Abstract
Notch is a transmembrane receptor that determines cell fates and pattern formation in all animal species. After specific ligand binding, the intracellular part of Notch is cleaved off and translocates to the nucleus, where it targets the DNA binding protein RBP-Jκ. In the absence of Notch, RBP-Jκ represses Notch target genes by recruiting a corepressor complex. We and others have previously identified SHARP as one component of this complex. Here, we show that the corepressor ETO as well as the leukemogenic fusion protein AML1/ETO directly interacts with SHARP, that ETO is part of the endogenous RBP-Jκ-containing corepressor complex, and that ETO is found at Notch target gene promoters. In functional assays, corepressor ETO, but not AML1/ETO, augments SHARP-mediated repression in an histone deacetylase-dependent manner. Furthermore, either the knockdown of ETO or the overexpression of AML1/ETO activates Notch target genes. Therefore, we propose that AML1/ETO can disturb the normal, repressive function of ETO at Notch target genes. This activating (or derepressing) effect of AML1/ETO may contribute to its oncogenic potential in myeloid leukemia.
A small number of signaling pathways are known to regulate gene expression and, hence, cell fates in many organ systems. Notch acts as the receptor in one of these pathways and is involved in regulating many cellular processes, such as stem cell maintenance or differentiation during the development and renewal of adult tissues (reviewed in references 5 and 17). In higher eukaryotes, well-studied examples of the influence of Notch on cell fate are neurogenesis and myogenesis in Drosophila (reviewed in reference 4) and hematopoiesis in mice (reviewed in references 27 and 38).
At the molecular level, the triggering of the Notch receptor by ligand binding leads to proteolytic processing within the transmembrane domain, which results in the release of the Notch intracellular domain Notch-IC. Notch-IC can translocate to the nucleus, where it targets the DNA binding protein RBP-Jκ, also known as CSL [CBF1, Su(H), Lag-1)], in order to activate Notch target genes. In mice and Drosophila melanogaster, the phenotypes that are produced upon depletion of RBP-Jκ are similar, but not identical, to those produced by the loss of Notch function. Subsequently, it became clear to us that many differences could be explained by the derepression of Notch target genes. Hence, we postulated that repression and activation via RBP-Jκ involve the recruitment of distinct corepressors and coactivator complexes (reviewed in references 5 and 23). Notch-IC binding to RBP-Jκ is crucial for switching from the repressed state to the activated state. When Notch-IC enters the nucleus, its binding to RBP-Jκ may trigger an allosteric change that facilitates the displacement of the transcriptional corepressor complex. Subsequently, Mastermind binds to Notch-IC/RBP-Jκ and the resulting ternary protein complex recruits coactivators, such as histone acetyltransferase p300 or PCAF (22, 33, 40, 44), chromatin remodeling factors, and the mediator complex, to activate transcription (12). When Notch-IC is absent from the nucleus, RBP-Jκ recruits a histone deacetylase (HDAC)-containing corepressor complex (15, 18, 32, 34).
We and others have previously described the protein SHARP (SMRT- and HDAC1-associated repressor protein) as part of the RBP-Jκ corepressor complex (21, 32). SHARP is a ubiquitously expressed, large protein of approximately 450 kDa, containing four RNA recognition motifs at its N terminus and a highly conserved SPOC domain at its C terminus (3, 30, 37). Its highly conserved repression domain has been described to interact with SMRT and N-CoR (37) and with CtIP/CtBP (34). Mice deficient for the murine SHARP homologue MINT die during late embryogenesis (21). Studies using fetal liver transfer and a conditional knockout approach have shown a SHARP deficiency to cause hematopoietic defects in marginal zone B-cell development and T-cell development (21, 39, 45).
ETO (also called myeloid translocational gene 8 protein [MTG8]) is best known as a fusion partner of AML1 in leukemias carrying the t(8;21) translocation (10, 29); in fact, the name of the protein is an acronym for the translocation (eight-twenty-one). ETO belongs to a family of conserved nuclear proteins whose members can be found from Drosophila to humans. It contains four evolutionarily conserved functional domains called nervy homology regions (NHRs). NHR2 is important for homodimerization and protein-protein interaction with other corepressors. Although ETO is not able to bind to DNA, it is reported to act as a negative transcriptional regulator. ETO can homodimerize and heterodimerize with other members of the ETO family as well as interact directly with the corepressors N-CoR, SMRT, and Sin3A (reviewed in reference 16) (13, 26, 41). The function of ETO as a corepressor depends also on recruitment of HDACs, especially HDAC1, -2, and -3 (2).
The t(8;21)(q22/q22) translocation, which fuses the ETO gene on human chromosome 8 with the AML1 gene on chromosome 21, is seen in approximately 12 to 15% of acute myeloid leukemia (AML) cases, and in about 40% of AML cases, it is seen with a French-American-British-classified M2 phenotype (9). AML1 (also known as Runx-1) is a transcription factor that forms a heterodimer with a non-DNA-binding protein, CBFβ (31, 41). The t(8;21) translocation fuses DNA encoding the N-terminal 177 amino acids of AML1, which includes the RUNT DNA-binding domain (which also interacts with CBFβ), in frame with the codons for amino acids 30 to 604 of ETO. The AML1/ETO fusion deletes the terminal activation domain of AML1 and acts as a dominant-negative form of AML1, which represses AML1 target genes. In contrast, AML1/ETO could also be found as an activator of transcription involving Bcl-2 (20) and enhanced the expression of p21WAF1 (35). However, the mechanism through which AML1/ETO can activate transcription remains unclear.
The focus of this study is to further characterize the RBP-Jκ/SHARP corepressor complex. In a yeast two-hybrid screen with the RBP-Jκ-interacting corepressor SHARP, ETO was identified as an interaction partner. SHARP-ETO interaction was confirmed in glutathione S-transferase (GST) pull-down and coimmunoprecipitation experiments. Furthermore, in chromatin immunoprecipitation (ChIP) experiments, the colocalization of RBP-Jκ/SHARP and ETO could be found at Notch target genes. Interestingly, the leukemogenic fusion protein AML1/ETO also interacts with SHARP and is present in the endogenous RBP-Jκ corepressor complex of Kasumi cells. However, in functional assays, ETO but not AML1/ETO augments SHARP-mediated repression. Moreover, AML1/ETO is able to disturb transcriptional repression at Notch target genes. Therefore, we propose that AML1/ETO deregulates not only AML1 target genes but also Notch target genes.
MATERIALS AND METHODS
Oligonucleotides.
For a list of all oligonucleotides used for the construction of plasmids and for real-time PCR, see the materials and methods section of the supplemental material.
Plasmids.
The ETO expression plasmid was made by inserting the SalI (blunted with Klenow fragment)-digested/NotI-digested ETO fragment from pBind-ETO (provided by J. Lausen) into the EcoRV-digested/NotI-digested pDNA3 vector. The AML1/ETO expression plasmid was produced from pGEX6P1-AML1/ETO (provided by J. Lausen) in a manner similar to that described above. For bacterial expression of GST fusion proteins, the KpnI (blunted with Klenow fragment)-digested/NotI-digested ETO fragment from pcDNA3-ETO was inserted into the SmaI-digested/NotI-digested pGEX6P1 vector (Amersham). The GST fusions of SHARP as well as the expression plasmids pcDNA3-Flag-SHARP-RD and pcDNA3-Flag2-SHARP have previously been described (32, 34). For mapping of the SHARP ETO interaction, the expression plasmids pcDNA3-Flag3-ETO-N (amino acids 1 to 305), pcDNA3-Flag3-ETO-C (amino acids 297 to 604), and pcDNA3-Flag3-ETO (amino acids 400 to 604) were constructed as follows. The N-terminal fragment of ETO (ETO-N) was amplified by PCR using ETO-N_001 and ETO-N_002 as primers, digested with EcoRI and XhoI, and inserted into the corresponding sites of pcDNA3-Flag3. The C terminus of ETO (ETO-C) was amplified using the primers ETO-C_001 and ETO-C_002, digested with EcoRV and XhoI, and inserted into the corresponding sites of pcDNA3-Flag3. The ETO deletion mutant (amino acids 400 to 604) was amplified using the primers ETO-del_001 and ETO-del_002 by PCR, digested with EcoRV and XhoI, and inserted into pcDNA3-Flag3. The pcDNA3-ETO-enhanced green fluorescent protein (eGFP) and pcDNA3-AML1/ETO-eGFP constructs were made as follows. ETO-C was amplified by PCR using the upstream primer ETO_001 and the downstream primer ETO_002, digested with EcoRI and XhoI, and inserted into the corresponding sites of pcDNA3-eGFP, resulting in pcDNA3-ETO-C-eGFP. For inserting the N-terminal part of ETO or AML1/ETO, the vectors pcDNA3-ETO and pcDNA3-AML1/ETO were digested with EcoRI and inserted into the corresponding site of pcDNA3-ETO-C-eGFP. The pcDNA3-Flag3-AML1/ETO(tr)-eGFP expression plasmid was produced as follows. The C-terminal part of AML1/ETO was deleted using the 5′ phosphorylated oligonucleotides TR_001 and TR_002. The oligonucleotides were annealed, and the resulting double-stranded DNA flanked by the restriction sites for BamHI and XhoI was inserted into the corresponding sites of pcDNA3-Flag3-AML1/ETO, resulting in pcDNA3-Flag3-AML1/ETO(tr). For eGFP fusion, pcDNA3-AML1/ETO-eGFP was digested with XhoI and XbaI and the eGFP sequence was inserted into the corresponding sites of pcDNA3-Flag3-AML1/ETO(tr). The RBP2N specific expression vector pcDNA3-RBP2N-d2EosFP (42) was digested with XhoI and XbaI, and the PCR-amplified eqFP611 cDNA (primers RBP_001 and RBP_002) was inserted, resulting in RBP2N-eqFP611. The reporter plasmids for the luciferase assays pFR-Luc, pGA981/6, and HES1-Luc as well as the expression plasmids pCMV-RBP-VP16, pCMV-Gal4-VP16-SHARP-RD, pCMV-mNotch-1-IC, and pcDNA3-Flag1-SHARP (amino acids 2770 to 3127) were described previously (32). The Hey1(−97/+85)-Luc reporter was a gift from M. Gessler. The ETO(xl) and XETOR expression plasmids were gifts from Ying Cao, University of Ulm.
