Abstract
Exonization of Alu elements creates primate-specific genomic diversity. Here we combine bioinformatic and experimental methodologies to reconstruct the molecular changes leading to exon selection. Our analyses revealed an intricate network involved in Alu exonization. A typical Alu element contains multiple sites with the potential to serve as 5′ splice sites (5′ss). First, we demonstrated the role of 5′ss strength in controlling exonization events. Second, we found that a cryptic 5′ss enhances the selection of a more upstream site and demonstrate that this is mediated by binding of U1 snRNA to the cryptic splice site, challenging the traditional role attributed to U1 snRNA of binding the 5′ss only. Third, we used a simple algorithm to identify specific sequences that determine splice site selection within specific Alu exons. Finally, by inserting identical exons within different sequences, we demonstrated the importance of flanking genomic sequences in determining whether an Alu exon will undergo exonization. Overall, our results demonstrate the complex interplay between at least four interacting layers that affect Alu exonization. These results shed light on the mechanism through which Alu elements enrich the primate transcriptome and allow a better understanding of the exonization process in general.
An average human mRNA precursor is 28,000 nucleotides (nt) long, comprising nine exons separated by eight introns. Internal exons are usually small (about 129 nt on average), and exons account for only ∼5% of human precursor mRNA (22). How does the splicing machinery find small exons embedded within long intronic sequences? Four splicing signals direct the splicing machinery to the correct exon-intron boundaries: the 5′ and 3′ splice sites (5′ss and 3′ss), located in the 5′ and 3′ ends of introns, respectively; the polypyrimidine tract; and the branch site sequence located upstream of the 3′ss. Intron removal is catalyzed by the spliceosome, a complex containing five snRNPs (U1, U2, U4/U6, and U5) and at least 150 non-snRNP proteins (4). In higher eukaryotes, the 5′ss is a region of 9 nt located across the exon-intron junction. The 5′ss sequence base pairs with a region of the U1 RNA. Most of the nucleotides in these positions are degenerate (7, 27, 34), and base pairing is not perfect between most pre-mRNAs and U1. High complementarity to U1 typically leads to constitutive splicing, whereas a low binding affinity of U1 for the 5′ss tends to result in alternative splicing (2, 35, 42).
Alternative splicing produces more than one isoform of mRNA from a single gene (14). Bioinformatic analysis indicates that >70% of all human genes are alternatively spliced. This contributes significantly to human proteome complexity and explains the numerical disparity between the number of genes in the human genome and the much larger number of proteins produced (14, 18, 31). There are five different types of alternative splicing; exon skipping is the most prevalent (20, 45, 50). In higher eukaryotes, the four consensus splicing signals are insufficient to define exon-intron boundaries (46). Exon selection is also controlled by exonic and intronic splicing regulatory elements (ESRs and ISRs, respectively). Proteins such as SR and hnRNPs bind to these sites and presumably assist the basal machinery in locating the correct splice junctions (6, 8, 44, 49). Such auxiliary elements can also promote the usage of cryptic (pseudo) splice sites, which are not selected under normal conditions (10, 12, 43). Thus, exon selection and alternative splicing regulation are outcomes of complex combinatorial effects that are largely uncharacterized (3, 5, 28, 39).
More than 5% of the alternatively spliced internal exons in the human genome are derived from Alu elements (41). Alu elements are short (∼280 nt), primate-specific retrotransposons. More than 1 million copies are dispersed throughout the human genome, with the majority located in introns (25, 36, 42). As far as we know, all alternatively spliced Alu exons were created via exonization of intronic elements. Most Alu elements have remained silent, while a small subset has undergone exonization, but there is high sequence similarity between exonized and nonexonized counterparts. These features render Alu elements an excellent platform for understanding the requirements of exonization. Indeed, previous studies of the sequences of these elements have determined the minimal conditions required for exonization and demonstrated that selective pressure is applied to maintain weak (suboptimal) splice sites that flank the alternatively spliced Alu exons (2, 25, 42). In addition, exonization of Alu sequences has been shown to be regulated not only by splice sites but also by splicing regulatory sequences within the Alu elements (24).
In this study, we have used bioinformatic and experimental analysis of splicing of Alu sequences to extend our understanding of the processes involved in selection of an exon. We began with the bioinformatic observation that Alu sequences tend to have multiple potential 5′ss. This led us to examine what determines the selection of a specific splice site. Our analyses revealed an intricate network. First, we demonstrated the role of 5′ss strength in governing exonization events. Second, we found that a cryptic 5′ss enhances the selection of a more upstream site and demonstrate that this enhancing effect is mediated by binding of U1 snRNA to the cryptic splice site. Third, we developed a simple algorithm for identifying specific ESRs within a given Alu exon. We subsequently showed that the identified ESRs cause certain splice sites to be selected. Finally, by inserting identical exons within different sequences, we demonstrated the importance of flanking sequences in determining whether a particular Alu exon will undergo exonization. Overall, our results shed light both on mechanistic aspects pertaining to exon recognition by the splicing machinery and on the evolutionary mechanisms allowing primate-specific transcriptomic enrichment by means of exonization events originating from Alu sequences.
