Abstract
Stable silencing of the INK4b-ARF-INK4a tumor suppressor locus occurs in a variety of human cancers, including malignant rhabdoid tumors (MRTs). MRTs are extremely aggressive cancers caused by the loss of the hSNF5 subunit of the SWI/SNF chromatin-remodeling complex. We found previously that, in MRT cells, hSNF5 is required for p16INK4a induction, mitotic checkpoint activation, and cellular senescence. Here, we investigated how the balance between Polycomb group (PcG) silencing and SWI/SNF activation affects epigenetic control of the INK4b-ARF-INK4a locus in MRT cells. hSNF5 reexpression in MRT cells caused SWI/SNF recruitment and activation of p15INK4b and p16INK4a, but not of p14ARF. Gene activation by hSNF5 is strictly dependent on the SWI/SNF motor subunit BRG1. SWI/SNF mediates eviction of the PRC1 and PRC2 PcG silencers and extensive chromatin reprogramming. Concomitant with PcG complex removal, the mixed lineage leukemia 1 (MLL1) protein is recruited and active histone marks supplant repressive ones. Strikingly, loss of PcG complexes is accompanied by DNA methyltransferase DNMT3B dissociation and reduced DNA methylation. Thus, various chromatin states can be modulated by SWI/SNF action. Collectively, these findings emphasize the close interconnectivity and dynamics of diverse chromatin modifications in cancer and gene control.
Epigenetic mechanisms confer inherited states of gene expression that involve neither alterations in the DNA sequence nor the continuous presence of the initiating signal (16, 28). The memory of gene expression status through cell divisions plays an important role in development and disease (8, 11, 17, 29, 32). For example, the stable silencing of tumor suppressor genes, such as p16INK4a, is believed to contribute to the development of human cancers. In a variety of tumors, p16INK4a is inactivated through epigenetic silencing, involving Polycomb group (PcG) proteins and DNA methylation (10, 17, 36). The principal function of p16INK4a is the induction of cellular senescence, a physiologically relevant state of permanent cell cycle arrest in response to aberrant proliferative signals (10, 25). p16INK4a acts mainly through inhibition of the Cyclin D1-CDK4 kinase, which phosphorylates and inactivates the retinoblastoma tumor suppressor protein (pRb). The p16INK4a gene is part of the INK4b-ARF-INK4a locus that encodes two other tumor suppressor proteins, p15INK4b and p14ARF (Fig. 1A) (10). p15INK4b is related to p16INK4a and also encodes a cyclin-dependent kinase inhibitor that activates pRb. Although the p14ARF transcription unit overlaps with the p16INK4a gene, it encodes a structurally unrelated protein that acts through activation of the p53 pathway.
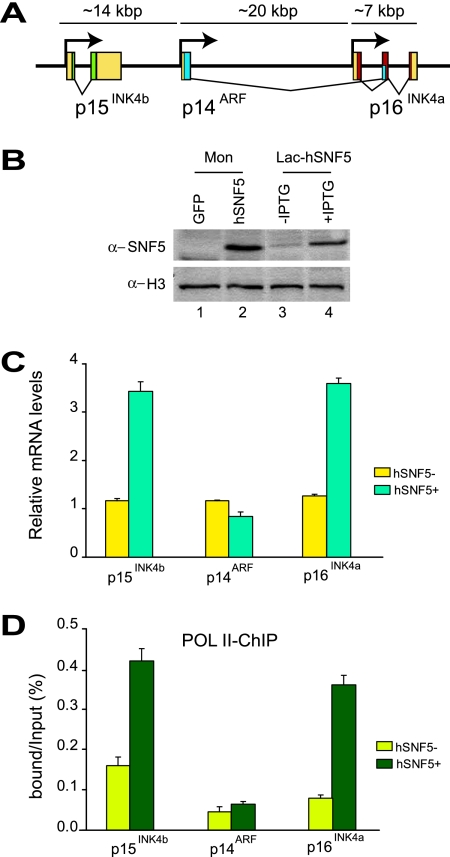
FIG. 1.
Reexpression of hSNF5 in MRT cells induces p15INK4b and p16INK4a but not p14ARF. (A) Organization of the human INK4b-ARF-INK4a locus (not drawn to scale). The genomic locus spans approximately 40 kbp of human chromosome 9 and encodes three distinct proteins: p15INK4b, p14ARF, and p16INK4a. The 5′ and 3′ untranslated regions (yellow boxes), the coding sequences of p15INK4b (green), p14ARF (blue), and p16INK4a (red) are indicated. (B) Western immunoblotting analysis of hSNF5 expression in MON cells transduced with lentiviruses expressing either GFP (lane1) or hSNF5 (lane2) and either noninduced (lane 3) or induced (lane 4) G401-derived Lac-hSNF5 cells. Cell lysates were resolved by SDS-PAGE and analyzed by Western immunoblotting with antibodies directed against hSNF5. Histone H3 serves as a loading control. (C) RT-qPCR analysis of gene expression in MRT cells reveals hSNF5-dependent induction of p15INK4b and p16INK4a, whereas p14ARF transcription remains unaffected. Cells were collected 48 h after transduction with lentiviruses expressing either GFP (yellow bars) or hSNF5 (blue bars). RT-qPCR analysis of isolated mRNA was used to determine the relative expression levels of p15INK4b, p16INK4a, p14ARF, and USP14 (a control gene that is independent of hSNF5). mRNA levels were plotted as percentage of USP14 mRNA × 0.006. The bar graphs represent the mean of three independent biological replicates, each analyzed by three separate qPCR reactions. The standard deviations are indicated. (D) RNA Pol II promoter binding was analyzed by ChIP-qPCR. Cross-linked chromatin was prepared from MRT cells lacking hSNF5 but expressing GFP (light green bars) or from cells expressing hSNF5 (dark green bars). All ChIP data presented here are the result of at least three independent experiments. The abundance of specific DNA sequences in the immunoprecipitates was determined by qPCR and corrected for the independently determined amplification curves for each primer set. Background levels were determined by ChIP using species and isotype-matched immunoglobulins directed against an unrelated protein (GST). ChIPs with antibodies directed against RNA Pol II were analyzed by qPCR using primer sets corresponding to the p15INK4b, p14ARF, and p16INK4a promoters. ChIP signal levels for each region are presented as a percentage of input chromatin.