Yeast two-hybrid screening.
Yeast two-hybrid screening was performed as described previously (32).
Cell culture and preparation of cell extracts.
The HEK293 (ATCC CRL 1573) and HeLa (ATCC CCL 2) cell lines were grown in Dulbecco's modified Eagle's medium (Gibco) supplemented with 10% fetal calf serum (FCS). The Kasumi cell line was grown in RPMI 1640 medium (Gibco) supplemented with 20% FCS, 5% HEPES, and 5% l-glutamine. The murine erythroleukemia (MEL) and human erythroleukemia (HEL) cell lines were grown in RPMI 1640 medium (Gibco) with 10% FCS, penicillin, and streptomycin. A pre-T-cell line called Beko was derived from mice deficient in T-cell receptor beta (kind gift from J. Kisielow). Beko cells were grown in Iscove's modified Dulbecco medium (Gibco) with 2% FCS, nonessential amino acids, 0.3 mg/liter Primatone, and 5 mg/liter insulin. All cell lines were maintained at 37°C under 5% CO2. Whole-cell lysates for Western blotting and immunoprecipitation experiments were prepared as follows. Cells were washed in phosphate-buffered saline (PBS) and suspended in ice-cold CHAPS lysis buffer (10 mM 3{3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 50 mM Tris-HCl, pH 7.8, 150 mM NaCl, 5 mM NaF, 0.5 mM phenylmethylsulfonyl fluoride and protease inhibitors; Complete mix; Roche) and incubated on ice for 1 h. The lysates were cleared by centrifugation at 80,000 × g for 30 min. Protein concentrations were determined by using the Bradford assay method (Bio-Rad). The extracts were used for Western blotting and immunoprecipitation experiments.
DNA transfection.
HEK293 cells were transfected using the calcium phosphate coprecipitation method (Promega), HeLa cells were transfected using the FuGENE transfection reagent (Roche), and Kasumi cells were transfected with Lipofectamine 2000 (Invitrogen). All transfections were performed according to the manufacturer's instructions.
In vitro protein translation.
Proteins were translated in vitro in the presence of [35S]methionine by using the reticulocyte lysate-coupled transcription/translation system (Promega). Translation and labeling quality were monitored by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
GST pull-down assay.
The GST fusion proteins were expressed in Escherichia coli strain BL21 (Stratagene) and stored as whole bacterial lysates at −80°C. Approximately 1 μg of GST protein and GST fusion protein were immobilized with glutathione-Sepharose beads (Amersham) and incubated together with the in vitro-translated proteins under rotation at 4°C for 1 h. Beads were washed four times with 600 μl buffer A (40 mM HEPES [pH 7.5], 5 mM MgCl2, 0.2 mM EDTA, 0.5% NP-40, and 100 mM KCl) and four times with 600 μl buffer B (equivalent to buffer A, but containing 300 mM KCl). After the washing steps, the beads were resuspended in SDS-PAGE loading buffer and the proteins were separated by SDS-PAGE. The gels were dried and exposed to X-ray films.
Immunoprecipitation.
Immunoprecipitation experiments were carried out using whole-cell extracts from HEK293 cells 24 h after cotransfection with Flag-SHARP-RD (3477 to 3664) and expression plasmids for ETO and AML1/ETO. Cells were lysed with 700 μl CHAPS lysis buffer. The extracts were incubated with 40 μl agarose-conjugated anti-Flag antibody (M2; Sigma) at 4°C overnight. The beads were washed five times with CHAPS lysis buffer and resuspended in SDS-polyacrylamide gel loading buffer. For the investigation of endogenous protein interactions, immunoprecipitation experiments were carried out using CHAPS cell extracts from Kasumi cells or HEL cells. The extracts were incubated with 4 μg of anti-ETO antibody (Santa Cruz) or 20 μl of an anti-SHARP antibody (αSHARP 2 [32]) at 4°C overnight, followed by a 2-h incubation with 40 μl Sepharose-conjugated protein G beads (Amersham). The beads were washed five times with CHAPS lysis buffer and resuspended in SDS-PAGE loading buffer.
Western blotting.
The proteins resolved in SDS-polyacrylamide gels (10%) were transferred electrophoretically at room temperature to polyvinylidene difluoride membranes (Millipore) for 1 h at 50 mA by using a Tris-glycine buffer system. The membranes were preblocked for 1 h in a solution of 2% or 3% milk powder in 0.1% Tween 20 in PBS (PBS-T) before the antibodies were added. The following antibodies were used: anti-ETO (goat polyclonal immunoglobulin G [IgG] raised against the C terminus of ETO; catalog no. sc-9737; Santa Cruz), secondary antibody peroxidase-conjugated rabbit anti-goat IgG (Jackson ImmunoResearch), anti-Flag (catalog no. M5; Sigma), secondary antibody peroxidase-conjugated sheep anti-mouse IgG (catalog no. NXA931; Amersham), anti-RBP-Jκ (rat monoclonal IgG2a [catalog no. T6709; Institute of Immunology Co., Ltd.] and secondary antibody peroxidase-conjugated goat anti-rat IgG [Dianova]), anti-VP16 (mouse monoclonal IgG directed against amino acids 456 to 490 of VP16 [catalog no. sc-7545; Santa Cruz] and secondary antibody peroxidase-conjugated sheep anti-mouse IgG [NXA931; Amersham]).
Luciferase assay.
HeLa cells (5 × 104) were transfected in 24-well plates (with 1 μg of reporter plasmid DNA), together with various amounts of expression plasmid. Luciferase activity was determined from at least three independent transfections with 20 μl of cleared lysate in an LB 9501 luminometer (Berthold) by using the luciferase assay system from Promega.
Purification of RBP-Jκ DNA-binding complexes.
Purification was performed as described previously (34). Whole-cell extracts from 109 Kasumi cells were used. The DNA-binding complexes were analyzed by electromobility shift assay (EMSA) and Western blotting.
EMSA.
The EMSA experiments were performed as described previously (34).
ChIP.
MEL and Kasumi cells were fixed for 1 h with 10 mM dimethyladipimate. For cross-linking, the cells were incubated in 1% formaldehyde for 10 min at room temperature. The cross-linking reaction was stopped with 125 mM glycine. Nuclear extracts were prepared by using a cell lysis buffer (50 mM Tris-HCl [pH 8.0], 2 mM EDTA, 0.1% NP-40, 10% glycerol, 2 mm dithiothreitol). The nuclei were resuspended in 600 μl resuspension buffer (50 mM Tris-HCl [pH 8.0], 1% SDS, 5 mM EDTA, 2 mM dithiothreitol) and sonicated. Chromatin was 10-fold diluted with dilution buffer (50 mM Tris-HCl [pH 8.0], 200 mM NaCl, 5 mM EDTA, 0.5% NP-40) and precleared. The precleared chromatin extract was incubated overnight with, in each case, 50 μg antibodies against ETO (rabbit polyclonal antibody), SHARP (rabbit polyclonal antibody), and RBP-Jκ (Chemicon). The immunoprecipitates were immobilized on protein A-Sepharose beads for 2 h. After washing the beads with 500 mM NaCl and 500 mM LiCl containing buffers, we eluted chromatin from the beads with elution buffer (Tris-EDTA, 2% SDS) at room temperature. The cross-linking of chromatin was reversed for 6 h at 65°C in the presence of 300 mM NaCl and RNase A. Chromatin was precipitated with ethanol, dissolved in Tris-EDTA-PK buffer (10 mM Tris-HCl [pH 7.5], 5 mM EDTA, 0.25% SDS) and treated with proteinase K for 2 h at 45°C. After purification by phenol-chloroform extraction, the chromatin was precipitated at −20°C in the presence of 300 mM NaCl, tRNA, glycogen, and ethanol overnight. The chromatin was analyzed by quantitative real-time PCR.
