Abstract
BUD23 was identified from a bioinformatics analysis of Saccharomyces cerevisiae genes involved in ribosome biogenesis. Deletion of BUD23 leads to severely impaired growth, reduced levels of the small (40S) ribosomal subunit, and a block in processing 20S rRNA to 18S rRNA, a late step in 40S maturation. Bud23 belongs to the S-adenosylmethionine-dependent Rossmann-fold methyltransferase superfamily and is related to small-molecule methyltransferases. Nevertheless, we considered that Bud23 methylates rRNA. Methylation of G1575 is the only mapped modification for which the methylase has not been assigned. Here, we show that this modification is lost in bud23 mutants. The nuclear accumulation of the small-subunit reporters Rps2-green fluorescent protein (GFP) and Rps3-GFP, as well as the rRNA processing intermediate, the 5′ internal transcribed spacer 1, indicate that bud23 mutants are defective for small-subunit export. Mutations in Bud23 that inactivated its methyltransferase activity complemented a bud23Δ mutant. In addition, mutant ribosomes in which G1575 was changed to adenosine supported growth comparable to that of cells with wild-type ribosomes. Thus, Bud23 protein, but not its methyltransferase activity, is important for biogenesis and export of the 40S subunit in yeast.
Ribosome biogenesis is a complex and highly ordered process in eukaryotic cells. Most of the events of ribosome assembly take place in the nuclear subcompartment, the nucleolus, which is organized around the transcription of the rRNAs. In Saccharomyces cerevisiae, the primary 35S precursor rRNA is transcribed by RNA polymerase I. Subsequent processing by endonucleases and exonucleases generates 18S RNA, the mature RNA of the small subunit, and 25S and 5.8S RNAs, two of the three RNAs of the large subunit. 5S RNA, the third RNA component of the large subunit is transcribed separately by RNA polymerase III. The processing pathway is well documented in S. cerevisiae (59), and more than 170 trans-acting factors that partake in ribosome biogenesis have been identified (10, 13, 56). The initial cleavage events at the A0, A1, and A2 sites are dependent on the U3 snoRNA and separate 20S pre-rRNA, the precursor for 18S RNA, from 27S RNA, the precursor for 5.8S and 25S rRNAs. In addition to nucleolytic processing, the ribosomal RNAs are highly modified by methylation and pseudouridylation. 2′-O-Ribose methylation and pseudouridylation are directed by box C/D and box H/ACA snoRNAs, respectively, that guide the modification enzymes to their targets (22). In addition, a small number of base methylations are carried out by specific methyltransferases (MTases) that recognize their substrates without the aid of guide RNAs. These base methylations differ from 2′-O methylation in that they typically occur at later steps in subunit biogenesis (6).
The two ribosomal subunits are exported out of the nucleus independent of each other. Nevertheless, both subunits use the importin-β-like export receptor Crm1 (reviewed in reference 64) that recognizes leucine-rich nuclear export sequences of cargo proteins (12, 49). In the case of the large subunit, Crm1 binds to the subunit indirectly through the export adapter Nmd3 (14, 18). An analogous adapter for the small subunit has not been identified, but it has been suggested that Ltv1 could provide this function (46). In addition to Crm1, the large subunit also requires the mRNA export adapter complex Mex67/Mtr2 (63) and the pre-60S RNA-associated factor Arx1 (5, 20). Consequently, multiple factors may assist export of the small subunit as well.
The small subunit is exported out of the nucleus as a pre-40S particle, containing 20S rRNA, its complement of small-subunit proteins, and a few trans-acting factors (45). Once in the cytoplasm, two further rRNA processing events are of note: the cleavage of 20S rRNA to 18S rRNA (57), liberating the D-A2 fragment of internal transcribed spacer 1 (ITS1), and dimethylation of A1781 and A1782 at the 3′ end of 18S rRNA (6). The dimethylation is a highly conserved rRNA processing event, found in all three domains of life. In yeast, it is carried out by Dim1 (28), whereas in Escherichia coli, the dimethylase is KsgA (51). Interestingly, Dim1 is essential, but the dimethylation event itself appears to have little consequence for cell growth.
Of the many rRNA methylations, only the m7G at position 1575 of the 18S rRNA (3) has an unknown MTase. The base numbering used here is the same as that used in the Saccharomyces Genome Database (http://www.yeastgenome.org/). G1575 was previously numbered as G1574 (3) and corresponds to G1338 in E. coli 16S rRNA (43). This modified guanine is in a position that may allow it to interact directly with the anticodon stem of tRNA in the P-site of the small subunit. Here we identify Bud23 as the MTase responsible for modification of G1575. As with the dimethylase Dim1, the MTase activity of Bud23 is dispensable for cell growth. However, deletion of BUD23 leads to a strong defect in subunit export. We suggest that Bud23 serves as a late checkpoint, monitoring the export competence of the small subunit.
MATERIALS AND METHODS
Strains and genetic procedures.
The S. cerevisiae strains used in this work were as follows: BY4741 (MATa his3Δ1 leu2Δ0 ura3Δ0 met15Δ0), AJY2161 (MATa bud23Δ::KanMX his3Δ1 leu2Δ0 ura3Δ0 lys2Δ0 met15Δ0), derived from sporulating the heterozygous deletion mutant (Research Genetics), and AJY1539 [MATa CRM1(T539C-HA) his3Δ1 leu2Δ0 ura3Δ0 met15Δ0] (16). Strain AJY1185 (MATa ade2-1 ura3-1 leu2-3 his3-11 trp1 can1-100 rdnΔΔ::HIS3 containing pAJ724), was made by replacing the ribosomal DNA (rDNA)-containing vector in reference 40.
Plasmids.
The plasmids and oligonucleotide primers used in this study are shown in Tables 1 and Table 2, respectively. Plasmids pAJ718 and pAJ724 were derived from pJD211.leu (47) by moving the rDNA locus as a SalI-NotI fragment into pRS425 and pRS424, respectively. pAJ1399 was made by moving RPS2-enhanced green fluorescent protein (eGFP) from pRS316-RPS2eGFP into pRS413. pAJ2151 was made by amplifying BUD23 using primers AJO1034 and AJO1036 and cloning into pAJ582 (16) as an EagI-BamHI fragment. pAJ2154 was made by amplifying BUD23 from genomic DNA by PCR using primers AJO1034 and AJO1035. The product was then ligated into pRS315 as an EagI-BamHI fragment. pAJ2155 was made by fusion PCR using primers AJO1034 and AJO1043 and primers AJO1042 and AJO1035. The second-round PCR product was cloned as described above for pAJ2154. pAJ2156 was made similarly, using the mutagenic primers AJO1048 and AJO1047. pAJ2157 was derived from pAJ718 by fusion PCR using the mutagenic primers AJO1049 and AJO1050 with the outside primers AJO214 and AJO1051. The product was cloned into pAJ718, replacing the corresponding wild-type sequence. pAJ2160 was constructed in a manner similar to that of pAJ2151 except AJO1034 and AJO1036 were used. pAJ2164 was made by fusion PCR using primers AJO1034 and AJO1067 and primers AJO1066 and AJO1035. The second-round PCR product was cloned into pAJ582 as an EagI-BamHI fragment. pAJ2175, expressing bud23ΔN from the NMD3 promoter, was made by amplifying BUD23 with primers AJO1068 and AJO1034. The truncated gene replaced the NMD3 coding sequence as an EcoRI-BamHI fragment in pAJ582. pAJ2176 was made as described above for pAJ2164, but using the outside primers AJO1034 and AJO1036. pLD50 (RPS3-GFP CEN HIS3) was from D. Lycan (46). All constructs were verified by sequencing.
TABLE 1.
Plasmids used in this study
| Plasmid | Relevant information | Reference or source |
|---|---|---|
| pAJ582 | NMD3-GFP LEU2 CEN | This study |
| pAJ718 | rDNA LEU2 CEN | This study |
| pAJ724 | rDNA URA3 2μm | This study |
| pAJ755 | NMD3-GFP URA3 CEN | This study |
| pAJ1399 | RPS2-eGFP HIS3 CEN | This study |
| pAJ1629 | SIK1-mRFP MET15 CEN | Z. Li and E. Marcotte, unpublished data |
| pAJ2151 | BUD23-GFP LEU2 CEN | This study |
| pAJ2154 | BUD23 LEU2 CEN | This study |
| pAJ2155 | bud23(D77K) LEU2 CEN | This study |
| pAJ2156 | bud23(G57E) LEU2 CEN | This study |
| pAJ2157 | rDNA(G1575A) LEU2 2μm | This study |
| pAJ2160 | bud23ΔC-GFP LEU2 CEN | This study |
| pAJ2164 | bud23ΔC(K4A)-GFP LEU2 CEN | This study |
| pAJ2175 | bud23ΔN-GFP LEU2 CEN | This study |
| pAJ2176 | bud23(K4A)-GFP LEU2 CEN | This study |
TABLE 2.