Malignant rhabdoid tumors (MRTs) are extremely aggressive cancers of early childhood that are associated with loss of the hSNF5 subunit of the SWI/SNF chromatin-remodeling complex (4, 19, 30, 34, 38). hSNF5 reexpression in MRT cells induces p16INK4a, but not p14ARF (3, 26). Accumulated evidence indicates that the failure to activate p16INK4a transcription due to the loss of hSNF5 (also known as INI1, BAF47, or SMARCB1) is an important oncogenic step in MRTs. First, in MRT cells induction of the epigenetically silenced p16INK4a gene is both necessary and sufficient for hSNF5-mediated mitotic checkpoint activation and cellular senescence (26, 40). Second, studies in mice have suggested that hSNF5 tumor suppression acts in parallel to p53 (14) but interacts functionally with the pRb pathway (13). In conclusion, MRT cells provide a physiologically relevant cell system to study the antagonistic effects of silencing and SWI/SNF action on the chromatin status of the multigene INK4b-ARF-INK4a tumor suppressor locus.
Genetic studies in Drosophila identified SWI/SNF as a trithorax group (trxG) activator, which counteracts PcG-mediated silencing (29, 32, 37). Significantly, the PcG protein BMI1 promotes oncogenesis in mice through silencing of the INK4a-ARF locus (15). Both the PRC1 and the PRC2 PcG complexes directly bind and silence the INK4a-ARF locus (5, 20). We therefore wondered whether this is also the case in MRT cells and, more interestingly, how SWI/SNF might overcome PcG silencing. Current models of PcG function favor the notion that binding of PcG silencing complexes create a chromatin structure that is refractory to remodeling by SWI/SNF (24, 29, 32, 33). This hypothesis is mainly based on results from in vitro experiments suggesting that PRC1-coated chromatin cannot be remodeled by SWI/SNF (35). However, this model raises a conundrum for genes that need to be reactivated after PcG silencing.
Here, we explored the molecular mechanism by which restoration of SWI/SNF functionality through hSNF5 reexpression overcomes epigenetic silencing and mediates p16INK4a transcriptional activation in MRT cells. Our results reveal that in vivo SWI/SNF activity can effectively overrule PcG complex-induced chromatin silencing. We suggest that the antagonistic interactions between SWI/SNF and PcG silencers involve a dynamic equilibrium rather than a static chromatin state.
MATERIALS AND METHODS
Cell culture and lentiviral procedures.
All tissue culture was performed according to standard protocols. The hSNF5-inducible MRT-derived cell lines have been described (26). The hSNF5 expressing lentiviral vector was generated by replacing the green fluorescent protein (GFP) encoding sequence of pRRLsin.sPPT.CMV.GFP.Wpre (9) with the hSNF5 cDNA (26). High-titer vector stocks were produced in 293T cells by cotransfection of transfer vector constructs with the packaging constructs by using standard transfection procedures (9). MON cells were transduced with the appropriate vector. To knock down BRG1, the cells were transduced with lentiviruses expressing short hairpin RNA (shRNA) directed against BRG1 (clone 15549; Expression Arrest-RNAi Consortium human shRNA library purchased from Open Biosystems) for 4 days. In a control experiment, the cells were transduced with GFP-expressing lentiviruses as described previously. Approximately 24 h after initial viral transduction, the lentivirus-mediated expression of hSNF5 or GFP controls was induced for additional 72 h. For 5-aza-2′-deoxycytidine (5-azadC) treatment, MON cells were incubated with 50 μmol of 5-azadC/liter, and the medium was refreshed every 24 h. After approximately 48 h, hSNF5 or GFP expression was induced for an additional 48 h, and then the cells were harvested. Protein, RNA extraction and reverse transcription-quantitative PCR (RT-qPCR) were performed as described below.
RNA purification and real-time RT-PCR analysis.
Total RNA was extracted from MRT cells by using the SV total RNA isolation system (Promega) 48 h after hSNF5 expression was induced. cDNA was synthesized from 1 μg of total RNA by using random hexamers and Superscript II RNase H-reverse transcriptase (Invitrogen). Quantitative real-time PCR (MyIQ; Bio-Rad) was performed with Sybr green I. PCR primers were designed by using Beacon designer (Premier Biosoft). A Q-PCR core kit (Invitrogen) was used with a 400 nM concentration of each primer under the following cycling conditions: 3 min at 95°C, followed by 40 cycles of 10 s at 95°C and 45 s at 60°C. The Usp14 gene, whose expression is hSNF5 independent, was used as an endogenous reference for normalization. Enrichment of specific DNA sequences was calculated by using the comparative threshold cycle (CT) method (22). PCR primer sequences are provided in Table 1.
TABLE 1.
Q-PCR primers used for ChIP, mRNA expression, or DIP assay along the INK4-ARF locus
| Technique and primer (location) | Primer set | Sequence (5′-3′) |
|---|---|---|
| ChIP | ||
| p15INK4b-Pm | A | GGCAGTGGTGAACATTCC |
| GCCCAAAGATGCTAGGAC | ||
| p14ARF-Pm | B | CGCCGTGTCCAGATGTCG |
| TGCTCTATCCGCCAATCAGG | ||
| p16INK4a (−8.7 kb) | C | ACTAGGCTTGTCCCACTTGC |
| TCAGTTCCTCTCTCCATTCTCC | ||
| p16INK4a (−0.6 kb) | D | CCCGTCCGTATTAAATAAACC |
| GACTGCTCTCTCCTTCCC | ||
| p16INK4a (−0.3 kb) | E | GGGCTCTCACAACTAGGAAAG |
| GGGTGTTTGGTGTCATAGGG | ||
| p16INK4a (+85 bp) | F | CCCCTTGCCTGGAAAGATAC |
| AGCCCCTCCTCTTTCTTCCT | ||
| p16INK4a (+0.5 kb) | G | CTGGAGGACGAAGTTTGC |
| AGGAGGAGGTCTGTGATTAC | ||
| p16INK4a (+1.5 kb) | H | GTGTTTCTCCTCTCCCTACTCC |
| CCGGGTTCAAGCTGTTGGC | ||
| p16INK4a (+5.6 kb) | I | ACCAAGACTTCGCTGACC |
| CAAGGAGGACCATAATTCTACC | ||
| mRNA expression | ||
| p15INK4b | TAGTGGAGAAGGTGCGACAG | |
| GCGCTGCCCATCATCATG | ||
| p14ARF | GGTTTTCGTGGTTCACATCC | |
| CCTAGACGCTGGCTCCTC | ||
| p16INK4a | CCCCTTGCCTGGAAAGATAC | |
| AGCCCCTCCTCTTTCTTCCT | ||
| USP14 | AACGCTAAAGGATGATGATTGGG | |
| TTTGGCTGAGGGTTCTTCTGG | ||
| DIP assay | ||
| p15INK4b | AGCAGCGTGGGAAAGAAGGG | |
| CTTGTTCTCCTCGCGCATTCC | ||
| p14ARF | CGCCGTGTCCAGATGTCG | |
| TGCTCTATCCGCCAATCAGG | ||
| p16INK4a | GGGCTCTCACAACTAGGAAAG | |
| GGGTGTTTGGTGTCATAGGG | ||
| H19 | GCGAGCCGTAAGCACAGC | |
| GCCGATTCCCATCCAGTTGAC |
Cell extracts and Western blotting.
