Abstract
Transcriptional regulation by the canonical Wnt pathway involves the stabilization and nuclear accumulation of β-catenin, which assembles with LEF1/TCF transcription factors and cofactors to activate Wnt target genes. Recently, the nuclear β-catenin complex has been shown to contain BCL9, which interacts with β-catenin and recruits Pygopus as a transcriptional coactivator. However, the presumed general functions of Pygopus and BCL9, which has been proposed to act as a scaffolding protein for Pygopus, have been challenged by the rather specific and modest developmental defects of targeted inactivations of both the Pygo1 and the Pygo2 genes. Here, we analyze the function of BCL9 in transcriptional activation by β-catenin. We find that BCL9 acts in a cell-type-specific manner and, in part, independent of Pygopus. We show that BCL9 itself contains a transcriptional activation domain in the C terminus, which functionally synergizes in lymphoid cells with the C-terminal transactivation domain of β-catenin. Finally, we identify amino acids in the transactivation domain of β-catenin that are important for its function and association with the histone acetyltransferases CBP/p300 and TRRAP/GCN5. Thus, BCL9 may serve to modulate and diversify the transcriptional responses to Wnt signaling in a cell-type-specific manner.
The canonical Wnt signaling pathway regulates multiple developmental processes, including cell proliferation and cell fate decisions (reviewed in references 14 and 17). In cells that receive a Wnt signal, the key effector of the pathway, β-catenin, is stabilized by the inhibition of a cytosolic destruction complex, consisting of the adenomatous polyposis coli (APC) protein, axin, casein kinase I, and GSK3β (reviewed in reference 33). Stabilized β-catenin accumulates in the cytoplasm and nucleus, where it associates with members of the LEF1/TCF family of transcription factors (reviewed in reference 63). LEF1/TCF transcription factors have no activation potential by themselves, but in association with β-catenin, they activate promoters containing multimerized LEF1/TCF-binding sites and natural promoters that respond to Wnt signals (6, 12, 22, 40, 44, 59, 61). Target gene activation depends on promoter architecture, cell type context, and the presence of specific LEF1/TCF family members. siamois and cdx1 are well-characterized Wnt target genes that are differentially activated by various LEF1/TCF proteins (8, 23, 28, 38). Diversification of the transcriptional response by LEF1/TCF proteins was found to involve a promoter-specific activation domain in the extended carboxy (C) termini, termed the E tails, of TCF1 and TCF4 proteins (3, 21, 23).
β-Catenin is a multidomain protein consisting of 12 armadillo repeats (arm) that mediate the alternative associations with the amino (N) termini of LEF1/TCF proteins, with components of the cytosolic APC/axin destruction complex and with the membrane-bound adhesion molecule E cadherin (reviewed in reference 51). In addition, β-catenin contains an amino-terminal domain that regulates protein stability and a C-terminal domain that confers transcription activation potential (27). The C-terminal domain of β-catenin has been found to interact with multiple proteins, including the histone acetyltransferases CBP/p300 and TRRAP/Tip60, the histone methylation complex MLL1, the MED12 component of the mediator complex, TBP, and Parafibromin, which is part of a transcription elongation complex (24, 32, 41, 48, 52). Transcription activation by β-catenin is also augmented by the association of Brg-1, a component of a nucleosome remodeling complex, with the armadillo repeats of β-catenin (5). However, the C terminus of β-catenin does not appear to be sufficient for transcriptional activation in response to Wnt signaling. First, β-catenin lacking the C-terminal domain retains an activation potential in mammalian transfection assays and in the fly (18, 27). In addition, a fusion of the C terminus of armadillo, the Drosophila orthologue of β-catenin, to the Drosophila TCF protein lacking the β-catenin interaction domain failed to restore signaling activity in transgenic flies (54). Additional activation functions of β-catenin could be accounted for by a transactivation function of the amino-terminal domain (27) and by the association of B-cell lymphoma 9 (BCL9)/Legless (Lgs) with the first armadillo repeats of β-catenin (34).
BCL9/Lgs was identified in a genetic screen for wingless/Wnt signal transducing components in Drosophila (34), and it had been previously found to be translocated and overexpressed in B-cell lymphomas (64). BCL9 has been shown to contain three homology domains (HD1, HD2, and HD3) that are highly conserved between Drosophila, zebrafish, and mammals (34). The HD2 domain of BCL9 mediates the interaction with β-catenin, whereas the HD1 domain associates with a nuclear protein, Pygopus (7, 34, 42, 53). Notably, the amino-terminal third of the Drosophila BCL9/Lgs protein, comprising all three homology domains, is necessary and sufficient to rescue wingless signal activity in BCL9/Lgs mutant flies (34). BCL9 by itself has not been found to mediate transcriptional activation, and the primary function of BCL9 in Wnt signaling has been proposed to consist of a “scaffolding” function for the recruitment of the transcriptional coactivator Pygopus (50). In mammalian cells, another BCL9 gene, termed BCL9-2 or BCL9L, has been identified and proposed to mediate a Wnt response in a Pygopus-independent manner by its association with a tyrosine-142-phosphorylated form of β-catenin, which impairs the association with α-catenin (1, 10). Additional analysis of BCL9L showed that it also collaborates with Pygopus and can functionally replace BCL9 both in cultured cells and in vivo (25).
The nuclear protein Pygopus has been shown to contain a C-terminal domain that mediates association with the HD1 domain of BCL9 and an amino-terminal PHD domain that has been implicated in recruiting a transcription coactivator (26, 34, 53). In addition, the exclusive nuclear localization of Pygopus has been shown to facilitate the nuclear localization of BCL9 protein, which by itself is found in both the cytoplasm and the nucleus (55). Based on these observations, a “chain of adaptors” model in which Pygopus is the most distal component has been proposed (50). Recently, Pygopus has also been found to be associated constitutively with TCF target genes in Drosophila salivary glands and tissue cultures in a BCL9/Lgs-independent manner (19). Analysis of the function of the Pygopus 1 (Pygo1) and Pygo2 genes in the mouse by the combined targeted gene inactivation revealed fairly specific developmental defects rather than general defects in Wnt signaling (47). Therefore, the question as to whether BCL9 can also function independently of Pygopus arises. Here, we study the function of BCL9 and find that it contains a transcriptional activation domain that synergizes with the activation domain of β-catenin and acts, at least in part, independently of Pygopus and in a cell-type-specific manner.
MATERIALS AND METHODS
Materials.
Anti-T7 (Novagen), anti-myc (Roche), and anti-β-catenin (Upstate) antibodies were used according to the manufacturer's instructions. Secondary antibodies were coupled to horseradish peroxidase (Jackson Immunoresearch). Monoclonal BCL9 antibodies were derived from rats immunized with amino acids (aa) 1 to 497 of human BCL9. Mouse anti-TRRAP and anti-GCN5 antibodies were a kind gift from L. Tora (Strasbourg). Rat anti-CBP was a kind gift from E. Kremmer (GSF Munich).
Cell culture and transfection.
Raji, Namalwa, Bjab, and Jurkat cells were grown in RPMI supplemented with 10% fetal calf serum and penicillin-streptomycin-glutamine (PSG). HeLa and HEK293 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum and PSG. Suspension cells were electroporated using the GenePulser II system (Bio-Rad). Lymphoid cells were electroporated using 1 μg reporter, 1 μg β-galactosidase, 0.5 μg TCF4E, and 3 μg β-catenin. BCL9 amounts are indicated in the figure legends. Pygopus1 (L-017154-00-005), Pygopus2 (L-013908-01-005), and control small interfering RNA (siRNA; D-001810-10-05) were transiently transfected at a concentration of 400 nM (Dharmacon). BCL9 and BCL9-2 knockdown Raji B cells were generated by stably integrating pSuper.neo+GFP (Oligoengine) with a BCL9-directed construct (5′GATCCCCCCTCAGAGCAGAGTATTAATTCAAGAGATTAATACTCTGCTCTGAGGTTTTTC) and pSuper.puro (Oligoengine) with a BCL9-2-directed construct (5′GATCCCCGGAACAGATTGGGCTGCATTTCAAGAGAATGCAGCCCAATCTGTTCCTTTTTC). Cells were selected with 1 mg/ml G418 and 1 μg/ml puromycin containing RPMI complete medium. HeLa cells were transiently transfected with Lipofectamine 2000 (Invitrogen). HEK293 cells were transiently transfected by the calcium phosphate method, using 0.1 μg reporter, 0.1 μg β-galactosidase, 0.05 μg TCF4E, 0.5 μg β-catenin, and 0.1 and 0.5 μg of BCL9.
Reporter gene assays.
The total amount of DNA was kept constant by addition of empty-vector plasmid DNA (pCI; Clontech). Cells were harvested at 36 h posttransfection in reporter lysis buffer (Promega), and luciferase assays were conducted according to the manufacturer's instructions (Promega). Firefly luciferase activities were normalized against the independently measured activity of cotransfected β-galactosidase, using chlorphenolred-β-d-galactopyranoside (CPRG) as a substrate.
Plasmids.
The hTCF4E, siamois, and cdx1 constructs have been described previously (23, 24). The LEF7-fos reporter, LEF1, and β-catenin constructs have been described previously (27). Gal4 VP16 was described previously (11). Gal45x luciferase was described previously (65). β-Catenin coding sequences were PCR amplified using Pfu polymerase (Stratagene) and cloned in pGEX3X (GE Healthcare). Point mutations in β-catenin have been introduced using QuikChange site-directed mutagenesis (Stratagene). CβS-BCL9-myc constructs under the control of a cytomegalovirus promoter have been cloned using standard cloning techniques. Bacterial BCL9 expression constructs were cloned into pGEX3X (GE Healthcare), yielding an N-terminal glutathione S-transferase (GST) fusion protein. Details are available upon request.