Overexpression in HEK293 cells and knockdown in Kasumi and pre-T cells.
HEK293 cells (3 × 106 in 10-cm dishes) were transfected with an empty pcDNA3 vector or expression plasmids pcDNA3-AML1/ETO, pcDNA3-AML1/ETO(tr), pcDNA3-mNotch-1-IC, and pCMV-RBP-VP16. The cells were harvested 24 h after transfection. After mRNA isolation and cDNA synthesis, the expression of Notch target genes was determined by real-time PCR. Kasumi cells were transfected with the pSIR-short interfering RNA (siRNA)-internal ribosome entry site-GFP vector, which expresses the specific AML1/ETO junction hairpin (CGA GAA CCU CGA AAU CGU ACT). GFP-positive and GFP-negative cells were sorted by fluorescence-activated cell sorting 3 days after transfection and subsequently analyzed via reverse transcription-PCR. The pre-T-cell line Beko was retrovirally infected with the pSIR-siRNA-histidinol vector, which expresses either hairpin ETO1.1 (GCA TGA GCA CCT TCT GCT CAA) or hairpin ETO1.2 (GAT CGC CTG TGG AAG TGA AGA). The retroviral packaging cell line Phoenix was transfected with calcium phosphate. Two days later, retroviral supernatants were taken and Beko cells were spin infected four times over 2 days. Cells were selected on 500 mM histidinol, and real-time PCR was performed 1 week after selection.
RESULTS
Corepressor SHARP physically interacts and colocalizes with ETO and AML1/ETO.
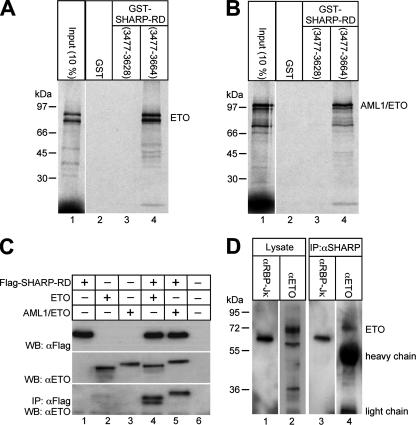
In order to identify proteins that interact with the RBP-Jκ-associated corepressor SHARP, we performed a yeast two-hybrid screen by using the repression domain of SHARP (SHARP-RD) as bait. A human embryonic brain library was screened, and the corepressor ETO was identified as an interactor. The association between SHARP-RD and ETO was verified in several independent assays (Fig. 1 and 2). First, in GST pull-down experiments, full-length ETO, radiolabeled in a cell-free system, interacted only with the bacterially expressed GST fusion protein GST-SHARP-RD (amino acids 3477 to 3664) (Fig. 1A, lane 4), not with a C-terminally truncated mutant of GST-SHARP-RD (amino acids 3477 to 3628) or GST alone (Fig. 1A, lanes 2 and 3). The fusion protein AML1/ETO, containing almost the whole ETO protein fused to the DNA-binding domain of AML1, also bound only to GST-SHARP-RD (amino acids 3477 to 3664) (Fig. 1B, lane 4), not to the truncated GST-SHARP-RD (amino acids 3477 to 3628) or GST alone (Fig. 1B, lanes 2 and 3).
FIG. 1.
ETO and AML1/ETO interact with SHARP in vitro (A and B) and in vivo (C and D). (A and B, lanes 4) Cell-free, synthesized, 35S-labeled ETO and AML1/ETO bind to GST-tagged SHARP-RD (amino acids 3477 to 3664) immobilized on glutathione-Sepharose beads. (A and B, lanes 3) They do not interact with a truncated version of immobilized GST-SHARP-RD (amino acids 3477 to 3628). (C) HEK293 cells were cotransfected with expression plasmids for ETO or AML1/ETO, together with Flag-tagged SHARP-RD. The expression of Flag-SHARP-RD, ETO, and AML1/ETO was detected by Western blotting (WB) using antibodies against the Flag tag (upper blot) and ETO (middle blot). ETO and AML1/ETO were coimmunoprecipitated only with SHARP-RD from lysates of cells cotransfected together with Flag-SHARP-RD (lower blot, lanes 4 and 5). (D) Endogenous RBP-Jκ and ETO could be coimmunoprecipitated from HEL cellular extracts by an anti-SHARP antibody (lanes 3 and 4). Coimmunoprecipitated ETO and AML1/ETO were detected on Western blots using the ETO antibody (αETO). Coimmunoprecipitated RBP-Jκ was detected using the RBP-Jκ antibody (αRBP-Jκ).
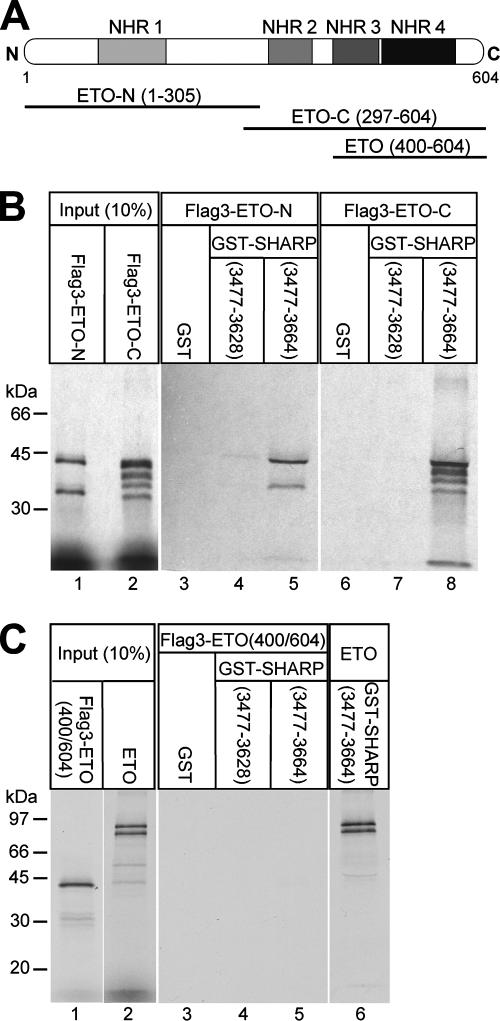
FIG. 2.
The first two functional domains of ETO (NHR1 and NHR2) are responsible for interaction with SHARP-RD. (A) Schematic representation of ETO and the truncated versions of ETO used in this study. (B) GST pull-down with the cell-free, synthesized, Flag-tagged, N-terminal (amino acids 1 to 305) and C-terminal (amino acids 297 to 604) halves of ETO. Both interact only with GST-SHARP-RD (amino acids 3477 to 3664) (lanes 5 and 8), not with the truncated GST-SHARP-RD (amino acids 3477 to 3628) (lanes 4 and 7) or GST alone (lanes 3 and 6). The GST fusions were immobilized on glutathione-Sepharose beads. (C) Cell-free synthesized ETO (amino acids 400 to 604) does not bind to immobilized GST-SHARP-RD any more (lanes 4 and 5). Cell-free synthesized full-length ETO was used as positive control (lane 6).
Secondly, coimmunoprecipitation experiments were performed in order to confirm the interaction of ETO (or AML1/ETO) and SHARP-RD in vivo. For this purpose, HEK293 cells were transiently transfected with expression plasmids for ETO or AML1/ETO alone or together with Flag-tagged SHARP-RD. Flag-SHARP-RD was immunoprecipitated with an anti-Flag antibody; ETO and AML1/ETO were detected in Western blot analyses by using an anti-ETO antibody (Fig. 1C). Immunoprecipitated ETO and AML1/ETO could be detected only if they were expressed together with Flag-SHARP-RD (Fig. 1C, bottom panel). In addition, after the incubation of cell lysates from ETO-expressing HEL cells (Fig. 1D, lane 2) with an antibody directed against endogenous SHARP protein (32), both RBP-Jκ (Fig. 1D, lane 3) and ETO (Fig. 1D, lane 4) were coimmunoprecipitated.