Cell extracts were prepared in radioimmunoprecipitation assay buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) and assayed for protein concentration, and 50 μg of extract was resolved by SDS-polyacrylamide gel electrophoresis (PAGE) and then analyzed by immunoblotting. Primary antibodies included SNF5 (ab12167 [Abcam]), BRG1 (ab4081 [Abcam]), SUZ12 (ab12073 [Abcam]), BMI1 (ab14389 [Abcam]), EZH2 (Sc-25383 [Santa Cruz]), DNMT3B (IMG-184A [IMGENEX]), and histone H3 (ab1791 [Abcam]). Western blots were developed by using an ECL detection kit (Pierce) according to the supplier's instructions.
ChIP and MeDIP assays.
Chromatin immunoprecipitations (ChIPs) were performed as described by the Upstate protocol. Cross-linked chromatin was prepared from ∼2 × 107 cells 48 h after transduction with lentiviruses expressing either hSNF5 or GFP as a control. Cells were treated with 1% formaldehyde for 20 min at room temperature. Chromatin isolation, sonication yielding fragments of 300 to 600 bp, and immunoprecipitations were performed according to previously established protocols. The following antibodies were used: SNF5 (ab12167 [Abcam]), BRG1 (ab4081 [Abcam]), SUZ12 (ab12073 [Abcam]), BMI1 (ab14389 [Abcam]), EZH2 (Sc-25383 [Santa Cruz]), DNMT3B (IMG-184A [IMGENEX]), POL II (Sc-899 [Santa Cruz]), mixed lineage leukemia 1 (MLL1; A300-086A [Bethyl Laboratories]), Histone H3 (ab1791 [Abcam]), H3-K4me3 (ab12209 [Abcam]), and H3-K27me3 (catalog no. 07-449 [Upstate]). The abundance of specific DNA sequences in the immunoprecipitates was determined by qPCR and corrected for the independently determined amplification curves of each primer set. ChIP using species- and isotype-matched immunoglobulins directed against an unrelated protein (glutathione S-transferase [GST]) were used to determine background levels. For methylated DNA immunoprecipitate (MeDIP) assays, genomic DNA was prepared by overnight proteinase K treatment, phenol-chloroform extraction, and ethanol precipitation. Genomic DNA was digested with MboI, and 4 μg of fragmented DNA was used for a standard MeDIP assay. After denaturation at 95°C for 10 min, the reaction was quenched on ice, and 500 μl of MeIP buffer (phosphate-buffered saline-0.05% Triton X-100), 50 μl of salmon sperm DNA-protein G-agarose beads (Upstate), and 10 μg of anti-5-methylcytosine monoclonal antibody (Eurogentec) was added. As a negative control, DNA fragments were incubated with nonspecific mouse immunoglobulin G. After an overnight incubation on a rotating wheel at 4°C, beads were washed with 700 μl of MeIP buffer and resuspended in 100 μl and treated with proteinase K for 3 h at 50°C. Finally, DNA was purified by conventional phenol-chloroform extraction, followed by ethanol precipitation and analysis by qPCR. The enrichment of specific DNA sequences was calculated by using the comparative CT method (22). PCR primer sequences are provided in Table 1. All data presented are the result of at least three independent ChIP or MeDIP experiments and triplicate qPCR reactions. The results were averaged, and standard errors were determined by using the R free software (http://www.r-project.org). The ChIP and MeDIP levels for each region are presented as the percentage of input chromatin. ChIPs using antibodies directed against H3-K27me3 or H3-K4me3 were normalized against histone H3 density.
RESULTS
hSNF5 is required for selective recruitment of BRG1 to the p15INK4b and p16INK4a promoters.
We utilized two distinct strategies to reexpress hSNF5 in MRT cells: the previously described Lac repressor-operator system (26) and transduction with hSNF5-expressing lentiviruses. Samples were taken 48 h after induction of hSNF5 expression in two different MRT-derived cell lines: MON and G401. We note that hSNF5 is not essential for SWI/SNF complex integrity (7, 23, 26). Lentiviral transduction resulted in expression levels that were comparable to that obtained using the Lac repressor operator system (Fig. 1B). Moreover, we previously showed that the levels of induced exogenous hSNF5 expression were comparable to the endogenous levels in a variety of cell lines (7, 23, 26). Below, we present results obtained for MON cells transduced with lentiviruses expressing either hSNF5 or GFP as a control. However, all results presented here were consistently observed for both cell lines and either delivery system.
To investigate the effect of hSNF5 on expression of the INK4b-ARF-INK4a locus, we quantified mRNA levels by RT-qPCR (Fig. 1C). In full agreement with our previous Western blotting results (26), we observed a clear induction of p16INK4a, but not p14ARF mRNA levels. In addition, we found that p15INK4b was upregulated. ChIP experiments revealed that hSNF5 expression triggers RNA polymerase II (RNA Pol II) recruitment to the p15INK4b and p16INK4a promoters, but not to p14ARF (Fig. 1D). We conclude that in the context of hSNF5 reexpression in MRT cells, p14ARF is not induced coordinately with p15INK4b and p16INK4a. Thus, the coregulation of the INK4b-ARF-INK4a locus observed in selective cell lines (12) is not a general phenomenon.
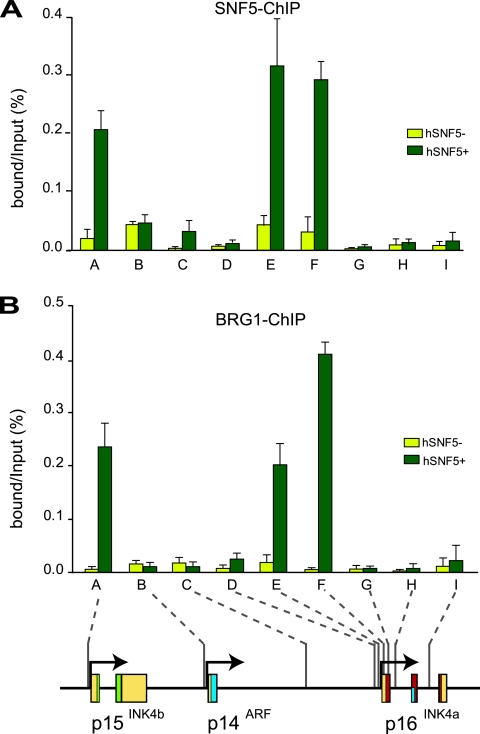
To gain insight into the mechanism of p15INK4b and p16INK4a activation by hSNF5, we analyzed the chromatin status at the INK4b-ARF-INK4a locus. For qPCR we used primer sets corresponding to the promoter areas of p15INK4b (primer set A) and p14ARF (primer set B), an intergenic control (primer set C), whereas we probed the p16INK4a locus at a higher resolution (primer sets D to I [Fig. 2, bottom]). ChIPs directed against hSNF5 revealed strong binding to the p15INK4b and p16INK4a promoters when hSNF5 is expressed (green bars; Fig. 2A). The p14ARF promoter was not bound by hSNF5, as reflected by the low background signal, also present in cells devoid of hSNF5 (yellow bars). Importantly, hSNF5 was strictly required to bring BRG1, the central SWI/SNF motor subunit, to the p15INK4b and p16INK4a promoters (Fig. 2B). We note that hSNF5 expression does not influence BRG1 levels in the cell (Fig. 3A) (26). Finally, these ChIPs demonstrated that SWI/SNF recruitment is limited to the p15INK4b and p16INK4a promoter areas and does not spread across the locus.