Immunoblot and coimmunoprecipitation assays.
Cells were lysed in coimmunoprecipitation buffer (CoIP buffer; 50 mM Tris-Cl, pH 7.5, 15 mM EGTA, 100 mM NaCl, 0.1% [wt/vol] Triton X-100) supplemented with protease inhibitor mix and 0.5 mM phenylmethylsulfonyl fluoride, using sonication. Cell debris was spun down and protein concentration determined according to the Bradford method (Bio-Rad). Five hundred micrograms to one milligram of total protein was used for one immunoprecipitation, using 1 μg of antibody. Precipitates bound to protein G beads (GE Healthcare) were washed several times with CoIP buffer and liberated with sodium dodecyl sulfate-polyacrylamide gel electrophoresis sample buffer. Proteins were resolved on 8 to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis gel, transferred to nitrocellulose membranes, and developed with the indicated primary and secondary antibodies. Immunoblots were incubated with enhanced chemiluminescence reagent (GE Healthcare) and exposed to X-ray films.
Recombinant protein purification.
Expression constructs were transformed into BL21 and induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at a temperature of 25°C. Upon sonication of the cell pellet, proteins were affinity purified over glutathione Sepharose (GE Healthcare) and extensively washed with a buffer containing 300 mM NaCl, 20 mM HEPES, pH 7.6, 10% glycerol, 0.5% Triton X-100, 1 mM EDTA, and 1 mM dithiothreitol. Proteins were eluted with a buffer containing 50 mM Tris-Cl, pH 8.0, and 20 mM glutathione, and for storage, the buffer was exchanged on a 25-ml desalting column (GE Healthcare) to phosphate-buffered saline, 10% glycerol, and 1 mM dithiothreitol. For circular dichroism (CD) measurements, GST-coupled proteins were cut with Factor Xa (NEB) according to the manufacturer's protocol. Cleaved GST was removed with glutathione Sepharose, and Factor Xa was removed using Factor Xa removal resin (Qiagen). CD protein samples were dialyzed against 25 mM sodium phosphate buffer, pH 7.5, without salt, and protein concentration was determined according to the theoretically determined extinction coefficients.
CD spectroscopy.
Spectra were accumulated on a Jasco J-720 spectrometer. Protein solutions were filtered prior measurement. Samples were collected at an ambient temperature in a cuvette with a 0.1-cm path length. Each CD spectrum consists of eight scans at 100 nm/min, with a 1-nm slit width. Data were collected from 190 to 300 nm. CD spectra were background corrected and scaled to mean molecular ellipticity.
GST pulldown assay.
Ten micrograms each of the indicated constructs was bound to glutathione beads and equilibrated in CoIP buffer. One microgram of recombinantly purified β-catenin was added, and after 1 h of incubation at 4°C on a rotary shaker, the beads were washed several times with CoIP buffer and analyzed as described for the coimmunoprecipitation assays. In the case of TRRAP interaction studies, recombinant β-catenin constructs were bound in the presence of 20 mM Tris-Cl, pH 7.5, 150 mM KCl, 0.2 mM EDTA, pH 8.0, 20% glycerol, and 0.1% Igepal CA-630. Bound proteins were incubated with 500 μg nuclear extract and essentially treated as described above, using the latter buffer instead of CoIP buffer.
Sybr green-based real-time PCR.
BCL9 and Pygopus mRNA expression levels were measured using Sybr green-based real-time PCR. Total RNA was isolated with TRIzol (Invitrogen), and 2 μg of RNA was transcribed to cDNA by using 200 U Superscript II (Invitrogen) and 0.1 nmol Oligo(dT)12-18 (Roche). PCR was performed with the AbiPrism 7500 sequence detection system (Applied Biosystems). Reverse transcriptase (RT) controls were done in parallel without adding enzyme. Obtained cycle numbers were normalized to β-actin levels. Primers were used at 100 nM, and the sequences were as follows: BCL9For, 5′-AGAGAGAAGCACAGCGCCTC; BCL9Rev, 5′-CTGCAGTCTGGTATTCTGGGAAG; BCL9-2For, 5′-CAGAACCCCCTGTCACTGATG; BCL9-2Rev, 5′-CGATGGTCTTGATGGCATTG; Pygo1For, 5′-CCAAACTCTG ACCATCTAGTGGC; Pygo1Rev, 5′-GGAACGTGAGGTGGCATTCT; Pygo2For, 5′-CAGCACCGGGAGGAAGC; Pygo2Rev, 5′-TCTGGACTCTTCATTTGCAGACC; β-ActinFor, 5′-TTGCCGACAGGATGCAGAA; β-ActinRev, 5′-CACGGAGTACTTGCGCTCAG; hAxin2For, 5′-GTGAAGGCCAATGGCCAA; hAxin2Rev, 5′-CAGGCGGTGGGTTCTCG; hEcadherinFor, 5′-GAACGCATTGCCACATACACTC; and hEcadherinRev, 5′-CATTCCCGTTGGATGACACAG.
Generation of β-catenin-deficient ES cells with stably integrated wild-type and M6 β-catenin constructs.
β-Cateninfloxdel/floxed embryonic stem (ES) cells (9) were cultured in Dulbecco's modified Eagle's medium supplemented with 15% fetal calf serum, PSG, nonessential amino acids, sodium pyruvate, and 1,000 U of leukemia inhibitory factor/ml. ES cells were stably transfected with either wild-type or M6 β-catenin constructs. Stable clones were selected with 1 μg/ml puromycin and identified by immunoblot analysis. Positive clones were transiently transfected with a modified Cre recombinase expression vector with an internal ribosome entry site-enhanced green fluorescent protein (EGFP; H. J. Fehling) to generate β-cateninfloxdel/floxdel ES cells. β-Cateninfloxdel/floxdel ES cell clones were identified due to EGFP expression. ES cells (β-cateninfloxdel/floxdel) were transiently transfected with Lipofectamine 2000 (Invitrogen), using 1.0 μg reporter, 1.0 μg β-galactosidase, 0.5 μg TCF4E, and 3.0 μg and 6.0 μg of wild-type β-catenin or 3.0 μg and 6.0 μg of M6 β-catenin. Further analysis was performed similarly to that for transfected cell lines.
RESULTS
Cell-type-specific function of BCL9 in β-catenin-dependent reporter gene activation.
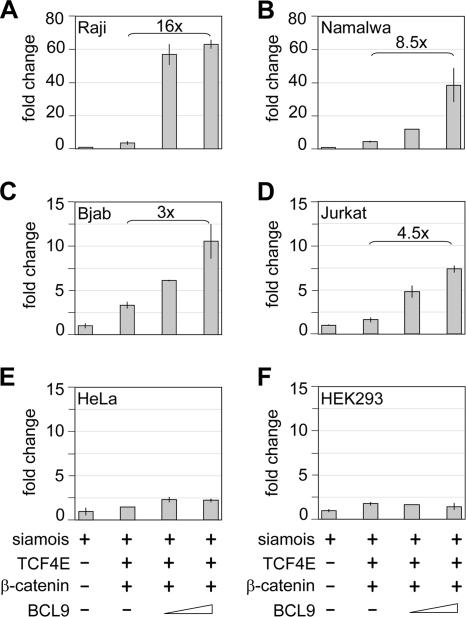
As an initial step in dissecting the function of BCL9 in transcriptional activation by β-catenin, we examined the potential of BCL9 to augment the activity of a natural Wnt-responsive promoter in different cell lines. To this end, we transfected a reporter construct, carrying the siamois promoter linked to the luciferase gene (8), together with expression plasmids for TCF4E, β-catenin, and BCL9, into cell lines representing the lymphoid B-cell lineage (Raji, Namalwa, and Bjab), the T-cell lineage (Jurkat), or fibroblastic cells (HeLa and HEK293). In lymphoid cells, we observed an up to 16-fold augmentation of promoter activity by BCL9, relative to the expression levels of TCF4E and β-catenin, whereas in fibroblastic cells, no significant enhancement by BCL9 was detected (Fig. 1A to F). In these experiments, we used the β-catenin expression plasmid at a concentration that allowed for a linear response to increasing amounts of the BCL9 expression plasmid. In principle, the observed differences in the response of the siamois promoter to the expression of exogenous BCL9 protein could be accounted for by cell-type-specific differences in the expression of endogenous BCL9. Therefore, we analyzed the expression of endogenous BCL9 protein by immunoblot analysis with a monoclonal antibody directed against BCL9 protein. No significant differences in protein expression were detected (data not shown), suggesting that the function of BCL9 differs in various cell types.
FIG. 1.
BCL9 mediates activation of a siamois reporter in a cell-type-specific manner. (A to D) BCL9 enhances transcriptional activity of the siamois luciferase reporter (1 μg) in B-cell (Raji, Namalwa, and Bjab) and T-cell (Jurkat) lines in transient transfection assays. Increasing amounts (0.5 and 1 μg) of the BCL9 expression plasmid were cotransfected with expression plasmids for TCF4E (0.5 μg) and wild-type β-catenin (3 μg). Luciferase values were normalized against the activity of a cotransfected β-galactosidase construct (1 μg). (E and F) Fibroblastic cell lines (HEK293 and HeLa) are unresponsive to BCL9 under similar conditions if transiently transfected with calcium phosphate or Lipofectamine. Duplicates of representative transfections are shown.
Promoter selectivity of BCL9 function.
