Abstract
The NF-κB signaling pathway regulates the activity of multiple dimeric transcription factors that are generated from five distinct monomers. The availabilities of specific dimers are regulated during cell differentiation and organ development and determine the cell's responsiveness to inflammatory or developmental signals. An altered dimer distribution is a hallmark of many chronic diseases. Here, we reveal that the cellular processes that generate different NF-κB dimers are highly connected through multiple cross-regulatory mechanisms. First, we find that steady-state expression of RelB is regulated by the canonical pathway and constitutive RelA activity. Indeed, synthesis control of RelB is the major determinant of noncanonical NF-κB dimer activation. Second, processing, not synthesis, of p100 and p105 is mechanistically linked via competitive dimerization with a limited pool of RelA and RelB. This homeostatic cross-regulatory mechanism determines the availability of the p50- and p52-containing dimers and also of the noncanonical IκB p100. Our results inform a wiring diagram to delineate NF-κB dimer formation that emphasizes that inflammatory and developmental signaling cannot be considered separately but are highly interconnected.
The innate and adaptive immune systems function on different timescales via distinct effector cells and are regulated by different cell surface receptors. However, the development and maintenance of the lymph nodes and Peyer's patches, which are indispensable for the adaptive immunity and provide microenvironments for lymphocyte maturation, require both the innate inflammatory signals mediated by tumor necrosis factor receptor (TNFR) as well as the developmental signals mediated by the lymphotoxin-β receptor (LTβR) (11, 22, 29, 32, 37, 39).
The transcription factor NF-κB has been implicated in both innate and adaptive immune responses. Inflammatory and developmental signals are thought to use discrete canonical and noncanonical pathways for NF-κB activation (31). The primary mediator of canonical NF-κB activity is the heterodimer RelA-p50 that consists of the RelA transcriptional activator and the nfkb1 protein p50, which is generated by constitutive proteasome-mediated processing of the precursor p105 (25). Three IκB proteins (IκB-α, -β, and -ɛ) retain the RelA-p50 dimer in the cytosol in an inactive state. The canonical pathway involves stimulus-responsive phosphorylation of the IκBs by IKK2/ΙΚΚβ, which tags them for degradation and allows nuclear translocation of the RelA-p50 dimer (13). In contrast, the noncanonical pathway regulates nuclear translocation of RelB dimers via a mechanism that involves NF-κB-inducing kinase and IKK1/IKKα-dependent phosphorylation of the nfkb2 protein p100. Subsequent proteasome-mediated removal of the C-terminal inhibitory domain from p100 generates p52, which then complexes with RelB to appear as nuclear RelB-p52 DNA binding activity (3, 35, 39, 41).
Previous biochemical studies revealed that the signaling through LTβR activates the NF-κB/RelB dimer via the noncanonical NF-κB pathway (7, 39). Lymphoid chemokine genes such as blc (Cxcl13) and slc (Ccl21), whose expression is important for lymph node development (33), were proposed to be RelB target genes. Genetic analyses also indicated that RelB plays an essential role in lymph node development, especially in maintaining lymph node architecture (38, 42). Homozygous knock-in mice expressing a nonactivatable IKKα variant (ikkαAA/AA) show impaired lymphoid organogenesis (35) due to stromal cell defects (4). Moreover, a mutant mouse strain with an inactivating mutation in the nik gene (nikaly/aly) (36) as well as nik−/− mice showed a complete lack of lymph nodes (24, 43), as was also observed in ltbr−/− mice (24).
However, we observed that nfkb2−/− mice (deficient in the primary regulator of noncanonical NF-κB signaling p100/p52) display defects only in the inguinal lymph node formation, with all other nodes present at wild-type frequencies (20). Residual p50-containing NF-κB activity was thought to be responsible for such incomplete penetrance of lymph node phenotypes in nfkb2−/− mice. Indeed, nfkb1−/− nfkb2−/− double knockout mice displayed a complete lack of lymph nodes, thus phenocopying LTβR-deficient mice (20). Interestingly, we observed that nfkb1−/− mice have inguinal lymph node defects, and derived murine embryonic fibroblasts (MEFs) showed attenuated RelB dimer activation in response to LTβR stimulation (20, 39). Collectively, these reports suggest that p105/p50 and p100/p52 have distinct yet overlapping roles downstream of LTβR. However, the biochemical mechanisms that allow for their coordinated functioning has remained elusive.
Strikingly, rela−/− mice exhibited an early organogenic defect with a complete absence of lymph nodes in newborn mice (1). LTβR signaling was shown to activate both RelA and RelB dimers, and it was postulated that a first phase of RelA activity is required to induce p100 synthesis and subsequent RelB-p52 activity (7, 28). However, this hypothesis has not been examined experimentally. Moreover, it remains unclear if the observed lymph node phenotype in rela−/− mice is due to a defect in LTβR-responsive RelB activation or whether RelA has a direct role in the lymphoid chemokine gene expression.
Therefore, secondary lymph node development and LTβR signaling appear to rely upon the constituents of both canonical (inflammatory) and noncanonical (developmental) NF-κB pathways. Here, we describe biochemical analyses that utilize a panel of knockout mouse cells to explore the functional interconnectedness of these two pathways in mediating signaling downstream of LTβR. Our studies reveal the existence of multiple distinct cross-regulatory mechanisms that control the generation of multiple NF-κB dimers and thereby determine their availability for stimulus-responsive activation. Our analyses allow us to present a comprehensive mechanistic model to describe a single NF-κB signaling system that integrates inflammatory and developmental signals.
MATERIALS AND METHODS
Cell culture and reagents.
MEFs were obtained from embryonic day 12.5 to 13.5 embryos grown in Dulbecco's modified Eagle's medium (Mediatech Inc.) supplemented with 10% bovine calf serum and used for experiments up to passage 5 (“primary”) or following immortalization by the 3T3 protocol (14). All experiments with NF-κB knockout cells were done at least once with primary MEFs, except when they were reconstituted with retroviral transgenes. The ikkβ− MEF line was a kind gift from Inder M. Verma. An agonistic monoclonal LTβR antibody (AF.H6) was kindly provided by Jeff Browning (Biogen, Inc.). Antibodies used for immunoprecipitation or immunoblotting and supershift analyses were obtained from Santa Cruz Biotechnology or were a kind gift from Mimi Ernst and Nancy Rice.
Gene expression analysis.
Growing primary or immortalized MEFs were stimulated with 0.3 to 0.5 μg/ml agonist antibodies or with 10 ng/ml of TNF. Immunoblotting was performed as described previously (2). In certain instances, band intensities were quantitated using ImageQuant software (version 5.2; GE Healthcare) and normalized to tubulin after background subtraction. Quantitative PCR (Q-PCR) analysis was performed as described earlier (20) using previously published primers (7). RNase protection assay (RPA) analysis was done accordingly (14) using radiolabeled probes specific for RelB and NF-κB2 mRNA. Band intensities were quantitated and normalized to L32 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA.
EMSA.
Electrophoretic mobility shift assays (EMSAs) were done by incubating nuclear extracts with radiolabeled DNA probe containing a κB site that binds to both RelA and RelB dimers (20). For supershift analysis, extracts were incubated with the indicated antibodies (see the figures) for 30 min prior to probe addition.
Retrovirus-mediated gene transduction.