ETO contains four highly conserved functional domains called nervy homology regions (NHR1 to NHR4). Previous investigations showed that these domains mediate interaction with other repressor proteins like N-CoR, SMRT, or Sin3A (2, 25, 26, 41). In order to identify which domains of ETO are responsible for SHARP interaction, several Flag-tagged, truncated versions of ETO were synthesized (Fig. 2A). Both the N-terminal part of ETO containing NHR1, Flag3-ETO-N (amino acids 1 to 305), and the C-terminal part, Flag3-ETO-C (amino acids 297 to 604), containing NHR2 to NHR4, associated with GST-SHARP-RD (amino acids 3477 to 3664) in GST pull-down experiments (Fig. 2B, lanes 5 and 8). Neither construct showed any interaction with GST alone (Fig. 2B, lanes 3 and 6) or with the truncated GST-SHARP-RD (amino acids 3477 to 3628) (Fig. 2B, lanes 4 and 7). In vitro-translated, Flag-tagged ETO (400 to 604) containing NHR3 and NHR4 was not able to bind to GST-SHARP-RD (amino acids 3477 to 3664) (Fig. 2C, lane 5) in a comparison with full-length ETO as a positive control (Fig. 2C, lane 6). These results demonstrate that SHARP interacts with ETO and AML1/ETO in vitro and in vivo. Furthermore, the first two functional domains of ETO contribute to the interaction with the full-length SHARP repression domain in a manner independent of each other.
Since, at the moment, it is not possible to detect SHARP by Western blotting (21, 32), we decided to use an immunofluorescence approach. To examine the subcellular localization of SHARP, ETO, and AML1/ETO, HEK293 cells were cotransfected with eGFP fusions of ETO, AML1/ETO, and a recently identified C-terminally truncated version of AML1/ETO (46), together with full-length SHARP (see Fig. S1A in the supplemental material). SHARP was immunostained using an anti-serum directed against SHARP. All three eGFP fusion proteins and SHARP locate to the nucleus (see Fig. S1A in the supplemental material). SHARP interacts with RBP-Jκ through a conserved interaction domain (32). To investigate the subcellular localization of SHARP, RBP-Jκ, and AML1/ETO triple-color fluorescence images were taken of the nuclei of HEK293 cells cotransfected with AML1/ETO-eGFP and a fusion of RBP2N with an optimized variant of the red fluorescent protein eqFP611 (42, 43). AML1/ETO-eGFP, RBP2N-eqFP611, and endogenous SHARP localized in the nuclei of HEK293 cells (see Fig. S1B in the supplemental material).
ETO and SHARP colocalize with RBP-Jκ at promoter regions of Notch target genes.
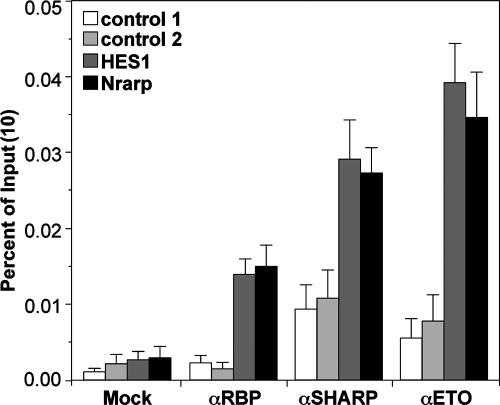
To investigate whether the ETO-SHARP interaction also takes place at Notch target genes in vivo, ChIP experiments were performed. We used ETO-expressing MEL cells and investigated whether ETO, SHARP, and RBP-Jκ can be found specifically at several Notch target gene promoters. After the immunoprecipitation of either RBP-Jκ, SHARP, or ETO, promoter regions of the HES1 gene and of the newly described Notch target Nrarp (24) could be specifically amplified, whereas several control regions (control 1, HES1 gene 3′ untranscribed region [UTR]; control 2, Hey1 gene 3′ UTR) remained at background levels (Fig. 3).
FIG. 3.
ETO, SHARP, and RBP-Jκ can be found associated at the promoter regions of the Notch target HES1 and Nrarp genes in MEL cells. ChIP experiments were performed as described in Materials and Methods. Mean values and standard deviations (error bars) of four independent experiments are shown. Control 1, HES1 gene 3′ UTR; control 2, Hey1 gene 3′ UTR.
AML1/ETO is part of the endogenous RBP-Jκ-complex.
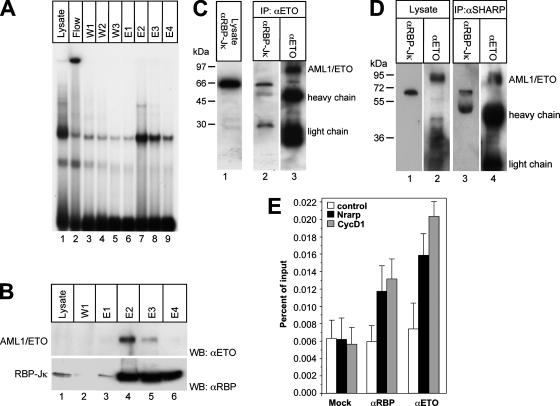
Previously, we isolated endogenous RBP-Jκ-containing complexes by DNA affinity chromatography (34). In order to investigate the formation of endogenous DNA-bound AML1/ETO-RBP-Jκ complexes, RBP-Jκ complexes were purified from Kasumi cells, which express AML1/ETO (11). Complex purification was verified by EMSA and Western blot analysis (Fig. 4A, lanes 7 to 9, and B, bottom panel, lanes 3 to 6). Western blot analyses further show that AML1/ETO coelutes with RBP-Jκ in fractions E2 and E3 (Fig. 4B, lanes 4 and 5). The presence of AML1/ETO within an endogenous RBP-Jκ protein complex was additionally confirmed by immunoprecipitation experiments with whole-cell lysates from Kasumi cells. For the precipitation of AML1/ETO, an anti-ETO antibody was used. RBP-Jκ could be coprecipitated with AML1/ETO, as shown by Western blots probed with an antibody against RBP-Jκ (Fig. 4C, lane 2). In addition, RBP-Jκ and AML1/ETO were also coimmunoprecipitated by using an antibody directed against endogenous SHARP (Fig. 4D, lanes 3 and 4). In order to investigate in vivo colocalization experiments with the fusion protein AML1/ETO and RBP-Jκ, ChIP experiments were performed using Kasumi cells. After precipitation with specific antibodies, RBP-Jκ and AML1/ETO were specifically found at the promoters of the Notch target Nrarp and cyclin D1 genes but not at the control (intron of the Hey1 gene) (Fig. 4E).
FIG. 4.
AML1/ETO is part of an endogenous RBP-Jκ complex in Kasumi cells. (A) Purification of endogenous RBP-Jκ complexes from Kasumi cells by DNA affinity chromatography. For purification, a DNA fragment containing 12 RBP-Jκ-binding sites was biotinylated and immobilized on a streptavidin-Sepharose column. The column was incubated with whole-cell lysates from Kasumi cells. After incubation and washing, the DNA-bound RBP-Jκ complexes were eluted with increasing concentrations of NaCl. (B) Western blot (WB) analysis of the lysate (lane 1), the first wash step (lane 2), and the eluted fractions (lanes 3 to 6) using antibodies against ETO (αETO) and RBP-Jκ (αRBP). AML1/ETO appears in the second and third elution steps, together with RBP-Jκ (lanes 4 and 5). (C) RBP-Jκ can be coimmunoprecipitated with AML1/ETO. Whole-cell lysates from Kasumi cells were incubated with an anti-ETO antibody overnight. The bound AML1/ETO complexes were immobilized on protein G-Sepharose beads. The cell lysates before (lane 1) and after precipitation (lane 2) were analyzed by Western blotting using an anti-RBP-Jκ antibody. (D) RBP-Jκ and AML1/ETO can be immunoprecipitated by using an antibody against SHARP. The cell lysates before (lanes 1 and 2) and after precipitation (lanes 3 and 4) were analyzed by using an anti-RBP-Jκ antibody or an anti-ETO antibody, respectively. (E) ETO and RBP-Jκ can be found associated at the promoter regions of the Notch target Nrarp and cyclin D1 genes in Kasumi cells. ChIP experiments were performed as described in Materials and Methods. Mean values and standard deviations (error bars) of four independent experiments are shown. As a control, an intron sequence of the Hey1 gene was used.
Taken together, these experiments demonstrate that AML1/ETO is a potential member of an endogenous DNA-bound RBP-Jκ complex in Kasumi cells. The association between AML1/ETO and RBP-Jκ is most probably mediated via SHARP, since we could not detect a direct interaction of AML1/ETO and RBP-Jκ.
ETO, but not AML1/ETO, augments SHARP-mediated repression.