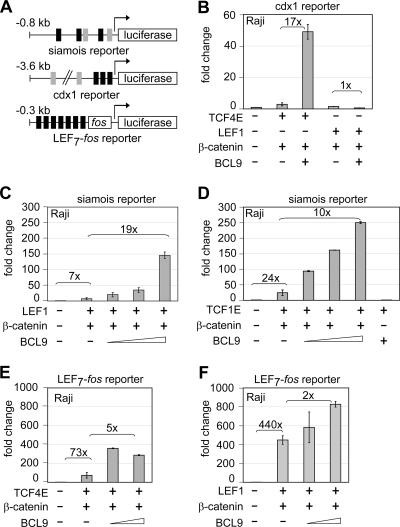
To address the question of whether the function of BCL9 can be observed with other Wnt-responsive promoters, we included the natural cdx1 promoter and the synthetic LEF7-fos promoter in our analysis (27, 38). The cdx1 promoter was chosen because of its differential regulation by LEF1 and TCF4E (23). Moreover, the LEF7-fos promoter, carrying multimerized LEF1/TCF-binding sites, provides a very strong response to β-catenin (27, 58). As anticipated, TCF4E and β-catenin augmented cdx1 promoter activity, whereas no significant activation was detected with LEF1 and β-catenin (Fig. 2B). Notably, BCL9 enhanced TCF4E-mediated promoter activity by a factor of 17, whereas no activation by BCL9 was observed with LEF1 (Fig. 2B). To determine whether LEF1 and β-catenin are responsive to BCL9 in a different promoter context, we used the siamois promoter and detected a 19-fold enhancement of promoter activity by BCL9 (Fig. 2C). Likewise, we found that TCF1E and also TCF4E conferred BCL9 responsiveness upon the siamois promoter (Fig. 1A to D and 2D).
FIG. 2.
Natural and synthetic reporter constructs, as well as different LEF1/TCF proteins, support BCL9-dependent reporter gene activation. (A) Schematic representation of reporter constructs used in this study. Strong and weak LEF1/TCF-binding sites are represented by black and gray boxes, respectively. Schematic representations of the siamois and cdx1 promoters were based on data from reference 23. (B) Transient transfection of BCL9 expression plasmid (0.5 μg), together with TCF4E or LEF1 expression plasmids and a cdx1-luciferase reporter construct. Ten times less of the LEF1 expression construct (0.05 μg) than of TCF4E (0.5 μg) was used to obtain equal protein expression levels (data not shown). (C, D) BCL9-dependent enhancement of siamois reporter activity in cells transfected with LEF1 (0.05 μg) or TCF1E (0.5 μg) expression plasmids. Increasing amounts of BCL9 expression plasmid (0.1, 0.3, and 1 μg) were supplemented with β-catenin expression plasmid (3 μg). (E, F) BCL9 modestly activates the synthetic LEF7-fos reporter in the presence of TCF4E or LEF1 under similar conditions.
BCL9 and BCL9L have been shown to activate a synthetic LEF1/TCF promoter only modestly (∼2-fold) in HEK293 cells (1, 34). To examine whether the function of BCL9 in this synthetic promoter context is more pronounced in Raji cells, we transfected these cells with the LEF7-fos luciferase construct, together with β-catenin and either TCF4E or LEF1. A modest two- to fivefold activation was observed upon expression of BCL9 (Fig. 2E and F). Likewise, no obvious BCL9 response was detected with the cdx1 and LEF7-fos promoters in HEK293 cells (Fig. 1F and data not shown). To examine whether Pygopus could enhance the BCL9 response of the LEF7-fos promoter, we also coexpressed Pygopus in HEK293 cells. Consistent with previous observations, we failed to detect an enhancement of the BCL9 response (56; data not shown). Moreover, we could not enhance the BCL9 effect by the expression of p300/CBP, which is limiting for Wnt-dependent gene activation in HEK293 cells (23; data not shown). These results suggest that BCL9 functions in a cell-type-specific manner whereby the contribution to β-catenin depends on the promoter context. A strong promoter activity correlates with a weak BCL9 response.
BCL9-dependent transactivation is, in part, independent of Pygopus.
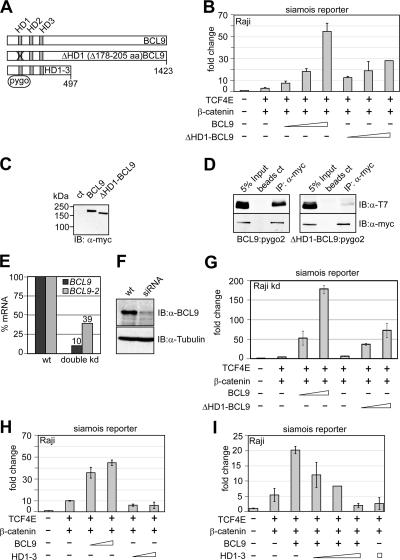
Currently, it is assumed that BCL9 has a scaffolding function for the recruitment of Pygopus as a transcriptional coactivator (50). To examine the relative contribution of Pygopus to the transactivation by BCL9, we generated and tested a mutant form of BCL9 (ΔHD1) in which the residues mediating the interaction with Pygopus (homology domain HD1) have been deleted (Fig. 3A). In transactivation assays with Raji cells, we assessed whether the observed activation potential of BCL9 is dependent on Pygopus. In comparison with the up to ∼18-fold enhancement of the activity of the siamois promoter by wild-type BCL9, the activation potential of ΔHD1-BCL9 was only 2-fold lower, suggesting that BCL9 itself may harbor a transactivation domain (Fig. 3B). Equal expression levels of the two constructs were confirmed by immunoblot analysis of extracts from HEK293 cells that have been transfected with the myc-tagged BCL9 or ΔHD1-BCL9 expression plasmid (Fig. 3C).
FIG. 3.
BCL9-dependent Wnt signaling is partly independent of Pygopus and requires the C terminus of BCL9. (A) Schematic representation of BCL9 constructs which were used in this study. Numbers underneath the bars indicate amino acids. Homology domains (HD) are depicted. (B) BCL9 expression plasmid (0.05, 0.15, and 0.5 μg) enhances siamois reporter activity in the absence of the Pygopus interaction domain (HD1). (C) BCL9 and ΔHD1-BCL9-myc are equally well expressed in HEK293 cells, as analyzed by immunoblotting (IB). ct, control; α-myc, anti-myc. (D) BCL9-myc but not ΔHD1-BCL9-myc interacts with T7-Pygopus2. HEK293 cells were transiently transfected with the indicated expression constructs (5 μg), and coimmunoprecipitation was performed as indicated. IP, immunoprecipitation. (E) Reduction of BCL9 and BCL9-2 mRNA levels in Raji double-knockdown (kd) cells as analyzed by real-time PCR. wt, wild type. (F) BCL9 protein levels are strongly reduced in Raji double-knockdown cells, as analyzed by immunoblotting with a BCL9 specific antibody. (G) BCL9 expression plasmid (0.05, 0.15, and 0.5 μg) enhances siamois reporter activity in the absence of the Pygopus interaction domain (HD1) in Raji BCL9 BCL9-2 double-knockdown cells. (H) C-terminally truncated BCL9 (HD1-3) (0.5 and 1 μg) fails to activate the siamois reporter in the presence of TCF4E (0.5 μg) and wild-type β-catenin (3 μg). (I) HD1-3 suppresses siamois promoter activation in a dose-dependent manner. Wild-type BCL9 expression plasmid (0.5 μg) was transfected alone or together with HD1-3 expression plasmid (0.3, 1, and 3 μg).
To confirm that deletion of the HD1 domain of BCL9 abrogates binding of Pygopus, we performed coimmunoprecipitations to detect, in extracts from transfected 293 cells, association of T7-tagged Pygo2 with myc-tagged BCL9 and myc-tagged ΔHD1-BCL9 (Fig. 3D). Efficient association of Pygo2 was observed with the wild-type but not the ΔHD1-BCL9 protein. The modest decrease of transcriptional activation by the ΔHD1 mutation could, in principle, also be due to a multimerization of the mutant BCL9 protein with endogenous wild-type BCL9. Therefore, we also examined the effect of Pygo2 on transcriptional activation by ΔHD1-BCL9 in Raji cells, in which the expression levels of endogenous BCL9 and BCL9-2 had been reduced by siRNA. We generated Raji cell clones by stable transfection of BCL9 and BCL9-2 siRNA and examined downregulation of mRNA and protein levels by quantitative RT-PCR and immunoblot analysis. In one clone, we observed 10-fold and ∼3-fold downregulations of BCL9 and BCL9-2 mRNA, respectively (Fig. 3E). We also observed a marked downregulation of BCL9 protein in immunoblot analysis (Fig. 3F). As no anti-BCL9-2 antibody is available, we could not assess the downregulation of BCL9-2 protein. Using the BCL9 BCL9-2 double-knockdown cells for transient transfection, we observed an approximately threefold increase of activation of β-catenin-dependent transcription by BCL9 compared to the level for wild-type Raji cells (Fig. 3G and B). However, activation by ΔHD1-BCL9 was also only modestly reduced in BCL9 BCL9-2 double-knockdown cells.
We also examined the effects of siRNA-mediated downregulation of Pygo1 and Pygo2 in Raji cells. Transient transfection of Pygo1 and Pygo2 siRNA into Raji cells reduced mRNA levels to 6% and 13%, respectively (data not shown). Cotransfection of Pygo1 and Pygo2 siRNA, individually or in combination, decreased BCL9-mediated transactivation of the siamois promoter by a factor of <2 (data not shown). Taken together, these experiments suggest that the transactivation function of BCL9 is, to a large extent, independent of Pygopus proteins.