MATERIALS AND METHODS
Compilation of exonizing and nonexonizing data sets.
We were interested in comparing typical Alu exons, which undergo exonization from the right arm in the antisense orientation, with nonexonizing Alu elements. Two initial data sets of exonizing and nonexonizing intronic Alu elements (exonic and intronic data sets) in the antisense orientation were obtained by querying the TranspoGene database (26). This yielded all Alu elements without and with an overlap of at least one expressed sequence tag (EST), in the case of the intronic and exonic data sets, respectively. Since a typical Alu exon is ∼300 nt in length, we next filtered out all Alu elements shorter than 250 nt.
To obtain instances in which the exonizations originated exclusively from the right arm, we performed pairwise global alignments between the Alu sequences in each of the two data sets and between the Alu-Jo consensus sequence, which was downloaded from Repbase (19) and is presented in Fig. 1B. These alignments were performed using the needle application with default parameters, which implements the Needleman-Wunsch global alignment algorithm (21, 32). We next filtered out all cases in the exonic data set in which the selected 3′ss and 5′ss were not located upstream of the poly(T) sequence which separates the two Alu arms. Based on these alignments, we also searched for putative 3′ss and 5′ss in the two data sets. For each of these two signals, we first empirically determined the sequence window in which they were located in the exonic data set. Specifically, we found that 97% of the 3′ss were located between positions 1 and 58 (relative to the consensus) and that >98% of the 5′ss were located between positions 105 and 181; the few atypical cases which did not conform to these rules were filtered out from the exonic data set. Within the 5′ss window, for each of the two data sets, we identified and assigned Senapathy (37) scores to four 5′ss sites, namely, each of sites A, B, and C (for the analyses presented in Fig. 2A, B, and C) and the highest-scoring site (for the analysis in Fig. 2D). In addition, in the case of the exonic data set, we also scored the biologically selected 5′ss (based on EST evidence). Following the various filtrations, we ended up with 323 Alu elements in the exonic data set and 177,410 Alu elements in the intronic data set.
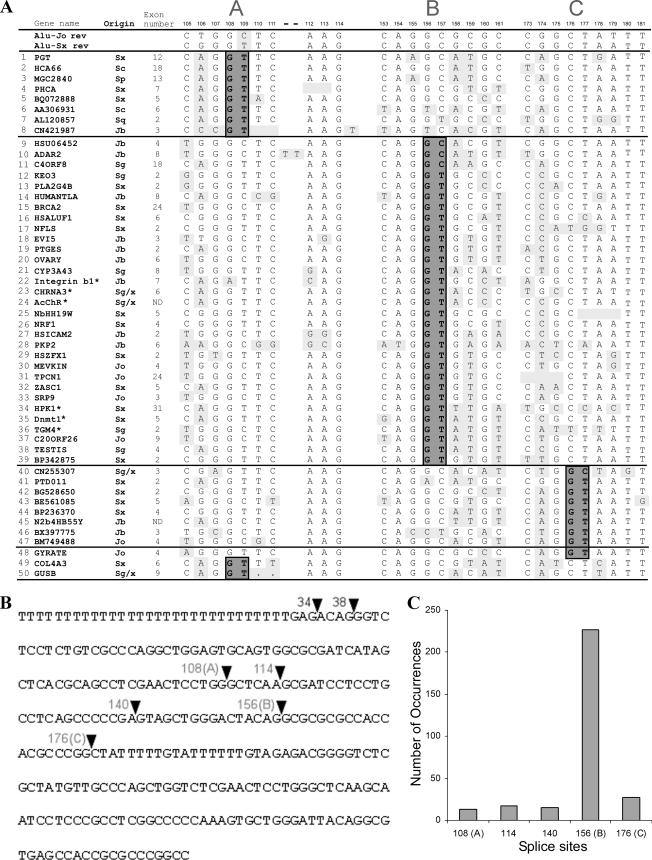
FIG. 1.