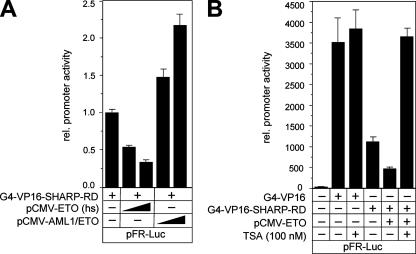
ETO has previously been described as a transcriptional corepressor (reviewed in reference 16). In order to investigate whether ETO is also able to augment SHARP-mediated repression, we performed several functional assays. HeLa cells were cotransfected with expression plasmids for Gal4-VP16-SHARP-RD, human ETO, or AML1/ETO, together with the Gal4-responsive reporter construct pFR-Luc. ETO enhanced the repression caused by Gal4-VP16-SHARP-RD in a dose-dependent manner up to fourfold (Fig. 5A). Surprisingly, the expression of AML1/ETO led to an up to twofold increase in promoter activity (Fig. 5A), suggesting that AML1/ETO interferes with the repressional activity of SHARP. Several control experiments were performed to probe the significance of these results (see Fig. S2 in the supplemental material). One, to exclude a possible deregulation of the activator by ETO or AML1/ETO, protein expression was analyzed after transfection of HEK293 cells with Gal4-VP16-SHARP-RD alone or together with ETO or AML1/ETO by Western blotting (see Fig. S2A in the supplemental material). Two, HeLa cells were cotransfected with expression plasmids for Gal4-VP16-SHARP-RD, ETO (xl) or the ETO-related XETOR from Xenopus laevis (6), together with the Gal4-responsive reporter construct pFR-Luc. ETO (xl), which showed a strong interaction with SHARP-RD (amino acids 3477 to 3664) in GST pull-down experiments (see Fig. S2B, lane 4, in the supplemental material), also enhanced the repression caused by Gal4-VP16-SHARP-RD in a dose-dependent manner (see Fig. S2C in the supplemental material). In contrast, XETOR, which failed to interact with SHARP (see Fig. S2B, lane 6, in the supplemental material), had no effect on promoter activity (see Fig. S2C in the supplemental material).
FIG. 5.
ETO, in contrast to AML1/ETO, acts as a corepressor for SHARP. (A) Human ETO increases, in a dose-dependent manner, the repression of promoter activity caused by SHARP-RD. In contrast, AML1/ETO increases the promoter activity. HeLa cells were transfected with the reporter construct pFR-Luc, the expression plasmid for the Gal4-VP16 fusion of SHARP-RD, and increasing amounts (100 ng and 150 ng) of the expression plasmids for ETO and AML1/ETO. (B) The repression caused by SHARP-RD and ETO is HDAC dependent. The reporter construct pFR-Luc was transfected alone or together with the expression plasmids for the Gal4-VP16 fusions and ETO into HeLa cells. The transfected cells were incubated for 6 h with TSA. The promoter activity was fully restored after the addition of TSA. Mean values and standard deviations (error bars) of at least four independent experiments are shown. rel., relative; −, absence of; +, presence of.
Previous studies have shown that ETO acts as an HDAC-dependent corepressor (28). To determine whether the repressional effect of ETO and SHARP is mediated by HDAC activity, HeLa cells were transfected with expression plasmids for Gal4-VP16 or Gal4-VP16-SHARP-RD and ETO, together with the Gal4-responsive reporter construct. Subsequently, the cells were treated with the HDAC inhibitor trichostatin A (TSA). The repression of the promoter activity caused by Gal4-VP16-SHARP-RD and ETO was completely abolished after TSA treatment (Fig. 5B). GST pull-down assays showed that cell-free synthesized HDAC1 directly interacts with GST-ETO. We also observed a direct interaction between HDAC1 and GST-AML1/ETO in GST pull-down assays (see Fig. S3 in the supplemental material). However, AML1/ETO did not act as a corepressor for SHARP.
ETO but not AML1/ETO represses RBP-VP16 mediated transcription.
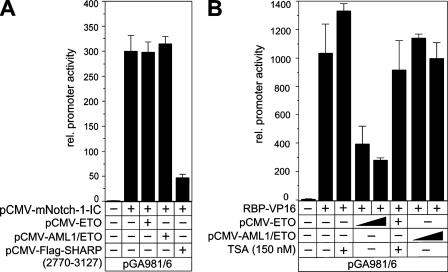
We have shown previously that RBP-Jκ/Notch-IC interaction is not compatible with RBP-Jκ/SHARP interaction due to competitive binding (32). The RBP-Jκ/Notch-IC-recruited coactivator complex is different from the RBP-Jκ/SHARP corepressor complex. To analyze the effect of ETO and AML1/ETO on Notch-1-IC activated transcription, we performed luciferase assays with the artificial promoter construct pGA981/6 containing several RBP-Jκ-binding sites. The promoter activity stimulated with Notch-1-IC was not changed by either ETO or AML1/ETO. In contrast, the cotransfection of a SHARP construct containing only the RBP-Jκ binding domain (amino acids 2770 to 3127) resulted in a clear reduction of promoter activity, most probably due to the displacement of the coactivator complex and not to repression (Fig. 6A). To analyze the effect of ETO and AML1/ETO in an RBP-Jκ/SHARP-dependent manner, the promoter construct was activated by cotransfection with RBP-VP16 (Fig. 6B), as shown previously (34). Promoter activity prestimulated with RBP-VP16 was reduced more than twofold by ETO, whereas AML1/ETO had no repressive effect on the promoter activity (Fig. 6B). In addition, treatment of the cells with the HDAC inhibitor TSA almost fully restored the promoter activity (Fig. 6B).
FIG. 6.
(A) ETO and AML1/ETO have no effect on the Notch-IC-recruited coactivator. The artificial reporter construct pGA981/6 was transfected into HeLa cells alone or together with a Notch-1-IC expression plasmid and an expression plasmid (100 ng) for ETO, AML1/ETO, or SHARP (amino acids 2779 to 3127). (B) ETO but not AML1/ETO acts as a corepressor for RBP-VP16-mediated transcription. The pGA981/6 reporter was transfected into HeLa cells alone or together with the RBP-VP16 expression plasmid and increasing amounts of expression plasmid (50 ng and 100 ng) for ETO and AML1/ETO. Where indicated, the HDAC inhibitor TSA (150 nM) was added to the cells 8 h prior to cell harvesting. Mean values and standard deviations (error bars) of at least three independent experiments are shown. rel., relative; −, absence of; +, presence of.
ETO but not AML1/ETO represses Hey1 and HES1 promoter activity.
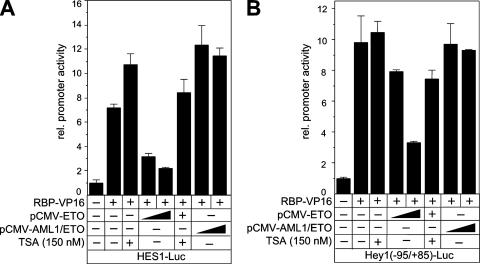
In humans, the HES1 and Hey1 genes had been identified as Notch target genes. In order to investigate the influence of ETO on these two Notch targets, luciferase assays were performed by using reporter constructs driven by either the HES1 or the Hey1 promoter. Cotransfection of RBP-VP16, together with the ETO expression plasmid, strongly reduced the promoter activity in a dose-dependent manner (Fig. 7A and B). Interestingly, cotransfection with AML1/ETO expression plasmid resulted in a slight activation of RBP-VP16-mediated transcriptional activity of the HES1 promoter construct (Fig. 7A), whereas AML1/ETO had no effect on the RBP-VP16-mediated transcriptional activity of the Hey1 reporter (Fig. 7B). The expression of the activator RBP-VP16 after the cotransfection of ETO or AML1/ETO was again tested by Western blotting (see Fig. S4 in the supplemental material). After treatment of the cells with TSA, the promoter activity was almost fully restored in all cases, suggesting that the ETO-mediated repression is indeed HDAC dependent (Fig. 7A and B).
FIG. 7.
ETO, but not AML1/ETO, acts as a corepressor for a RBP-Jκ-mediated transcription of HES1 (A) and Hey1 (B) genes. The reporter constructs were transfected into HeLa cells alone or together with the RBP-VP16 expression plasmid and increasing amounts of expression plasmid (50 ng and 100 ng) for ETO and AML1/ETO. Where indicated, the HDAC inhibitor TSA (150 nM) was added to the cells 8 h prior to cell harvesting. Mean values and standard deviations (error bars) of at least four independent experiments are shown. rel., relative; −, absence of; +, presence of.