Previous experiments with Drosophila melanogaster indicated that a BCL9 deletion construct comprising only homology domains 1 to 3 (HD1-3) was able to rescue development in a BCL9/Lgs mutant background (34). In contrast, the amino-terminal third of BCL9, containing HD1-3, was unable to augment β-catenin-dependent transactivation, despite a level of expression similar to that of wild-type BCL9 (Fig. 3H and data not shown). Moreover, we found that overexpression of the HD1-3 fragment of BCL9 reduced transactivation by wild-type BCL9 to a level that is even lower than that observed without expression of exogenous BCL9 (Fig. 3I). This “squelching” effect of HD1-3, below the level observed with TCF4E-β-catenin alone, suggests that endogenous BCL9 contributes to reporter activity.
Delineation of a transactivation domain in the C terminus of BCL9.
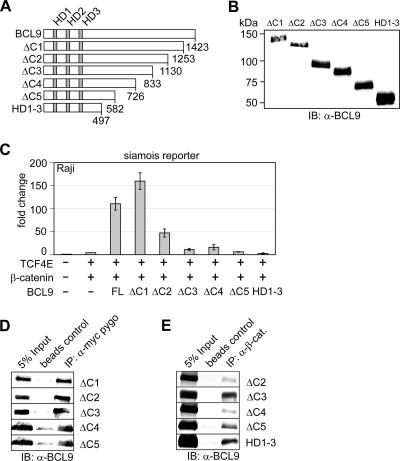
To delineate the putative transactivation domain in BCL9, we generated a series of C-terminally truncated proteins (Fig. 4A). Immunoblot analysis with an anti-BCL9 antibody indicated that the expression levels of these constructs were similar (Fig. 4B). Truncation of amino acids residing C-terminal of residue 833 (ΔC3) markedly impaired BCL9-dependent transactivation (Fig. 4C). To confirm that this mutant BCL9 protein and other C-terminally truncated proteins are properly folded, we analyzed their potential for interaction with Pygopus and β-catenin. To this end, we cotransfected expression plasmids for BCL9 and epitope-tagged Pygopus or β-catenin and performed a coimmunoprecipitation experiment (Fig. 4D and E). As anticipated, this analysis confirmed that all C-terminally truncated BCL9 proteins were able to interact with both Pygopus and β-catenin. ClustalW analysis of the region C-terminal of residue 833 revealed two domains (HD4 and HD5) that are conserved between vertebrate BCL9 and BCL9-2 proteins but are not conserved in Drosophila Legless (not shown).
FIG. 4.
Delineation of a transactivation domain in the C terminus of BCL9. (A) Schematic representation of BCL9 deletion constructs. Numbers indicate amino acids. (B) Immunoblot analysis (IB) of C-terminal BCL9 truncations reveals similar expression levels. α-BCL9, anti-BCL9. (C) BCL9-dependent activation is lost upon progressive deletion of the BCL9 C terminus. BCL9 deletion constructs (1 μg) were transiently expressed in the presence of TCF4E (0.5 μg) and wild-type β-catenin (3 μg). (D) BCL9 deletion constructs interact with the known interaction partners β-catenin and Pygopus. HEK293 cells were transiently transfected with the indicated expression constructs (5 μg), and coimmunoprecipitation was performed as indicated. IP, immunoprecipitation.
The HD4-5 region of BCL9 contains a transactivation domain.
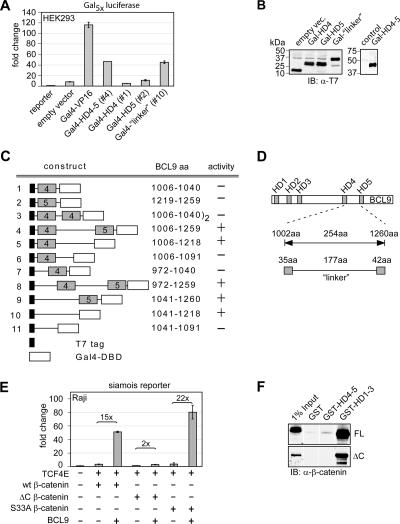
To assess whether the region of BCL9 comprising HD4 and HD5 acts as a transactivation domain, we fused the domain to the Gal4 DNA-binding domain (DBD) and analyzed its activity in reporter assays, using a reporter carrying multimerized Gal4-binding consensus sites (Fig. 5A). Indeed, Gal4 HD4-5 of BCL9 was found to transactivate the reporter at 50% of the level observed with the strong transactivation domain of VP16. All constructs were expressed to similar extents (Fig. 5B). Reporter activation was solely dependent on the presence of a region between the conserved HD4 and HD5 domains, which we termed the “linker” region (Fig. 5A and C). In summary, we were able to identify a transactivation domain in the C terminus of BCL9 which comprises about 170 aa (Fig. 5D).
FIG. 5.
The BCL9 C terminus is sufficient to activate a synthetic Gal4 reporter and requires the C-terminal transactivation domain of β-catenin. (A) Part of the BCL9 C terminus fused to the Gal4 DBD suffices to activate a synthetic reporter consisting of multimerized Gal4 DBD-binding sites upstream of luciferase and thereby defines a transactivation domain. The indicated gene constructs (0.1 μg each) were transiently transfected into HEK293 cells. (B) Immunoblot analysis (IB) of T7 epitope-labeled fusion proteins confirms similar expression levels. Indicated constructs were transiently expressed in HEK293 cells, and whole-cell extract was analyzed. α-T7, anti-T7. (C) Delineation of the transactivation domain of BCL9 in fusions with the Gal4 DBD. Numbers indicate the amino acid sequences of BCL9. “Activity” indicates the ability of the respective construct to activate a synthetic Gal4 reporter in transfection experiments. (D) Schematic representation of the BCL9 transactivation domain. Homology domains (HD) 4 and 5 are marked according to their homology to BCL9-2. (E) BCL9-mediated transactivation depends on the presence of the C-terminal transactivation domain of β-catenin. To allow for similar protein accumulations of wild-type and mutant β-catenin (data not shown), 3 μg of the wild-type β-catenin gene construct or 0.3 μg of the S33A or ΔC β-catenin gene constructs was transfected, together with 0.5 μg of the BCL9 construct as indicated. (F) Recombinantly purified full-length (FL) and ΔC-β-catenin interact with GST-HD1-3 of BCL9 but not HD4-5 in a GST pulldown assay. β-Catenin was visualized by immunoblot analysis.
BCL9 synergizes with the C-terminal transactivation domain of β-catenin.
The presence of an activation domain in BCL9 that accounted for much of the activity of BCL9 in β-catenin-dependent transcription raised the question of whether the activation domains of BCL9 and β-catenin synergize. To this end, we compared the BCL9-mediated transactivations in combination with wild-type and ΔC β-catenin, which lacks the C-terminal transactivation domain. Consistent with previous observations, ΔC β-catenin alone activated the siamois promoter at a threefold-lower level than wild-type β-catenin (Fig. 5E) (27). In combination with BCL9, the activation of the siamois promoter by ΔC β-catenin was augmented only 2-fold, whereas a 15-fold enhancement was observed with wild-type β-catenin (Fig. 5E). As ΔC β-catenin accumulates at a higher level than wild-type β-catenin, we used S33A β-catenin, which accumulates at a level similar to that for ΔC β-catenin (27; data not shown). With S33A β-catenin, we observed a 22-fold enhancement of transactivation by BCL9. The marked dependence of BCL9-mediated transactivation on the C-terminal region of β-catenin raised the question of whether the C terminus of β-catenin has a role in the physical interaction between BCL9 and β-catenin. To examine this possibility, we performed a GST pulldown experiment with GST-HD1-3 and wild-type β-catenin or ΔC β-catenin. With both forms of β-catenin, we observed similar efficiencies of interaction (Fig. 5F). Thus, the dependence of the transactivation function of BCL9 on the C-terminal domain of β-catenin may reflect a functional synergy between both activation domains.
Mutational analysis of the β-catenin C terminus identifies amino acids that influence coactivator binding and transactivation.
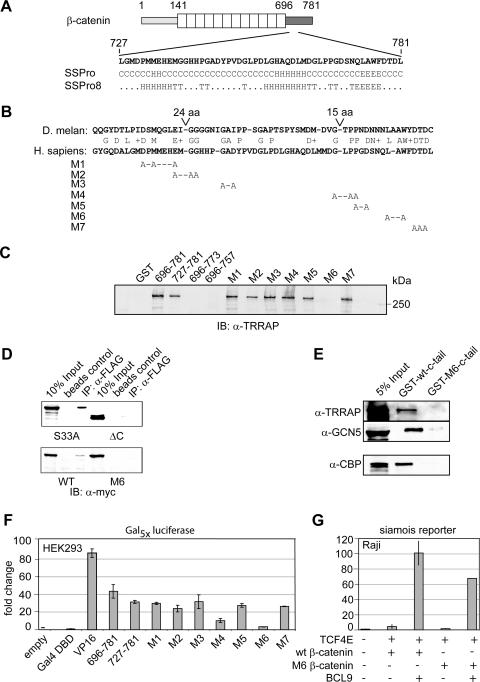
To gain further insight into the function of the C-terminal activation domain of β-catenin, we performed a mutational analysis. Secondary structure analysis of the C-terminal region predicted two short α-helices and a β-sheet motif (Fig. 6A). Moreover, a comparison of the amino acid sequences of human β-catenin and D. melanogaster armadillo allowed for the identification of conserved residues (Fig. 6B). We introduced clustered point mutations that changed conserved residues to alanines (constructs M1 to M7), and we examined the effects of these mutations on the physical interaction with coactivators. Consistent with a recent report, a mass spectrometric analysis of proteins that bind to the C-terminal region of β-catenin allowed us to identify TRRAP (transformation/transcription domain-associated protein) as an interaction partner (48; data not shown). To define the minimal domain of β-catenin that interacts with TRRAP, we performed a GST pulldown experiment in which GST fusions of different domains of β-catenin were incubated with a nuclear extract from HeLa cells. Immunoblot analysis with an anti-TRRAP antibody indicated that the amino acids between positions 727 and 781 of β-catenin comprise the minimal domain that interacts with TRRAP (Fig. 6C). Further truncations of the β-catenin C terminus markedly impaired the interaction with TRRAP (Fig. 6C). Analysis of the set of point mutations indicated that the M6 mutation (L774A/W776A) significantly reduced the interaction with TRRAP (Fig. 6C). We also confirmed the effects of the M6 mutation on the interaction with TRRAP in coimmunoprecipitations with extracts from HEK293 cells that have been transfected with myc-tagged β-catenin and FLAG-tagged TRRAP constructs. Efficient coimmunoprecipitation was detected with wild-type and S33A β-catenin but not with mutant β-catenin lacking the C terminus (ΔC) or carrying the M6 mutation, although in long exposures of the autoradiogram, a weak signal could be detected (Fig. 6D and data not shown). The immunoblot analysis for detection of myc-tagged β-catenin, whose results are shown in the lower panel of Fig. 6D, also demonstrates that wild-type and M6 β-catenin constructs are expressed at equal levels.