5′ss selection of Alu-derived exons. (A) Alignment of 50 exonized Alu elements in the antisense orientation with respect to the pre-mRNA. This data set is based on previous studies (25, 36, 41) as well as on further literature (marked by asterisks; see Table S1 in the supplemental material for references). The 26 nt presented contain three possible 5′ss selected during Alu exonization. The first two intronic positions at each site are highlighted in dark gray and marked 5′ss A, B, and C. Consensus sequences of subfamilies S and Jo appear in the first two rows. Single mutations differing from the ancestral S and Jo subfamilies are highlighted in light gray. Rows 48 to 50 represent the 5′ss of Alu sequences whose constitutive exonization was shown to cause a genetic disease, either OAT deficiency (GYRATE), Alport syndrome (COL4A3), or Sly syndrome (GUSB). The mutation that causes Alport syndrome is in the 3′ss region (−7G to T), as shown previously (25). The mutations causing Sly syndrome and OAT deficiency are both in 5′ss regions: the mutation causing OAT syndrome is in row 48, position 176, and the mutations resulting in Sly deficiency are in row 50, positions 110 and 111. See Table S1 in the supplemental material for references for these three genetic diseases. Gene names are given according to RefSeq conventions, and the Alu exon number is the exon serial number for each gene. (B) Most frequently used splice sites in Alu right-arm exonizations in the antisense orientation, mapped onto the Alu-Jo consensus sequence (19). The 3′ss at positions 34 and 38 and the 5′ss at positions 108, 114, 140, 156, and 176 are indicated with arrowheads marking the exon-intron junctions at these sites. (C) Frequencies of selection of the five main 5′ss resulting in Alu exonization.
FIG. 2.
Overview of sites A, B, and C in Alu elements. (A) Pictograms of sites A, B, and C in intronic Alu elements in the intronic data set compared to pictograms for these three sites used as splice sites in sequences from the exonic data set. (B) Percentages of Alu elements in which a minimal 5′ss (“GT” or “GC”) exists at sites A, B, and C in the exonic and intronic data sets. (C) Mean strengths of the three sites selected from sequences in the exonic data set compared with those regions in sequences in the intronic data set. (D) Sensitivity of splice site strength as a predictor of selection of the three sites. The y axis represents the number of cases in which a given splice site was strongest divided by the number of cases in which it was selected.
Plasmid constructs.
Minigenes were inserted into the pEGFP-C1 vector (Clontech), which contains green fluorescent protein (GFP), as described previously (13). Human genomic sequences were amplified using primers containing additional sequences for restriction enzymes. The PCR products were restriction digested and inserted into the vector, followed by DNA sequencing. We used the following three minigenes: (i) the ADAR2 minigene (adenosine deaminase) contains the human genomic sequence from the beginning of exon 7 through the end of exon 9 (2.2 kb); (ii) the PGT minigene (putative glucosyltransferase) contains the human genomic sequence from the beginning of exon 11 through the end of exon 13 (1.8 kb); and (iii) IKBKAP, referenced also as the IKAP minigene (inhibitor of κ-light polypeptide gene enhancer in B cell kinase complex-associated protein), contains three constitutive exons, i.e., exons 19 to 21 (1.87 kb). To prevent amplification of the endogenous mRNA and to achieve amplification of the product originating solely from the plasmid, we used a forward primer directed against the multiple cloning site of the pEGFP-C3 plasmid and a reverse primer directed against the transcription termination site of the plasmid.
Site-directed mutagenesis.
A supercoiled double-stranded DNA vector (pEGFP-C3) with an insert of the desired minigene and two synthetic oligonucleotide primers containing the desired mutations were extended by PCR analysis using Pfu Turbo DNA polymerase (Stratagene). The PCR machine was programmed for 18 cycles, and the elongation time corresponded to 2 min for each 1 kb. The PCR products were treated with 12 U DpnI (New England Biolabs) for 1 h at 37°C. The resulting DNA (1 to 3 μl) was transformed into Escherichia coli strain XL1. After transformation, the XL1 competent cells repaired the nicks in the mutated plasmids to generate the full-length plasmids. We then performed a colony-picking and miniprep extraction (Gibco/BRL). All plasmids were confirmed by sequencing (25). Plasmids containing deletions/insertions were amplified using 5′ phosphate-oligonucleotide primers complementary to flanking regions of the desired deletion/insertion sequences. The PCR products were treated as described previously (13).
Transfection and RT-PCR.
