the methyltransferase activity of PRMT5 is required.
DISCUSSION
Herein, we have identified PRMT5 as a repressor recruited to the SNAIL complex via interaction with the AJUBA corepressor. We demonstrated that PRMT5 can form multiprotein complexes containing SNAIL and AJUBA which function to repress the canonical SNAIL target gene E-cadherin. We showed that treatment of the p19 cells with the methyltransferase inhibitor MTA or with siRNA targeting PRMT5 stimulates E-cadherin expression. Further, PRMT5 was shown to specifically bind to the proximal promoter of the E-cadherin gene, and concomitantly, the methylation status of histones at this locus, represented by H4R3, is increased in the presence of the SNAIL. Together, our data suggest that PRMT5 is a key mediator for the regulation of the expression of the E-cadherin gene and that the methyltransferase activity of PRMT5 is involved in the transcriptional repression of the SNAIL complex.
The PRMT5 protein is a member of the type II protein arginine methyltransferases and can methylate transcription factors and histones on specific arginine residues to regulate gene expression (2, 13, 37, 38). For example, PRMT5 was found to interact with BRG1 and BRM, components of the human SWI/SNF chromatin-remodeling complex, to methylate histones H2A and H4 on arginine 3 and H3 on arginine 8. These activities of PRMT5 result in the repression of genes, such as ST7 and NM23, and the promotion of a tumorigenic state in NIH 3T3 cells (37, 38). PRMT5 can interact with Blimp1, a zinc finger transcriptional repressor, and suppresses the expression of Dhx38 by the methylation of histones H2A and H4 on arginine 3 (2). The PRMT5 protein has also been found to be part of the E2F complex in the cyclin E1 promoter, correlating with the repression of the transcription of the cyclin E1 gene (13).
The evidence described above suggests that PRMT5 is involved in the transcriptional repression. Paradoxically, the majority of the PRMT5 protein at steady state is found in the cytoplasm. How PRMT5 is translocated, retained, and targeted to specific genes in the nucleus is not clear. We demonstrate that PRMT5 can be translocated into the nucleus via the formation of a complex with AJUBA and SNAIL and that SNAIL may function as a nuclear anchor to target PRMT5 to its target genes.
The MEP50 protein is also found in the AJUBA-PRMT5 complex. The MEP50 protein contains WD motifs and is constitutively associated with the PRMT5 protein (17, 18). Several WD motif-containing proteins were recently shown to be essential for global histone methylation and the regulation of gene transcription (47). The MEP50 protein was shown to direct PRMT5 to specific histones and is indispensable for PRMT5-dependent histone modification (18). Further work will be necessary to establish the role of MEP50 in SNAIL-mediated gene repression and PRMT5 function.
The SNAIL family of proteins play key roles in the regulation of EMT events during development and metastasis and also serve as early markers for the malignant phenotype and prognosis (4, 5, 7, 11, 30, 34). Recently, Snail was shown to be spontaneously upregulated during the process of tumor recurrence in mice, and high levels of Snail expression strongly predict decreased relapse-free survival in women with breast cancer (34). These observations strongly imply a critical role of Snail in the process of breast cancer recurrence. Therefore, the identification of proteins involved in SNAIL-dependent repression will not only shed new light for understanding the mechanisms of SNAIL in EMT and tumor recurrence but also provide new targets for potential drug development and diagnostics.
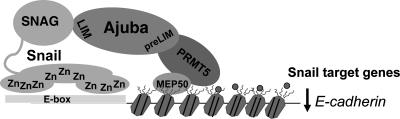
Previous studies have demonstrated the association of SNAIL with potential coregulators through its SNAG domain. These include HDAC1, HDAC2, and the corepressor mSIN3A (39). HDACs are commonly found in large protein complexes in vivo both in the cytoplasm and in the nucleus and may direct the regulation of gene expression, the cell cycle, differentiation, and DNA repair. HDAC1 and HDAC2 have been shown to associate with SMRT, the CoREST complex, mSIN3, N-CoR, and Mi-2/NuRD and play essential roles in gene silencing (12, 41). However, how SNAIL mediates the complex assembly remains elusive. Here, we showed that AJUBA recruits PRMT5 via its PreLIM region to the SNAG domain of SNAIL (Fig. 9). Since both HDAC1 and HDAC2 and AJUBA-PRMT5 interact with the SNAG domain of SNAIL, it will be interesting to examine the complementary roles of these enzymes in SNAIL-mediated repression.