FIG. 6.
Mutational analysis of the β-catenin C terminus identifies residues that are important for coactivator binding and transactivation. (A) The C terminus (aa 727 to 781) of β-catenin is predicted to consist of helical (H) and β-sheet (E) motifs (T, turn; C, unstructured). Secondary structure contribution was calculated from the primary amino acid sequence (16). (B) Comparison of armadillo and β-catenin reveals conserved amino acids. Combinations of these amino acids were mutated to alanine as indicated. (C) L774A/W776A (M6) mutation of the β-catenin C terminus abolishes in vitro TRRAP binding. Mutant constructs were fused to GST and examined for binding to TRRAP in a GST pulldown assay with HeLa nuclear extract. Bound and subsequently eluted TRRAP was detected by immunoblot analysis (IB) with an anti-TRRAP (α-TRRAP) antibody. (D) L774A/W776A (M6) β-catenin displays reduced TRRAP binding in the context of the full-length protein. myc-tagged β-catenin constructs and Flag-TRRAP were transiently expressed in HEK293 cells and analyzed for interaction in coimmunoprecipitation assays. IP, immunoprecipitation; WT, wild type. (E) L774A/W776A (M6) β-catenin displays also reduced binding toward CBP. GST pulldown assays with wild-type and M6 mutant β-catenin C-terminal peptides were performed using Pd36 nuclear extract. (F) Transient transfections for determination of the transactivation potentials of various mutant β-catenin constructs that had been fused to the Gal4 DBD. The indicated gene constructs (0.1 μg) were transfected, together with a luciferase reporter carrying multimerized Gal4-binding sites, into HEK293 cells. (G) BCL9 (0.5 μg) activates the siamois reporter (1 μg) independently of the M6 β-catenin (3 μg) mutation in a transient transfection assay with Raji cells.
We also examined which histone acetyltransferase activity was associated with the TRRAP complex that interacts with the C-terminal domain of β-catenin. In a GST pulldown assay, GCN5 was found to interact with the wild-type C tail of β-catenin but not the M6 mutant C tail (Fig. 6E). In this assay, we also observed a similar effect of the M6 mutation on the interaction with CBP/p300, a bona fide coactivator of β-catenin, raising the possibility that the point mutations affect the overall structure of this transactivation domain (see below). Finally, we examined the effects of the truncations and point mutations in the context of a fusion with the Gal4 DBD. In transfection assays of HEK293 cells with a Gal4 reporter construct and expression plasmids for the various Gal4 fusion proteins, the M4 and M6 constructs reduced reporter activity 3- and 10-fold, respectively (Fig. 6F). We also examined the effects of the M6 mutation on the BCL9-mediated transactivation of the siamois reporter. In this experiment, we observed a modest decrease of the responsiveness to BCL9 relative to the level for wild-type β-catenin (Fig. 6G). Taken together, these results indicate that amino acids at the very C terminus of β-catenin are involved in binding of TRRAP and CBP/p300 and in mediating the activation potential of the transactivation domain.
L774A/W776A mutant β-catenin displays differences in secondary structure formation and reporter gene activation.
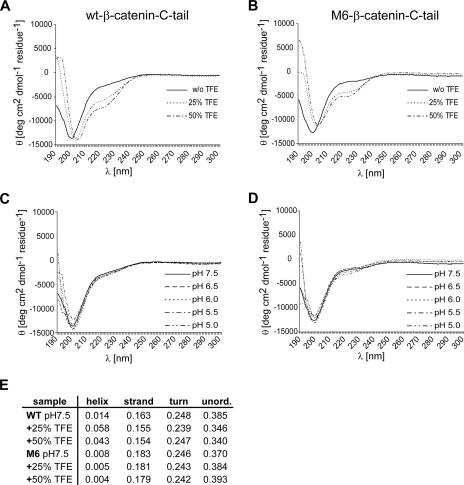
Mutation of the hydrophobic amino acids L774 and W776 virtually abrogated the putative hydrophobic β-sheet at the extreme C terminus of β-catenin (Fig. 6A). To assess the effect of the mutation on the secondary structure, we adopted the CD approach (31, 49). Purified recombinant wild-type and mutant (M6) β-catenin peptides (aa 727 to 781) were analyzed in the absence or presence of trifluoroethanol (TFE) and at different pH concentrations. Both parameters were shown to facilitate the formation of secondary structure, which are observed during the interaction of unfolded protein domains with protein partners (13). With the wild-type β-catenin peptide, the addition of TFE resulted in a new shoulder at 220 nm, suggesting the formation of some secondary structure (Fig. 7A). The shoulder was less pronounced with the M6 β-catenin peptide (Fig. 7B). Variations of pH had no significant effect on the CD spectra of both wild-type and M6 β-catenin peptides (Fig. 7C and D). Quantitative analysis of the spectra by use of the CONTIN algorithm revealed a modest induction of secondary structures in the wild-type β-catenin peptide upon addition of TFE (Fig. 7E). This analysis suggested that the M6 mutation impairs the ability of the C-terminal β-catenin peptide to adopt some secondary structure.
FIG. 7.
L774A/W776A (M6) β-catenin displays modest differences in secondary structure. (A to D) CD analysis of purified wild-type (wt) and mutant (M6) β-catenin C termini. (A and B) Addition of TFE modestly induces secondary structure formation. Wild-type and L774A/W776A (M6) β-catenin C termini in 25 mM sodium phosphate were measured in the absence and presence of increasing amounts of TFE. (C and D) pH variation does not affect secondary structure formation. Wild-type and M6 β-catenin levels were measured in 25 mM sodium phosphate at different pHs. (E) Computational analysis of CD data reveals differences in helix formation upon TFE treatment. Secondary structure contributions were calculated using the CONTIN algorithm. Computational analysis was conducted with Dichroweb software (62).
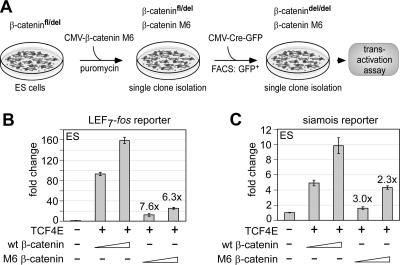
The reduction of activation potential of the Gal4 M6 C tail versus the level for the Gal4 wild-type C tail raised the question of the effects of the M6 mutation in the context of the full-length protein. In transfection assays, the effects of mutations in β-catenin have been proposed to be obscured by the presence of endogenous wild-type β-catenin (27). Therefore, we stably transfected an expression plasmid encoding M6 β-catenin into ES cells in which the endogenous β-catenin protein could be conditionally deleted (Fig. 8A). This strategy allowed for the maintenance of the adhesive function of β-catenin, which is important for the propagation of ES cells (9). Clones that expressed myc-tagged M6 β-catenin were identified by immunoblot analysis and used for the inactivation of endogenous β-catenin by transfection of a cytomegalovirus-Cre-internal ribosome entry site-green fluorescent protein (GFP) plasmid, whereby transfected cells were enriched by sorting for GFP-expressing cells. Cre-mediated inactivation of the endogenous β-catenin gene was confirmed by PCR analysis (data not shown). To assess the effects of the M6 mutation relative to wild-type β-catenin, we transfected the modified ES cells with the siamois or the LEF7-fos reporter, together with M6 or wild-type β-catenin (Fig. 8B and C). The level of reporter gene activation by M6 β-catenin was in the case of the LEF7-fos reporter eightfold lower than that observed with wild-type β-catenin but somewhat modestly reduced (threefold) in the presence of the siamois promoter. We also examined the effects of the M6 mutation on the β-catenin-dependent activation of an endogenous Wnt target gene. As we did not observe a robust Wnt response in the ES cell clone, we compared the expression of endogenous Axin2, a well-characterized Wnt target gene (30, 35, 39), in HEK293 cells that have been transfected with LEF1 and β-catenin expression plasmids and treated with the Wnt signaling surrogate lithium chloride or (as a control) sodium chloride. In mock-transfected cells, the number of Axin2 transcripts, detected by quantitative RT-PCR, was increased sixfold by lithium chloride treatment (data not shown). Transfection of wild-type β-catenin further increased expression of Axin2 by a factor of 2, whereas this increase was reduced by 50% in cells transfected with the M6 β-catenin expression plasmid. In summary, these results suggest that the mutation impairs, in part, the transactivation potential of full-length β-catenin, which contains an additional transactivation domain in the amino terminus that may compensate for the M6 mutation in vivo (27).
FIG. 8.