Retroviral constructs (pBabe.puro or its derivatives) were cotransfected with pCL.Eco into 293T cells using the calcium-phosphate method. At ∼40 h posttransfection the supernatant was filtered through a 0.45-μm-pore-size cellulose acetate filter and used to infect MEFs. Transduced cells were selected with puromycin hydrochloride. Retroviral constructs expressing wild-type IKKβ (IKKβ-wt), IKKβ-AA (an activation-defective mutant), and IKKβ-EE (a constitutively active mutant) were kindly provided by D. Shultz and M. Karin. RelB- and p100-expressing retroviral constructs have been described elsewhere (2). RelA.pBabe.puro was constructed by cloning the murine RelA gene into the EcoR1 and Sal1 sites of pBabe.puro.
RESULTS
NF-κB/RelA and RelB dimers in LTβR-induced expression of lymphoid chemokine genes.
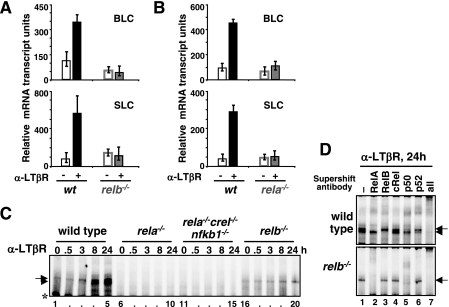
Previous studies have shown that the expression of organogenic lymphoid chemokines is required for lymph node development (33). It was proposed that the RelB-p52 dimer activated upon LTβR engagement mediates expression of the lymphoid chemokines (4). However, rela−/− mice (rescued with the combined deletion of the tnfr1 gene) also exhibited defects in lymph node development due to the requirement of RelA in the nonhematopoietic stromal cells (1). To genetically dissect the signaling pathways downstream of LTβR, we utilized an agonist LTβR antibody that induces NF-κB DNA binding activity in a cell culture system (2, 32). Total RNA was isolated from LTβR-stimulated MEFs and subjected to Q-PCR analysis to measure gene expression. Both blc and slc genes were activated in response to LTβR stimulation in wild-type MEFs, but induction of both genes was defective in relb−/− MEFs (Fig. 1A). These data provide genetic evidence that the previously reported RelB recruitment to a variant κB site in these promoters (4) plays a critical role for the expression of lymphoid chemokine genes. However, stimulation of rela−/− MEFs with LTβR agonist revealed similar defects in the activation of lymphoid chemokines (Fig. 1B). Therefore, our genetic analyses suggest not only that RelA may be required for the expression of RelA target genes that are induced upon LTβR stimulation, such as those encoding VCAM-1 (7, 33) but also that it appears to be important for the expression of lymphoid chemokine genes that have been classified as RelB targets (4).
FIG. 1.
Requirement of RelA for noncanonical NF-κB signaling induced by LTβR. (A and B) BLC and SLC mRNA levels in resting cells (open bars) and in response to LTβR stimulation as measured by real-time Q-PCR) in early-passage fibroblasts of the indicated genotypes. mRNA levels relative to the GAPDH control are plotted. (C) NF-κB DNA binding activity in MEF lines of the indicated genotypes during LTβR signaling. Specific κB-DNA binding complexes are indicated by arrows (RelA dimers) and arrowheads (RelB dimers). The asterisk denotes a nonspecific DNA-protein complex. (D) The composition of NF-κB DNA binding activity induced upon LTβR stimulation in wild-type or relb−/− MEFs was examined by supershift analysis using the indicated antibodies. At 24 h poststimulation RelA-p50 and RelB-p52 dimers appear as the major NF-κB DNA binding activity in the nucleus, with accompanying RelA-p52 and RelB-p50 dimers as minor DNA binding activity. α, anti.
To understand the role of RelA in the expression of RelB target genes, we examined NF-κB DNA binding activity in wild-type and rela−/− MEFs in response to LTβR stimulation by EMSA. In wild-type MEFs, LTβR stimulation activated both RelA and RelB dimers with sustained temporal profiles (Fig. 1C, lanes 1 to 5, and D, top panel) (2, 42). The majority of the induced RelA DNA binding activity was composed of RelA-p50 dimer with only a minor amount of RelA-p52 activity (Fig. 1D). In contrast, the majority of the induced RelB DNA binding activity in MEFs was composed of RelB-p52 dimer, with a smaller fraction of RelB-p50 dimer (Fig. 1D). RelA dimer activation was found to be intact in relb−/− MEFs, thus implying a RelB-independent activation mechanism for RelA downstream of LTβR (Fig. 1C, lanes 16 to 20, and D, bottom panel). However, activation of the RelB dimer was much reduced in rela−/− MEFs even at late time points (Fig. 1C, lanes 6 to 10). Similarly, rela−/− c-rel−/− nfkb1−/− MEFs, which are genetically devoid of all three canonical NF-κB dimer-forming subunits, RelA, c-Rel, and p50, did not show detectable noncanonical RelB-p52 activity upon LTβR stimulation (lanes 11 to 15).
Our results in a rela−/− MEF cell line contrast with those previously published (9, 42), which showed RelB activation in an immortalized rela−/− MEF cell line that has a partially transformed phenotype (12). In a comparative study, we utilized early passage (or primary) rela−/− and rela−/− c-rel−/− nfkb1−/− MEFs and two independently immortalized rela−/− MEF cell lines. In contrast to the previously used rela−/− cell line, attenuated activation of RelB dimers was observed upon LTβR stimulation in this rela−/− cell line or in early-passage rela−/− and rela−/− c-rel−/− nfkb1−/− MEFs (data not shown).
RelA is required for noncanonical NF-κB activation by controlling homeostatic RelB expression.
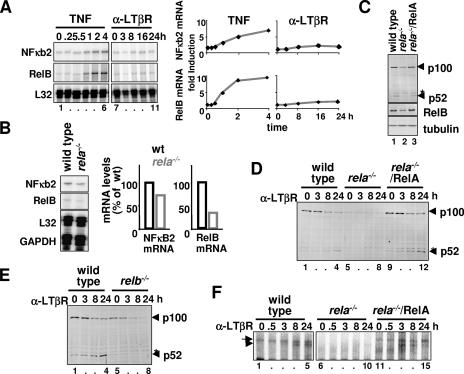
The genes encoding RelB and p100/p52 are known NF-κB target genes (6, 10, 19). Stimuli that activate the canonical NF-κB/RelA dimer were shown to induce p100 protein synthesis (2, 7, 26, 42). On that basis, it was suggested that LTβR-mediated sequential activation of RelA-p50 and RelB-p52 dimers is potentiated by RelA-induced transcription of the nfkb2 gene (7). Inducibly synthesized p100 was proposed to be processed into p52, allowing for RelB-p52 dimer activation (26, 34). To test the proposed model, we quantitatively examined transcriptional activation of relb and nfkb2 genes in wild-type MEFs upon LTβR stimulation. Using an RPA, we found robust induction of both RelB and NF-κB2 mRNA in response to TNF stimulation but could detect only a small increase in the level of those mRNAs upon LTβR stimulation (Fig. 2A). This observation does not sufficiently explain the quantitative dependence of RelB dimer activation on RelA (Fig. 1C).
FIG. 2.
RelA is required for LTβR signaling and controls steady-state RelB expression. (A) RPA to examine NF-κB2 and RelB mRNA levels in wild-type MEFs stimulated with TNF or LTβR agonist (left panel). Relative induction levels of NF-κB2 and RelB mRNA in response to TNFR or LTβR stimulation are plotted after normalizing to L32 mRNA (right panel). (B) Steady-state level of NF-κB2 and RelB mRNA in wild-type or rela−/− MEFs was examined by RPA (left panel). Respective mRNA levels were quantified and normalized to L32 and GAPDH mRNA and plotted (right panel). (C) Immunoblotting for p100 and RelB in wild-type, rela−/−, or rela−/− MEFs reconstituted with a retroviral RelA transgene. Presence of a nonspecific band is denoted with an asterisk. (D and E) Immunoblotting to examine p100 processing into p52 during LTβR signaling in MEFs of the indicated genotypes. (F) NF-κB-DNA binding activities induced upon LTβR stimulation in rela−/− MEFs that express the RelA transgene were analyzed by EMSA. α, anti.