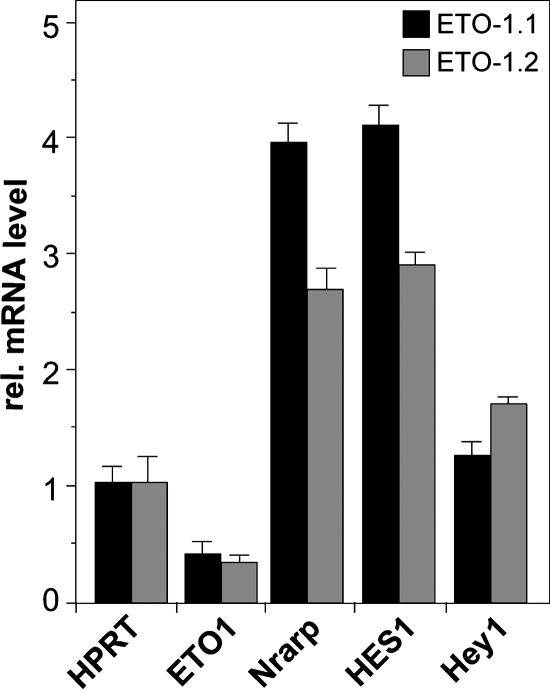
Either knockdown of ETO1 or overexpression of AML1/ETO activates Notch target genes.
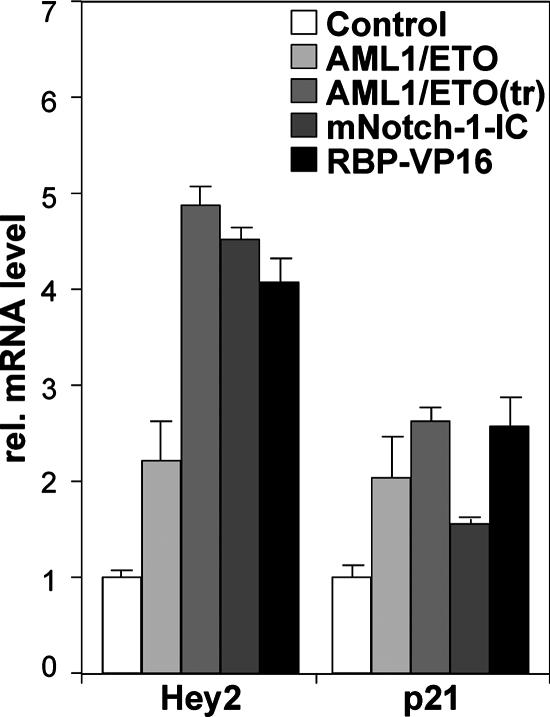
In order to investigate the loss of function of ETO at endogenous Notch target genes, we performed knockdown experiments in a pre-T-cell line derived from T-cell receptor beta knockout mice (Beko) that exclusively express ETO1 and not ETO2. Retroviral infection with two independent ETO1 siRNAs led to an upregulation of the Notch target HES1 and Nrarp genes and to a very minor upregulation of the Hey1 gene (Fig. 8). To analyze the effect of AML1/ETO expression on endogenous Notch target genes, we transfected HEK293 cells with expression plasmids for either AML1/ETO or the C-terminally truncated AML1/ETO(tr) that has previously been described (46) to be an even more potent oncogene. As a control, empty vector, Notch-IC, or RBP-VP16 was transfected. After mRNA isolation and cDNA synthesis, the expression of Hey2, p21WAF1, and actin for normalization was studied by real-time PCR. AML1/ETO, like Notch-IC or RBP-VP16, stimulates transcription by up to sixfold in the case of Hey2 and two- to threefold in the case of p21WAF1 (Fig. 9).
FIG. 8.
Knockdown of ETO1 activates the transcription of Notch target HES1 and Nrarp genes in pre-T cells. Two independent hairpins (ETO-1.1 and ETO-1.2) were used to infect a mouse pre-T-cell line. The expression of Notch target HES1 and Nrarp genes is upregulated, whereas the Hey1 gene level is only slightly affected. The values shown have been normalized with the expression of hypoxanthine phosphoribosyltransferase (HPRT). Mean values and standard deviations (error bars) of at least four independent measurements are shown. rel., relative.
FIG. 9.
AML1/ETO and AML1/ETO(tr) activate transcription of the Notch target Hey2 and p21Waf1 genes in HEK293 cells. Cells were transiently transfected with empty expression plasmid pcDNA3 or expression plasmid for AML1/ETO, AML1/ETO(tr), Notch-1-IC, or RBP-VP16. The mRNA levels of the Notch targets Hey2 and p21Waf1 were determined by reverse transcription-PCR. As a control, the empty pcDNA3 vector was used. The values have been normalized with actin expression. Mean values and standard deviations (error bars) of at least four independent measurements are shown. rel., relative.
Specific knockdown of AML1/ETO leads to downregulation of Notch target genes.
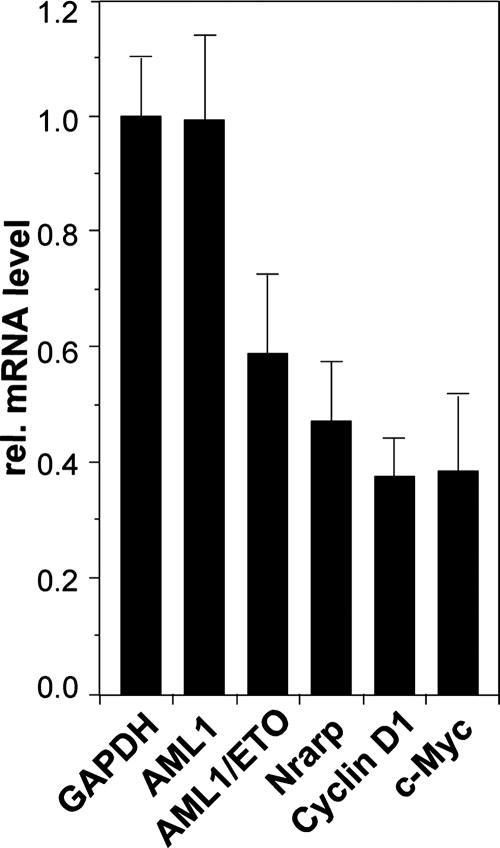
To analyze the effect of AML1/ETO on Notch target gene expression in the leukemic Kasumi cell line expressing AML1/ETO due to the t(8;21) translocation, we designed an siRNA that targets a 25-nucleotide region spanning the fusion of AML1 and ETO. Kasumi cells were transfected with the pSIR-siRNA-internal ribosome entry site-GFP vector expressing the specific AML1/ETO junction hairpin. GFP-positive and GFP-negative cells were sorted by fluorescence-activated cell sorting and subsequently analyzed for the expression of AML1, AML1/ETO, GAPDH (glyceraldehyde-3-phosphate dehydrogenase) for normalization, and several Notch target genes (Fig. 10). Specific downregulation of AML1/ETO mRNA, but not that of AML1, was observed. The twofold knockdown effect was also described previously by two other groups (14, 19). In a manner similar to that described above, Notch target Nrarp, cyclin D1, and c-Myc genes were significantly downregulated upon AML1/ETO knockdown (Fig. 10).
FIG. 10.
Downregulation of the Notch target Nrarp, cyclin D1, and c-Myc genes after specific knockdown of AML1/ETO in Kasumi cells. Cells were transfected with a construct expressing a hairpin RNA directed against the AML1/ETO fusion region and GFP. After GFP-positive cells were sorted, relative (rel.) mRNA levels were determined by real-time PCR. The values have been normalized with the expression of GAPDH. Mean values and standard deviations (error bars) of four independent measurements are shown. rel., relative.
DISCUSSION
RBP-Jκ is the central player in the transcriptional regulation of Notch target genes and functions in both transcriptional repression and activation. Notch-IC enters the nucleus, binds to RBP-Jκ, and activates target genes. In the absence of Notch-IC, RBP-Jκ recruits a corepressor complex that keeps Notch target genes inactive. Previously, we described SHARP as an RBP-Jκ-interacting corepressor (32). We could subsequently show that corepressors CtIP and CtBP are recruited by RBP-Jκ/SHARP to repress transcription of the Notch target gene Hey1 (34). Here, we have investigated the role of ETO and the leukemogenic fusion protein AML1/ETO in RBP-Jκ/SHARP-mediated repression of Notch target genes and could demonstrate that ETO, but not AML1/ETO, is able to augment RBP-Jκ/SHARP-mediated repression of Notch target genes.
The repression domain of SHARP functions as a platform for different corepressors.
It has previously been shown that the SHARP repression domain can interact with SMRT and N-CoR (37) as well as with CtIP/CtBP (34). In the present study, we show the functional interaction of SHARP and ETO. These observations suggest a redundancy in the sense that different repressor complexes act at different Notch target genes or in different cell types. Alternatively, the RBP-Jκ/SHARP corepressor complexes might cross talk with other repressor complexes.
Mechanism of AML1/ETO-mediated disruption of repression.