Transactivation defect of the β-catenin M6 mutation. (A) Scheme of the generation of β-catenindel/del/β-catenin M6 mutant ES cells. L774A/W776A (M6) β-catenin expression plasmid was stably integrated in ES cells, and the endogenous β-catenin gene was inactivated by transient transfection of a Cre-GFP expression plasmid. CMV, cytomegalovirus. (B and C) Transient transfection of mutant ES cells with expression plasmids encoding TCF4E or wild-type (wt) or M6 β-catenin shows a reduced activation potential of the M6 β-catenin on the LEF7-fos reporter (B) and the siamois reporter (C). The reduction of transactivation by M6 β-catenin relative to the level for wt β-catenin is indicated.
DISCUSSION
Transcriptional activation by β-catenin in response to Wnt signals involves both a C-terminal transactivation domain that binds multiple cofactors, including chromatin-modifying complexes, and BCL9, which has been proposed to act as a scaffold for the recruitment of Pygopus. Despite the identification of multiple proteins that interact with the C-terminal domain of β-catenin, it remains relatively unclear whether and how these proteins synergize with BCL9 to mediate the regulation of Wnt target genes. In our study, we found that BCL9 has three unexpected properties in the activation of Wnt target genes.
First, we observed cell type specificity in the function of BCL9. Although BCL9 mRNA is ubiquitously expressed and immunoblot analysis with a monoclonal anti-BCL9 antibody revealed similar protein expression levels in different cell lines, BCL9 augments β-catenin-dependent transcription primarily in lymphoid cells. In transfection assays, we observed a 5-fold activation of a siamois reporter construct in Jurkat T cells and an up to 16-fold activation in Raji B cells. In contrast, no significant promoter activation by BCL9 was detected in fibroblastic HeLa and HEK293 cells. Given the similar BCL9 protein expression levels in these cells, the cell type specificity of BCL9 function is rather unexpected and raises the possibility that posttranslational modifications or the interactions with partner proteins differ in lymphoid versus nonlymphoid cells. BCL9 has been initially identified as a gene that is translocated (t1;14)(q21;q32) in human B-cell lymphomas (64). The translocation and presumed overexpression of BCL9 protein have been proposed to contribute to the transformation of B-lymphoid cells. Moreover, a significant number of B-cell non-Hodgkin lymphomas have been correlated with alterations in the genomic organization and expression of the BCL9 locus (29). The effect of BCL9 overexpression on leukemogenesis may also be related to the cell-type-specific function of BCL9. Up-regulation of the BCL9-related protein BCL9L has also been detected in invasive carcinomas of colonic mucosa cells, raising the possibility that dysregulation of BCL9 and BCL9L contributes to aberrant activation of a nuclear Wnt response (46).
A second aspect of our findings concerns the promoter specificity of BCL9 function. In our transfection experiments, we found that the natural cdx1 and siamois promoters, which contain multiple LEF1/TCF-binding sites, respond much more strongly to BCL9 than the synthetic LEF7-fos promoter. In these experiments, we noted that the efficiency of activation by β-catenin is significantly lower with natural promoters than with the synthetic promoter. In particular, the activation of the LEF7-fos promoter by LEF1 and β-catenin is ∼440-fold, and it is enhanced only 2-fold by the coexpression of BCL9. In contrast, the activation of the siamois promoter by LEF1 and β-catenin is 7-fold, but it can be augmented 19-fold by BCL9. Moreover, the replacement of LEF1 with TCF1E, which increases β-catenin-dependent activation of the siamois promoter by a factor of 24, results in an additional 10-fold activation by BCL9. However, reporter activation via BCL9 is independent of the E tail, which has been recently identified as an auxiliary DBD in TCF1E and TCF4E (4; data not shown). Nevertheless, the increased activity of the TCF1E/β-catenin complex, relative to the level for LEF1/β-catenin, may reflect differences in binding of Wnt-responsive elements by LEF1 and TCF1E (3, 4). The more pronounced effect of BCL9 in the context of natural promoters raises the interesting possibility that the promoter strength might influence the dependence on BCL9. Similar promoter selectivity has been previously observed for the coactivator OcaB, which interacts with Oct transcription factors. In B-lymphoid cells, a subset of immunoglobulin kappa promoters with “suboptimal” Oct-binding sites shows a pronounced dependence on OcaB, whereas kappa promoters with “optimal” Oct-binding sites are independent of OcaB (15).
A third surprising result of our study was the identification of a potent transactivation function of BCL9. Previous genetic and functional analysis of Drosophila indicated that the amino-terminal third of BCL9/Lgs protein, including homology domains 1 to 3, is necessary and sufficient for the transcriptional regulation of Wnt target genes by BCL9/Lgs (34). Moreover, the forced expression of the HD1-3 part of BCL9 was found to complement mutations in BCL9/Lgs, and the function of BCL9 was attributed to the recruitment of Pygopus via the HD1 domain to DNA-bound TCF/β-catenin complexes (26, 34). According to these findings, a “chain of adaptors” model in which BCL9/Lgs acts solely as a “scaffolding” protein has been proposed (50). However, our transactivation experiments with lymphoid cell lines revealed a strong transactivation domain in the C-terminal half of BCL9. We found that truncations of the C terminus virtually abrogate the transcriptional activation potential of BCL9. In addition, we showed that the activation by BCL9 is, at least in part, independent of the recruitment of Pygopus, as the deletion of the HD1 domain or siRNA-mediated downregulation of Pygo1 and Pygo2 had a relatively modest effect on transactivation by BCL9.
The C-terminal region of BCL9 includes two homology domains (HD4 and HD5) that are highly conserved between mice, humans, and zebrafish. Interestingly, these homology domains are not found in the Drosophila Legless protein. HD4 and HD5 are also highly conserved between BCL9 and BCL9L/BCL9-2 (1). However, our functional analysis indicated that transactivation is mediated by a proline-rich domain residing between HD4 and HD5. Proline-rich domains have been identified as transactivation domains in several transcription factors (reviewed in references 20 and 57). A modest twofold contribution of the C-terminal half of BCL9L/BCL9-2 to transcriptional activation has been observed in transfection assays of HEK293 and SW480 cells, whereas a fourfold effect was observed in S33Y-β-catenin-induced transformation (1). Although these effects are modest compared to the function of the transactivation domain of BCL9 in Raji lymphoid cells, we consider it likely that the effects are mediated by the equivalent region in BCL9L.
Despite the cell-type-specific function of the transactivation domain in the context of full-length BCL9 protein, the domain shows similar activities in lymphoid and nonlymphoid cell lines when fused to a Gal4 DBD. This difference raises the interesting possibility that the lymphoid specificity of BCL9 function may involve the “unmasking” of a constitutive transactivation domain. Such changes in protein structure may involve cell-type-specific protein modifications or the differential interaction of protein partners. For example, the function of the C-terminal transactivation domain of β-catenin is regulated by modification of the tyrosine 654 residue in armadillo repeat 12, which has been proposed to modulate its juxtaposition with the C-terminal transactivation domain (43).
The proteins or complexes that determine the function of the C-terminal transactivation domain of BCL9 are still unknown. The apparent functional synergy with the transactivation domain of β-catenin raised the possibility that one of the known interaction partners of the C terminus of β-catenin also binds to the transactivation domain of BCL9. To this end, we performed coimmunoprecipitations of overexpressed epitope-tagged BCL9 to detect interactions with coexpressed CBP/p300, TRRAP, or parafibromin/Hyx. However, no interactions were detected, suggesting that another protein is involved in mediating the functional synergy. A functional collaboration between BCL9/Lgs and the C-terminal transactivation domain of β-catenin has also been observed in experiments in which a mutant BCL9/Lgs protein, carrying a D164A mutation that abrogates the interaction with β-catenin, was used in a transactivation experiment with β-catenin and parafibromin (41). In this experiment, the mutant form of β-catenin was found not to respond to parafibromin, suggesting that the function of Parafibromin is dependent on both the C terminus of β-catenin and BCL9. Parafibromin is a component of the PAF complex which is involved in transcriptional initiation and elongation by RNA polymerase II (2, 45, 66). Based on the interactions of CBP/p300, Brg1, and parafibromin with distinct but overlapping domains of β-catenin, it has been suggested that the recruitment of these cofactors results in a sequential or concerted histone acetylation, chromatin remodeling, and polymerase activation (41). Indeed, a careful kinetic study of the recruitment of β-catenin and β-catenin-associated proteins by Sierra and coworkers, in which the association of proteins with the c-Myc promoter was determined by chromatin immunoprecipitations, allowed for insight into the dynamics of target gene activation (48). This study showed that LEF1/TCF proteins are bound to the Wnt-responsive promoter prior to stimulation, whereas β-catenin and BCL9, Pygopus, p300 and RNA polymerase II are recruited 30′ after stimulation (48). Notably, β-catenin and its protein partners dissociated 60′ after stimulation and a cycling reassociation and dissociation of these proteins were found to be regulated by casein kinase II-mediated phosphorylation of the β-catenin interaction domain of LEF1 (60).
In addition to the Brg1 and CBP/p300 chromatin-modifying complexes that associate with the C terminus of β-catenin, the TRRAP/Tip60 histone acetyltransferase complex and the mixed-lineage-leukemia 1 (MLL-1) histone methyltransferase complex, which promotes H3K4 trimethylation, have been identified as cofactors of β-catenin (37, 48). Consistent with these observations, we have also identified the TRRAP/GCN5 (SAGA) complex as an interaction partner of the C terminus of β-catenin.