Next, we asked if RelA is required for the constitutive synthesis of NF-κB2 or RelB mRNA in resting cells. Comparison of the relative abundance revealed that the NF-κB2 mRNA level was reduced to 70% and that the RelB mRNA level was reduced to 30% in rela−/− MEFs (Fig. 2B). Immunoblotting of wild-type and rela−/− MEF extracts showed a proportional reduction of RelB protein but more significantly reduced levels of p100 (Fig. 2C). Consistent with the observed defect in RelB-p52 dimer activation (Fig. 1C), LTβR-responsive generation of p52 was also significantly reduced in rela−/− cells (Fig. 2D, lanes 5 to 8). Interestingly, in relb−/− MEFs, p100 was completely degraded upon LTβR engagement (Fig. 2E, lanes 5 to 8), indicating that RelB may stabilize and protect de novo generated p52 from proteolysis. Thus, limited availability of RelB in rela−/− MEFs may in part be responsible for the observed defect in p52 generation upon receptor stimulation. As discussed later (see Fig. 6F), the availability of RelB is limited not only because its reduced synthesis (Fig. 2B) but also because RelB protein is sequestered in dimers with p50 made available by the loss of RelA protein in these cells. Consistent with this hypothesis, retroviral transduction of a RelA transgene into rela−/− MEFs not only rescued the steady-state level of p100 and RelB proteins (Fig. 2C) but also restored p52 production upon LTβR stimulation (Fig. 2D, lanes 9 to 12) and the activation of both RelA and RelB DNA binding dimers (Fig. 2F, lanes 11 to 15). Collectively, our analyses indicate that the relb gene and, to a lesser extent, the nfkb2/p100 gene are transcriptionally regulated by RelA in resting cells; this constitutive, or homeostatic, level of transcription of RelB mRNA is important for p52 generation and the activation of the RelB-p52 dimer in response to noncanonical stimuli such as those transduced through LTβR.
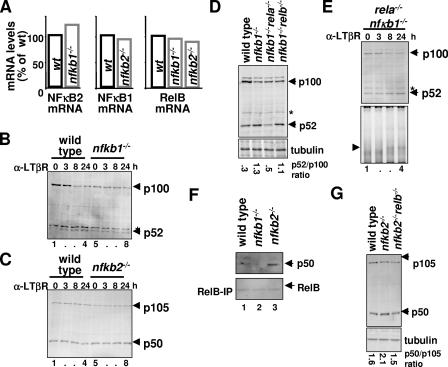
FIG. 6.
Cross-regulation between p105/p50 and p100/p52 at the level of protein processing. (A) Steady-state levels of RelB, NF-κB2, and NF-κB1 mRNAs were measured in nfkb1−/− and nfkb2−/− primary MEFs by RPA and compared with wild-type MEFs in a bar diagram. (B) Immunoblotting to reveal constitutive and LTβR-stimulated processing of p100 to p52 in wild-type and nfkb1−/− primary MEFs. (C) The levels of p105 and p50 proteins in resting and LTβR-stimulated wild-type and nfkb2−/− MEFs were examined by immunoblot analysis. (D) Homeostatic level of p52 and p100 in primary MEFs of the indicated genotypes were examined by immunoblot analysis. Band intensities were quantified using ImageQuant software, normalized to tubulin, and expressed as a ratio of processed product to precursor. (E) Immunoblot (top) and EMSA (bottom) analysis to reveal p52 accumulation and RelB-p52 dimer activation, respectively, in nfkb1−/− rela−/− MEFs in response to LTβR stimulation. (F) The levels of p50 (top) associated with RelB (bottom) in wild-type and nfkb2−/− MEFs were examined by immunoblot analysis of the RelB immunoprecipitate obtained from the indicated cell extracts. (G) Homeostatic level of p50 and p105 in primary MEFs of the indicated genotypes were examined by immunoblot analysis, and the ratios of processed product to precursor were quantified. α, anti.
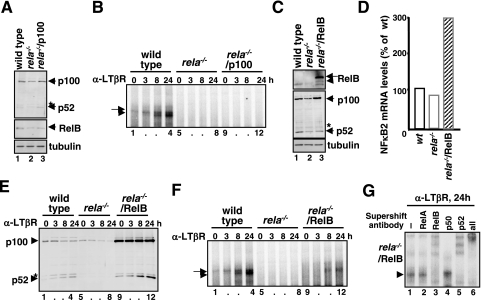
To test whether constitutive transcription of p100 or RelB may be sufficient, we first expressed p100 from an ectopic retroviral promoter (27) in rela−/− MEFs. Although such constitutive expression rescued the p100 protein level in these cells (Fig. 3A), it did not restore RelB dimer activation (Fig. 3B, lanes 9 to 12). Similarly, we expressed RelB in rela−/− MEFs and found an elevated level of p100 protein (Fig. 3C) due to transcriptional upregulation of the nfkb2 gene by constitutive RelB activity (Fig. 3D) in the nucleus and RelB-dependent stabilization of p100. LTβR stimulation of these cells resulted in the gradual appearance of p52 (Fig. 3E, lanes 9 to 12), which complexes with the constitutively expressed (transgenic) RelB to appear as RelB-p52 DNA binding activity (Fig. 3F, lanes 9 to 12, and G). However, rescue of RelB-p52 DNA binding activity was only partial in the rela−/−/RelB cell line as overexpression of the RelB transgene led to elevated synthesis of p100 protein (Fig. 3C) that sequestered RelB even in the stimulated cells (data not shown). Despite the incomplete functional reconstitution, our analysis indicated that the control of RelB synthesis, but not of p100, is a primary determinant for RelB-p52 dimer activation via noncanonical signaling. Interestingly, we noticed an increased RelB level (data not shown) in the previously utilized rela−/− (1) MEF line (9, 42), which provides a plausible explanation of why noncanonical NF-κB activity is intact in these cells.
FIG. 3.
Constitutive expression of RelB is sufficient to restore LTβR-induced RelB activation in rela−/− MEFs. (A and C) Immunoblotting to reveal p100 (A) or RelB (C) expression from transgene in rela−/− MEFs. (B and F) EMSA to examine NF-κB DNA binding activity in response to LTβR stimulation in rela−/− MEFs that expresses p100 (B) or RelB (F) from transgene. (D) Steady-state level of NF-κB2 mRNA in rela−/− MEFs in the absence or presence of the retroviral RelB transgene was analyzed by RPA. (E) Immunoblotting to examine processing of p100 and generation of p52 upon LTβR stimulation in rela−/− MEFs in the absence or presence of the retroviral RelB transgene. (G) Supershift analysis to examine the composition NF-κB DNA binding activity induced upon LTβR stimulation in rela−/− MEFs that expresses RelB transgene. α, anti.
A regulatory role of IKKβ on RelB dimer activation.