FIG. 9.
Model for the roles of AJUBA and PRMT5 in SNAIL-mediated gene repression. Small circles, methyl groups; hatched oblongs, nucleosomes; wiggly lines, histone tails.
It is a reasonable assumption that, in addition to PRMT5, other factors are recruited to the SNAIL complex via interactions with AJUBA. Like many other LIM proteins, AJUBA has been shown to function as a scaffold protein and can interact with a variety of proteins, including transcription regulators, kinases, and cytoskeleton proteins (14, 19, 23, 26-28, 33, 42). Therefore, an understanding of the spatial and temporal regulation of these protein complexes should shed new light on the roles of these important and clinically relevant transcription factors.
Acknowledgments
We thank Gideon Dreyfuss for the gift of the Prmt5 plasmid and Jae-Gahb Park for the E-cadherin promoter reporter plasmid. We also thank David E. White, Dmitri Negorev, David C. Schultz, and Alexey V. Ivanov for helpful discussion.
F.J.R. is supported, in part, by NIH grants (CA095561 and CA092088) and grant DAMD17-02-1-0631 from the Pardee foundation, by the Emerald foundation, and by the Samuel Waxman Cancer Research Foundation. Z.H. is supported by an NIH training grant (T32, CA09171-31A1). G.D.L. is supported by NIH grants (CA75315 and CA106496) and the Washington University/Pfizer Biomedical Research Program. E.M.L. is an HHMI predoctoral fellow. We acknowledge the National Cancer Institute-supported Wistar Institute Cancer Center shared facilities for genomics, protein expression, proteomics, and hybridoma and the Commonwealth Universal Research Enhancement Program, Pennsylvania Department of Health. K.A. is supported by NIH grant KO1-CA095620.
Footnotes
Published ahead of print on 17 March 2008.
REFERENCES
- 1.Aguilera, A., L. S. Aroeira, M. Ramirez-Huesca, M. L. Perez-Lozano, A. Cirugeda, M. A. Bajo, G. Del Peso, A. Valenzuela-Fernandez, J. A. Sanchez-Tomero, M. Lopez-Cabrera, and R. Selgas. 2005. Effects of rapamycin on the epithelial-to-mesenchymal transition of human peritoneal mesothelial cells. Int. J. Artif. Organs 28164-169. [DOI] [PubMed] [Google Scholar]
- 2.Ancelin, K., U. C. Lange, P. Hajkova, R. Schneider, A. J. Bannister, T. Kouzarides, and M. A. Surani. 2006. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 8623-630. [DOI] [PubMed] [Google Scholar]
- 3.Ayyanathan, K., H. Peng, Z. Hou, W. J. Fredericks, R. K. Goyal, E. M. Langer, G. D. Longmore, and F. J. Rauscher III. 2007. The Ajuba LIM domain protein is a corepressor for SNAG domain mediated repression and participates in nucleocytoplasmic shuttling. Cancer Res. 679097-9106. [DOI] [PubMed] [Google Scholar]
- 4.Bagnato, A., and L. Rosano. 2007. Epithelial-mesenchymal transition in ovarian cancer progression: a crucial role for the endothelin axis. Cells Tissues Organs 18585-94. [DOI] [PubMed] [Google Scholar]
- 5.Barrallo-Gimeno, A., and M. A. Nieto. 2005. The Snail genes as inducers of cell movement and survival: implications in development and cancer. Development 1323151-3161. [DOI] [PubMed] [Google Scholar]
- 6.Cano, A., M. A. Perez-Moreno, I. Rodrigo, A. Locascio, M. J. Blanco, M. G. del Barrio, F. Portillo, and M. A. Nieto. 2000. The transcription factor snail controls epithelial-mesenchymal transitions by repressing E-cadherin expression. Nat. Cell Biol. 276-83. [DOI] [PubMed] [Google Scholar]
- 7.Castro Alves, C., E. Rosivatz, C. Schott, R. Hollweck, I. Becker, M. Sarbia, F. Carneiro, and K. F. Becker. 2007. Slug is overexpressed in gastric carcinomas and may act synergistically with SIP1 and Snail in the down-regulation of E-cadherin. J. Pathol. 211507-515. [DOI] [PubMed] [Google Scholar]