Although the function of BCL9 and its cofactor Pygopus in the nuclear response of Wnt signaling has been established both genetically and biochemically, the question as to whether they are obligatory cofactors of β-catenin or whether they act in a more specific context remains. For example, targeted inactivation of the Pygo2 gene or both Pygo1 and Pygo2 genes in the mouse results in relatively modest phenotypic defects (36, 47). In particular, Pygo1 Pygo2 double-deficient mice showed defects in the development of the lens of the eye and defects in the branching morphogenesis of the kidney. In addition, the crossing of Pygo1 Pygo2 double-deficient mice with transgenic mice carrying a lacZ reporter gene under the control of multimerized LEF1/TCF-binding sites indicated that Wnt signaling activity is modestly impaired (36, 47). Our observation that BCL9 can, at least in part, also act independently of Pygopus raises the interesting possibility that distinct cofactor complexes are recruited at different promoters and/or cell types. Conversely, Pygopus has also been found to directly interact with the Drosophila TCF protein, which may allow for bypassing the adaptor function of BCL9/Lgs (19). BCL9 has three conserved protein domains (HD3 to HD5), for which no interaction partners have yet been identified. These protein domains of BCL9 may allow for interactions with other DNA-binding proteins, providing some dependence on promoter context. In conclusion, our experiments indicated that BCL9 harbors a strong activation domain that synergizes with β-catenin and functions in a cell-type-specific and promoter-selective manner, which may help to augment and diversify the nuclear responses to Wnt signaling.
Acknowledgments
We are grateful to K. Basler for sharing a BCL9 cDNA construct. We thank R. Kemler for providing floxed β-catenin ES cells. We thank A. Hecht for donating several plasmids. We thank L. Tora for anti-TRRAP and anti-GCN5 antibodies. E. Kremmer we thank for expert help in the generation of BCL9 antibodies. We thank T. Stehle for the opportunity to use his spectropolarimeter. We thank H. J. Fehling for the supply with a Cre-EGFP construct.
This work was supported by the funds of the Max Planck Society and a grant from the German Research Foundation (DFG).
Footnotes
Published ahead of print on 17 March 2008.
REFERENCES
- 1.Adachi, S., T. Jigami, T. Yasui, T. Nakano, S. Ohwada, Y. Omori, S. Sugano, B. Ohkawara, H. Shibuya, T. Nakamura, and T. Akiyama. 2004. Role of a BCL9-related beta-catenin-binding protein, B9L, in tumorigenesis induced by aberrant activation of Wnt signaling. Cancer Res. 648496-8501. [DOI] [PubMed] [Google Scholar]
- 2.Adelman, K., W. Wei, M. B. Ardehali, J. Werner, B. Zhu, D. Reinberg, and J. T. Lis. 2006. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol. Cell. Biol. 26250-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atcha, F. A., J. E. Munguia, T. W. Li, K. Hovanes, and M. L. Waterman. 2003. A new beta-catenin-dependent activation domain in T cell factor. J. Biol. Chem. 27816169-16175. [DOI] [PubMed] [Google Scholar]
- 4.Atcha, F. A., A. Syed, B. Wu, N. Hoverter, N. N. Yokoyama, J. H. Ting, J. E. Munguia, H. J. Mangalam, J. L. Marsh, and M. L. Waterman. 2007. A unique DNA binding domain converts TCFs into strong Wnt effectors. Mol. Cell. Biol. 278352-8363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, N., A. Hurlstone, H. Musisi, A. Miles, M. Bienz, and H. Clevers. 2001. The chromatin remodelling factor Brg-1 interacts with beta-catenin to promote target gene activation. EMBO J. 204935-4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behrens, J., J. P. von Kries, M. Kuhl, L. Bruhn, D. Wedlich, R. Grosschedl, and W. Birchmeier. 1996. Functional interaction of beta-catenin with the transcription factor LEF-1. Nature 382638-642. [DOI] [PubMed] [Google Scholar]
- 7.Belenkaya, T. Y., C. Han, H. J. Standley, X. Lin, D. W. Houston, and J. Heasman. 2002. pygopus encodes a nuclear protein essential for wingless/Wnt signaling. Development 1294089-4101. [DOI] [PubMed] [Google Scholar]
- 8.Brannon, M., M. Gomperts, L. Sumoy, R. T. Moon, and D. Kimelman. 1997. A beta-catenin/XTcf-3 complex binds to the siamois promoter to regulate dorsal axis specification in Xenopus. Genes Dev. 112359-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brault, V., R. Moore, S. Kutsch, M. Ishibashi, D. H. Rowitch, A. P. McMahon, L. Sommer, O. Boussadia, and R. Kemler. 2001. Inactivation of the beta-catenin gene by Wnt1-Cre-mediated deletion results in dramatic brain malformation and failure of craniofacial development. Development 1281253-1264. [DOI] [PubMed] [Google Scholar]
- 10.Brembeck, F. H., T. Schwarz-Romond, J. Bakkers, S. Wilhelm, M. Hammerschmidt, and W. Birchmeier. 2004. Essential role of BCL9-2 in the switch between beta-catenin's adhesive and transcriptional functions. Genes Dev. 182225-2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruhn, L., A. Munnerlyn, and R. Grosschedl. 1997. ALY, a context-dependent coactivator of LEF-1 and AML-1, is required for TCRalpha enhancer function. Genes Dev. 11640-653. [DOI] [PubMed] [Google Scholar]
- 12.Brunner, E., O. Peter, L. Schweizer, and K. Basler. 1997. pangolin encodes a Lef-1 homologue that acts downstream of Armadillo to transduce the Wingless signal in Drosophila. Nature 385829-833. [DOI] [PubMed] [Google Scholar]
- 13.Buck, M. 1998. Trifluoroethanol and colleagues: cosolvents come of age. Recent studies with peptides and proteins. Q. Rev. Biophys. 31297-355. [DOI] [PubMed] [Google Scholar]
- 14.Cadigan, K. M., and R. Nusse. 1997. Wnt signaling: a common theme in animal development. Genes Dev. 113286-3305. [DOI] [PubMed] [Google Scholar]
- 15.Casellas, R., M. Jankovic, G. Meyer, A. Gazumyan, Y. Luo, R. Roeder, and M. Nussenzweig. 2002. OcaB is required for normal transcription and V(D)J recombination of a subset of immunoglobulin kappa genes. Cell 110575-585. [DOI] [PubMed] [Google Scholar]
- 16.Cheng, J., A. Z. Randall, M. J. Sweredoski, and P. Baldi. 2005. SCRATCH: a protein structure and structural feature prediction server. Nucleic Acids Res. 33W72-W76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Clevers, H. 2006. Wnt/beta-catenin signaling in development and disease. Cell 127469-480. [DOI] [PubMed] [Google Scholar]
- 18.Cox, R. T., L.-M. Pai, C. Kirkpatrick, J. Stein, and M. Pfeifer. 1999. Roles of the C terminus of armadillo in wingless signaling in drosophila. Genetics 153319-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de la Roche, M., and M. Bienz. 2007. Wingless-independent association of Pygopus with dTCF target genes. Curr. Biol. 17556-561. [DOI] [PubMed] [Google Scholar]
- 20.Dyson, H. J., and P. E. Wright. 2005. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell Biol. 6197-208. [DOI] [PubMed] [Google Scholar]
- 21.Fang, M., J. Li, T. Blauwkamp, C. Bhambhani, N. Campbell, and K. M. Cadigan. 2006. C-terminal-binding protein directly activates and represses Wnt transcriptional targets in Drosophila. EMBO J. 252735-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giese, K., A. Amsterdam, and R. Grosschedl. 1991. DNA-binding properties of the HMG domain of the lymphoid-specific transcriptional regulator LEF-1. Genes Dev. 52567-2578. [DOI] [PubMed] [Google Scholar]
- 23.Hecht, A., and M. P. Stemmler. 2003. Identification of a promoter-specific transcriptional activation domain at the C terminus of the Wnt effector protein T-cell factor 4. J. Biol. Chem. 2783776-3785. [DOI] [PubMed] [Google Scholar]
- 24.Hecht, A., K. Vleminckx, M. P. Stemmler, F. van Roy, and R. Kemler. 2000. The p300/CBP acetyltransferases function as transcriptional coactivators of beta-catenin in vertebrates. EMBO J. 191839-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hoffmans, R., and K. Basler. 2007. BCL9-2 binds Arm/beta-catenin in a Tyr142-independent manner and requires Pygopus for its function in Wg/Wnt signaling. Mech. Dev. 12459-67. [DOI] [PubMed] [Google Scholar]
- 26.Hoffmans, R., R. Stadeli, and K. Basler. 2005. Pygopus and legless provide essential transcriptional coactivator functions to armadillo/beta-catenin. Curr. Biol. 151207-1211. [DOI] [PubMed] [Google Scholar]
- 27.Hsu, S. C., J. Galceran, and R. Grosschedl. 1998. Modulation of transcriptional regulation by LEF-1 in response to Wnt-1 signaling and association with beta-catenin. Mol. Cell. Biol. 184807-4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu, Y., J. Kazenwadel, and R. James. 1993. Isolation and characterization of the murine homeobox gene cdx-1. J. Biol. Chem. 26827214-27225. [PubMed] [Google Scholar]