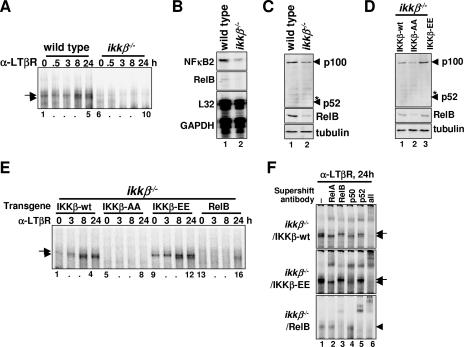
LTβR signaling involves IKKα-mediated phosphorylation and subsequent proteasomal processing of p100 into p52 to allow for NF-κB activation (9, 28, 42). In contrast, p100 processing upon LTβR engagement was shown to be qualitatively intact in ikkβ−/− MEFs (7, 28), leading to the notion of two separate NF-κB signaling pathways, the canonical IKKβ-mediated and the noncanonical IKKα-mediated pathways (31). However, our EMSA analyses with ikkβ−/− MEFs indicated a complete absence of NF-κB/RelB activation in response to LTβR stimulation (Fig. 4A, lanes 6 to 10).
FIG. 4.
Constitutive IKKβ activity determines LTβR-induced activation of RelB/NF-κB. (A) NF-κB DNA binding activities induced upon LTβR stimulation in MEFs of the indicated genotypes were analyzed by EMSA. (B) Steady-state levels of NF-κB2 and RelB mRNA in wild-type and ikkβ−/− MEFs was examined by RPA. (C and D) Immunoblotting to reveal the steady-state level of p100 and RelB protein in MEFs of the indicated genotypes. (E) NF-κB DNA binding activity induced by LTβR stimulation in ikkβ−/− MEFs expressing transgenic wild-type (lanes 1 to 4), inactive (lanes 5 to 8), or constitutively active forms (lanes 9 to 12) of IKKβ or ectopic RelB protein (lanes 13 to 16). (F) Supershift analysis to examine the composition of NF-κB DNA binding activity induced upon LTβR stimulation in the ikkβ−/− MEFs reconstituted with the indicated retroviral transgene. α, anti.
Previously, we have reported that the three canonical IκBs, IκB-α, -β, and -ɛ, are important for regulating the constitutive DNA binding activity of RelA-p50 dimer in the nucleus (2). Steady-state activity of IKKβ was shown to control IκB homeostasis and thus constitutive RelA-p50 activity in resting cells (30). Given that ikkβ−/− MEFs lack constitutive NF-κB activity (Fig. 4A), we asked if the steady-state levels of the constituents of the noncanonical pathway are reduced in these cells. Our analysis revealed that the NF-κB2/p100 mRNA and protein levels were reduced in ikkβ−/− MEFs, while RelB mRNA and protein were almost undetectable (Fig. 4B and C).
By mutating the serines present in the activation loop of IKKβ (S177 and S181), the activation potential of the kinase can be modulated (8, 23). Retroviral reconstitution of ikkβ−/− MEFs with the IKKβ-wt transgene, but not the activation-defective mutant IKKβ-AA, restored the constitutive level of RelB and p100 protein (Fig. 4D, lanes 1 and 2). Moreover, reconstitution of ikkβ−/− cells with the constitutively active mutant IKKβ-EE resulted in an increase in RelB and p100 levels above wild type (Fig. 4D). Thus, our analyses indicated that the activation state of IKKβ in resting cells determines the homeostatic levels of cellular RelB and p100 proteins. To explore the functional consequences of IKKβ-dependent constitutive synthesis, we examined RelB dimer activation in these reconstituted cells. IKKβ-AA-reconstituted MEFs, similar to the parental ikkβ−/− cells, lack constitutive RelA-p50 DNA binding and showed no RelB activation upon LTβR stimulation (Fig. 4E, lanes 5 to 8), while expression of IKKβ-wt rescued both constitutive RelA-p50 activity and LTβR-responsive RelB activation (Fig. 4E, lanes 1 to 4, and F, top panel). Strikingly, expression of the IKKβ-EE transgene in ikkβ−/− MEFs not only resulted in increased constitutive RelA-p50 DNA binding (Fig. 4E, lane 9) but also rescued RelB dimer activation in response to LTβR engagement (Fig. 4E, lanes 9 to 12, and F, middle panel). Further, transgenic RelB expression was sufficient to restore RelB-p52 dimer activation defects in ikkβ−/− MEFs (Fig. 4E, lanes 13 to 16, and F, bottom panel) as shown for rela−/− MEFs (Fig. 3).
In sum, we have revealed that the constitutive nuclear RelA-p50 activity plays a critical role for LTβR-mediated activation of the noncanonical RelB-p52 dimer by ensuring RelB expression. Furthermore, constitutive IKKβ activity is required for RelB expression, although IKKβ does not participate in noncanonical signaling per se, such as p100 processing and RelB nuclear localization. By considering the functional mechanisms that control cellular homeostasis, we have identified cross talk within the resting cells, even prior to stimulation, between the canonical and the noncanonical NF-κB signaling pathways that determines the cellular responsiveness to developmental stimuli.
Genetic redundancy between nfkb1 and nfkb2 in lymph node development involves RelB.
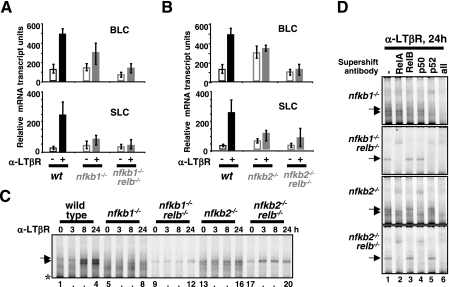
Previously, we have reported that nfkb2−/− mice show only partial defects in lymph node development and also that nfkb1−/− mice lack inguinal lymph nodes (20). A complete defect, similar to that seen in the ltβr−/− mice (32), was observed upon combined deletion of both the nfkb1 and nfkb2 genes (20). These results indicated that there is genetic redundancy between p50 and p52 in lymph node development and that both proteins may be coordinately required. To address the underlying molecular mechanism, we examined the expression of lymphoid chemokines, blc and slc, in a variety of knockout cells. We found that nfkb1−/− MEFs are partially defective in the expression of these RelB target genes in response to LTβR stimulation (Fig. 5A). However, nfkb1−/− relb−/− MEFs showed a complete lack of blc or slc gene expression upon LTβR stimulation, confirming that the remaining lymphoid chemokine expression was RelB dependent. In nfkb2−/− MEFs, we found not only that LTβR-induced gene activation was abrogated, confirming that nfkb2/p100 is the primary regulator of the noncanonical pathway, but also that the constitutive level of lymphoid chemokine mRNA was elevated (Fig. 5B). By using nfkb2−/− relb−/− MEFs that are deficient in both p100 and RelB, we found that constitutive levels of BLC and SLC mRNA were restored to wild-type levels. Thus, we were able to determine that the elevated basal expression of these chemokines in nfkb2−/− MEFs was in fact due to RelB. These results suggest that misregulation of RelB is responsible for masking the defective lymph node development phenotype in nfkb2−/− mice.
FIG. 5.
Requirement of both nfkb1 and nfkb2 gene products for induction of RelB target genes and NF-κB/RelB DNA binding activity in response to LTβR signaling. (A and B) Expression of BLC and SLC genes upon LTβR stimulation in primary MEFs of the indicated genotypes. (C) NF-κB DNA binding activities induced by LTβR stimulation in primary MEFs of the indicated genotypes were examined by EMSA. The presence of RelB in single nfkb1−/− and nfkb2−/− knockout cells perturbs NF-κB RelA activation. (D) Supershift analysis to reveal the composition of NF-κB DNA binding activity induced upon LTβR stimulation in the MEFs of the indicated genotypes. α, anti.