Oligonucleotides used in this study
| Oligonucleotide | Sequence |
|---|---|
| AJO214 | GTTCGCCTAGACGCTCTCTTC |
| AJO1024 | ATGCTCTTGCCAAAACAAAAAAATCCATTTTCAAAATTATTAAATTTCTT |
| AJO1027 | TGTGTACAAAGGGCAGGGACGTA |
| AJO1034 | CGCTAAACGAGTCCAACC |
| AJO1035 | CTTTTTACTGATGAGCTTCC |
| AJO1036 | CGAGGATCCGGAACCTGTGTCTTCTTTTCC |
| AJO1042 | CATGTGTGGTGTGGTTTGAAGATATCGCCCAGC |
| AJO1043 | CAAACCACACCACACATG |
| AJO1047 | GTCCGGACTGTCTGGGGA |
| AJO1048 | TCCCCAGACAGTCCGGACTCGCACCCGATATC |
| AJO1049 | TTATTGCTCTTCAACGAGAAATTCCTAGTAAGC |
| AJO1050 | CTCGTTGAAGAGCAATAA |
| AJO1051 | TGCTATGGTATGGTGACG |
| AJO1065 | GCTGGATCCGGCTAGCGTCCATGGTCACACCGTC |
| AJO1066 | GTGGACGACCCAGAGTCTGCTGCCAATGCCGCTTACTACCTTGTGTTGAGC |
| AJO1067 | AGACTCTGGGTCGTCCAC |
| AJO1068 | GCTGAATTCGAGAACGTCAACTTGAAG |
RNA analysis.
All RNAs were prepared using MasterPure yeast RNA purification kit (Epicenter). Northern blotting was done as described elsewhere (Z. Li and E. Marcotte, unpublished data). Twelve micrograms of total RNA was used for primer extension with 32P-labeled primer AJO1027 as described previously (2) except that denaturation was done in water.
l-[3H-methyl]methionine pulse-chase analysis was carried out as described previously (17). Strains were transformed with pRS411 and grown in 20 ml methionine-deficient (Met−) medium to an optical density at 600 nm (OD600) of ∼0.2. Cells were collected, resuspended in 3 ml of Met− medium, incubated at 30°C for 25 min, and then labeled for 2 minutes with 0.3 mCi [3H-methyl]methionine. Unlabeled methionine was then added to a final concentration of 0.6 mM, and aliquots were collected over time. Six micrograms of total RNA was separated on a 1.2% formaldehyde agarose gel transferred to nylon membrane. The membrane was sprayed with En3Hance (Dupont) and exposed to film.
Sucrose gradient sedimentation.
Yeast strains BY4741 and AJY2161 were grown in yeast extract-peptone-dextrose (YPD) at 30°C to a density of OD600 of 0.3. For polysome analysis, cycloheximide was added to 150 μg/ml, cultures were immediately poured onto ice, and cells were collected by centrifugation. All the steps were carried out on ice or at 4οC. Cell pellets were washed with lysis buffer (10 mM Tris-HCl [pH 7.5], 10 mM MgCl2, 100 mM KCl, 6 mM β-mercaptoethanol, 150 μg/ml cycloheximide, 1 mM phenylmethylsulfonyl fluoride [PMSF], 1 μM leupeptin, and 1 μM pepstatin), resuspended in an equal volume of lysis buffer, and broken open by vortexing in the presence of glass beads. The extract was centrifuged for 10 min at 15,000 × g at 4°C, and the supernatant was recovered. Eleven OD260 units were loaded onto a continuous 7% to 47% sucrose gradient in lysis buffer and centrifuged for 2.5 h at 40,000 rpm (Beckman SW40 rotor). Gradients were continuously monitored for absorbance at 254 nm. For measuring subunit ratios, cycloheximide was omitted, and a Mg2+-free lysis buffer was used (50 mM Tris-HCl [pH 7.4], 50 mM NaCl, 1 mM dithiothreitol, 1 mM PMSF, 1 μM leupeptin, and 1 μM pepstatin). Scion Image (NIH) software was used to quantify subunit levels.
Microscopy.
Overnight cultures of cells were diluted to an OD600 of ∼0.1 in fresh medium and then incubated for 5 to 6 h at 30°C and either visualized directly or prepared for fluorescence in situ hybridization using a procedure modified from http://singerlab.aecom.yu.edu/protocols/. Cells were fixed in 4.5% formaldehyde and 11% acetic acid for 30 min at room temperature. Cells were washed twice in 0.1 M potassium phosphate buffer (pH 7.5) and then washed with 0.1 M potassium phosphate (pH 7.5) and 1.2 M sorbitol. Spheroplasting was done in the presence of 20 mM vanadyl-RNase complex, 28 mM β-mercaptoethanol, 0.06 mg/ml PMSF, and 50 μg/ml Zymolyase T-20 for 10 min at 37°C. Cells were allowed to adhere to polylysine-coated slides and placed in 70% ethanol. After rehydration with 4× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), cells were incubated in 50% formamide, 10% dextran sulfate, 4× SSC, 1× Denhardt's solution, 20 mM vanadyl-RNase complex. After 1 h at 37°C, 1 ng/μl Cy3-labeled oligonucleotide AJO1024 was added, and incubation was continued for 20 h. Cells were then washed successively with 2× SSC, 1× SSC, and 1× phosphate-buffered saline (PBS). DNA was stained for 5 min with 4′,6′-diamidino-2-phenylindole (DAPI) at 1 μg/ml in PBS. Cells were washed once in PBS and then mounted. Fluorescence was visualized on a Nikon E800 microscope fitted with an 100× objective and a Photometrics CoolSNAP ES digital camera controlled with NIS-Elements AR 2.10.
Bioinformatics analyses.
Searches of the nonredundant version of current sequence databases were carried out at the NCBI using PSI-BLAST (1) with default parameters, except for using a stringent expectation (E) value threshold of 1e−30 to restrict the analysis to Bud23 orthologs and prevent “explosion” of the search by inclusion of all members of the large Rossmann-fold methyltransferase (RFM) superfamily. Multiple-sequence alignment of the Bud23 protein family was calculated using PROMALS (41) with default parameters.
Secondary structure prediction, identification of ordered and disordered regions, and tertiary fold recognition (FR) was carried out via the GeneSilico metaserver (27), which is a gateway for >30 third-party methods (for references to original methods, see http://genesilico.pl/meta2/). On the basis of the FR alignments to the top-scoring templates (1xxl, 1kpi, and 2avn), we constructed the homology model of Bud23 using the so-called FRankenstein's Monster method (23, 24), which has been validated as one of the best approaches for template-based modeling in CASP5 and CASP6. Model quality has been assessed using PROQ (61) and MetaMQAP (M. Pawlowski, R. Matlak, M. Gajda, and J. M. Bujnicki, unpublished data, and https://genesilico.pl/toolkit/). Mapping of sequence conservation onto the model was done via the COLORADO3D server (44), using the Rate4Site method, with the JTT substitution matrix and ML model for rate inference.
Comparisons between the Bud23 family and families in the Clusters of Orthologous Groups (COG) database were done with HHpred (48). Based on the Bud23 family alignment, a phylogenetic tree was calculated with MEGA 3.1 (26), using the neighbor-joining method, with the JTT model of substitutions and pairwise deletions. The stability of individual nodes was calculated using the bootstrap test (1,000 replicates).
RESULTS
Bud23 is required for efficient rRNA processing and nuclear export of 40S ribosomal subunits.
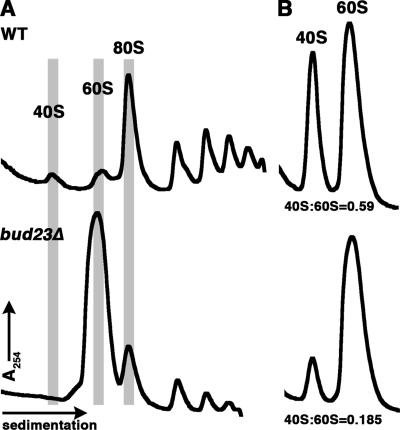
We initially identified BUD23 as encoding a potential ribosome biogenesis factor from a comprehensive bioinformatics analysis of protein function in yeast in which BUD23 was observed to associate predominantly with known ribosomal biogenesis factors in a functional gene network (31). BUD23 is not essential, but its deletion causes a severe slow growth phenotype. Sucrose gradient analysis showed that deletion of BUD23 (bud23Δ) results in a significant depletion of 40S subunits and loss of polysomes (Fig. 1A). The high free 60S peak in this profile is typical of a 40S biogenesis defect. To quantify the subunit imbalance, we measured the ratio of small to large subunits under conditions that dissociate all 40S from 60S subunits (Fig. 1B). Normalizing to the subunit ratio from wild-type cells, the ratio of 40S to 60S in the bud23Δ mutant was 0.31, indicating a 70% reduction in the level of 40S subunits in the bud23Δ mutant compared to the wild type.