293T cells were cultured in Dulbecco's modified Eagle medium supplemented with 4.5 g/ml glucose (Biological Industries) and 10% fetal calf serum in a six-well dish under standard conditions at 37°C with 5% CO2. Cells were grown to 50% confluence, and transfection was performed using Fugene 6 (Roche) with 1 μg of plasmid DNA. RNA was harvested after 48 h. Total cytoplasmic RNA was extracted using TriReagent (Sigma), followed by treatment with 1 U RNase-free DNase (Promega). Reverse transcription-PCR (RT-PCR) amplification was performed for 1 h at 42°C, using an oligo(dT) reverse primer and 2 U reverse transcriptase from avian myeloblastosis virus (Roche). The spliced cDNA products derived from the expressed minigenes were detected by PCR. For amplification of ADAR2 and PGT, we used a pEGFP-specific reverse primer (CGCTTCTAACATTCCTATCCAAGCGT). For the forward primers, we used an ADAR2 exon 7 primer (CCCAAGCTTTTGTATGTGGTCTTTCTGTTCTGAAG) and a PGT exon 9 primer (AATCTTACTCATGTTACTA). Amplification was performed for 30 cycles, consisting of 30 s at 94°C, 50 s at 61°C, and 1 min at 72°C. The products were resolved in a 2% agarose gel and confirmed by sequencing. Cytoplasmic RNAs from three different transfection experiments were measured by gel electrophoresis, and the intensities of the bands were quantified with Image-J 1.36 (pixel detection software). The PCR remained in an exponential phase throughout 30 cycles, as demonstrated by loading products of 18, 21, 25, 30, and 32 PCR cycles. In addition, Image-J quantification of ADAR2 and PGT RT-PCR products correlates with real-time RT-PCR quantification produced by the Roche LightCycler PCR and detection system (13). The level of mRNA of the housekeeping gene encoding glyceraldehyde-3-phosphate dehydrogenase was used as the internal control for each transfection. Each RT-PCR experiment was performed at least three times, and the fluctuation was <10% among the experiments.
RESULTS
Each Alu element is composed of two related but distinct monomers, the left and right arms. Exonization of Alu sequences tends to occur predominantly from the right arm, in the antisense orientation relative to the mRNA precursor (25). We began by visually inspecting a multiple alignment of exonizing Alu sequences (a sample can be viewed in Fig. 1A). Despite the high degree of similarity between the sequences of Alu monomers, the locations of the selected 3′ss and 5′ss were found to be variable. Three of the main 5′ss sites, which we termed sites A, B, and C, are shown in Fig. 1A; the site labeled B was predominantly selected.
This raises intriguing questions. How does the splicing machinery “decide” which 5′ss to select? Do adjacent splicing signals compete for selection by the splicing machinery? If so, are evolutionary pressures exerted on adjacent splicing signals? The last question was prompted by an observation, apparent in Fig. 1A, that whenever site B or A was selected, there was not a splice consensus sequence at site C. This may reflect a negative selective pressure that prevents the appearance of competing adjacent splicing signals.
To obtain an overview of splice site selection within Alu sequences, we began with a bioinformatic approach. We compiled two data sets, including a data set of 323 Alu elements in the antisense orientation with EST evidence supporting exonization from the right arm (exonic data set) and a data set of ∼177,000 intronic Alu elements in the antisense orientation that lack any EST evidence of exonization (intronic data set). To allow direct comparison between specific positions within different Alu sequences, we performed pairwise global alignments of each of the sequences from each of the two data sets (intronic and exonic) against the Alu consensus. We next determined which positions (relative to the consensus) within the exonic data set served as splice sites. We then searched for putative splicing signals within the right arms of the two data sets (see Materials and Methods).
We found that within the exonic data set, the most frequently selected 5′ss was at position 156 (referring to the first intronic position of the 5′ss); this site was selected in 227 of the 323 (70%) right-arm Alu exons (labeled site B in Fig. 1A). The second most frequently selected 5′ss was at position 176 (selected in 27 cases; site C). Three other positions, positions 108 (site A), 114, and 140, were selected at approximately equal frequencies (13 to 17 instances of each). The locations of these five sites on the Alu consensus sequence are shown in Fig. 1B, and their frequencies of selection are presented in Fig. 1C. There were two main 3′ss, at positions 34 and 38 relative to the consensus. These positions have been analyzed previously (25).
In our subsequent analyses, we decided to focus on sites A, B, and C. Sites B and C were selected because they are the most frequently selected splice sites. We focused on site A as well, since aberrant selection of this site is implicated in the pathogenesis of Alport syndrome and Sly syndrome (Fig. 1A, rows 49 and 50; a deletion in row 50 is marked with dots) (33, 48). Notably, aberrant selection of site C is involved in the pathogenesis of OAT deficiency (Fig. 1A, row 48) (29).