- 29.Itoyama, T., G. Nanjungud, W. Chen, V. G. Dyomin, J. Teruya-Feldstein, S. C. Jhanwar, A. D. Zelenetz, and R. S. Chaganti. 2002. Molecular cytogenetic analysis of genomic instability at the 1q12-22 chromosomal site in B-cell non-Hodgkin lymphoma. Genes Chromosomes Cancer 35318-328. [DOI] [PubMed] [Google Scholar]
- 30.Jho, E.-H., T. Zhang, C. Domon, C.-K. Joo, J.-N. Freund, and F. Constantini. 2002. Wnt/β-catenin/TCF signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol. Cell. Biol. 221172-1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelly, S. M., T. J. Jess, and N. C. Price. 2005. How to study proteins by circular dichroism. Biochim. Biophys. Acta 1751119-139. [DOI] [PubMed] [Google Scholar]
- 32.Kim, S., X. Xu, A. Hecht, and T. G. Boyer. 2006. Mediator is a transducer of Wnt/beta-catenin signaling. J. Biol. Chem. 28114066-14075. [DOI] [PubMed] [Google Scholar]
- 33.Kimelman, D., and W. Xu. 2006. beta-Catenin destruction complex: insights and questions from a structural perspective. Oncogene 257482-7491. [DOI] [PubMed] [Google Scholar]
- 34.Kramps, T., O. Peter, E. Brunner, D. Nellen, B. Froesch, S. Chatterjee, M. Murone, S. Zullig, and K. Basler. 2002. Wnt/wingless signaling requires BCL9/legless-mediated recruitment of pygopus to the nuclear beta-catenin-TCF complex. Cell 10947-60. [DOI] [PubMed] [Google Scholar]
- 35.Leung, J. Y., F. T. Kolligs, R. Wu, Y. Zhai, R. Kuick, S. Hanash, K. R. Cho, and E. R. Fearon. 2002. Activation of Axin2 expression by β-catenin-T cell factor. A feedback repressor pathway regulating Wnt signaling. J. Biol. Chem. 27721657-21665. [DOI] [PubMed] [Google Scholar]
- 36.Li, B., C. Rheaume, A. Teng, V. Bilanchone, J. E. Munguia, M. Hu, S. Jessen, S. Piccolo, M. L. Waterman, and X. Dai. 2007. Developmental phenotypes and reduced Wnt signaling in mice deficient for pygopus 2. Genesis 45318-325. [DOI] [PubMed] [Google Scholar]
- 37.Li, J., C. Sutter, D. S. Parker, T. Blauwkamp, M. Fang, and K. M. Cadigan. 2007. CBP/p300 are bimodal regulators of Wnt signaling. EMBO J. 262284-2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lickert, H., C. Domon, G. Huls, C. Wehrle, I. Duluc, H. Clevers, B. I. Meyer, J.-N. Freund, and R. Kemler. 2000. Wnt/β-catenin signaling regulates the expression of the homeobox gene cdx1 in embryonic intestine. Development 1273805-3813. [DOI] [PubMed] [Google Scholar]
- 39.Lustig, B., B. Jerchow, M. Sachs, S. Weiler, T. Pietsch, U. Karsten, M. van de Wetering, H. Clevers, P. M. Schlag, W. Birchmeier, and J. Behrens. 2002. Negative feedback loop of Wnt signaling through upregulation of Conductin/Axin2 in colorectal cancer and liver tumors. Mol. Cell. Biol. 221184-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Molenaar, M., M. van de Wetering, M. Oosterwegel, J. Peterson-Maduro, S. Godsave, V. Korinek, J. Roose, O. Destree, and H. Clevers. 1996. XTcf-3 transcription factor mediates beta-catenin-induced axis formation in Xenopus embryos. Cell 86391-399. [DOI] [PubMed] [Google Scholar]
- 41.Mosimann, C., G. Hausmann, and K. Basler. 2006. Parafibromin/Hyrax activates Wnt/Wg target gene transcription by direct association with beta-catenin/Armadillo. Cell 125327-341. [DOI] [PubMed] [Google Scholar]
- 42.Parker, D. S., J. Jemison, and K. M. Cadigan. 2002. Pygopus, a nuclear PHD-finger protein required for Wingless signaling in Drosophila. Development 1292565-2576. [DOI] [PubMed] [Google Scholar]
- 43.Piedra, J., D. Martinez, J. Castano, S. Miravet, M. Dunach, and A. G. de Herreros. 2001. Regulation of beta-catenin structure and activity by tyrosine phosphorylation. J. Biol. Chem. 27620436-20443. [DOI] [PubMed] [Google Scholar]
- 44.Riese, J., X. Yu, A. Munnerlyn, S. Eresh, S. C. Hsu, R. Grosschedl, and M. Bienz. 1997. LEF-1, a nuclear factor coordinating signaling inputs from wingless and decapentaplegic. Cell 88777-787. [DOI] [PubMed] [Google Scholar]
- 45.Rozenblatt-Rosen, O., C. M. Hughes, S. J. Nannepaga, K. S. Shanmugam, T. D. Copeland, T. Guszczynski, J. H. Resau, and M. Meyerson. 2005. The parafibromin tumor suppressor protein is part of a human Paf1 complex. Mol. Cell. Biol. 25612-620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakamoto, I., S. Ohwada, H. Toya, N. Togo, K. Kashiwabara, T. Oyama, T. Nakajima, H. Ito, S. Adachi, T. Jigami, and T. Akiyama. 2007. Up-regulation of a BCL9-related beta-catenin-binding protein, B9L, in different stages of sporadic colorectal adenoma. Cancer Sci. 9883-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwab, K. R., L. T. Patterson, H. A. Hartman, N. Song, R. A. Lang, X. Lin, and S. S. Potter. 2007. Pygo1 and Pygo2 roles in Wnt signaling in mammalian kidney development. BMC Biol. 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sierra, J., T. Yoshida, C. A. Joazeiro, and K. A. Jones. 2006. The APC tumor suppressor counteracts beta-catenin activation and H3K4 methylation at Wnt target genes. Genes Dev. 20586-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sreerama, N., and R. W. Woody. 2000. Estimation of protein secondary structure from circular dichroism spectra: inclusion of denatured proteins with native proteins in the analysis. Anal. Biochem. 287243-251. [DOI] [PubMed] [Google Scholar]
- 50.Stadeli, R., and K. Basler. 2005. Dissecting nuclear Wingless signalling: recruitment of the transcriptional co-activator Pygopus by a chain of adaptor proteins. Mech. Dev. 1221171-1182. [DOI] [PubMed] [Google Scholar]
- 51.Stadeli, R., R. Hoffmans, and K. Basler. 2006. Transcription under the control of nuclear Arm/beta-catenin. Curr. Biol. 16R378-R385. [DOI] [PubMed] [Google Scholar]
- 52.Takemaru, K. I., and R. T. Moon. 2000. The transcriptional coactivator CBP interacts with beta-catenin to activate gene expression. J. Cell Biol. 149249-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, B., F. Townsley, R. Rosin-Arbesfeld, H. Musisi, and M. Bienz. 2002. A new nuclear component of the Wnt signalling pathway. Nat. Cell Biol. 4367-373. [DOI] [PubMed] [Google Scholar]
- 54.Thompson, B. J. 2004. A complex of Armadillo, Legless, and Pygopus coactivates dTCF to activate wingless target genes. Curr. Biol. 14458-466. [DOI] [PubMed] [Google Scholar]
- 55.Townsley, F. M., A. Cliffe, and M. Bienz. 2004. Pygopus and Legless target Armadillo/beta-catenin to the nucleus to enable its transcriptional co-activator function. Nat. Cell Biol. 6626-633. [DOI] [PubMed] [Google Scholar]
- 56.Townsley, F. M., B. Thompson, and M. Bienz. 2004. Pygopus residues required for its binding to Legless are critical for transcription and development. J. Biol. Chem. 2795177-5183. [DOI] [PubMed] [Google Scholar]
- 57.Triezenberg, S. J. 1995. Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5190-196. [DOI] [PubMed] [Google Scholar]
- 58.van de Wetering, M., R. Cavallo, D. Dooijes, M. van Beest, J. van Es, J. Loureiro, A. Ypma, D. Hursh, T. Jones, A. Bejsovec, M. Peifer, M. Mortin, and H. Clevers. 1997. Armadillo coactivates transcription driven by the product of the Drosophila segment polarity gene dTCF. Cell 88789-799. [DOI] [PubMed] [Google Scholar]
- 59.van de Wetering, M., M. Oosterwegel, D. Dooijes, and H. Clevers. 1991. Identification and cloning of TCF-1, a T lymphocyte-specific transcription factor containing a sequence-specific HMG box. EMBO J. 10123-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang, S., and K. A. Jones. 2006. CK2 controls the recruitment of Wnt regulators to target genes in vivo. Curr. Biol. 162239-2244. [DOI] [PubMed] [Google Scholar]
- 61.Waterman, M. L., W. H. Fischer, and K. A. Jones. 1991. A thymus-specific member of the HMG protein family regulates the human T cell receptor C alpha enhancer. Genes Dev. 5656-669. [DOI] [PubMed] [Google Scholar]
- 62.Whitmore, L., and B. A. Wallace. 2004. DICHROWEB: an online server for protein secondary structure analyses from circular dichroism spectroscopic data. Nucleic Acids Res. 32W668-W673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Willert, K., and K. A. Jones. 2006. Wnt signaling: is the party in the nucleus? Genes Dev. 201394-1404. [DOI] [PubMed] [Google Scholar]
- 64.Willis, T. G., I. R. Zalcberg, L. J. A. Coignet, I. Wlodarska, M. Stul, D. M. Jadayel, C. Bastard, J. G. Treleaven, D. Catovsky, M. L. M. Silva, and M. J. S. Dyer. 1998. Molecular cloning of translocation t(1;14)(q21;q32) defines a novel gene (BCL9) at chromosome 1q21. Blood 911873-1881. [PubMed] [Google Scholar]
- 65.Yang, W. M., S. C. Tsai, Y. D. Wen, G. Fejer, and E. Seto. 2002. Functional domains of histone deacetylase-3. J. Biol. Chem. 2779447-9454. [DOI] [PubMed] [Google Scholar]
- 66.Yart, A., M. Gstaiger, C. Wirbelauer, M. Pecnik, D. Anastasiou, D. Hess, and W. Krek. 2005. The HRPT2 tumor suppressor gene product parafibromin associates with human PAF1 and RNA polymerase II. Mol. Cell. Biol. 255052-5060. [DOI] [PMC free article] [PubMed] [Google Scholar]