FIG. 1.
Deletion of BUD23 results in reduced levels of 40S subunits. (A) Whole-cell extracts were prepared from wild-type (WT) and bud23Δ cells and fractionated by ultracentrifugation through 7 to 47% sucrose gradients. The positions of free 40S monosomes and free 60S and 80S monosomes are shaded. Note that in the bud23Δ sample, the largest peak corresponds to free 60S. (B) The ratio of small to large subunits was determined by making extracts in the absence of cycloheximide, to allow ribosome runoff, and in the absence of Mg2+ to dissociate 40S from 60S subunits. Extracts were fractionated as described above for panel A.
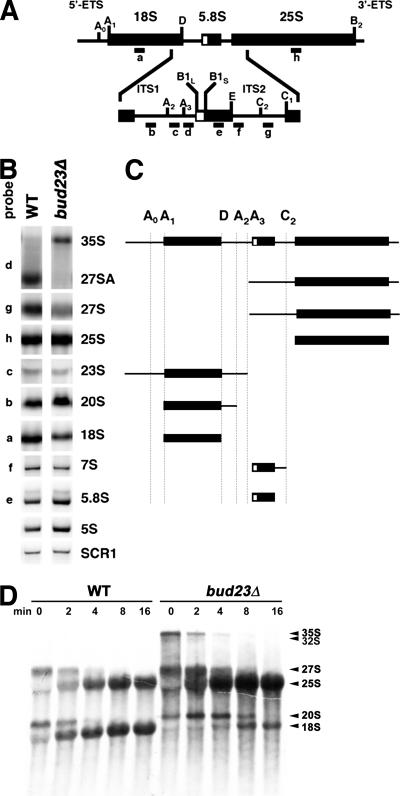
We next examined rRNA processing by Northern blotting for different rRNA intermediate species in the steady-state total RNA isolated from bud23Δ and wild-type cells. rRNA processing was clearly perturbed in the bud23Δ mutant: there was accumulation of 35S RNA and strong depletion of 27SA, indicating a defect in cleavage at site A2. Nevertheless, the overall levels of 25S and 5.8S RNAs along with the level of 7S, a precursor of 5.8S, were not affected, indicating that subsequent processing of the rRNA transcript downstream of site A2 was not affected. Notably, 20S RNA showed a slight accumulation (1.2-fold the level in wild-type cells), but we did not detect increased levels of 23S or 22S species (Fig. 2B; also data not shown), while the levels of 18S RNA were clearly reduced. These results suggest that although the bud23Δ mutant is strongly affected for cleavage at A2, the protein is also needed for an event downstream of producing 20S RNA.
FIG. 2.
Northern blot analysis of pre-rRNA species in bud23 mutant cells. (A) Cartoon of the processing pathway of rRNA in yeast. The positions of oligonucleotide probes (probes a to h) used for Northern blotting are shown. 5′-ETS, external transcribed spacer. (B) Ten micrograms of total RNA prepared from wild-type (WT) (BY4741) and bud23Δ (AJY2161) cells was separated on a 1% agarose formaldehyde gel, transferred to a membrane, and probed with the indicated probes. (C) Cartoon depicting the indicated pre-rRNA species detected in panel B. (D) Pulse-chase labeling of rRNA was carried out as described in Materials and Methods. Wild-type (BY4741) and bud23Δ (AJY2161) (both containing pRS411, MET15) were labeled with [methyl-3H] methionine for 2 minutes and then chased with excess unlabeled methionine. Aliquots were collected and quickly frozen at the indicated times after labeling. Total RNA was separated on a formaldehyde agarose gel, transferred to a membrane, and exposed to X-ray film after spraying with En3Hance (Dupont). The positions of RNA species are identified to the right of the gel.
To examine the kinetics of rRNA processing, we used pulse-labeling of rRNA with [3H-methyl]methionine. In wild-type cells, processing to mature 18S and 25S rRNAs was nearly complete within 4 minutes after labeling (Fig. 2D, WT cells). In contrast, bud23Δ cells showed a significant delay in conversion of 20S to 18S rRNA, and the overall level of 18S rRNA at 16 min was significantly reduced compared to that of 25S RNA. This is most likely accounted for by rapid turnover of the 20S RNA-containing particle. There was also a modest increase in 35S RNA and a slight delay in processing 27S to 25S rRNA; however, accumulation of 25S rRNA was not impaired. The apparently greater amount of 25S rRNA in bud23Δ cells versus wild-type cells is due to loading equivalent amounts of total RNA, which in the case of the mutant contains relatively less 18S and consequently more 25S RNA. The defect in processing 20S to 18S rRNA observed by Northern blotting and by pulse-chase analysis could reflect a defect in export or processing 20S rRNA or could be due to instability of 20S rRNA in the cytoplasm.
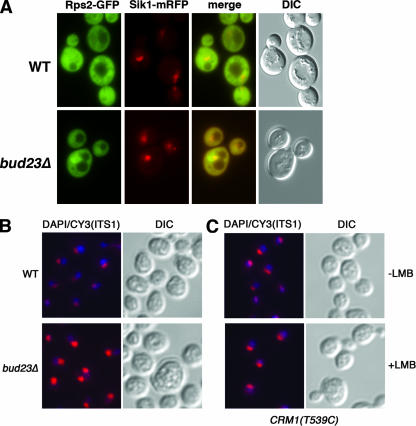
To examine small-subunit export, we took advantage of a GFP fusion to the small ribosomal subunit protein Rps2 that can serve as a reporter of the small subunit to monitor export (36). Rps2-GFP accumulated in the nuclei of bud23Δ cells but did not accumulate in wild-type cells (Fig. 3A). Comparison of the localization of Rps2-GFP with Sik1-monomeric red fluorescent protein (mRFP), a nucleolar reporter (19), indicated that the pre-40S particles were accumulating in the nucleolus as well as the nucleoplasm. Because the nuclear Rps2-GFP fluorescence could represent free protein rather than ribosome-bound protein, we also assayed the localization of a second small-subunit reporter, Rps3-GFP (46). We observed nuclear accumulation of Rps3-GFP as well (data not shown).
FIG. 3.
Rps2 and ITS1 accumulate in the nucleus in bud23Δ mutant cells. (A) Wild-type (WT) (BY4741) and bud23Δ cells (AJY2161) were transformed with pAJ1399 (RPS2-eGFP) and pAJ1629 (SIK1-mRFP). Fluorescence was visualized in green (GFP) and red (RFP) channels. The merged images and differential interference contrast (DIC) images are also shown. (B) Wild-type and bud23Δ cells were prepared for fluorescence in situ hybridization using a Cy3-labeled probe complementary to the 5′ end of ITS1. (C) For comparison, the localization of ITS1 was examined in AJY1539 [CRM1(T539C)] mutants cells not treated with LMB (−LMB) or treated with 0.1 μg/ml LMB (+LMB) for 30 min to block ribosomal subunit export.
The processing of rRNA to 20S rRNA and the accumulation of both Rps2 and Rps3 reporters in the nucleus suggest that the pre-40S particles are blocked for export in bud23Δ mutant cells. To examine this more directly, we monitored the localization of 5′ internal transcribed spacer 1 (ITS1) (Fig. 2A) by fluorescence in situ hybridization (Fig. 3B). In wild-type cells, ITS1 is abundant in the nucleolus, but because of the rapid export of pre-40S particles, it is not readily detected in the nucleoplasm (Fig. 3B, top panel). A low level of ITS1 is also observed in the cytoplasm (37), where it is degraded by Xrn1 after cleavage from 18S rRNA (50). bud23Δ cells showed strong enrichment of ITS1 in the nucleolus compared to wild-type cells and increased nucleoplasmic signal for ITS1, evident from the merge of DAPI and Cy3 signals in the nucleoplasm (Fig. 3B, bottom panel). Export of ribosomal subunits in yeast requires the export receptor Crm1 (14, 18, 37). Consequently, as a control for a block in export, we used leptomycin B (LMB) to inhibit the function of Crm1 in the LMB-sensitive mutant crm1(T539C). In the presence of LMB, ITS1 accumulated in the nucleolus and nucleoplasm (Fig. 3C, bottom panel), similar to what we observed for in bud23Δ cells. These results indicate that bud23 mutants are defective for nuclear export of the 40S subunit. From this, we also conclude that the lack of processing of 20S rRNA to 18S rRNA results from a block in export, rather than instability of 18S rRNA in the cytoplasm.