We sought to determine whether splicing signals at these three sites formed part of the ancestral Alu sequence or whether these regions became splice sites as a result of specific mutations that subsequently led to exonization. We created pictograms of sites A, B, and C, based on the sequences in the exonic data set. In parallel, we created pictograms of these three sites based on the sequences in the intronic data set. These two sets of pictograms are presented in Fig. 2A. For all three splice sites, various changes were observed between the selected splice sites in the exonizing data set and those in the nonexonizing one. All changes across the 9-nt 5′ss reflect a strengthening of the splice sites. For site A, the predominant changes were an increase in A at position −2 relative to the same position in the nonexonized sequences (i.e., the second-to-last position with respect to the exon-intron boundary) and an increase in T at position +2; for site B, there was an increase in T at position +2 and in G at position +5; and for site C, there was an increase in A at position −2 and in G at position +1. All of these changes result in stronger binding between the 5′ss and U1 snRNA (7).
To complement this analysis, we next compared the frequencies of occurrence of splice sites A, B, and C in the exonic and intronic data sets (Fig. 2B). For a splice site to be defined as existent, we demanded that there be a “GT” or “GC” at the two intronic positions potentially serving as 5′ss. For sites A and B, a 5′ss was found in the vast majority of Alu sequences in both the exonic and intronic data sets. For site C, however, the two data sets differed: a 5′ss was found in only 1.4% of the Alu elements in the intronic data set but was found in 10.5% of the Alu elements in the exonic data set. Thus, there was an ∼8-fold increase in the number of splice sites at site C in the exonic data set with respect to the intronic data set. This indicates that once a mutation, from “C” to “G” at position +1 of site C (as can be inferred from Fig. 1A), creates a functional splice site at site C, there is a tendency for the Alu element containing this newly formed splice site to undergo exonization. Notably, the scarcity of existent splice sites at site C explains our observation (Fig. 1A) that in all instances in which site B was selected, site C lacked a potential splice site. This phenomenon, therefore, does not reflect a selection against the coappearance of two splice sites but merely appears to reflect the general nucleotide composition at site C.
To understand the balance of power among the three sites, we compared the mean strengths of the selected splice sites in the exonic data set to the mean splice site strengths for the intronic data set in cases in which a splice site at the relevant position was existent (Fig. 2C). Two phenomena were observed. First, the selected splice sites were consistently stronger, in all three sites, than their counterparts in the intronic group, reflecting the fact that exonization of Alu sequences is more likely to occur from stronger splice sites. Second, considerable differences were found in terms of splice site strength, as follows: site A is by far the weakest site, with a mean Senapathy score, in cases in which it was selected, of 61.5; site B is of intermediate strength, with a mean score of 78.4; and site C is the strongest site, with a mean 5′ss score of 83.6. The differences in strengths among the three splice sites are due to the nucleotide compositions of the three sites, which can also be observed directly in Fig. 1A. The “basal” nucleotide composition of existent 5′ss at site A comprises five positions optimal for base pairing with U1 snRNA (positions −3 through +2), site B comprises six or seven optimal pairs (positions −3 through +2, position +5, and position +6), and site C comprises eight such positions (all positions except for position +5).
How does the splicing machinery decide which splice site to select? Are considerations of splice site strength sufficient to explain and to predict splice site selection? To test this, we examined the sensitivity of splice site strength as a predictor of selection. For each of the three sites, we calculated the fraction of instances in which a splice site was strongest within the sequence window in which 5′ss of Alu elements tended to appear (see Materials and Methods) divided by the total number of instances in which it was selected (Fig. 2D). Splice site strength was found to be a very sensitive measure for sites B and C. In all 27 cases in which site C was selected, it was the strongest splice site (sensitivity = 100%). Of 227 cases in which site B was selected, it was the strongest site in 219 cases (sensitivity = 96.5%). Splice site A, however, differed from these two sites. Of the 13 instances in which it was selected, it was strongest in only 4 (sensitivity = 30.7%), indicating that in ∼70% of the cases in which site A was selected, there was a stronger candidate present that was not selected. These results indicate that whereas selection of sites B and C is primarily a function of their strength, selection of site A presumably involves further factors.
Cross talk between different 5′ss.
We next set out to analyze 5′ss selection in two ex vivo systems. For this purpose, we cloned two minigenes, containing three exons separated by two introns, in which the first and third exons are constitutively spliced and the second exon is an alternatively spliced exon originating from an Alu element. The first minigene is derived from exons 7 to 9 of the human ADAR2 gene (Fig. 3A), and the second minigene is derived from exons 9 to 11 of the human PGT gene (Fig. 3C). The minigenes were transfected into 293T cells. RNA was collected, and the splicing pattern of the minigene mRNAs was examined by RT-PCR analysis, using specific primers for the exogenous mRNAs. The inclusion/skipping ratio was measured by a densitometry program and showed fluctuations of <10% in independent experiments (see Materials and Methods).