FIG. 2.
hSNF5 mediates BRG1 recruitment to the p15INK4b and p16INK4a promoters. (A) ChIP-qPCR analysis of hSNF5 binding to the INK4b-ARF-INK4a locus revealed that hSNF5 binds directly to the p15INK4b and p16INKa promoters, but not to p14ARF. Cross-linked chromatin was isolated from MRT cells that either lack (light green bars) or express (dark green bars) hSNF5. qPCR primer sets correspond to the p15INK4b promoter (A), the p14ARF promoter (B), an intergenic control region (C), and various regions of the p16INK4a locus (sets D to I). Primer sets E and F cover the p16INK4a promoter. The positions of the amplified regions on the INK4b-ARF-INK4a locus are indicated at the bottom. (B) BRG-1 binding to the p15INK4b and p16INK4a promoters is hSNF5 dependent, as revealed by ChIP-qPCR with antibodies directed against BRG-1. The procedures were as described in the legend to Fig. 1.
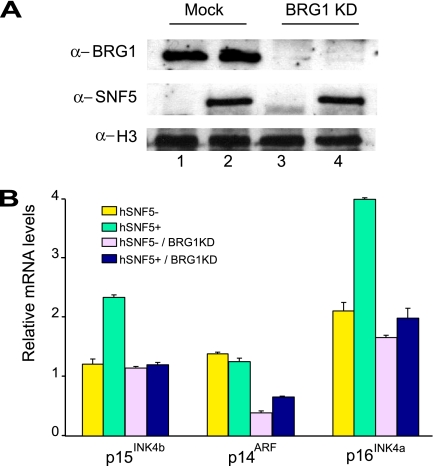
FIG. 3.
BRG1 is required for hSNF5-mediated induction of p15INK4b and p16INK4a. (A) Western blot analysis of the BRG1 protein levels in MON cell extracts prepared 4 days after BRG1 knockdown using lentiviral transduction with viruses expressing shRNA targeting BRG1 mRNA (clone 15549; Open Biosystems; top panel, lanes 3 and 4). As a control, cells were transduced with lentiviruses expressing GFP. One day after transduction with either GFP- or shRNA-expressing lentiviruses, cells were transduced again with viruses expressing either GFP (middle panel, lanes 1 and 3) or hSNF5 (lanes 2 and 3). Cell extracts were prepared 72 h after hSNF5 expression. Histone H3 serves as a loading control. (B) Loss of BRG1 abrogates transcriptional activation of p15INK4b and p16INK4a by hSNF5. Relative expression levels of p15INK4b, p14ARF, and p16INK4a in these cells were determined by RT-qPCR of isolated mRNA, 72 h after hSNF5 expression. The bar graphs represent the mean of three independent experiments, each analyzed in triplicate by RT-qPCR.
Does hSNF5 mediate gene activation by itself, or does its function depend on the chromatin remodeling activity of SWI/SNF? To investigate this question, we used lentiviral expressed shRNAs to deplete MON cells for BRG1, the central ATPase that is critical for chromatin remodeling (Fig. 3A). Western immunoblot analysis of extracts from mock-treated cells (lanes 1 and 2) or cells transduced with lentiviruses expressing shRNAs directed against BRG1 (lanes 3 and 4) revealed a dramatic shRNA-mediated loss of BRG1. Next, we used lentiviral transduction of viruses expressing either GFP (lanes 1 and 3) or hSNF5 (lanes 2 and 4) to test the effect of hSNF5 reexpression in cells depleted of BRG1. Western immunoblotting confirmed our earlier observation (26) that hSNF5 reexpression did not influence BRG1 levels (Fig. 3A, compare lanes 1 and 2). Antibodies directed against histone H3 were used as a loading control. In agreement with our earlier results, hSNF5 expression in mock-treated cells led to a robust induction of both 15INK4b and p16INK4a, but not of p14ARF, as revealed by RT-qPCR (Fig. 3B). However, in BRG1-depleted cells hSNF5 could no longer activate 15INK4b and p16INK4a expression. Surprisingly, the low level of p14ARF expression was reduced further due to the loss of BRG1, indicating a potential role for BRG1, but not hSNF5. We conclude that hSNF5-mediated induction of 15INK4b and p16INK4a is critically dependent on BRG1, the key catalytic subunit of SWI/SNF.
SWI/SNF displaces PcG silencing complexes.