To address the biochemical basis for these gene expression phenotypes, we measured LTβR-induced κB-DNA binding activity in these knockout cells. In nfkb1−/− MEFs, p52-containing NF-κB dimers were only weakly activated during signaling at late time points (Fig. 5C, lanes 5 to 8, and D), whereas nfkb1−/− relb−/− double knockout MEFs revealed some RelA-p52 activation but no RelB activity (Fig. 5C, lanes 9 to 12, and D). In contrast, we observed constitutive RelB-p50 activity in nfkb2−/− MEFs (Fig. 5C, lanes 13 to 16, and D), which was ablated in nfkb2−/− relb−/− doubly deficient cells (Fig. 5C, lanes 17 to 20, and D). These results suggest that the four gene products of nfkb1 and nfkb2 (p105/p50 and p100/p52, respectively) function in concert (i) to control the availability of RelB-containing dimers in the cell, (ii) to ensure their inhibition in resting cells, and thus (iii) to mediate their stimulus-responsive activation.
Cross-regulation via processing of p105/p50 and p100/p52 proteins.
Next, we examined the potential mechanisms underlying deregulated RelB activity in single knockout cells. To this end, we first investigated the possibility that relb and nfkb2 or nfkb1 transcription may be perturbed in nfkb1−/− or in nfkb2−/− MEFs, respectively. Instead, our quantitative analysis indicated that the steady-state levels of RelB and NF-κB2 mRNA in nfkb1−/− MEFs or of RelB and NF-κB1 mRNA in nfkb2−/− MEFs are similar to the levels of wild-type cells (Fig. 6A). However, immunoblot analysis of nfkb1−/− cell extracts indicated a lower level of p100 protein due to elevated constitutive processing of p100 into p52; LTβR signaling did not further increase the level of p52 in these mutant cells (Fig. 6B, lanes 1 to 8). Similarly, we observed an increase in the steady-state level of p50 in nfkb2−/− MEFs compared to the wild-type cells, although p50 generation was not responsive to LTβR signaling (Fig. 6C, lanes 1 to 8).
Based on our analyses, we propose that the absence of the major RelA-interacting partner p50 leads to the elevated level of RelA-p52 complexes in nfkb1−/− MEFs, thereby promoting steady-state p100 processing. Cellular IκB proteins sequester the RelA-p52 dimer in the cytosol and release bound NF-κB dimer as nuclear activity upon TNF stimulation of nfkb1−/− MEFs (14). However, elevated constitutive processing depletes the cellular pool of p100 complexes that are available for LTβR-responsive processing, thereby attenuating activation of p52-containing dimer during LTβR stimulation (Fig. 5C).
To address the hypothesis that the availability of RelA and RelB regulates the degree of constitutive processing of p105 and p100, we examined the ratio of precursor to mature processing product in double knockout cells. Indeed, we found that the p52-to-p100 ratio, which was increased in nfkb1−/− MEFs, could be restored to the wild-type level in nfkb1−/− rela−/− MEFs but not in nfkb1−/− relb−/− MEFs (Fig. 6D). These results confirm that the availability of RelA in the absence of p50 potentiates constitutive p100 processing that generates p52.
Furthermore, p100 restored in nfkb1−/− rela−/− MEFs was still subject to inducible processing and generated p52 upon LTβR signaling (Fig. 6E, top panel). Interestingly, these cells also rescued the defect in LTβR-responsive p52 generation in rela−/− MEFs (Fig. 2D). We suggest that p50 made available by the absence of RelA (in rela−/− MEFs) sequesters RelB and thereby interferes with RelB-dependent stabilization of p52 during LTβR signaling. Once such p50 competition was relieved in nfkb1−/− rela−/− MEFs, RelB-p52 DNA binding activity was again induced by LTβR stimulation (Fig. 6E, bottom panel). Due to the reduced RelB mRNA synthesis in the absence of RelA, however, the observed DNA binding activity was weaker than in wild-type cells.
In contrast, the absence of the primary RelB-interacting partner p52/p100 in nfkb2−/− MEFs resulted in increased RelB-p50 association (Fig. 6F), and this finding accounts for the increased level of p50 in these cells. Indeed, the nfkb2 and relb compound deficiency restored the p50/p105 ratio to that observed for wild-type MEFs (Fig. 6G). In sum, by utilizing a panel of knockout cells devoid of various NF-κB proteins, we reveal cross-regulation between p105/p50 and p100/p52 at the level of precursor processing that is based on competition for the dimerization partners, RelA and RelB.
DISCUSSION
The molecular mechanisms by which NF-κB is activated has been studied in some detail. In the present study we examined the mechanisms that regulate the formation of several important NF-κB dimers via the expression of monomer constituents, their proteolytic processing to mature subunits, and dimerization. Our results allow us to chart the NF-κB dimer generation pathways in a wiring diagram that integrates the previously described activation mechanisms, as discussed below.
The single NF-κB dimer signaling module.
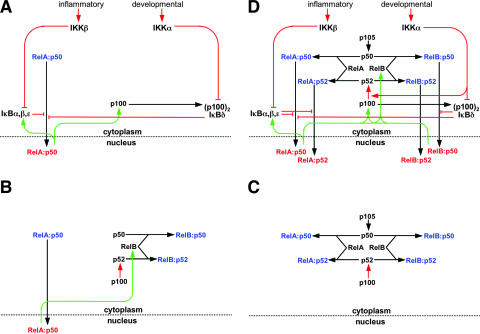
During inflammatory signaling the RelA-p50 heterodimer acts as the primary mediator of NF-κB activity in MEFs. Three canonical IκBs, IκBα, -β and -ɛ, were shown to regulate the RelA-p50 dimer activation in the inflammatory pathway (13, 16). A detailed understanding of the biochemical events has permitted the construction of a mathematical model that recapitulates the experimentally observed NF-κB activation dynamics in inflammatory settings (15, 17, 40). However, this model, which comprises three IκBs, proved inadequate to describe RelA-p50 activation during developmental signaling. Subsequent biochemical analyses revealed the asymmetric (p100)2 homodimer IκBδ as a fourth IκB molecule, which we included in the mathematical model that then recapitulated RelA-p50 activation in response to both inflammatory and developmental signals (2). The functional connectivity between these components is presented in the form of a schematic wiring diagram in Fig. 7A. In this diagram, canonical IκBs and IκBδ/(p100)2 inhibit RelA-p50 dimer nuclear translocation. Inducible processes (red lines), such as IKKβ-mediated IκB degradation or IKKα-mediated (p100)2 degradation, relieve this inhibition. Constitutive processes (black lines), such as RelA-p50 dimer assembly from the monomer subunits or RelA-p50 dimer nuclear import, also regulate NF-κB DNA binding activity. Inducible resynthesis of the four IκBs by NF-κB-dependent transcription (green lines) mediates feedback within this module.
FIG. 7.
Wiring diagrams to depict functional relationships within the NF-κB signaling system. NF-κB monomers are shown in black, cytoplasmic dimers are in blue, and nuclear dimers are in red. Constitutive processes are shown in black, regulated processes are in red, and feedback processes are in green. (A) A wiring diagram of the regulation of the NF-κB/RelA-p50 dimer in response to inflammatory or developmental stimuli. A mathematical model of these functional connections has been constructed and shown to recapitulate signaling cross talk between these pathways in the control of RelA-p50 activity (2). (B) A wiring diagram to summarize our findings (Fig. 2 and 3) that steady-state RelB synthesis and the availability of latent RelB dimers are dependent on constitutive RelA-p50 activity. In Fig. 4, we also showed that constitutive RelA-p50 activity is controlled by constitutive IKKβ activity. (C) A wiring diagram to describe the molecular competition between p50 and p52 for their dimerization partners RelA and RelB. This competition determines the rate of p50 and p52 generation via processing from their p105 and p100 precursors. Thus, p105/p50 and p100/p52 processing is interdependent through competition for common interaction partners. (D) A wiring diagram of the NF-κB signaling system that accounts for the generation of four NF-κB dimers that are detected in response to LTβR signaling, namely, RelA-p50, RelA-p52, RelB-p50, and RelB-p52. This diagram is based on previously established connectivity (A) and incorporates the insights from the current study summarized in panels B and C. The NF-κB signaling system receives signals from both inflammatory and developmental cues through IKK2/IKKβ and IKK1/IKKα kinases, respectively, to activate distinct NF-κB dimers. For the sake of clarity, we have depicted only signal-responsive protein complexes, omitting dimers such as p50-p50, p105-p50, p105-RelA, p100-p52, or p100-RelB that do not change in abundance during LTβR induced signaling.