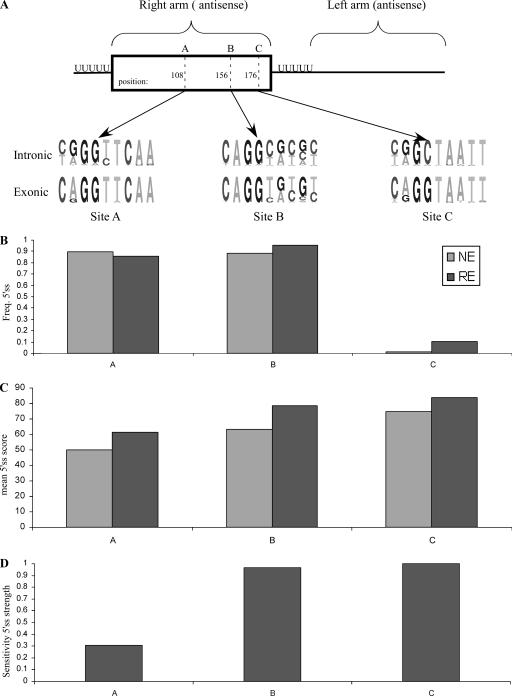
FIG. 3.
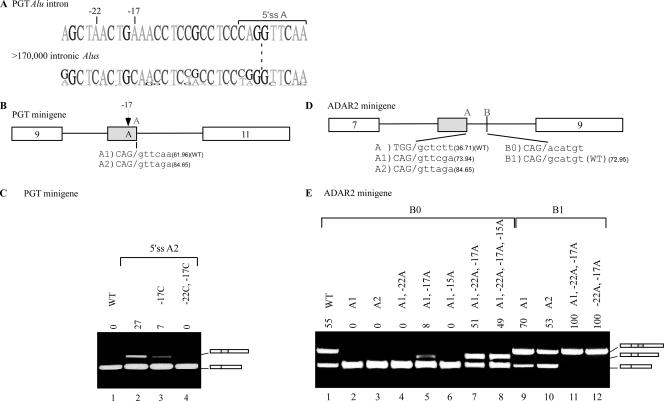
5′ss selection in Alu exons. (A) Diagram of the ADAR2 minigene containing exons 7, 8, and 9. Exon 8 is an Alu-derived exon; the three shades of gray represent the three potential 5′ss regions A, B, and C. The last three exonic and first six intronic nucleotides of the wt and mutated 5′ss are shown, marked as A to A2, B0 to B3, and C to C2. Senapathy scores of the 5′ss are given in parentheses. (B) Plasmids containing the indicated mutants were introduced into 293T cells by transfection. Total cytoplasmic RNA was extracted, and splicing products were separated in a 2% agarose gel after RT-PCR. Lane 1, splicing products of wt ADAR2 (marked B1 in panel A); lanes 2 to 11, splicing products of the indicated mutants. The numbers above the lanes indicate the percentages of inclusion of exon 8. The three minigene mRNA products are shown on the right. Empty boxes define constitutive exons, and the shaded boxes indicate the selection of either of the three 5′ss. (C) Diagram showing the PGT minigene, containing exons 9, 10, and 11. Exon 10 does not exonize under endogenous conditions but constitutes an intronic Alu element on the verge of exonization. The three shades of gray in exon 10 represent three potential 5′ss (sites A, B, and C). Exons 9 and 11, shown by empty boxes, are constitutively spliced. The wt 5′ss (marked A1) and six mutated 5′ss are shown in the lower part, marked as A2, B to B3, and C to C2. Senapathy scores of the 5′ss are presented as described above. (D) Analysis of in vivo splicing essentially as described for panel B. Lane 1, splicing products of wt PGT (marked A1 in panel C); lane 2, splicing products of a 3′ss mutant (from GAG to TAG); lanes 3 to 12, splicing products of the indicated mutants. The three minigene mRNA products are shown on the right. Empty boxes indicate constitutively spliced exons, whereas the boxes colored in three shades of gray indicate selection of 5′ss A, B, or C. All PCR products were verified by sequencing.
The 5′ss of the ADAR2 Alu exon is of the GC type (Fig. 1A, row 10) and is included in 55% of the minigene mRNAs (Fig. 3B, lane 1; the minigene and the endogenous ADAR2 mRNA gave similar inclusion levels). In addition to the wild-type (wt) 5′ss B (marked B1), we created two different 5′ss at position C, which differed in strength but were both considerably stronger than site B1 (marked 5′ss C1 and C2; see Fig. 3A for the sequences of these sites). The presence of either of the two stronger 5′ss at position C resulted in a shift from alternative selection of 5′ss B to constitutive selection of 5′ss C (Fig. 3B, compare lane 1 with lanes 2 and 3), compatible with our previous conclusion that splice site strength strongly determines splice site selection.