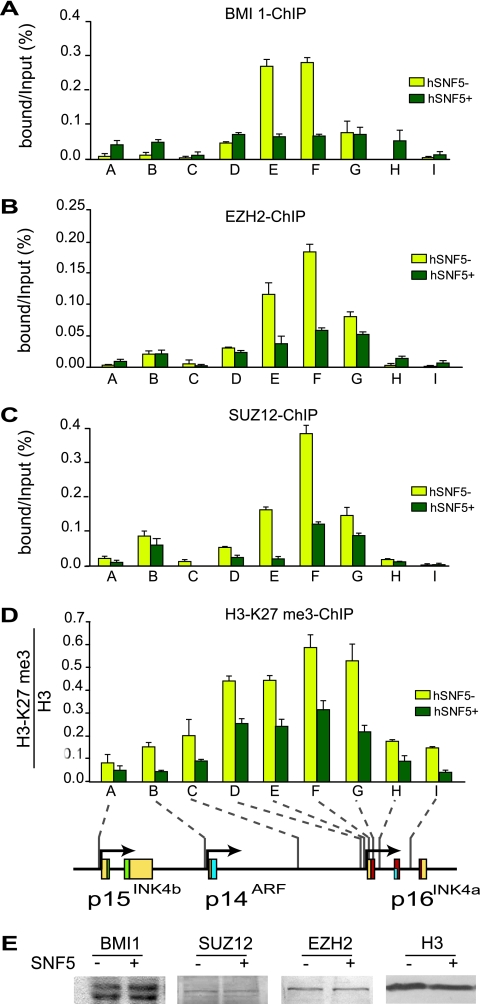
Overexpression of the PcG protein BMI1 promotes oncogenesis in mice through silencing of the INK4a-ARF locus (15). Growth of normal and cancerous prostate cells is controlled by PcG protein CBX7-dependent repression of INK4a-ARF (2). Recently, it was observed that both PcG complexes PRC1 and PRC2 can directly bind and silence the INK4a-ARF locus (5, 20). Because hSNF5 expression in MRT cells leads to SWI/SNF recruitment to the p15INK4b and p16INK4a promoters, we tested its effect on PcG complex binding. First, we established whether in MRT cells PcG silencers occupy the INK4b-ARF-INK4a locus by ChIPs directed against the PRC1 subunit BMI1 and the PRC2 subunits EZH2 and Suz12 (Fig. 4A to C). We observed strong binding of PcG silencers to the p16INK4a promoter region (primer sets E and F) in the absence of hSNF5. Outside this domain of ∼0.8 kb, PcG protein binding tapers off across the locus. Relatively low amounts of PcG proteins were detected at the p14ARF and p15INK4b promoters. Strikingly, hSNF5 expression strongly reduces the binding of both PRC1 and PRC2. We conclude that during p16INK4a activation, SWI/SNF mediates the eviction of PcG silencing complexes. The PRC2 subunit EZH2 catalyzes histone H3 lysine 27 trimethylation (H3-K27me3), a chromatin mark associated with gene repression (29, 32). Although this could be due to ChIP efficiency, the H3-K27me3 histone mark appears to spread more broadly than PRC2 (Fig. 4D). Clearly, H3-K27me3 is not a stable mark but is reduced upon hSNF5 expression and removal of PRC2. This is likely to be the result of the action of the recently identified H3-K27me3 demethylases (1, 21). Finally, we note that the loss of PcG protein binding was not due to a change in their expression, as revealed by Western blotting (Fig. 4E). In conclusion, our results demonstrated that SWI/SNF can effectively counteract prebound PcG complexes during regulation of an endogenous gene. Thus, unlike previously published in vitro experiments (35), PcG binding does not necessarily block SWI/SNF action in vivo. We suggest that the antagonistic interactions between SWI/SNF and PcG silencers involve a dynamic equilibrium rather than a static chromatin state.
FIG. 4.
Restoration of SWI/SNF causes eviction of PcG silencers and loss of H3-K27 methylation. ChIPs using antibodies directed against BMI1 (A), EZH2 (B), SUZ12 (C), and H3-K27me3 (D) were performed. Cross-linked chromatin was isolated from MRT cells that either lack (yellow bars) or express (dark green bars) hSNF5. ChIPs were analyzed by qPCR using primer sets specific for the regions indicated by A to I along the INK4b-ARF-INK4a locus, revealing that PcG silencer binding peaks at the p16INK4a promoter. After hSNF5 induction both PRC1 (BMI1) and PRC2 (EZH2 and SUZ12) were removed, and H3-K27me3 was strongly reduced. H3-K27me3 ChIPs were normalized to H3 ChIP. Procedures were as described in the legend to Fig. 1. (E) hSNF5 expression does not affect BMI1, SUZ12, and EZH2 levels. Western immunoblotting analysis of BMI1, SUZ12, and EZH2 expression in MON cells transduced by either GFP- or SNF5-expressing lentiviruses. Cell lysates were resolved by SDS-PAGE and analyzed by Western immunoblotting with antibodies to BMI1, SUZ12, and EZH2, respectively. Histone H3 serves as a loading control.
SWI/SNF induced recruitment of MLL1.
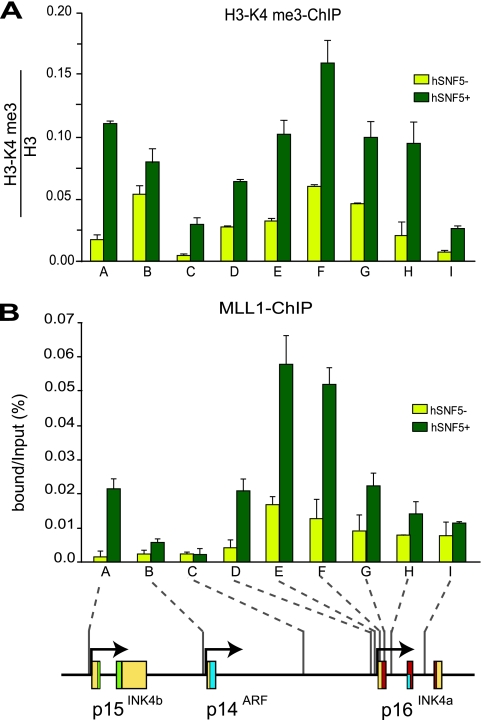
The loss of the H3-K27me3 repressive histone mark after PcG silencer eviction by SWI/SNF prompted us to investigate whether this is accompanied by the appearance of positive marks. We therefore determined the effect of hSNF5 expression on H3-K4me3 by ChIP. We found that concomitant with the decrease in H3-K27me3, the active H3-K4me3 mark is strongly induced at p16INK4a and p15INK4b (Fig. 5A). At the p14ARF promoter H3-K4me3 levels are at an intermediate level but not significantly affected by hSNF5 expression. The prominent H3-K4me3 methyltransferase MLL1 is the human homologue of Drosophila TRX, the founding member of the trxG (6). ChIPs revealed that hSNF5 expression triggers MLL1 binding to the INK4b and INK4a loci (Fig. 5B). Unlike SWI/SNF, which is targeted mainly to the promoter regions, MLL1 appears to be more broadly distributed on the p16INK4a gene. Because MLL1 translocations are associated with highly aggressive lymphoid and myeloid infant leukemia (6), it will be of interest to investigate whether the oncogenic MLL1 fusion proteins are defective in p16INK4a activation. Collectively, our results show that during hSNF5-mediated p16INK4a activation, the trxG activators SWI/SNF and MLL1 replace the PcG silencers PRC1 and PRC2. We conclude that the antagonism between PcG and trxG proteins is not limited to fly development but also operates in human cancer cells.
FIG. 5.
hSNF5 induced recruitment of H3-K4 methylase MLL1. ChIPs with antibodies directed against H3-K4me3 (A) and MLL1 (B) reveal increased H3-K4me3 and MLL1 binding at p15INK4b and p16INK4a after hSNF5 expression. Cross-linked chromatin was isolated from MRT cells that either lack (yellow bars) or express (dark green bars) hSNF5. ChIPs were analyzed by qPCR using the primer sets specific for the regions indicated by A to I along the INK4b-ARF-INK4a locus. Histone H3-K4me3 ChIPs were normalized to histone H3 ChIP. The procedures were as described in the legend to Fig. 1.
SWI/SNF recruitment leads to reduced DNA methylation at the p16INK4a promoter.