A multi-NF-κB dimer signaling system.
LTβR signaling activates multiple NF-κB dimers (3, 5, 20, 39) that may have distinct functions in gene expression (4, 14). However, a comprehensive description of their coordinated activation is lacking. The prevalence of various NF-κB dimers in resting and in activated cells is modulated by the synthesis, degradation, and processing mechanisms that generate constituent monomers as well as by association and dissociation rates of the monomer subunits to generate dimers. Here, we present wiring diagrams to recapitulate both generation of these NF-κB dimers and their activation during signaling. For the sake of clarity, we have depicted only signal-responsive protein and protein complexes, omitting pools of dimers, such as p50-p50, p105-p50, p105-RelA, p100-p52, or p100-RelB, or c-Rel-containing dimers that do not appear to change in abundance during LTβR signaling in fibroblast cells.
As depicted in Fig. 7B, RelB heterodimerizes with either p50 or p52. The level of RelB synthesis is determined by constitutive activity of the canonical pathway via RelA-dependent transcription of relb (Fig. 2 and 3). This cross-regulation mechanism controls the cellular abundance of the RelB monomer subunit and thus latent RelB dimer (Fig. 7B). Our data showed that homeostatic control of RelB expression is more important to noncanonical dimer activation than the previously proposed RelA-inducible expression of p100 (7) during LTβR signaling.
Similarly, RelA monomer is capable of dimerizing with p50 or p52. Indeed, p50 and p52 compete for binding to RelA and RelB (Fig. 7C). Constitutive p105 processing generates p50 in the resting cell. In contrast, p100 processing to generate p52 is inhibited by RelB but is responsive to developmental stimuli such as LT. Thus, the competition between p50 and p52 for binding partners RelA and RelB regulates the ratio of p100/p52 and therefore the availability of the noncanonical regulator p100 (Fig. 5 and 6). Alterations in the expression of RelA and p105/p50 will therefore (but unintuitively) affect the strength of the noncanonical signaling, potentially providing signaling cross talk between inflammatory and developmental signaling pathways.
Finally, we combined these proposed homeostatic cross-regulatory mechanisms (Fig. 7B and C) with the canonical and noncanonical signaling inputs, mediated typically (but not exclusively) by IKKβ and IKKα, respectively (Fig. 7A), to construct a wiring diagram to describe an NF-κB signaling system that accounts for the generation and activation of four distinct dimers (Fig. 7D). In this diagram, inflammatory signals activate the RelA-p50 dimer through IKKβ-mediated IκB degradation, while IκBδ/(p100)2-inhibited RelA-p50 and RelB-p50 complexes are released into the nucleus upon IKKα-mediated developmental signaling. Furthermore, p52 is cotranslationally generated from p100 in response to IKKα signaling and dimerizes with RelB and RelA to appear as RelB-p52 and RelA-p52 DNA binding complexes. The RelA-p50 dimer controls RelB synthesis and thus modulates stimulus-responsive activation of the RelB dimers via the noncanonical pathway. On the other hand, feedback inhibition mediated by the resynthesized IκBs is capable of terminating RelA responses (15). In addition, we found that RelB-containing dimers may transcriptionally upregulate nfkb2 and relb, forming a positive feedback loop that due to its potential cancer relevance deserves further study.
Perturbations in the NF-κB signaling system in gene knockouts.
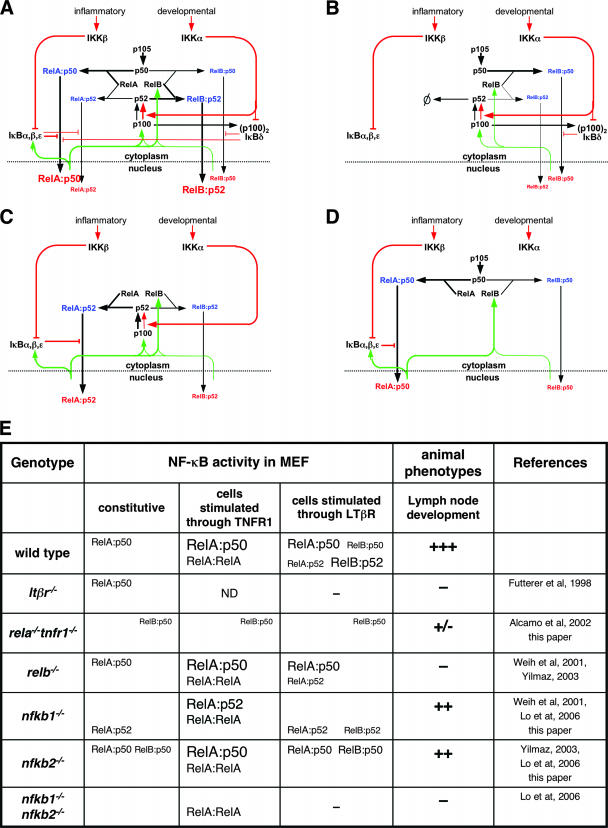
We next examined whether our newly constructed wiring diagram helps explain misregulated dimer activation in various NF-κB knockouts (Fig. 8E). The wiring diagram in Fig. 7D qualitatively depicted functional connectivities but did not reflect the relative strengths of biochemical reactions. For example, p50's preferred binding partner is RelA, whereas p52-RelB dimers are more abundant than p52-RelA dimers. To appropriately reflect such reaction preferences, we have indicated relative reaction preferences by the line width of the arrows in a modified wiring diagram (Fig. 8A). This wiring diagram serves as a starting point for examining the effects of gene knockouts of NF-κB family members.
FIG. 8.
(A) A modified version of the wiring diagram that now indicates the preferred biochemical reactions with bold lines and lesser reactions with thin lines to more quantitatively mirror the NF-κB signaling system in wild-type cells that responds to both inflammatory and developmental signaling. (B to D) Wiring diagrams that describe mutant NF-κB signaling systems in rela−/−, nfkb1−/−, or nfkb2−/− cells, respectively. (E) A comprehensive description of the NF-κB dimers that are activated during signaling through TNFR1 or LTβR in MEFs of the indicated genotypes. Relative DNA binding activities of a given NF-κB dimer under various conditions are represented with different font sizes. ND, not determined. Observed lymph node phenotypes of various NF-κB gene knockout mice are shown: +++, normal development; −, severe defect in lymph node development; ++ and +/−, intermediate phenotypes.
The lack of RelA activity not only resulted in the complete absence of the NF-κB activity during inflammatory signaling but also abrogated the noncanonical pathway (Fig. 8E). Two distinct mechanisms are responsible for such defective RelB activation (Fig. 8B). First, RelA deficiency reduces RelB mRNA synthesis. Second, excess p50 (resulting from the lack of RelA) competes with p52 for RelB binding and thus interferes with p52 generation and RelB-p52 dimer formation. Therefore, it is possible that the observed phenotype in rela−/− tnfr1−/− mice (1) could be in part due to the attenuated RelB activity during lymph node development (Fig. 8E).