Bud23 is predicted to be a methyltransferase.
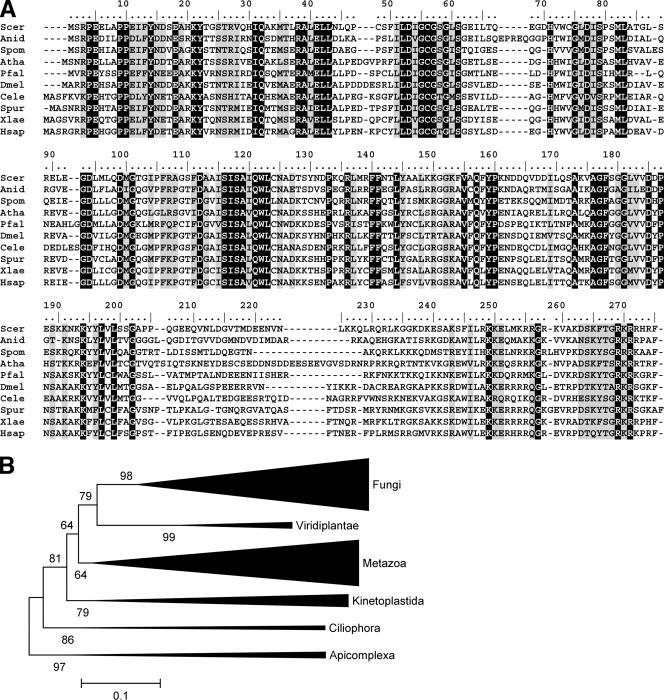
Based on sequence comparisons, Bud23 was previously predicted to be a MTase, but without any indication of its possible substrate specificity (39). Our searches of the nonredundant sequence database at the NCBI using PSI-BLAST revealed a family of Bud23 orthologs conserved in members of the domain Eukarya, but absent from members of the domains Archaea and Bacteria (Fig. 4). Comparisons at the family level, carried out using the HHpred method for detection of remote homology, revealed that Bud23 proteins are most closely related to a large superfamily of RFM enzymes (proteins whose catalytic domain comprises a seven-stranded β-sheet surrounded on both sides by α-helices, similar to Rossmann-fold dehydrogenases) (7). These enzymes are involved in modification of small molecules, including the following: the UbiE family of MTases involved in ubiquinone/menaquinone biosynthesis (COG2226; E value of 9.2e−26), cyclopropane fatty acid (Cfa) synthase and related MTases (COG2230; E value of 3e−22), and the UbiG family of 2-polyprenyl-3-methyl-5-hydroxy-6-metoxy-1,4-benzoquinol MTases (COG2227; E value of 3.7e−20). Closer inspection of the family-family alignments revealed potential conservation of the common cofactor-binding site (sequence motifs I to III) between Bud23 and small-molecule MTases. However, significant differences between their putative catalytic centers (sequence motifs IV, VI, and VIII) are consistent with different substrate specificities of these enzymes.
FIG. 4.
Bud23 is highly conserved but restricted to eukaryotes. (A) BLAST sequence alignment of Bud23 orthologs. Bud23 orthologs were from the following species (the accession numbersof these orthologs shown in brackets): Saccharomyces cerevisiae (Scer) [6319895], Aspergillus nidulans (Anid) [67539238], Schizosaccharomyces pombe (Spom) [19115061], Arabidopsis thaliana (Athal) [15242087], Plasmodium falciparum (Pfal) [124506365], Drosophila melanogaster (Dmel) [21355093], Caenorhabditis elegans (Cele) [17552330], Strongylocentrotus purpuratus (Spur) [72006191]), Xenopus laevis (Xlae) [148234441], and Homo sapiens (Hsap) [23199995]. Gaps introduced to maximize alignment are indicated by dashes. (B) Phylogenetic tree of Bud23 orthologs calculated using the neighbor-joining method as described in Materials and Methods. Lengths of branches correspond to the estimated evolutionary distances between Bud23 variants from different taxons (the bar below the tree indicates the distance of 0.1 unit according to the JTT matrix). Numbers at the nodes indicate the statistical support for grouping of the corresponding taxons, calculated from 1,000 bootstrap resamplings of the alignment. Values close to 100 indicate strong support.
Bioinformatics analysis of the Bud23 sequence via the GeneSilico metaserver (see Materials and Methods for details) revealed that residues ∼10 to 210 show a propensity to form a well-folded globular domain rich in α-helices and β-strands, while the C-terminal 65 residues are most likely intrinsically disordered, with a tendency to form two α-helices in the regions from residues 220 to 233 and 239 to 255). According to the FR analysis, the globular part of Bud23 exhibits very strong similarity to the RFM fold (PCONS [62] score 3.4, significantly above the threshold of 1, indicating a robust prediction). We constructed a three-dimensional model of Bud23 with the N-terminal region folded as the α/β globular domain with a topology characteristic for the RFM superfamily (7) and with the C-terminal region modeled as a random coil containing two loose α-helices. The methyl group donor S-adenosylmethionine (AdoMet) has been docked into the predicted active site in the globular domain by analogy to the known structures of RFM proteins. The model of the RFM domain (amino acids [aa] 1 to 210) has been evaluated by the PROQ method as potentially “very good” (predicted LG score of 3.675), suggesting that the predicted structure can be used to make functional predictions. On the other hand, the conformation of the C-terminal tail (aa 211 to 275) should be regarded as undefined; the current model presents a single tentative variant out of a large number of possible conformations.
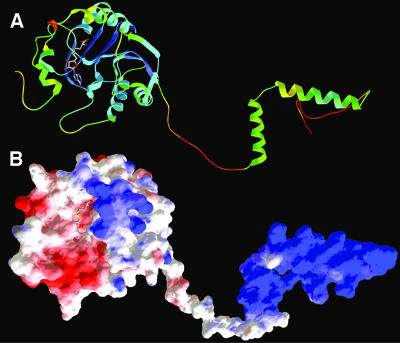
Figure 5 shows the ribbon and surface representations of the Bud23 model. Mapping of the sequence conservation onto the protein structure reveals concentration of conserved residues in the predicted catalytic pocket of the N-terminal domain, and in the helical part of the “loopy” C terminus (data not shown). Interestingly, mapping of the electrostatic potential onto the surface reveals that the part of the RFM domain predicted to bind AdoMet is negatively charged (compatible with the positive charge on the cofactor molecule), while a protrusion formed mostly by a well-structured loop comprising aa 189 to 194 and the C-terminal disordered tail (aa 211 to 275) is positively charged, suggesting potential involvement in binding of the negatively charged RNA backbone. Indeed, the positively charged loop in the RFM domain corresponds to a region frequently involved in protein-substrate interactions in other members of the RFM superfamily, e.g., rRNA methyltransferase ErmC′ (34). By analogy to other members of the RFM superfamily and on the basis of the analysis of sequence conservation among Bud23 homologs, we predict that residues D53, D77, and D99 are involved in coordination of the methionine, ribose, and adenine moieties of AdoMet, respectively. Although catalytic residues are not conserved between different families of RFM enzymes, our model of Bud23 predicts that invariant (or nearly invariant) residues S118, Q121, Q157, Y159, and K194 line a pocket adjacent to the AdoMet-binding site, which suggests their involvement in the binding of the target base and catalysis of the methyl transfer reaction. Finally, we predict that conserved positively charged residues R3, K190, K191, and K193 on the surface of the RFM domain and K243, K249, K250, K265, R269, K270, and R271 (and possibly their other, less-conserved neighbors) in the C-terminal tail are involved in rRNA binding. Additionally, aa 255 to 274 are predicted to contain a nuclear localization signal (NLS) (PSORT II [http://psort.nibb.ac.jp/]). It is likely that the C-terminal tail of Bud23 is disordered in the absence of the substrate (as predicted by bioinformatics methods) but may assume a folded structure upon formation of the protein-RNA complex.
FIG. 5.