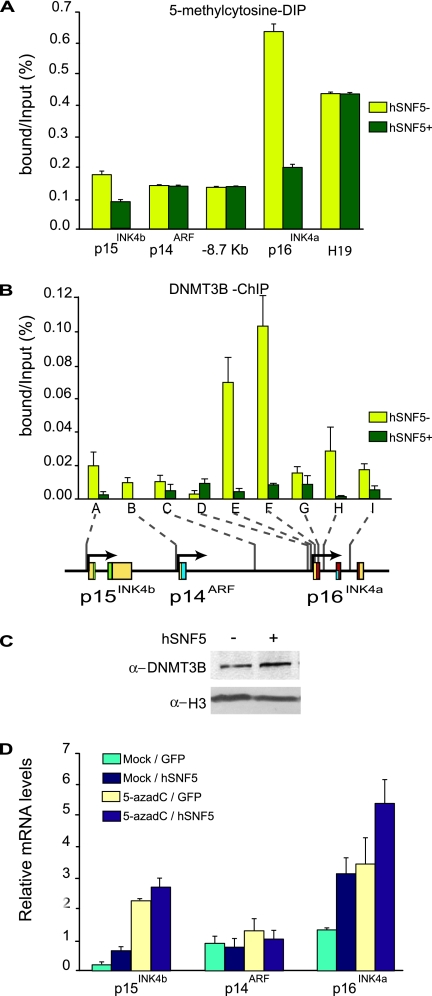
Our results thus far emphasized the dynamic rather than static nature of chromatin states. We therefore wondered what the effect of SWI/SNF promoter binding might be on CpG island methylation, a mark that is usually considered to be highly stable. A multitude of studies has implicated DNA hyper-methylation of a promoter-proximal CpG island in the epigenetic inactivation of p16INK4a in a wide variety of human cancers (17). We investigated the DNA methylation status of the p16INK4a promoter in MRT cells using antibodies directed against methylated CpG for immunoprecipitation (MeDIP). We detected DNA methylation at the p16INK4a promoter, which was dramatically reduced after hSNF5 expression (Fig. 6A). Although CpG methylation levels at the p15INK4b promoter were comparably low, again we observed a clear reduction. These effects were gene selective, because CpG methylation at p14ARF or the imprinted H19 locus remained unchanged. Next, we examined the distribution of the DNA methyltransferase DNMT3B, which has previously been associated with PcG silencing (31, 39). ChIPs revealed that, similar to the PcG silencers, DNMT3B binding peaks at the p16INK4a promoter region and tapers off to a low level across the locus. However, concomitant with the drop in CpG methylation, DNMT3B is evicted from the locus after SWI/SNF binding (Fig. 6B). Western immunoblotting demonstrated that the loss of DNMT3B binding is not caused by a reduced expression (Fig. 6C). We conclude that, similar to the chromatin association of PcG silencers, DNA methylation can be highly dynamic and modulated by SWI/SNF recruitment.
FIG. 6.
Loss of DNA methylation at the p16INK4a promoter after SWI/SNF restoration. (A) MeDIP analysis of changes in CpG DNA methylation at the p16INK4a promoter, harboring a CpG island. A number of other regions (devoid of CpG islands) within the INK4b-ARF-INK4a locus were amplified as controls. The imprinted and hypermethylated H19 gene was used as a reference. Genomic DNA was isolated from MRT cells that either lack (yellow bars) or express (dark green bars) hSNF5. After MboI digestion and denaturation, MeDIP was performed with antibodies directed against 5-methylcytosine and quantified by using qPCR. Binding is expressed as a percentage of the input DNA. The bar graphs represent the mean of three independent MeDIP experiments, each analyzed in triplicate by qPCR. (B) Reactivation of SWI/SNF leads to loss of the DNA methyltransferase DNMT3B, as revealed by ChIPs on chromatin isolated from MRT cells that either lack (yellow bars) or express (dark green bars) hSNF5. The ChIPs were analyzed by qPCR using primer sets specific for the regions indicated by A-I along the INK4b-ARF-INK4a locus. The procedures were as described in the legend to Fig. 1. (C) hSNF5 expression does not affect DNMT3B expression levels. Western immunoblot analysis of DNMT3B in MON cells transduced by either GFP- or SNF5-expressing lentiviruses. Histone H3 serves as a loading control. (D) Treatment with a DNA methylation inhibitor and hSNF5 expression has additive effects on p15INK4b and p16INK4a induction. MON cells were either mock treated or incubated with 50 μmol of 5-azadC/liter. Approximately 48 h later the cells were transduced with lentiviruses expressing either GFP or hSNF5. Relative expression levels of p15INK4b, p14ARF, and p16INK4a were determined by the RT-qPCR of isolated mRNA. The bar graphs represent the mean of three independent experiments each analyzed in triplicate by RT-qPCR.
Next, we tested the effect of the DNA methylation inhibitor 5-azadC on the expression of the INK4b-ARF-INK4a locus in MRT cells. We incubated MON MRT cells with 5-azadC, followed by transduction with either GFP- or hSNF5-expressing lentiviruses. Expression analysis by RT-qPCR revealed that the addition of 5-azadC resulted in a robust induction of p15INK4b and p16INK4a, but not of p14ARF. Thus, treatment with a DNA methylation inhibitor leads to transcriptional derepression, supporting the functional importance of CpG methylation in p15INK4b and p16INK4a silencing. When 5-azadC treatment was combined with hSNF5 reexpression, we observed modestly additive stimulation of p15INK4b and p16INK4a transcription, whereas p14ARF remained unaffected. The absence of synergism could be interpreted as a suggestion that removal of CpG methylation and SWI/SNF action ultimately have similar consequences for the local chromatin structure. Again, these findings emphasize the close interconnectivity of various chromatin structure modulating activities.
DISCUSSION
The loss of hSNF5 in MRT cells precludes SWI/SNF binding to the p15INK4b and p16INK4a promoters, which are both stably silenced in these cells. By reexpressing hSNF5 in MRT cells, we created a switch allowing us to study the effect of SWI/SNF recruitment to a silenced locus bound by PcG silencers. Reexpression of hSNF5 in MRT cells overcomes epigenetic silencing and mediates transcriptional activation of p15INK4b and p16INK4a. In contrast, p14ARF is not induced, demonstrating that the INK4b-ARF-INK4a locus is not coordinately regulated in MRT cells. Because SWI/SNF itself lacks sequence-specific DNA-binding activity, an important future goal is the identification of promoter binding recruiters. An issue that has remained unresolved for a considerable time is whether hSNF5 functions as part of the SWI/SNF complex or if it acts independently. shRNA-mediated depletion of BRG1 revealed that hSNF5 was completely dependent on the motor subunit of the SWI/SNF complex to activate either p15INK4b or p16INK4a. Thus, it appears that gene activation by hSNF5 is dependent on BRG1-mediated chromatin remodeling. These results dovetail well with our functional dissection of Drosophila SWI/SNF remodelers in genome wide expression control (7, 23, 26). Both studies suggest that SWI/SNF subunits do not function outside the remodeling complexes.