It has been suggested that RelB binding to p100 inhibits its stimulus-responsive processing G. Ghosh, personal communication. We propose here that RelA, in contrast, allows constitutive p100 processing, which results in elevated processing in nfkb1−/− MEFs (Fig. 6) and functional compensation at the level of gene expression during TNF signaling in nfkb1−/− MEFs (14). However, elevated p100 processing also depletes the cellular pool of p100 that can be inducibly processed into p52 during noncanonical signaling (Fig. 8C). As such, functional compensation in inflammatory signaling impairs the system's responsiveness to developmental signals. Therefore, our system wiring diagram illustrates an explanation for the observed NF-κB activation defect in nfkb1−/− mice that may impair lymph node development (Fig. 8E) (20). Similarly, the wiring diagram (Fig. 8D) illustrates that in nfkb2−/− MEFs, all of the RelB protein is available to bind p50, thereby, depleting the pool of constitutive p105. As RelB-p50 is not subject to inhibition by canonical IκB proteins, it appears as a constitutive DNA binding activity in the nucleus. We postulate that the transcriptional activity of the constitutive RelB-p50 dimer partly masks the phenotype that is otherwise expected in nfkb2−/− mice (Fig. 8E) (20).
Therefore, genetic and biochemical analyses indicated that lymph node development and its maintenance require both components of the canonical (rela and nfkb1) and the noncanonical (relb and nfkb2) pathways. Understanding LTβR signaling requires integration of both pathways into a single NF-κB signaling system. The system wiring diagram describes mechanisms that function in the resting cell, prior to stimulation, to control the homeostasis of various NF-κB dimers. Regulation of the homeostatic state emerged as an important factor underlying dimer activation during LTβR signaling. Hence, our results emphasized not only the functional interdependence of the canonical and the noncanonical pathways but also the functional interconnectedness of the homeostatic mechanisms that are responsible for NF-κB dimer generation and the stimulus-induced, dynamic mechanisms that control dimer activation (Fig. 8A).
Such interdependencies and functional interconnectedness are not limited to the NF-κB/IκB proteins but also exist at the level of the kinases that regulate them. The noncanonical signal transducer IKKα was shown to be involved in terminating canonical NF-κB/RelA activity induced in macrophages upon lipopolysaccharide stimulation (18). Furthermore, the absence of IKKβ in hepatocytes was compensated by other kinase(s), presumably by IKKα, to allow NF-κB activation in response to TNF (21). Our understanding of the cross-regulatory mechanisms between IKK subunits is currently limited by an incomplete biochemical characterization of the multiple kinase complexes that they form.
In sum, our analysis of the NF-κB system reveals interesting systems properties such as complexity (four dimers, nine proteins, and two kinases), robustness (partial penetrance of the phenotype in nfkb2−/− mice), sensitivity (unexpected phenotype in nfkb1−/− mice), and cross-regulation (RelB activation defects in rela−/− mice). The NF-κB signaling system consists of 15 different potential homo- or heterodimers comprising five homologous proteins, RelA, c-Rel, RelB, p50, and p52 (16). Our current system wiring diagram describes four of them that are activated during LTβR signaling, but it does not convey the dynamics of activation. Construction of a mathematical model that recapitulates NF-κB monomer expression, dimer formation, and LTβR-responsive activation may enable further understanding of the emergent network properties in homeostatic and dynamic control.
Acknowledgments
We thank J. Browning for LTβR agonist antibody, N. Rice for p50/p105 and p52/p100 antibodies, and Santa Cruz Biotechnology for a variety of other antibodies; we thank C. Lynch for help with the experiments. We are grateful to G. Ghosh and J. D. Kearns for helpful discussions and M. Karin for critical reading of the manuscript.
This study was supported by the following grants: NIH GM071573 and NIH GM071862 to A.H.
Footnotes
Published ahead of print on 25 February 2008.
REFERENCES
- 1.Alcamo, E., N. Hacohen, L. C. Schulte, P. D. Rennert, R. O. Hynes, and D. Baltimore. 2002. Requirement for the NF-κB family member RelA in the development of secondary lymphoid organs. J. Exp. Med. 195233-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Basak, S., H. Kim, J. D. Kearns, V. Tergaonkar, E. O'Dea, S. L. Werner, C. A. Benedict, C. F. Ware, G. Ghosh, I. M. Verma, and A. Hoffmann. 2007. A fourth IκB protein within the NF-κB signaling module. Cell 128369-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beinke, S., and S. C. Ley. 2004. Functions of NF-κB1 and NF-κB2 in immune cell biology. Biochem. J. 382393-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bonizzi, G., M. Bebien, D. C. Otero, K. E. Johnson-Vroom, Y. Cao, D. Vu, A. G. Jegga, B. J. Aronow, G. Ghosh, R. C. Rickert, and M. Karin. 2004. Activation of IKKαtarget genes depends on recognition of specific κB binding sites by RelB:p52 dimers. EMBO J. 234202-4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonizzi, G., and M. Karin. 2004. The two NF-kappaB activation pathways and their role in innate and adaptive immunity. Trends Immunol. 25280-288. [DOI] [PubMed] [Google Scholar]
- 6.Bren, G. D., N. J. Solan, H. Miyoshi, K. N. Pennington, L. J. Pobst, and C. V. Paya. 2001. Transcription of the RelB gene is regulated by NF-κB. Oncogene 207722-7733. [DOI] [PubMed] [Google Scholar]
- 7.Dejardin, E., N. M. Droin, M. Delhase, E. Haas, Y. Cao, C. Makris, Z. W. Li, M. Karin, C. F. Ware, and D. R. Green. 2002. The lymphotoxin-beta receptor induces different patterns of gene expression via two NF-κB pathways. Immunity 17525-535. [DOI] [PubMed] [Google Scholar]
- 8.Delhase, M., M. Hayakawa, Y. Chen, and M. Karin. 1999. Positive and negative regulation of IκB kinase activity through IKKbeta subunit phosphorylation. Science 284309-313. [DOI] [PubMed] [Google Scholar]
- 9.Derudder, E., E. Dejardin, L. L. Pritchard, D. R. Green, M. Korner, and V. Baud. 2003. RelB/p50 dimers are differentially regulated by tumor necrosis factor-alpha and lymphotoxin-beta receptor activation: critical roles for p100. J. Biol. Chem. 27823278-23284. [DOI] [PubMed] [Google Scholar]
- 10.de Wit, H., W. H. Dokter, S. B. Koopmans, C. Lummen, M. van der Leij, J. W. Smit, and E. Vellenga. 1998. Regulation of p100 (NFKB2) expression in human monocytes in response to inflammatory mediators and lymphokines. Leukemia 12363-370. [DOI] [PubMed] [Google Scholar]
- 11.Fu, Y. X., and D. D. Chaplin. 1999. Development and maturation of secondary lymphoid tissues. Annu. Rev. Immunol. 17399-433. [DOI] [PubMed] [Google Scholar]