Structural model of Bud23. (A) Bud23 model shown in ribbon representation, colored according to the predicted accuracy (estimated agreement with the native structure). Ribbons are colored blue (highly confident, predicted error of ∼1 Å) to green (predicted low accuracy, expected differences between the model and the native structure of up to ∼5 Å) to red (predicted very low accuracy, error difficult to estimate). The orientation of two helices in the C terminus (right) with respect to each other and with respect to the catalytic domain (left) is completely arbitrary, as this region is likely to be disordered. (B) Bud23 model colored according to the distribution of electrostatic potential, from red (−3 kT) to blue (+3 kT).
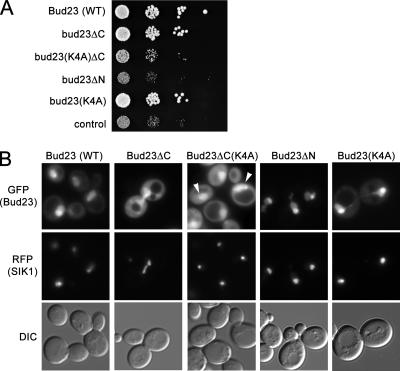
On the basis of sequence alignments and our structural prediction (Fig. 4 and 5), we made several mutations in Bud23 and tested them for function by complementation in growth assays and for cellular localization. Deletion of the highly basic C terminus (aa 220 to 275), Bud23ΔC, encompassing the predicted NLS, resulted in no obvious defect in growth (Fig. 6A) and a modest relocalization of the protein to the cytoplasm (Fig. 6B). Thus, this region is important for normal nuclear localization but must be redundant with other nuclear import signals. We changed the four lysines at positions 190, 191, 193, and 194, forming a basic cluster adjacent to the Ado-Met binding pocket (Fig. 5B), to alanines. Surprisingly, this mutant (Bud23-K4A) showed wild-type growth and localization (Fig. 6A and B). However, when combined with the C-terminal deletion (Bud23ΔC-K4A), we observed significantly reduced growth and loss of localization to the nucleolus (Fig. 6), suggesting that this basic cluster contributes to nucleolar localization. However, considering that Bud23-K4A was not apparently excluded from the nucleolus, the basic C-terminal tail must also contribute to nucleolar localization. Finally, deletion of the entire MTase domain (aa 1 to 219), Bud23ΔN, resulted in a severe growth defect (Fig. 6A), comparable to that of the vector control, indicating that the MTase domain is necessary for Bud23 function. Bud23ΔN was localized exclusively to the nucleolus and nucleoplasm, with no cytoplasmic localization evident (Fig. 6B), consistent with the notion that the C terminus contains an NLS.
FIG. 6.
Localization of mutant Bud23 proteins. (A) Wild-type (WT) and mutant Bud23 proteins were expressed as GFP fusions from plasmids in the bud23Δ mutant AJY2161. The constructs were as follows: WT (pAJ2151); Bud23ΔC, deleted of amino acids 220 to 275 (pAJ2160); Bud23ΔC(K4A), Bud23ΔC in which the lysines at positions 190, 191, 193 and 194 were changed to alanines (pAJ2164); Bud23ΔN, deleted of amino acids 1 to 219 (pAJ2175); and Bud23(K4A) (pAJ2176). Serial dilutions of cultures were spotted onto rich growth medium and incubated at 30°C for 3 days. (B) GFP fusions of wild-type and mutant Bud23 proteins, as in panel A, were expressed in a BUD23 wild-type strain that also expressed the nucleolar marker Sik1 as a fusion to RFP (19). Cells were fixed with formaldehyde before visualization in green (GFP) and red (RFP) channels. Differential interference contrast (DIC) images are shown for reference. The white arrowheads in the Bud23ΔC(K4A) GFP panel indicate the position of the nucleolus, where the GFP signal is diminished.
Bud23 methylates G1575 of 18S rRNA.
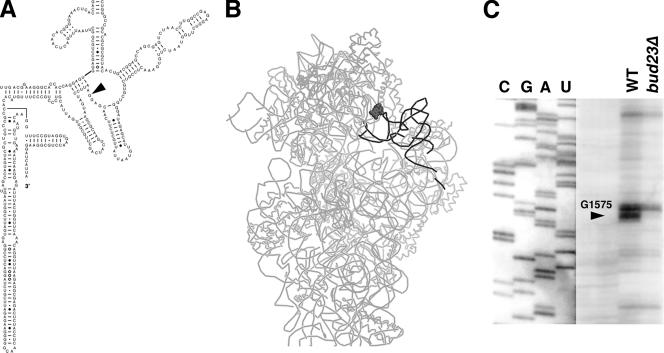
Considering that rRNA is highly methylated, an obvious question was whether 18S rRNA is a substrate for Bud23. The only known methylation for which the enzyme had not been identified was m7 of guanosine at position 1575. G1575 is a highly conserved residue, corresponding to G1338 in E. coli 16S rRNA, that contacts the P-site tRNA in the decoding center (42) (Fig. 7A and B). To test whether Bud23 was responsible for methylation at G1575, we prepared total RNA from wild-type and bud23Δ cells and carried out primer extension through the methylation site. Base modification hinders the progress of reverse transcriptase, resulting in a transcription stop product. The reverse transcription products were compared to a DNA sequencing ladder of DNA corresponding to this region of the rRNA. As seen in Fig. 7C, the expected stop observed in wild-type cells at position G1575 was absent in the RNA prepared from bud23Δ cells. The band 3 nucleotides above G1575 that is present in both wild-type and mutant samples arises from a separate methylation event at G1572, directed by snoRNA57 (15, 33).
FIG. 7.
Bud23 methylates G1575 of 18S rRNA. (A) Secondary structure of the 3′ portion of yeast 18S rRNA (URL http://www.rna.ccbb.utexas.edu/). The black arrowhead indicates the position of G1575. (B) Structure of the 30S subunit from E. coli (PDB structure 29B0) (42) showing the position of G1338 (dark gray, filled), corresponding to G1575 in yeast and the position of the P-site tRNA (dark gray). (C) Total RNA was prepared from wild-type (WT) and bud23Δ mutant cells. Primer extension was carried out with a primer that would yield a 61-nucleotide product if extended to G1575. Products were resolved on a denaturing 8% polyacrylamide gel. A DNA sequence ladder using the same primer was run to identify the position of G1575.
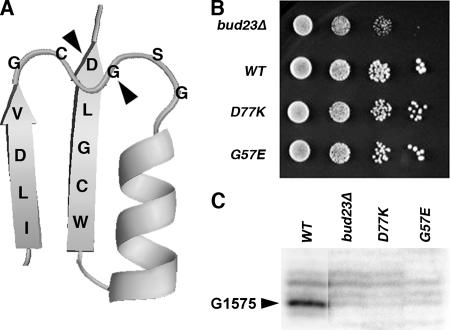
These results strongly suggest that Bud23 is the MTase that modifies G1575. To examine this more directly and to determine the in vivo consequence(s) of this methylation, we mutated the catalytic site of Bud23. Mutational analyses of other MTases have shown that changes in the AdoMet binding pocket commonly inactivate these enzymes. On the basis of modeling studies and inactivating mutations in other MTases (9), we decided to change glycine 57 to glutamate and aspartate 77 to lysine (Fig. 8A). These mutants nearly fully complemented the slow growth phenotype of a bud23 deletion mutant (Fig. 8B). The slight growth defect of a D77K mutant may be due to perturbation of protein structure by the change of charge. The growth rate of the double G57E D77K mutant was indistinguishable from the D77K single mutant (data not shown.) Although these mutants complemented the slow growth of a bud23Δ mutant, primer extension analysis of rRNA prepared from G57E and D77K mutant cells showed no detectable methylation at G1575 similar to what was observed for the bud23Δ mutant (Fig. 8C).
FIG. 8.
Catalytically inactive bud23 mutants do not methylate G1575 but do complement the growth defect of a bud23 deletion. (A) Cartoon of Bud23 structure, showing mutated residues G57E and D77K that are part of the AdoMet binding pocket. The relative positions of the two β-strands and α-helix have been altered slightly from the structure modeled in Fig. 5. (B) Growth test of bud23 mutants. Cultures of a bud23Δ strain (AJY2161) carrying an empty vector (pRS315) (bud23Δ), plasmid-borne wild-type BUD23 (pAJ2154) (WT), and bud23(D77K) (pAJ2155) (D77K), and bud23(G57E) (pAJ2156) (G57E) mutants were standardized for cell density, and 10-fold serial dilutions were plated onto selective medium. Plates were incubated for 3 days at 30°C. (C) Total RNA was prepared from the strains in panel B. Primer extension was carried out as described in the legend to Fig. 7. The gray scale was adjusted in the three right lanes to equalize the signal for background primer extension stops.