Although the whole INK4b-ARF-INK4a locus is inactive, it is not evenly coated by PcG silencing complexes. Instead, we found that PRC1 and PRC2 binding peaks at the repressed p16INK4a promoter. These results indicate that the ∼0.8-kb p16INK4a promoter area might function as a nucleation site for PcG complex recruitment. The H3-K27me3 mark appears to spread more broadly than the PcG proteins, indicative of a “kiss-and-go” mechanism of histone methylation by PRC2. A similar difference in the pattern of distribution of H3-K27me3 and PRC2 has been observed for Drosophila PREs and associated genes (18, 27). In contrast to the purported primacy of PRC1 prebinding over SWI/SNF action (35), we found that in MRT cells PRC1 could be effectively displaced by SWI/SNF. In fact, SWI/SNF binding initiates a cascade of chromatin reprogramming, which completely resets the epigenetic status of p15INK4b and p16INK4a but leaves p14ARF largely unaffected. The trxG activators SWI/SNF and MLL1 supplant their antagonists PRC1, PRC2, and the DNA methyltransferase DNMT3B. Concomitant with RNA Pol II recruitment and gene transcription, active histone marks replace repressive ones, and DNA methylation at the p16INK4a promoter is strongly reduced. We speculate that during stem cell differentiation, SWI/SNF remodelers might play a comparable role in removal of PcG silencers from genes that need to be expressed.
Previous studies have suggested a functional association between PcG silencing and CpG methylation by DNA methyltransferases such as DNMT3B (31, 39). Here, we observed that DNMT3B was displaced by SWI/SNF action concomitantly with the PcG silencing complexes. Moreover, we found that addition of the DNA methylation inhibitor 5-azadC leads to the transcriptional derepression of p15INK4b and p16INK4a, whereas p14ARF remained unaffected. These results support the functional importance of CpG methylation for the silencing of these genes in MRT cells and might have therapeutic implications. Combining the 5-azadC treatment with hSNF5 expression in MRT cells resulted in modestly additive stimulation of p15INK4b and p16INK4a transcription. We interpret this absence of synergistic activation as an indication that the removal of CpG methylation and SWI/SNF action start a cascade that results in similar changes in chromatin structure. Collectively, these findings emphasize the intertwined dynamics of diverse chromatin marks and SWI/SNF function during transcriptional regulation.
Acknowledgments
We thank D. Trono for the gift of lentiviral vectors; T. van Dijk for advice on lentiviral procedures; Parham Solaimani for the Brg1 knockdown experiment, and J. Svejstrup, R. Vries, S. Philipsen, and A. Bracken for valuable comments on the manuscript.
This study was supported by grants from the Dutch Cancer Society KWF (EMCR2006-3583) and NWO Chemical Sciences (TOP700.52.312) to C.P.V.
Footnotes
Published ahead of print on 10 March 2008.
REFERENCES
- 1.Agger, K., P. A. C. Cloos, J. Christensen, D. Pasini, S. Rose, J. Rappsilber, I. Issaeva, E. Canaani, A. E. Salcini, and K. Helin. 2007. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature 449731-734. [DOI] [PubMed] [Google Scholar]
- 2.Bernard, D., J. F. Martinez-Leal, S. Rizzo, D. Martinez, D. Hudson, T. Visakorpi, G. Peters, A. Carnero, D. Beach, and J. Gil. 2005. CBX7 controls the growth of normal and tumor-derived prostate cells by repressing the Ink4a/Arf locus. Oncogene 245543-5551. [DOI] [PubMed] [Google Scholar]
- 3.Betz, B. L., M. W. Strobeck, D. N. Reisman, E. S. Knudsen, and B. E. Weissman. 2002. Re-expression of hSNF5/INI1/BAF47 in pediatric tumor cells leads to G1 arrest associated with induction of p16ink4a and activation of RB. Oncogene 215193-5203. [DOI] [PubMed] [Google Scholar]
- 4.Biegel, J. A., J. Y. Zhou, L. B. Rorke, C. Stenstrom, L. M. Wainwright, and B. Fogelgren. 1999. Germ-line and acquired mutations of INI1 in atypical teratoid and rhabdoid tumors. Cancer Res. 5974-79. [PubMed] [Google Scholar]
- 5.Bracken, A. P., D. Kleine-Kohlbrecher, N. Dietrich, D. Pasini, G. Gargiulo, C. Beekman, K. Theilgaard-Monch, S. Minucci, B. T. Porse, J.-C. Marine, K. H. Hansen, and K. Helin. 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 21525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canaani, E., T. Nakamura, T. Rozovskaia, S. T. Smith, T. Mori, C. M. Croce, and A. Mazo. 2004. ALL-1/MLL1, a homologue of Drosophila TRITHORAX, modifies chromatin and is directly involved in infant acute leukaemia. Br. J. Cancer 90:Br J. Cancer 2004 Feb 23;. 90756-760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Doan, D., T. M. Veal, Z. Yan, W. Wang, S. N. Jones, and A. N. Imbalzano. 2004. Loss of the INI1 tumor suppressor does not impair the expression of multiple BRG1-dependent genes or the assembly of SWI/SNF enzymes. Oncogene 233462-3473. [DOI] [PubMed] [Google Scholar]
- 8.Feinberg, A. P., R. Ohlsson, and S. Henikoff. 2006. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 721-33. [DOI] [PubMed] [Google Scholar]
- 9.Follenzi, A., G. Sabatino, A. Lombardo, C. Boccaccio, and L. Naldini. 2002. Efficient gene delivery and targeted expression to hepatocytes in vivo by improved lentiviral vectors. Hum. Gene Ther. 13243-260. [DOI] [PubMed] [Google Scholar]
- 10.Gil, J., and G. Peters. 2006. Regulation of the INK4b-ARF-INK4a tumour suppressor locus: all for one or one for all. Nat. Rev. Mol. Cell. Biol. 7667-677. [DOI] [PubMed] [Google Scholar]
- 11.Goldberg, A. D., C. D. Allis, and E. Bernstein. 2007. Epigenetics: a landscape takes shape. Cell 128635-638. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez, S., P. Klatt, S. Delgado, E. Conde, F. Lopez-Rios, M. Sanchez-Cespedes, J. Mendez, F. Antequera, and M. Serrano. 2006. Oncogenic activity of Cdc6 through repression of the INK4/ARF locus. Nature 440702-706. [DOI] [PubMed] [Google Scholar]