Mutations that considerably strengthened site B relative to the wt 5′ss B1 (Fig. 3A) resulted in a shift from alternative to constitutive selection of 5′ss B, suggesting that the inclusion level of the Alu exon may be regulated by the strength of base pairing between site B and U1 snRNA (Fig. 3B, lane 4) (42). Under these conditions of constitutive selection of splice site B, we examined whether splice sites at position C could compete with site B. We added splice sites C1 and C2, both of which are strong splice sites, to a transcript containing B3. Although C1 and C2 are both stronger than B3, site B was selected constitutively in both cases (Fig. 3B, lanes 5 and 6), indicating that a medium or strong 5′ss at position C does not affect the constitutive selection of a strong 5′ss at position B.
We next generated an intermediate 5′ss, site B2, which is stronger than B1 but weaker than B3. Changing the wt site to site B2 resulted in a 95% inclusion level of the Alu exon (Fig. 3B, lane 7). Surprisingly, when a potential site was added at position C (5′ss C1), no skipping was observed, and B2 was selected in a constitutive manner (Fig. 3B, lane 8). This suggests that selection of site B was enhanced by the presence of a downstream cryptic 5′ss at site C. In the presence of an even stronger splice site (5′ss C2), site B was selected as the main splice site in approximately 83% of cases, and site C was selected as a minor site, with approximately 17% selection (Fig. 3B, lane 9). Taken together, these results suggest the following. First, a strong predisposition to select splice site B exists in the ADAR exon. Once the strength of the 5′ss at this position is over a certain threshold, it is almost invariably selected, regardless of the strength of 5′ss at downstream positions. This predisposition may explain why, in the vast majority of right-arm Alu exonizations, site B is selected as the 5′ss. Second, a cryptic splice site downstream of site B (at site C) enhances the selection of site B.
We then determined whether we could shift the selected 5′ss from site B to site A. When the relatively weak wt 5′ss B1 was present, the addition of a 5′ss at position A did not shift selection toward 5′ss A1 or A2 (not shown). Even a point mutation that eliminated 5′ss B (G→A at position +1, indicated as 5′ss B0 in Fig. 3A) did not result in the selection of 5′ss A1 or A2 (Fig. 3B, lanes 10 and 11), despite the fact that both are stronger splice sites than B1. These results confirm that there is a strong predisposition for the selection of site B in the ADAR exon.
To confirm our observations regarding the interplay between sites B and C, we examined conditions leading to exonization of the Alu exon in the PGT minigene (Fig. 3C). The PGT Alu element is not exonized in its minigene environment (Fig. 3D, lane 1). However, mutating the third-to-last position within the intron upstream of the PGT Alu exon from G to T (Fig. 3C), thereby strengthening the 3′ss, results in exonization of the Alu sequence, with site A being selected (Fig. 3D, lane 2) (25). We found that such exonization can also be achieved by a mutation that strengthens site A (mutation A2) (Fig. 3D, lane 3). Notably, this 5′ss A2 is identical to the 5′ss A2 in the ADAR2 gene, yet in the PGT gene it was alternatively selected and in ADAR2 it was invariably skipped.
Is site A influenced by other potential splice sites? To test this, we created further mutations in sites B and C. Mutations that created weak and medium-strength 5′ss at site B (sites B1 and B2, respectively) (Fig. 3B) were not sufficient to activate exonization of the PGT Alu exon (Fig. 3D, lanes 4 and 6), even when they were stronger than the selected A2 site (site B2 is stronger than site A2). The same splice site sequences were sufficient to cause alternative selection of site B in the ADAR gene. However, these two potential 5′ss at position B had a silencing effect on the selection of 5′ss A2 (Fig. 3D; note the gradual decrease in inclusion level apparent in lanes 3, 5, and 7, correlating with the gradual increase in strength of the B site). This demonstrates that a weak or medium-strength putative 5′ss at site B reduced selection of site A in the PGT gene. However, further strengthening of site B (to B3) resulted in a shift from full skipping to 70% inclusion (Fig. 3D, lane 8), with site B being selected. Again, these findings demonstrate the multiple factors underlying splice site selection; splice site strength determines the outcome only up to a certain threshold, and beneath it, interplay between the two sites is apparent. In addition, the apparent predisposition for selection of site A is indicative of the role played by surrounding sequences.