- 8.Cicchini, C., D. Filippini, S. Coen, A. Marchetti, C. Cavallari, I. Laudadio, F. M. Spagnoli, T. Alonzi, and M. Tripodi. 2006. Snail controls differentiation of hepatocytes by repressing HNF4alpha expression. J. Cell. Physiol. 209230-238. [DOI] [PubMed] [Google Scholar]
- 9.Collins, T., J. R. Stone, and A. J. Williams. 2001. All in the family: the BTB/POZ, KRAB, and SCAN domains. Mol. Cell. Biol. 213609-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Boer, T. P., T. A. van Veen, M. F. Bierhuizen, B. Kok, M. B. Rook, K. J. Boonen, M. A. Vos, P. A. Doevendans, J. M. de Bakker, and M. A. van der Heyden. 2007. Connexin43 repression following epithelium-to-mesenchyme transition in embryonal carcinoma cells requires Snail1 transcription factor. Differentiation 75208-218. [DOI] [PubMed] [Google Scholar]
- 11.Ding, J. X., Y. J. Feng, L. Q. Yao, M. Yu, H. Y. Jin, and L. H. Yin. 2006. The reinforcement of invasion in epithelial ovarian cancer cells by 17 beta-estradiol is associated with up-regulation of Snail. Gynecol. Oncol. 103623-630. [DOI] [PubMed] [Google Scholar]
- 12.Ekwall, K. 2005. Genome-wide analysis of HDAC function. Trends Genet. 21608-615. [DOI] [PubMed] [Google Scholar]
- 13.Fabbrizio, E., S. El Messaoudi, J. Polanowska, C. Paul, J. R. Cook, J. H. Lee, V. Negre, M. Rousset, S. Pestka, A. Le Cam, and C. Sardet. 2002. Negative regulation of transcription by the type II arginine methyltransferase PRMT5. EMBO Rep. 3641-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feng, Y., and G. D. Longmore. 2005. The LIM protein Ajuba influences interleukin-1-induced NF-κB activation by affecting the assembly and activity of the protein kinase Cζ/p62/TRAF6 signaling complex. Mol. Cell. Biol. 254010-4022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Friedman, J. R., W. J. Fredericks, D. E. Jensen, D. W. Speicher, X. P. Huang, E. G. Neilson, and F. J. Rauscher III. 1996. KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 102067-2078. [DOI] [PubMed] [Google Scholar]
- 16.Friesen, W. J., S. Paushkin, A. Wyce, S. Massenet, G. S. Pesiridis, G. Van Duyne, J. Rappsilber, M. Mann, and G. Dreyfuss. 2001. The methylosome, a 20S complex containing JBP1 and pICln, produces dimethylarginine-modified Sm proteins. Mol. Cell. Biol. 218289-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friesen, W. J., A. Wyce, S. Paushkin, L. Abel, J. Rappsilber, M. Mann, and G. Dreyfuss. 2002. A novel WD repeat protein component of the methylosome binds Sm proteins. J. Biol. Chem. 2778243-8247. [DOI] [PubMed] [Google Scholar]
- 18.Furuno, K., T. Masatsugu, M. Sonoda, T. Sasazuki, and K. Yamamoto. 2006. Association of Polycomb group SUZ12 with WD-repeat protein MEP50 that binds to histone H2A selectively in vitro. Biochem. Biophys. Res. Commun. 3451051-1058. [DOI] [PubMed] [Google Scholar]
- 19.Goyal, R. K., P. Lin, J. Kanungo, A. S. Payne, A. J. Muslin, and G. D. Longmore. 1999. Ajuba, a novel LIM protein, interacts with Grb2, augments mitogen-activated protein kinase activity in fibroblasts, and promotes meiotic maturation of Xenopus oocytes in a Grb2- and Ras-dependent manner. Mol. Cell. Biol. 194379-4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gregory, R. I., T. P. Chendrimada, and R. Shiekhattar. 2006. MicroRNA biogenesis: isolation and characterization of the microprocessor complex. Methods Mol. Biol. 34233-47. [DOI] [PubMed] [Google Scholar]
- 21.Gregory, R. I., K. P. Yan, G. Amuthan, T. Chendrimada, B. Doratotaj, N. Cooch, and R. Shiekhattar. 2004. The microprocessor complex mediates the genesis of microRNAs. Nature 432235-240. [DOI] [PubMed] [Google Scholar]