The phenotype of the bud23 catalytic mutants suggests that methylation of G1575 has little impact on cell growth. As another means of examining the significance of this G1575 methylation, we mutated G1575 to A in 18S rRNA in a plasmid-borne copy of the 35S rDNA transcriptional unit under control of its native promoter. The mutant rDNA was then introduced into a yeast strain in which the genomic rDNA array had been completely deleted (60), and instead, rRNA was expressed from a URA3 plasmid. After introduction of the plasmid containing mutant rDNA, the preexisting URA3 plasmid containing wild-type rDNA was selected against on medium containing 5-fluoroorotic acid. For controls, we separately introduced a plasmid bearing the wild-type rDNA locus and an empty vector. The mutant rDNA fully complemented growth, indicated by its growth on 5-FOA, whereas the empty vector was unable to complement growth (Fig. 9A). In similar experiments in which we exchanged plasmids containing mutant rDNA for ones with wild-type rDNA, we have noted high levels of interplasmid recombination that could lead to misinterpretation of the effects of mutant rRNA. Fortuitously, G1575 occurs within an EcoRI site in the rDNA. Thus, to demonstrate that the plasmid with G1575A mutation was the sole source of rDNA in these cells, we amplified the plasmid DNA of the region surrounding G1575 by PCR and then tested for the sensitivity of the PCR product to cleavage by EcoRI. Whereas the PCR product from wild-type cells was fully cleaved by EcoRI, the product from G1575A-containing cells was completely resistant to EcoRI cleavage (Fig. 9B). Together, these results indicate that Bud23 is the MTase for G1575 but that the m7G1575 modification itself does not have a significant impact on cell growth. Rather, it is the Bud23 protein and not its methylation activity that is necessary for efficient biogenesis and export of the small subunit.
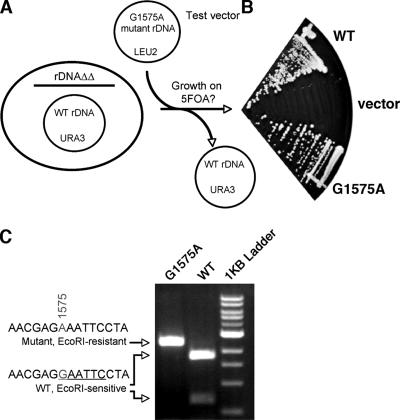
FIG. 9.
Changing guanosine at position 1575 to adenosine has no discernible effect on cell growth. (A) Schematic of strategy to introduce mutant ribosomes into yeast. 5-Fluoroorotic acid (5FOA)-containing media selects against the URA3 vector, forcing cells to utilize the rDNA locus introduced on the LEU2 vector. WT, wild type. (B) LEU2 plasmids expressing wild-type rRNA (pAJ718) (WT), G1575A mutant (pAJ2157), or no rRNA (pRS425) (vector) were transformed into strain AJY1185 and tested for growth on 5FOA-containing medium. (C) A 2.7-kb fragment spanning the G1575A mutation of the plasmid-borne rDNA locus was amplified by PCR using primers AJO214 and AJO1051. The PCR product was treated with EcoRI and analyzed on an agarose gel. The brightest band of the 1-kb ladder is 3 kb.
DISCUSSION
Bud23 is required for the eukaryote-specific methylation of G1575.
We have shown here that the previously uncharacterized yeast protein Bud23 is responsible for the methylation of the N7 position of G1575 in 18S rRNA. To our knowledge, this is the last known methylation of 18S rRNA in yeast for which the MTase was not known. Bud23 is highly conserved throughout the domain Eukarya but orthologs are not detectable in the domain Bacteria (see below). Similarly, the methylation of G1575 is a conserved rRNA modification in eukaryotes that is not observed in prokaryotes (35). Although Bud23 is required for methylation of G1575, point mutations that inactivate the protein's MTase activity cause little phenotype. On the other hand, deletion of the protein leads to a significant impact on cell growth and a 70% reduction in 40S subunit levels. Thus, the protein, but not its MTase activity, is important for the production of 40S subunits.
G1575 is a highly conserved residue in the ribosome. In the bacterial small subunit, G1338 (corresponding to G1575 in yeast) is positioned immediately above and contacting the anticodon loop of the P-site tRNA, with the N7 position projecting away from the tRNA and into the body of the subunit (42). Considering the conservation of sequence surrounding this residue, we imagine a similar architecture for the eukaryotic ribosome as well. Despite the fact that this residue is in a functional center of the ribosome, methylation of G1575 does not appear to be important for normal cell growth. This conclusion comes from the demonstration that mutations in Bud23 that disrupt its MTase activity or mutation of the substrate guanosine at 1575 to adenosine has no obvious impact on cell growth. This lack of phenotype is not uncommon for loss of rRNA methylation events. In yeast and bacteria, many 2′-O methyl modifications seem to have little consequence for cell growth (8, 30). Similarly, 16S rRNA in bacteria and 18S rRNA in eukaryotes undergoes a highly conserved base dimethylation near the 3′ end. In E. coli, this is carried out by KsgA, a nonessential protein. ksgA mutants display little phenotype other than resistance to the antibiotic kasugamycin; however, extracts prepared from ksgA mutants showed altered translation fidelity and requirements for translation initiation factors (51). Dim1, the yeast ortholog of KsgA, methylates A1781 and A1782, in the 3′ end of 18S rRNA. Although DIM1 is essential, conditional dim1-2 mutants are completely defective for dimethylation but do not significantly affect cell growth. Nevertheless, extracts prepared from dim1-2 mutants do not support translation in vitro (29). Thus, dimethylation has a subtle effect on ribosome function that is enhanced in vitro. Considering the position of G1575 in the P-site, it seems likely that methylation of G1575 is important for translation fidelity by constraining the binding of the P-site tRNA in the small subunit. This may be particularly important for frameshifting, which depends on repositioning of the P-site tRNA (4). Additional work is required to determine whether there is an underlying defect in translation in the absence of methylation at G1575.
Phylogenetic analysis of the Bud23 family reveals that it is ubiquitously present in Eukarya, but absent from Bacteria and Archaea. Searches of the COG database revealed that the closest homologs (paralogs) of the Bud23 family belong to small-molecule MTases, in particular, the UbiE, Cfa, and UbiG families. Likewise, protein structure prediction methods revealed that Bud23 sequence is most compatible with the structures of small-molecule MTases rather than with MTases acting on nucleic acids or proteins. This relationship suggests that the ancestor of the Bud23 family originated by duplication of a small-molecule MTase followed by development of a novel substrate specificity (i.e., toward rRNA, possibly by acquisition of positively charged loops) and minor adjustment of the catalytic site, rather than by duplication of some existing rRNA MTase. A similar scenario has been proposed for HEN1 MTase involved in methylation of 2′-OH groups in micro-RNAs (54). HEN1 nonetheless seems to be a product of an independent duplication, as its similarity to Bud23 is relatively low, in the order of similarity between all different MTase families (E value of 4.5e−12). Bud23 and HEN1 also exhibit completely different patterns of conserved residues in motifs IV, VI, and VIII that are typically associated with the MTase active site and vary between MTase families involved in different types of reaction (e.g., also between UbiE/Cfa/UbiG MTases).
Nuclear export of 40S subunits.
In the pathway for assembly and export of the small ribosomal subunit, the pre-40S particle that is exported out of the nucleus contains the 20S pre-rRNA intermediate. Once in the cytoplasm, 20S rRNA is cleaved to generate mature 18S rRNA (57, 58), releasing the 5′ fragment of ITS1 that is then degraded. Thus, the substrate for export contains 20S rRNA. In screening for novel ribosome biogenesis factors (Li and Marcotte, unpublished), we observed a nuclear export defect for bud23Δ mutant cells. This was confirmed using both Rps2-GFP and Rps3-GFP reporters and by monitoring the localization of ITS1. In bud23Δ cells, both Rps2 and Rps3 reporters accumulated in the nucleolus as well as the nucleoplasm. ITS1 also strongly accumulated in the nucleolus and nucleoplasm, consistent with the slightly elevated overall levels of 20S rRNA in bud23Δ cells. Crm1 is an export receptor that is required for export of the both the 40S and 60S subunits (14, 18, 37, 53, 55). The accumulation of ITS1 in bud23Δ cells was similar to what we observed in the presence of LMB, an inhibitor of Crm1 (25). The nuclear export phenotype of a bud23 mutant was specific because mutations in other 40S biogenesis factors, such as Bud22 (Li and Marcotte, unpublished), did not accumulate 20S rRNA, due to an earlier block in 18S processing (Li and Marcotte, unpublished). We suggest that Bud23 protein is required for efficient export of the small subunit.