- 13.Guidi, C. J., R. Mudhasani, K. Hoover, A. Koff, I. Leav, A. N. Imbalzano, and S. N. Jones. 2006. Functional interaction of the retinoblastoma and Ini1/Snf5 tumor suppressors in cell growth and pituitary tumorigenesis. Cancer Res. 668076-8082. [DOI] [PubMed] [Google Scholar]
- 14.Isakoff, M. S., C. G. Sansam, P. Tamayo, A. Subramanian, J. A. Evans, C. M. Fillmore, X. Wang, J. A. Biegel, S. L. Pomeroy, J. P. Mesirov, and C. W. Roberts. 2005. Inactivation of the Snf5 tumor suppressor stimulates cell cycle progression and cooperates with p53 loss in oncogenic transformation. Proc. Natl. Acad. Sci. USA 10217745-17750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs, J. J. L., K. Kieboom, S. Marino, R. A. DePinho, and M. van Lohuizen. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397164-168. [DOI] [PubMed] [Google Scholar]
- 16.Jenuwein, T., and C. D. Allis. 2001. Translating the histone code. Science 2931074-1080. [DOI] [PubMed] [Google Scholar]
- 17.Jones, P. A., and S. B. Baylin. 2007. The epigenomics of cancer. Cell 128683-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kahn, T. G., Y. B. Schwartz, G. I. Dellino, and V. Pirrotta. 2006. Polycomb complexes and the propagation of the methylation mark at the Drosophila ubx gene. J. Biol. Chem. 28129064-29075. [DOI] [PubMed] [Google Scholar]
- 19.Klochendler-Yeivin, A., C. Muchardt, and M. Yaniv. 2002. SWI/SNF chromatin remodeling and cancer. Curr. Opin. Genet. Dev. 1273-79. [DOI] [PubMed] [Google Scholar]
- 20.Kotake, Y., R. Cao, P. Viatour, J. Sage, Y. Zhang, and Y. Xiong. 2007. pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4a tumor suppressor gene. Genes Dev. 2149-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, M. G., R. Villa, P. Trojer, J. Norman, K.-P. Yan, D. Reinberg, L. Di Croce, and R. Shiekhattar. 2007. Demethylation of H3K27 regulates Polycomb recruitment and H2A ubiquitination. Science 318447-450. [DOI] [PubMed] [Google Scholar]
- 22.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25402-408. [DOI] [PubMed] [Google Scholar]
- 23.Moshkin, Y. M., L. Mohrmann, W. F. J. van Ijcken, and C. P. Verrijzer. 2007. Functional differentiation of SWI/SNF remodelers in transcription and cell cycle control. Mol. Cell. Biol. 27651-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muller, J., and J. A. Kassis. 2006. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr. Opin. Genet. Dev. 16476-484. [DOI] [PubMed] [Google Scholar]
- 25.Narita, M., and S. W. Lowe. 2005. Senescence comes of age. Nat. Med. 11920-922. [DOI] [PubMed] [Google Scholar]
- 26.Oruetxebarria, I., F. Venturini, T. Kekarainen, A. Houweling, L. M. Zuijderduijn, A. Mohd-Sarip, R. G. Vries, R. C. Hoeben, and C. P. Verrijzer. 2004. p16INK4a is required for hSNF5 chromatin remodeler-induced cellular senescence in malignant rhabdoid tumor cells. J. Biol. Chem. 2793807-3816. [DOI] [PubMed] [Google Scholar]
- 27.Papp, B., and J. Muller. 2006. Histone trimethylation and the maintenance of transcriptional ON and OFF states by TrxG and PcG proteins. Genes Dev. 202041-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ptashne, M. 2007. On the use of the word “epigenetic”. Curr. Biol. 17R233-R236. [DOI] [PubMed] [Google Scholar]
- 29.Ringrose, L., and R. Paro. 2007. Polycomb/Trithorax response elements and epigenetic memory of cell identity. Development 134223-232. [DOI] [PubMed] [Google Scholar]
- 30.Roberts, C. W., and S. H. Orkin. 2004. The SWI/SNF complex: chromatin and cancer. Nat. Rev. Cancer 4133-142. [DOI] [PubMed] [Google Scholar]
- 31.Schlesinger, Y., R. Straussman, I. Keshet, S. Farkash, M. Hecht, J. Zimmerman, E. Eden, Z. Yakhini, E. Ben-Shushan, B. E. Reubinoff, Y. Bergman, I. Simon, and H. Cedar. 2007. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39232-236. [DOI] [PubMed] [Google Scholar]
- 32.Schuettengruber, B., D. Chourrout, M. Vervoort, B. Leblanc, and G. Cavalli. 2007. Genome regulation by Polycomb and Trithorax proteins. Cell 128735-745. [DOI] [PubMed] [Google Scholar]
- 33.Schwartz, Y. B., and V. Pirrotta. 2007. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 89-22. [DOI] [PubMed] [Google Scholar]
- 34.Sevenet, N., E. Sheridan, D. Amram, P. Schneider, R. Handgretinger, and O. Delattre. 1999. Constitutional mutations of the hSNF5/INI1 gene predispose to a variety of cancers. Am. J. Hum. Genet. 651342-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shao, Z., F. Raible, R. Mollaaghababa, J. R. Guyon, C.-t. Wu, W. Bender, and R. E. Kingston. 1999. Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 9837-46. [DOI] [PubMed] [Google Scholar]
- 36.Sparmann, A., and M. van Lohuizen. 2006. Polycomb silencers control cell fate, development and cancer. Nat. Rev. Cancer 6846-856. [DOI] [PubMed] [Google Scholar]
- 37.Tamkun, J. W., R. Deuring, M. P. Scott, M. Kissinger, A. M. Pattatucci, T. C. Kaufman, and J. A. Kennison. 1992. brahma: a regulator of Drosophila homeotic genes structurally related to the yeast transcriptional activator SNF2/SWI2. Cell 68561-572. [DOI] [PubMed] [Google Scholar]
- 38.Versteege, I., N. Sevenet, J. Lange, M. F. Rousseau-Merck, P. Ambros, R. Handgretinger, A. Aurias, and O. Delattre. 1998. Truncating mutations of hSNF5/INI1 in aggressive paediatric cancer. Nature 394203-206. [DOI] [PubMed] [Google Scholar]
- 39.Vire, E., C. Brenner, R. Deplus, L. Blanchon, M. Fraga, C. Didelot, L. Morey, A. Van Eynde, D. Bernard, J.-M. Vanderwinden, M. Bollen, M. Esteller, L. Di Croce, Y. de Launoit, and F. Fuks. 2006. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 439:871-874. [DOI] [PubMed] [Google Scholar]
- 40.Vries, R. G., V. Bezrookove, L. M. Zuijderduijn, S. K. Kia, A. Houweling, I. Oruetxebarria, A. K. Raap, and C. P. Verrijzer. 2005. Cancer-associated mutations in chromatin remodeler hSNF5 promote chromosomal instability by compromising the mitotic checkpoint. Genes Dev. 19665-670. [DOI] [PMC free article] [PubMed] [Google Scholar]