- 22.Grimes, H. L., T. O. Chan, P. A. Zweidler-McKay, B. Tong, and P. N. Tsichlis. 1996. The Gfi-1 proto-oncoprotein contains a novel transcriptional repressor domain, SNAG, and inhibits G1 arrest induced by interleukin-2 withdrawal. Mol. Cell. Biol. 166263-6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirota, T., N. Kunitoku, T. Sasayama, T. Marumoto, D. Zhang, M. Nitta, K. Hatakeyama, and H. Saya. 2003. Aurora-A and an interacting activator, the LIM protein Ajuba, are required for mitotic commitment in human cells. Cell 114585-598. [DOI] [PubMed] [Google Scholar]
- 24.Hou, Z., S. Srivastava, M. J. Mistry, M. P. Herbst, J. P. Bailey, and N. D. Horseman. 2003. Two tandemly linked interferon-gamma-activated sequence elements in the promoter of glycosylation-dependent cell adhesion molecule 1 gene synergistically respond to prolactin in mouse mammary epithelial cells. Mol. Endocrinol. 171910-1920. [DOI] [PubMed] [Google Scholar]
- 25.Julien, S., I. Puig, E. Caretti, J. Bonaventure, L. Nelles, F. van Roy, C. Dargemont, A. G. de Herreros, A. Bellacosa, and L. Larue. 2007. Activation of NF-kappaB by Akt upregulates Snail expression and induces epithelium mesenchyme transition. Oncogene 267445-7456. [DOI] [PubMed] [Google Scholar]
- 26.Kadrmas, J. L., and M. C. Beckerle. 2004. The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5920-931. [DOI] [PubMed] [Google Scholar]
- 27.Kanungo, J., S. J. Pratt, H. Marie, and G. D. Longmore. 2000. Ajuba, a cytosolic LIM protein, shuttles into the nucleus and affects embryonal cell proliferation and fate decisions. Mol. Biol. Cell 113299-3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kisseleva, M., Y. Feng, M. Ward, C. Song, R. A. Anderson, and G. D. Longmore. 2005. The LIM protein Ajuba regulates phosphatidylinositol 4,5-bisphosphate levels in migrating cells through an interaction with and activation of PIPKIα. Mol. Cell. Biol. 253956-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krause, C. D., Z. H. Yang, Y. S. Kim, J. H. Lee, J. R. Cook, and S. Pestka. 2007. Protein arginine methyltransferases: evolution and assessment of their pharmacological and therapeutic potential. Pharmacol. Ther. 11350-87. [DOI] [PubMed] [Google Scholar]
- 30.Kurrey, N. K., A. K, and S. A. Bapat. 2005. Snail and Slug are major determinants of ovarian cancer invasiveness at the transcription level. Gynecol. Oncol. 97155-165. [DOI] [PubMed] [Google Scholar]
- 31.Langer, E. M., Y. Feng, Z. Hou, F. J. Rauscher III, K. L. Kroll, and G. D. Longmore. 2008. Ajuba LIM proteins are Snail corepressors required for neural crest development in Xenopus. Dev. Cell 14424-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, M. G., R. Villa, P. Trojer, J. Norman, K. P. Yan, D. Reinberg, L. Di Croce, and R. Shiekhattar. 2007. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318447-450. [DOI] [PubMed] [Google Scholar]
- 33.Marie, H., S. J. Pratt, M. Betson, H. Epple, J. T. Kittler, L. Meek, S. J. Moss, S. Troyanovsky, D. Attwell, G. D. Longmore, and V. M. Braga. 2003. The LIM protein Ajuba is recruited to cadherin-dependent cell junctions through an association with alpha-catenin. J. Biol. Chem. 2781220-1228. [DOI] [PubMed] [Google Scholar]
- 34.Moody, S. E., D. Perez, T. C. Pan, C. J. Sarkisian, C. P. Portocarrero, C. J. Sterner, K. L. Notorfrancesco, R. D. Cardiff, and L. A. Chodosh. 2005. The transcriptional repressor Snail promotes mammary tumor recurrence. Cancer Cell 8197-209. [DOI] [PubMed] [Google Scholar]