- 12.Gapuzan, M. E., O. Schmah, A. D. Pollock, A. Hoffmann, and T. D. Gilmore. 2005. Immortalized fibroblasts from NF-κB RelA knockout mice show phenotypic heterogeneity and maintain increased sensitivity to tumor necrosis factor alpha after transformation by v-Ras. Oncogene 246574-6583. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh, S., and M. Karin. 2002. Missing pieces in the NF-κB puzzle. Cell 109(Suppl.)S81-S96. [DOI] [PubMed] [Google Scholar]
- 14.Hoffmann, A., T. H. Leung, and D. Baltimore. 2003. Genetic analysis of NF-κB/Rel transcription factors defines functional specificities. EMBO J. 225530-5539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hoffmann, A., A. Levchenko, M. L. Scott, and D. Baltimore. 2002. The IκB-NF-κB signaling module: temporal control and selective gene activation. Science 2981241-1245. [DOI] [PubMed] [Google Scholar]
- 16.Hoffmann, A., G. Natoli, and G. Ghosh. 2006. Transcriptional regulation via the NF-κB signaling module. Oncogene 256706-6716. [DOI] [PubMed] [Google Scholar]
- 17.Kearns, J. D., S. Basak, S. L. Werner, C. S. Huang, and A. Hoffmann. 2006. IκBɛ provides negative feedback to control NF-κB oscillations, signaling dynamics, and inflammatory gene expression. J. Cell Biol. 173659-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lawrence, T., M. Bebien, G. Y. Liu, V. Nizet, and M. Karin. 2005. IKKα limits macrophage NF-κB activation and contributes to the resolution of inflammation. Nature 4341138-1143. [DOI] [PubMed] [Google Scholar]
- 19.Liptay, S., R. M. Schmid, E. G. Nabel, and G. J. Nabel. 1994. Transcriptional regulation of NF-κB2: evidence for κB-mediated positive and negative autoregulation. Mol. Cell. Biol. 147695-7703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lo, J. C., S. Basak, E. S. James, R. S. Quiambo, M. C. Kinsella, M. L. Alegre, F. Weih, G. Franzoso, A. Hoffmann, and Y. X. Fu. 2006. Coordination between NF-κB family members p50 and p52 is essential for mediating LTbetaR signals in the development and organization of secondary lymphoid tissues. Blood 1071048-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Luedde, T., U. Assmus, T. Wustefeld, A. Meyer zu Vilsendorf, T. Roskams, M. Schmidt-Supprian, K. Rajewsky, D. A. Brenner, M. P. Manns, M. Pasparakis, and C. Trautwein. 2005. Deletion of IKK2 in hepatocytes does not sensitize these cells to TNF-induced apoptosis but protects from ischemia/reperfusion injury. J. Clin. Investig. 115849-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mebius, R. E. 2003. Organogenesis of lymphoid tissues. Nat. Rev. Immunol. 3292-303. [DOI] [PubMed] [Google Scholar]
- 23.Mercurio, F., H. Zhu, B. W. Murray, A. Shevchenko, B. L. Bennett, J. Li, D. B. Young, M. Barbosa, M. Mann, A. Manning, and A. Rao. 1997. IKK-1 and IKK-2: cytokine-activated IκB kinases essential for NF-κB activation. Science 278860-866. [DOI] [PubMed] [Google Scholar]
- 24.Miyawaki, S., Y. Nakamura, H. Suzuka, M. Koba, R. Yasumizu, S. Ikehara, and Y. Shibata. 1994. A new mutation, aly, that induces a generalized lack of lymph nodes accompanied by immunodeficiency in mice. Eur. J. Immunol. 24429-434. [DOI] [PubMed] [Google Scholar]
- 25.Moorthy, A. K., O. V. Savinova, J. Q. Ho, V. Y. Wang, D. Vu, and G. Ghosh. 2006. The 20S proteasome processes NF-κB1 p105 into p50 in a translation-independent manner. EMBO J. 251945-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mordmuller, B., D. Krappmann, M. Esen, E. Wegener, and C. Scheidereit. 2003. Lymphotoxin and lipopolysaccharide induce NF-κB-p52 generation by a co-translational mechanism. EMBO Rep. 482-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morgenstern, J. P., and H. Land. 1990. Advanced mammalian gene transfer: high titre retroviral vectors with multiple drug selection markers and a complementary helper-free packaging cell line. Nucleic Acids Res. 183587-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller, J. R., and U. Siebenlist. 2003. Lymphotoxin beta receptor induces sequential activation of distinct NF-κB factors via separate signaling pathways. J. Biol. Chem. 27812006-12012. [DOI] [PubMed] [Google Scholar]
- 29.Neumann, B., A. Luz, K. Pfeffer, and B. Holzmann. 1996. Defective Peyer's patch organogenesis in mice lacking the 55-kD receptor for tumor necrosis factor. J. Exp. Med. 184259-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.O'Dea, E. L., D. Barken, R. Q. Peralta, K. T. Tran, S. L. Werner, J. D. Kearns, A. Levchenko, and A. Hoffmann. 2007. A homeostatic model of IκB metabolism to control constitutive NF-κB activity. Mol. Syst. Biol. 3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pomerantz, J. L., and D. Baltimore. 2002. Two pathways to NF-κB. Mol. Cell 10693-695. [DOI] [PubMed] [Google Scholar]
- 32.Rennert, P. D., D. James, F. Mackay, J. L. Browning, and P. S. Hochman. 1998. Lymph node genesis is induced by signaling through the lymphotoxin beta receptor. Immunity 971-79. [DOI] [PubMed] [Google Scholar]
- 33.Schneider, K., K. G. Potter, and C. F. Ware. 2004. Lymphotoxin and LIGHT signaling pathways and target genes. Immunol. Rev. 20249-66. [DOI] [PubMed] [Google Scholar]
- 34.Sen, R. 2006. Control of B lymphocyte apoptosis by the transcription factor NF-κB. Immunity 25871-883. [DOI] [PubMed] [Google Scholar]
- 35.Senftleben, U., Y. Cao, G. Xiao, F. R. Greten, G. Krahn, G. Bonizzi, Y. Chen, Y. Hu, A. Fong, S. C. Sun, and M. Karin. 2001. Activation by IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science 2931495-1499. [DOI] [PubMed] [Google Scholar]
- 36.Shinkura, R., K. Kitada, F. Matsuda, K. Tashiro, K. Ikuta, M. Suzuki, K. Kogishi, T. Serikawa, and T. Honjo. 1999. Alymphoplasia is caused by a point mutation in the mouse gene encoding NF-κB-inducing kinase. Nat. Genet. 2274-77. [DOI] [PubMed] [Google Scholar]
- 37.Ware, C. F. 2005. Network communications: lymphotoxins, LIGHT, and TNF. Annu. Rev. Immunol. 23787-819. [DOI] [PubMed] [Google Scholar]
- 38.Weih, D. S., Z. B. Yilmaz, and F. Weih. 2001. Essential role of RelB in germinal center and marginal zone formation and proper expression of homing chemokines. J. Immunol. 1671909-1919. [DOI] [PubMed] [Google Scholar]
- 39.Weih, F., and J. Caamano. 2003. Regulation of secondary lymphoid organ development by the nuclear factor-kappaB signal transduction pathway. Immunol. Rev. 19591-105. [DOI] [PubMed] [Google Scholar]
- 40.Werner, S. L., D. Barken, and A. Hoffmann. 2005. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science 3091857-1861. [DOI] [PubMed] [Google Scholar]
- 41.Xiao, G., E. W. Harhaj, and S. C. Sun. 2001. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell 7401-409. [DOI] [PubMed] [Google Scholar]
- 42.Yilmaz, Z. B., D. S. Weih, V. Sivakumar, and F. Weih. 2003. RelB is required for Peyer's patch development: differential regulation of p52-RelB by lymphotoxin and TNF. EMBO J. 22121-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yin, L., L. Wu, H. Wesche, C. D. Arthur, J. M. White, D. V. Goeddel, and R. D. Schreiber. 2001. Defective lymphotoxin-beta receptor-induced NF-κB transcriptional activity in NF-κB-inducing kinase-deficient mice. Science 2912162-2165. [DOI] [PubMed] [Google Scholar]