It is tempting to speculate that the conservation of Bud23 throughout eukaryotes and its absence from prokaryotes reflects a function that evolved because of the acquisition of a nuclear envelope. Evolution of the nuclear envelope necessitated a mechanism for movement of macromolecules across the envelope. Although Bud23 itself does not appear to contain a nuclear export signal and thus is not likely to be an export adapter, Bud23 may be necessary for the efficient recruitment of an export adapter or other protein important for recruitment of an adapter.
In addition to Crm1, export of the small subunit depends on Yrb2 (38), a factor required for efficient Crm1-mediated export (52), as well as Ltv1, a potential Crm1-dependent adapter for the small subunit in yeast (46). Several small-subunit proteins are also involved in export of the small subunit in yeast. These include the small-subunit proteins Rps0, Rps2, Rps3, Rps15, and Rps20 (11, 32). Thus, Bud23 could be required for the correct assembly of one or more of these proteins into the pre-40S particle.
We have previously presented the idea of “structural proofreading” with respect to recruitment of the large-subunit adapter Nmd3 (21). Similar considerations have been presented for the role of rRNA modifications, which cluster around the functional centers of the ribosome (8). Considering the position of G1575 in the P-site of the small subunit and the apparent role of Bud23 in regulating export, it is possible that the Bud23 protein recognizes its substrate only after correct folding and assembly of the small subunit around the decoding center. The binding of Bud23 could then represent a mechanism of structural proofreading for the small subunit, serving as a check point in ribosome assembly that is coupled to export. Despite the fact that the MTase activity of Bud23 is dispensable, the protein is highly conserved, suggesting that its activity is under selective pressure. The methylation event itself may serve as an “inspection sticker,” marking the subunit as having been recognized by Bud23, possibly preventing Bud23 from rebinding to the subunit or allowing its release from the subunit. However, because the methylation activity of Bud23 is not critical for its function, such a mark must not be a strict requirement for releasing Bud23.
Acknowledgments
This work was supported by grant N301 2396 33 from the Polish Ministry of Science and Higher Education to J. M. Bujnicki, by NIH RO1 GM53655 to A. W. Johnson, and by grants from the NSF (IIS-0325116 and EIA-0219061), NIH (GM06779-01, GM076536-01), Welch (F1515), and a Packard Fellowship (E.M.M.) to E. Marcotte.
We thank S. Stevens, D. Lycan, and J. Dinman for yeast strains and plasmids.
Footnotes
Published ahead of print on 10 March 2008.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 253389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1988. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
- 3.Bakin, A., and J. Ofengand. 1995. Mapping of the 13 pseudouridine residues in Saccharomyces cerevisiae small subunit ribosomal RNA to nucleotide resolution. Nucleic Acids Res. 233290-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baranov, P. V., R. F. Gesteland, and J. F. Atkins. 2004. P-site tRNA is a crucial initiator of ribosomal frameshifting. RNA 10221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradatsch, B., J. Katahira, E. Kowalinski, G. Bange, W. Yao, T. Sekimoto, V. Baumgartel, G. Boese, J. Bassler, K. Wild, R. Peters, Y. Yoneda, I. Sinning, and E. Hurt. 2007. Arx1 functions as an unorthodox nuclear export receptor for the 60S preribosomal subunit. Mol. Cell 27767-779. [DOI] [PubMed] [Google Scholar]
- 6.Brand, R. C., J. Klootwijk, T. J. Van Steenbergen, A. J. De Kok, and R. J. Planta. 1977. Secondary methylation of yeast ribosomal precursor RNA. Eur. J. Biochem. 75311-318. [DOI] [PubMed] [Google Scholar]
- 7.Bujnicki, J. M. 1999. Comparison of protein structures reveals monophyletic origin of the AdoMet-dependent methyltransferase family and mechanistic convergence rather than recent differentiation of N4-cytosine and N6-adenine DNA methylation. In Silico Biol. 1175-182. [PubMed] [Google Scholar]
- 8.Decatur, W. A., and M. J. Fournier. 2002. rRNA modifications and ribosome function. Trends Biochem. Sci. 27344-351. [DOI] [PubMed] [Google Scholar]
- 9.Farrow, K. A., D. Lyras, G. Polekhina, K. Koutsis, M. W. Parker, and J. I. Rood. 2002. Identification of essential residues in the Erm(B) rRNA methyltransferase of Clostridium perfringens. Antimicrob. Agents Chemother. 461253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fatica, A., and D. Tollervey. 2002. Making ribosomes. Curr. Opin. Cell Biol. 14313-318. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira-Cerca, S., G. Poll, P. E. Gleizes, H. Tschochner, and P. Milkereit. 2005. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol. Cell 20263-275. [DOI] [PubMed] [Google Scholar]
- 12.Fornerod, M., M. Ohno, M. Yoshida, and I. W. Mattaj. 1997. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell 901051-1060. [DOI] [PubMed] [Google Scholar]
- 13.Fromont-Racine, M., B. Senger, C. Saveanu, and F. Fasiolo. 2003. Ribosome assembly in eukaryotes. Gene 31317-42. [DOI] [PubMed] [Google Scholar]
- 14.Gadal, O., D. Strauss, J. Kessl, B. Trumpower, D. Tollervey, and E. Hurt. 2001. Nuclear export of 60S ribosomal subunits depends on Xpo1p and requires a nuclear export sequence-containing factor, Nmd3p that associates with the large subunit protein Rpl10p. Mol. Cell. Biol. 213405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ghazal, G., D. Ge, J. Gervais-Bird, J. Gagnon, and S. Abou Elela. 2005. Genome-wide prediction and analysis of yeast RNase III-dependent snoRNA processing signals. Mol. Cell. Biol. 252981-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hedges, J., M. West, and A. W. Johnson. 2005. Release of the export adapter, Nmd3p, from the 60S ribosomal subunit requires Rpl10p and the cytoplasmic GTPase Lsg1p. EMBO J. 24567-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho, J., and A. W. Johnson. 1999. NMD3 encodes an essential cytoplasmic protein required for stable 60S ribosomal subunits in Saccharomyces cerevisiae. Mol. Cell. Biol. 192389-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho, J. H. N., G. Kallstrom, and A. W. Johnson. 2000. Nmd3p is a Crm1p-dependent adapter protein for nuclear export of the large ribosomal subunit. J. Cell Biol. 1511057-1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huh, W. K., J. V. Falvo, L. C. Gerke, A. S. Carroll, R. W. Howson, J. S. Weissman, and E. K. O'Shea. 2003. Global analysis of protein localization in budding yeast. Nature 425686-691. [DOI] [PubMed] [Google Scholar]
- 20.Hung, N. J., K. Y. Lo, S. S. Patel, K. Helmke, and A. W. Johnson. 2008. Arx1 is a nuclear export receptor for the 60S ribosomal subunit in yeast. Mol. Biol. Cell 19735-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson, A. W. 2003. Nuclear export of ribosomal subunits. In M. Olson (ed.), Nucleolus. Landes Biosciences, Georgetown, TX.
- 22.Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 203617-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kosinski, J., I. A. Cymerman, M. Feder, M. A. Kurowski, J. M. Sasin, and J. M. Bujnicki. 2003. A “FRankenstein's monster” approach to comparative modeling: merging the finest fragments of Fold-Recognition models and iterative model refinement aided by 3D structure evaluation. Proteins 53(Suppl. 6)369-379. [DOI] [PubMed] [Google Scholar]
- 24.Kosinski, J., M. J. Gajda, I. A. Cymerman, M. A. Kurowski, M. Pawlowski, M. Boniecki, A. Obarska, G. Papaj, P. Sroczynska-Obuchowicz, K. L. Tkaczuk, P. Sniezynska, J. M. Sasin, A. Augustyn, J. M. Bujnicki, and M. Feder. 2005. FRankenstein becomes a cyborg: the automatic recombination and realignment of fold recognition models in CASP6. Proteins 61(Suppl. 7)106-113. [DOI] [PubMed] [Google Scholar]
- 25.Kudo, N., B. Wolff, T. Sekimoto, E. P. Schreiner, Y. Yoneda, M. Yanagida, S. Horinouchi, and M. Yoshida. 1998. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp. Cell Res. 242540-547. [DOI] [PubMed] [Google Scholar]
- 26.Kumar, S., K. Tamura, and M. Nei. 2004. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 5150-163. [DOI] [PubMed] [Google Scholar]
- 27.Kurowski, M. A., and J. M. Bujnicki. 2003. GeneSilico protein structure prediction meta-server. Nucleic Acids Res. 313305-3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lafontaine, D., J. Delcour, A. L. Glasser, J. Desgres, and J. Vandenhaute. 1994. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol. 241492-497. [DOI] [PubMed] [Google Scholar]
- 29.Lafontaine, D., T. Preiss, and D. Tollervey. 1998. Yeast 18S rRNA dimethylase Dim1p: a quality control mechanism in ribosome synthesis? Mol. Cell. Biol. 182360-2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lane, B. G. 1998. Historical perspectives on RNA nucleoside modifications, p. 1-20. In H. Grosjean and R. Benne (ed.), Modification and editing of RNA. ASM Press, Washington, DC.