- 35.Nakakura, E. K., D. N. Watkins, V. Sriuranpong, M. W. Borges, B. D. Nelkin, and D. W. Ball. 2001. Mammalian Scratch participates in neuronal differentiation in P19 embryonal carcinoma cells. Brain Res. Mol. Brain Res. 95162-166. [DOI] [PubMed] [Google Scholar]
- 36.Nieto, M. A. 2002. The snail superfamily of zinc-finger transcription factors. Nat. Rev. Mol. Cell Biol. 3155-166. [DOI] [PubMed] [Google Scholar]
- 37.Pal, S., S. N. Vishwanath, H. Erdjument-Bromage, P. Tempst, and S. Sif. 2004. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol. Cell. Biol. 249630-9645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pal, S., R. Yun, A. Datta, L. Lacomis, H. Erdjument-Bromage, J. Kumar, P. Tempst, and S. Sif. 2003. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol. Cell. Biol. 237475-7487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peinado, H., E. Ballestar, M. Esteller, and A. Cano. 2004. Snail mediates E-cadherin repression by the recruitment of the Sin3A/histone deacetylase 1 (HDAC1)/HDAC2 complex. Mol. Cell. Biol. 24306-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peinado, H., M. Del Carmen Iglesias-de la Cruz, D. Olmeda, K. Csiszar, K. S. Fong, S. Vega, M. A. Nieto, A. Cano, and F. Portillo. 2005. A molecular role for lysyl oxidase-like 2 enzyme in snail regulation and tumor progression. EMBO J. 243446-3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peterson, C. L. 2002. HDAC's at work: everyone doing their part. Mol. Cell 9921-922. [DOI] [PubMed] [Google Scholar]
- 42.Pratt, S. J., H. Epple, M. Ward, Y. Feng, V. M. Braga, and G. D. Longmore. 2005. The LIM protein Ajuba influences p130Cas localization and Rac1 activity during cell migration. J. Cell Biol. 168813-824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Richard, S., M. Morel, and P. Cleroux. 2005. Arginine methylation regulates IL-2 gene expression: a role for protein arginine methyltransferase 5 (PRMT5). Biochem. J. 388379-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shin, Y., I. J. Kim, H. C. Kang, J. H. Park, H. R. Park, H. W. Park, M. A. Park, J. S. Lee, K. A. Yoon, J. L. Ku, and J. G. Park. 2004. The E-cadherin −347G->GA promoter polymorphism and its effect on transcriptional regulation. Carcinogenesis 25895-899. [DOI] [PubMed] [Google Scholar]
- 45.Shin, Y., I. J. Kim, H. C. Kang, J. H. Park, H. W. Park, S. G. Jang, M. R. Lee, S. Y. Jeong, H. J. Chang, J. L. Ku, and J. G. Park. 2004. A functional polymorphism (−347 G->GA) in the E-cadherin gene is associated with colorectal cancer. Carcinogenesis 252173-2176. [DOI] [PubMed] [Google Scholar]
- 46.White, D. E., D. Negorev, H. Peng, A. V. Ivanov, G. G. Maul, and F. J. Rauscher III. 2006. KAP1, a novel substrate for PIKK family members, colocalizes with numerous damage response factors at DNA lesions. Cancer Res. 6611594-11599. [DOI] [PubMed] [Google Scholar]
- 47.Wysocka, J., T. Swigut, T. A. Milne, Y. Dou, X. Zhang, A. L. Burlingame, R. G. Roeder, A. H. Brivanlou, and C. D. Allis. 2005. WDR5 associates with histone H3 methylated at K4 and is essential for H3 K4 methylation and vertebrate development. Cell 121859-872. [DOI] [PubMed] [Google Scholar]
- 48.Zhou, B. P., J. Deng, W. Xia, J. Xu, Y. M. Li, M. Gunduz, and M. C. Hung. 2004. Dual regulation of Snail by GSK-3beta-mediated phosphorylation in control of epithelial-mesenchymal transition. Nat. Cell Biol. 6931-940. [DOI] [PubMed] [Google Scholar]
- 49.Zweidler-Mckay, P. A., H. L. Grimes, M. M. Flubacher, and P. N. Tsichlis. 1996. Gfi-1 encodes a nuclear zinc finger protein that binds DNA and functions as a transcriptional repressor. Mol. Cell. Biol. 164024-4034. [DOI] [PMC free article] [PubMed] [Google Scholar]