It has previously been described that AML1/ETO acts in a dominant manner to inhibit AML1-mediated transcription (reviewed in reference 7). However, there are reports which indicate that AML1/ETO can be an activator regarding Bcl-2 (20) and enhance the expression of p21Waf1 (35) and HES1 (1). Our finding that ETO, but not AML1/ETO, can act in concert with RBP-Jκ/SHARP to repress transcription at Notch target genes was surprising. Different potential scenarios might explain our results at the molecular level. (i) The Runx domain of AML1 is the binding domain of CBFβ, and this heterodimer has been characterized extensively (47). The heterodimer of AML1/ETO and CBFβ could potentially activate Notch target genes. (ii) The Runx domain of AML has previously been described to interact with other transcriptional activators, including Ets family factors, c-Jun/c-fos, and C-EBP family factors (reviewed in reference 36). Potentially, such an interactor causes transcriptional activation at Notch target genes. (iii) Alternatively, AML1/ETO and ETO localization in the nucleus could be affected in a way that cannot be resolved by standard microscopic techniques. Both proteins have been found in the nucleus, but more careful analysis might reveal differences in localization. An alternative splice variant of AML1/ETO that promotes leukemogenesis more efficiently has previously been described (46). As shown in Fig. 9, the truncated form of AML1/ETO, AML1/ETO(tr), upregulates Notch target genes in a manner stronger than that of full-length AML1/ETO. The C-terminal region that is truncated contains NHR3 and NHR4, which are also the binding sites for SMRT. It is possible that the lack of ETO repression in the AML1/ETO(tr) domain makes it an even better activator (“derepressor”).
Upregulation of Jagged1 by AML1/ETO.
It has previously been reported that the expression of AML1/ETO causes the upregulation of the Notch ligand Jagged1 and thereby results in the specific activation of Notch signaling through the Jagged1 ligand (1). Our results are in line with the upregulation of certain Notch target genes by AML1/ETO (see Fig. S5 in the supplemental material). To test whether this upregulation is mediated by Notch processing due to ligand binding, we added the presenilin inhibitor N-[N-(3,5-difluorophenacetyl-l-alanyl)]-S-phenylglycine t-butyl ester (DAPT) to the cells expressing AML1/ETO or mNotch-1-IC-ΔE as a control. As expected, in mNotch-1-IC-ΔE-expressing cells, DAPT downregulates the expression of luciferase from the HES1-specific reporter construct. In contrast, in AML1/ETO-expressing cells, DAPT has no effect on HES1 promoter activity (see Fig. S5 in the supplemental material). Therefore, HES1 transactivation by AML1/ETO seems to be independent of Jagged1. These results argue against a positive autofeedback loop via Jagged1-mediated Notch processing. Alternatively, we propose that AML1/ETO upregulates Notch target genes by directly disrupting the normal corepressor function of SHARP/ETO.
Does derepression of Notch target genes by AML1/ETO contribute to leukemogenesis?
The activation of Notch signaling favors the expansion of hematopoietic stem cells (reviewed in reference 27). These findings would be in line with the observation that the leukemogenic fusion protein AML1/ETO promotes the expression of certain Notch target genes. Retroviral transduction of hematopoietic stem cells of AML1/ETO recapitulates many of the hematopoietic developmental abnormalities seen in humans, but animals do not develop leukemia for several months (8). This observation suggests that a principal contribution of AML1/ETO to AML is to maintain the undifferentiated state or even expand these progenitor cells. The activation of Notch target genes might be crucial in this respect. We suggest that endogenous ETO is a novel corepressor to keep Notch target gene expression silent. The leukemogenic fusion protein AML1/ETO can disrupt the repressive ETO function and activate certain Notch target genes. This in turn might favor the expansion of hematopoietic progenitor cells and possibly contribute to leukemogenesis.
In summary, our results indicate that corepressor ETO is a novel component of the RBP-Jκ/SHARP corepressor complex and that ETO, but not AML1/ETO, augments repression at Notch target genes. It will be interesting to further investigate the significance of the disruption of the repressive function ETO by AML1/ETO in myeloid leukemia.
Supplementary Material
Acknowledgments
We thank J. Lausen, D. van Essen, M. Gessler, and Ying Cao for providing us with plasmids and J. Kisielow for the pre-T-cell line Beko. We thank M. Krötschel, E. Rüber, R. Rittelmann, and S. Schirmer for excellent technical assistance.
This study was supported by the Deutsche Forschungsgemeinschaft Emmy-Noether fellowship (BO-1639/3-2) to T.B., DFG grants Wi-1990/2-1 to J.W. and SFB497/B9 and SFB518/A18 to F.O., and by funds from the Max Planck Society (to T.B.).
Footnotes
Published ahead of print on 10 March 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alcalay, M., N. Meani, V. Gelmetti, A. Fantozzi, M. Fagioli, A. Orleth, D. Riganelli, C. Sebastiani, E. Cappelli, C. Casciari, M. T. Sciurpi, A. R. Mariano, S. P. Minardi, L. Luzi, H. Muller, P. P. Di Fiore, G. Frosina, and P. G. Pelicci. 2003. Acute myeloid leukemia fusion proteins deregulate genes involved in stem cell maintenance and DNA repair. J. Clin. Investig. 1121751-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann, J. M., J. Nip, D. K. Strom, B. Lutterbach, H. Harada, N. Lenny, J. R. Downing, S. Meyers, and S. W. Hiebert. 2001. ETO, a target of t(8;21) in acute leukemia, makes distinct contacts with multiple histone deacetylases and binds mSin3A through its oligomerization domain. Mol. Cell. Biol. 216470-6483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ariyoshi, M., and J. W. Schwabe. 2003. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev. 171909-1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Artavanis-Tsakonas, S., M. D. Rand, and R. J. Lake. 1999. Notch signaling: cell fate control and signal integration in development. Science 284770-776. [DOI] [PubMed] [Google Scholar]
- 5.Bray, S. J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7678-689. [DOI] [PubMed] [Google Scholar]
- 6.Cao, Y., H. Zhao, and H. Grunz. 2002. XETOR regulates the size of the proneural domain during primary neurogenesis in Xenopus laevis. Mech. Dev. 11935-44. [DOI] [PubMed] [Google Scholar]
- 7.Davis, J. N., L. McGhee, and S. Meyers. 2003. The ETO (MTG8) gene family. Gene 3031-10. [DOI] [PubMed] [Google Scholar]
- 8.de Guzman, C. G., A. J. Warren, Z. Zhang, L. Gartland, P. Erickson, H. Drabkin, S. W. Hiebert, and C. A. Klug. 2002. Hematopoietic stem cell expansion and distinct myeloid developmental abnormalities in a murine model of the AML1-ETO translocation. Mol. Cell. Biol. 225506-5517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Downing, J. R. 1999. The AML1-ETO chimaeric transcription factor in acute myeloid leukaemia: biology and clinical significance. Br. J. Haematol. 106296-308. [DOI] [PubMed] [Google Scholar]
- 10.Downing, J. R., D. R. Head, A. M. Curcio-Brint, M. G. Hulshof, T. A. Motroni, S. C. Raimondi, A. J. Carroll, H. A. Drabkin, C. Willman, K. S. Theil, et al. 1993. An AML1/ETO fusion transcript is consistently detected by RNA-based polymerase chain reaction in acute myelogenous leukemia containing the (8;21)(q22;q22) translocation. Blood 812860-2865. [PubMed] [Google Scholar]
- 11.Erickson, P. F., G. Dessev, R. S. Lasher, G. Philips, M. Robinson, and H. A. Drabkin. 1996. ETO and AML1 phosphoproteins are expressed in CD34+ hematopoietic progenitors: implications for t(8;21) leukemogenesis and monitoring residual disease. Blood 881813-1823. [PubMed] [Google Scholar]
- 12.Fryer, C. J., J. B. White, and K. A. Jones. 2004. Mastermind recruits CycC:CDK8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 16509-520. [DOI] [PubMed] [Google Scholar]