- 31.Lee, I., Z. Li, and E. M. Marcotte. 2007. An improved, bias-reduced probabilistic functional gene network of baker's yeast, Saccharomyces cerevisiae. PLoS ONE 2e988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leger-Silvestre, I., P. Milkereit, S. Ferreira-Cerca, C. Saveanu, J. C. Rousselle, V. Choesmel, C. Guinefoleau, N. Gas, and P. E. Gleizes. 2004. The ribosomal protein Rps15p is required for nuclear exit of the 40S subunit precursors in yeast. EMBO J. 232336-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lowe, T. M., and S. R. Eddy. 1999. A computational screen for methylation guide snoRNAs in yeast. Science 2831168-1171. [DOI] [PubMed] [Google Scholar]
- 34.Maravic, G., M. Feder, S. Pongor, M. Flogel, and J. M. Bujnicki. 2003. Mutational analysis defines the roles of conserved amino acid residues in the predicted catalytic pocket of the rRNA:m6A methyltransferase ErmC′. J. Mol. Biol. 33299-109. [DOI] [PubMed] [Google Scholar]
- 35.McCloskey, J. A., and J. Rozenski. 2005. The small subunit rRNA modification database. Nucleic Acids Res. 33D135-D138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milkereit, P., D. Strauss, J. Bassler, O. Gadal, H. Kühn, S. Schütz, N. Gas, J. Lechner, E. Hurt, and H. Tschochner. 2003. A Noc complex specifically involved in the formation and nuclear export of ribosomal 40 S subunits. J. Biol. Chem. 2784072-4081. [DOI] [PubMed] [Google Scholar]
- 37.Moy, T. I., and P. A. Silver. 1999. Nuclear export of the small ribosomal subunit requires the ran-GTPase cycle and certain nucleoporins. Genes Dev. 132118-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moy, T. I., and P. A. Silver. 2002. Requirements for the nuclear export of the small ribosomal subunit. J. Cell Sci. 1152985-2995. [DOI] [PubMed] [Google Scholar]
- 39.Niewmierzycka, A., and S. Clarke. 1999. S-Adenosylmethionine-dependent methylation in Saccharomyces cerevisiae. Identification of a novel protein arginine methyltransferase. J. Biol. Chem. 274814-824. [DOI] [PubMed] [Google Scholar]
- 40.Oakes, M., J. P. Aris, J. S. Brockenbrough, H. Wai, L. Vu, and M. Nomura. 1998. Mutational analysis of the structure and localization of the nucleolus in the yeast Saccharomyces cerevisiae. J. Cell Biol. 14323-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pei, J., and N. V. Grishin. 2007. PROMALS: towards accurate multiple sequence alignments of distantly related proteins. Bioinformatics 23802-808. [DOI] [PubMed] [Google Scholar]
- 42.Petry, S., D. E. Brodersen, F. Murphy IV, C. M. Dunham, M. Selmer, M. J. Tarry, A. C. Kelley, and V. Ramakrishnan. 2005. Crystal structures of the ribosome in complex with release factors RF1 and RF2 bound to a cognate stop codon. Cell 1231255-1266. [DOI] [PubMed] [Google Scholar]
- 43.Piekna-Przybylska, D., W. A. Decatur, and M. J. Fournier. 2007. New bioinformatic tools for analysis of nucleotide modifications in eukaryotic rRNA. RNA 13305-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasin, J. M., and J. M. Bujnicki. 2004. COLORADO3D, a web server for the visual analysis of protein structures. Nucleic Acids Res. 32W586-W589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schäfer, T., D. Strauss, E. Petfalski, D. Tollervey, and E. Hurt. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J. 221370-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seiser, R. M., A. E. Sundberg, B. J. Wollam, P. Zobel-Thropp, K. Baldwin, M. D. Spector, and D. E. Lycan. 2006. Ltv1 is required for efficient nuclear export of the ribosomal small subunit in Saccharomyces cerevisiae. Genetics 174679-691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smith, M. W., A. Meskauskas, P. Wang, P. V. Sergiev, and J. D. Dinman. 2001. Saturation mutagenesis of 5S rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 218264-8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Soding, J. 2005. Protein homology detection by HMM-HMM comparison. Bioinformatics 21951-960. [DOI] [PubMed] [Google Scholar]
- 49.Stade, K., C. S. Ford, C. Guthrie, and K. Weis. 1997. Exportin 1 (Crm1p) is an essential nuclear export factor. Cell 901041-1050. [DOI] [PubMed] [Google Scholar]
- 50.Stevens, A., C. L. Hsu, K. R. Isham, and F. W. Larimer. 1991. Fragments of the internal transcribed spacer 1 of pre-rRNA accumulate in Saccharomyces cerevisiae lacking 5′→3′ exoribonuclease 1. J. Bacteriol. 1737024-7028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suvorov, A. N., B. van Gemen, and P. H. van Knippenberg. 1988. Increased kasugamycin sensitivity in Escherichia coli caused by the presence of an inducible erythromycin resistance (erm) gene of Streptococcus pyogenes. Mol. Gen. Genet. 215152-155. [DOI] [PubMed] [Google Scholar]
- 52.Taura, T., H. Krebber, and P. A. Silver. 1998. A member of the Ran-binding protein family, Yrb2p, is involved in nuclear protein export. Proc. Natl. Acad. Sci. USA 957427-7432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thomas, F., and U. Kutay. 2003. Biogenesis and nuclear export of ribosomal subunits in higher eukaryotes depend on the CRM1 export pathway. J. Cell Sci. 1162409-2419. [DOI] [PubMed] [Google Scholar]
- 54.Tkaczuk, K. L., A. Obarska, and J. M. Bujnicki. 2006. Molecular phylogenetics and comparative modeling of HEN1, a methyltransferase involved in plant microRNA biogenesis. BMC Evol. Biol. 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trotta, C. R., E. Lund, L. Kahan, A. W. Johnson, and J. E. Dahlberg. 2003. Coordinated nuclear export of 60S ribosomal subunits and NMD3 in vertebrates. EMBO J. 222841-2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tschochner, H., and E. Hurt. 2003. Pre-ribosomes on the road from the nucleolus to the cytoplasm. Trends Cell Biol. 13255-263. [DOI] [PubMed] [Google Scholar]
- 57.Udem, S. A., and J. R. Warner. 1973. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 2481412-1416. [PubMed] [Google Scholar]
- 58.Vanrobays, E., P. E. Gleizes, C. Bousquet-Antonelli, J. Noaillac-Depeyre, M. Caizergues-Ferrer, and J. P. Gelugne. 2001. Processing of 20S pre-rRNA to 18S ribosomal RNA in yeast requires Rrp10p, an essential non-ribosomal cytoplasmic protein. EMBO J. 204204-4213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Venema, J., and D. Tollervey. 1999. Ribosome synthesis in Saccharomyces cerevisiae. Annu. Rev. Genet. 33261-311. [DOI] [PubMed] [Google Scholar]
- 60.Wai, H. H., L. Vu, M. Oakes, and M. Nomura. 2000. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 283524-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wallner, B., and A. Elofsson. 2003. Can correct protein models be identified? Protein Sci. 121073-1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wallner, B., and A. Elofsson. 2005. Pcons5: combining consensus, structural evaluation and fold recognition scores. Bioinformatics 214248-4254. [DOI] [PubMed] [Google Scholar]
- 63.Yao, W., D. Roser, A. Kohler, B. Bradatsch, J. Bassler, and E. Hurt. 2007. Nuclear export of ribosomal 60S subunits by the general mRNA export receptor Mex67-Mtr2. Mol. Cell 2651-62. [DOI] [PubMed] [Google Scholar]
- 64.Zemp, I., and U. Kutay. 2007. Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 5812783-2793. [DOI] [PubMed] [Google Scholar]