- 13.Gelmetti, V., J. Zhang, M. Fanelli, S. Minucci, P. G. Pelicci, and M. A. Lazar. 1998. Aberrant recruitment of the nuclear receptor corepressor-histone deacetylase complex by the acute myeloid leukemia fusion partner ETO. Mol. Cell. Biol. 187185-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heidenreich, O., J. Krauter, H. Riehle, P. Hadwiger, M. John, G. Heil, H. P. Vornlocher, and A. Nordheim. 2003. AML1/MTG8 oncogene suppression by small interfering RNAs supports myeloid differentiation of t(8;21)-positive leukemic cells. Blood 1013157-3163. [DOI] [PubMed] [Google Scholar]
- 15.Hsieh, J. J., S. Zhou, L. Chen, D. B. Young, and S. D. Hayward. 1999. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc. Natl. Acad. Sci. USA 9623-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hug, B. A., and M. A. Lazar. 2004. ETO interacting proteins. Oncogene 234270-4274. [DOI] [PubMed] [Google Scholar]
- 17.Ilagan, M. X., and R. Kopan. 2007. SnapShot: notch signaling pathway. Cell 1281246. [DOI] [PubMed] [Google Scholar]
- 18.Kao, H. Y., P. Ordentlich, N. Koyano-Nakagawa, Z. Tang, M. Downes, C. R. Kintner, R. M. Evans, and T. Kadesch. 1998. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 122269-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasashima, K., E. Sakota, and T. Kozu. 2004. Discrimination of target by siRNA: designing of AML1-MTG8 fusion mRNA-specific siRNA sequences. Biochimie 86713-721. [DOI] [PubMed] [Google Scholar]
- 20.Klampfer, L., J. Zhang, A. O. Zelenetz, H. Uchida, and S. D. Nimer. 1996. The AML1/ETO fusion protein activates transcription of BCL-2. Proc. Natl. Acad. Sci. USA 9314059-14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuroda, K., H. Han, S. Tani, K. Tanigaki, T. Tun, T. Furukawa, Y. Taniguchi, H. Kurooka, Y. Hamada, S. Toyokuni, and T. Honjo. 2003. Regulation of marginal zone B cell development by MINT, a suppressor of Notch/RBP-J signaling pathway. Immunity 18301-312. [DOI] [PubMed] [Google Scholar]
- 22.Kurooka, H., and T. Honjo. 2000. Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J. Biol. Chem. 27517211-17220. [DOI] [PubMed] [Google Scholar]
- 23.Lai, E. C. 2004. Notch signaling: control of cell communication and cell fate. Development 131965-973. [DOI] [PubMed] [Google Scholar]
- 24.Lamar, E., G. Deblandre, D. Wettstein, V. Gawantka, N. Pollet, C. Niehrs, and C. Kintner. 2001. Nrarp is a novel intracellular component of the Notch signaling pathway. Genes Dev. 151885-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lausen, J., S. Cho, S. Liu, and M. H. Werner. 2004. The nuclear receptor co-repressor (N-CoR) utilizes repression domains I and III for interaction and co-repression with ETO. J. Biol. Chem. 27949281-49288. [DOI] [PubMed] [Google Scholar]
- 26.Lutterbach, B., J. J. Westendorf, B. Linggi, A. Patten, M. Moniwa, J. R. Davie, K. D. Huynh, V. J. Bardwell, R. M. Lavinsky, M. G. Rosenfeld, C. Glass, E. Seto, and S. W. Hiebert. 1998. ETO, a target of t(8;21) in acute leukemia, interacts with the N-CoR and mSin3 corepressors. Mol. Cell. Biol. 187176-7184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maillard, I., T. Fang, and W. S. Pear. 2005. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23945-974. [DOI] [PubMed] [Google Scholar]
- 28.Melnick, A. M., J. J. Westendorf, A. Polinger, G. W. Carlile, S. Arai, H. J. Ball, B. Lutterbach, S. W. Hiebert, and J. D. Licht. 2000. The ETO protein disrupted in t(8;21)-associated acute myeloid leukemia is a corepressor for the promyelocytic leukemia zinc finger protein. Mol. Cell. Biol. 202075-2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyoshi, H., K. Shimizu, T. Kozu, N. Maseki, Y. Kaneko, and M. Ohki. 1991. t(8;21) breakpoints on chromosome 21 in acute myeloid leukemia are clustered within a limited region of a single gene, AML1. Proc. Natl. Acad. Sci. USA 8810431-10434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Newberry, E. P., T. Latifi, and D. A. Towler. 1999. The RRM domain of MINT, a novel Msx2 binding protein, recognizes and regulates the rat osteocalcin promoter. Biochemistry 3810678-10690. [DOI] [PubMed] [Google Scholar]
- 31.Ogawa, E., M. Inuzuka, M. Maruyama, M. Satake, M. Naito-Fujimoto, Y. Ito, and K. Shigesada. 1993. Molecular cloning and characterization of PEBP2 beta, the heterodimeric partner of a novel Drosophila runt-related DNA binding protein PEBP2 alpha. Virology 194314-331. [DOI] [PubMed] [Google Scholar]
- 32.Oswald, F., U. Kostezka, K. Astrahantseff, S. Bourteele, K. Dillinger, U. Zechner, L. Ludwig, M. Wilda, H. Hameister, W. Knochel, S. Liptay, and R. M. Schmid. 2002. SHARP is a novel component of the Notch/RBP-Jkappa signalling pathway. EMBO J. 215417-5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oswald, F., B. Tauber, T. Dobner, S. Bourteele, U. Kostezka, G. Adler, S. Liptay, and R. M. Schmid. 2001. p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 217761-7774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oswald, F., M. Winkler, Y. Cao, K. Astrahantseff, S. Bourteele, W. Knochel, and T. Borggrefe. 2005. RBP-Jκ/SHARP recruits CtIP/CtBP corepressors to silence Notch target genes. Mol. Cell. Biol. 2510379-10390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peterson, L. F., M. Yan, and D. E. Zhang. 2007. The p21Waf1 pathway is involved in blocking leukemogenesis by the t(8;21) fusion protein AML1-ETO. Blood 1094392-4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peterson, L. F., and D. E. Zhang. 2004. The 8;21 translocation in leukemogenesis. Oncogene 234255-4262. [DOI] [PubMed] [Google Scholar]
- 37.Shi, Y., M. Downes, W. Xie, H. Y. Kao, P. Ordentlich, C. C. Tsai, M. Hon, and R. M. Evans. 2001. Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 151140-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tanigaki, K., and T. Honjo. 2007. Regulation of lymphocyte development by Notch signaling. Nat. Immunol. 8451-456. [DOI] [PubMed] [Google Scholar]
- 39.Tsuji, M., R. Shinkura, K. Kuroda, D. Yabe, and T. Honjo. 2007. Msx2-interacting nuclear target protein (Mint) deficiency reveals negative regulation of early thymocyte differentiation by Notch/RBP-J signaling. Proc. Natl. Acad. Sci. USA 1041610-1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wallberg, A. E., K. Pedersen, U. Lendahl, and R. G. Roeder. 2002. p300 and PCAF act cooperatively to mediate transcriptional activation from chromatin templates by notch intracellular domains in vitro. Mol. Cell. Biol. 227812-7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang, J., T. Hoshino, R. L. Redner, S. Kajigaya, and J. M. Liu. 1998. ETO, fusion partner in t(8;21) acute myeloid leukemia, represses transcription by interaction with the human N-CoR/mSin3/HDAC1 complex. Proc. Natl. Acad. Sci. USA 9510860-10865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wiedenmann, J., S. Ivanchenko, F. Oswald, F. Schmitt, C. Rocker, A. Salih, K. D. Spindler, and G. U. Nienhaus. 2004. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc. Natl. Acad. Sci. USA 10115905-15910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wiedenmann, J., A. Schenk, C. Rocker, A. Girod, K. D. Spindler, and G. U. Nienhaus. 2002. A far-red fluorescent protein with fast maturation and reduced oligomerization tendency from Entacmaea quadricolor (Anthozoa, Actinaria). Proc. Natl. Acad. Sci. USA 9911646-11651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wu, L., J. C. Aster, S. C. Blacklow, R. Lake, S. Artavanis-Tsakonas, and J. D. Griffin. 2000. MAML1, a human homologue of Drosophila mastermind, is a transcriptional co-activator for NOTCH receptors. Nat. Genet. 26484-489. [DOI] [PubMed] [Google Scholar]
- 45.Yabe, D., H. Fukuda, M. Aoki, S. Yamada, S. Takebayashi, R. Shinkura, N. Yamamoto, and T. Honjo. 2007. Generation of a conditional knockout allele for mammalian Spen protein Mint/SHARP. Genesis 45300-306. [DOI] [PubMed] [Google Scholar]
- 46.Yan, M., E. Kanbe, L. F. Peterson, A. Boyapati, Y. Miao, Y. Wang, I. M. Chen, Z. Chen, J. D. Rowley, C. L. Willman, and D. E. Zhang. 2006. A previously unidentified alternatively spliced isoform of t(8;21) transcript promotes leukemogenesis. Nat. Med. 12945-949. [DOI] [PubMed] [Google Scholar]
- 47.Zhang, L., Z. Li, J. Yan, P. Pradhan, T. Corpora, M. D. Cheney, J. Bravo, A. J. Warren, J. H. Bushweller, and N. A. Speck. 2003. Mutagenesis of the Runt domain defines two energetic hot spots for heterodimerization with the core binding factor beta subunit. J. Biol. Chem. 27833097-33104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.