Under conditions where site B was alternatively selected (using B3), we sought to determine whether in the PGT gene, as in the ADAR gene, a putative splice site at site C would increase the selection of site B. At site C, we created two mutations, C1 and C2 (see Fig. 3B for details), that were identical to the ones used in the ADAR minigene. Insertion of C1 and C2 mutants resulted in 52% and 100% inclusion levels in PGT Alu elements, respectively, with site C being selected (Fig. 3D, lanes 11 and 12). However, a combination of 5′ss B3 and either C1 or C2 resulted in the selection of site B only (Fig. 3D, compare lane 8 with lanes 9 and 10). This indicates, as in the case of the ADAR exon, that the presence of a downstream 5′ss at site C functions as an enhancer for the selection of site B (compare with Fig. 3B, lanes 7 and 8).
Factors underlying predisposition for selection of site B in ADAR.
We next sought to determine what underlies the clear predisposition for selection of site B in the ADAR minigene, even when it is weak and in the presence of stronger candidates. To find putative regions that enhance the selection of site B, we compared the positions upstream of splice site B to the corresponding positions in the ∼177,000 introns in the intronic data set. We reasoned that a position differing considerably in the exonizing ADAR gene with respect to nonexonizing counterparts may be involved in the exonization of the former. We found that position −15 upstream of site B in the Alu exon presented the greatest change in comparison to nonexonizing counterparts. The overwhelming majority of nonexonizing Alu elements contain a T at this position, whereas in the ADAR2 gene this position has a G (as illustrated in Fig. 4A). Further bioinformatic support for the potential importance of this mutation was that analysis of this site using ESRsearch revealed that without the mutation, namely, as in the intronic Alu elements, position −15 overlaps with only three predicted ESRs. However, with the T→ G change found in the ADAR gene, this position overlaps with eight putative ESRs.
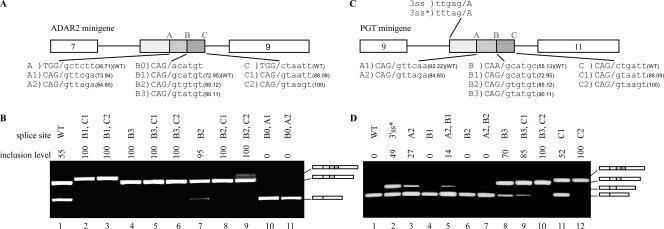
FIG. 4.
Site B regulation and U1 complementation in ADAR2 Alu exon. (A) The upper sequence shows the last 20 nt and first 6 nt of the exon-intron junction of site B of the ADAR2 Alu exon. Position −15 is marked above the exon sequence. The lower sequence is a pictogram of the sequences at this site from an alignment of ∼177,000 intronic Alu elements in the intronic data set. (B) Diagram of ADAR2 minigene. 5′ss B and C are marked above the box. wt and mutated 5′ss are shown in the lower part, marked as B1, B2, B4, C, and C2 to C5; Senapathy scores are given in parentheses. Position −15 upstream of 5′ss B is marked above the box. (C) Mutated 5′ss with matching U1 snRNAs used for complementation assays. Four complementary U1 snRNAs were created, to match sites C3, C4, B4, and C5. The highlighted nucleotides represent complementary mutations. (D) In vivo splicing and complementation assay. Lane 1, splicing products of wt ADAR2 gene; lanes 2 to 11, splicing products of exon 8 mutants; lanes 7, 9, and 11 are splicing products obtained after complementation with the indicated U1 snRNA. The three boxes colored in shades of gray indicate the selection of either of the two potential 5′ss.
Indeed, combining the wt 5′ss B1 (Fig. 4B) with a −15 G→T mutation, the ADAR exon was fully skipped (Fig. 4D, lane 2), validating our bioinformatic prediction. The same mutation combined with the stronger 5′ss B2 also decreased the selection of this exon (Fig. 4D, compare lanes 3 and 4). Thus, this position acts as an enhancer of site B. However, examination of the 227 Alu exons resulting from selection of site B revealed that only three cases shared this particular T→G mutation at position −15; this was not different in a statistically significant manner from the proportion of occurrences of this mutation in the intronic data set. This suggests that in the course of evolution, different mechanisms have independently developed among different Alu elements, governing splice site selection.
U1 snRNA binding to site C enhances selection of site B.
Thus far, we have established that the existence of a splice site at site C enhances selection of the more upstream site B. We now sought to determine whether this enhancing effect is mediated by binding of U1 snRNA to site C. For this purpose, we used a complementation assay with mutated U1 snRNAs that have a higher affinity for site C than for site B (Fig. 4C). In order to focus on cross talk between sites B and C without ESR interference, we used an ADAR2 Alu exon, with 5′ss B2 and a position −15 G→T point mutation. Although this mutation lowered the inclusion level of the Alu exon from almost 100% to 50%, combining it with C2 at site C restored a high inclusion level (Fig. 4D, compare lanes 3, 4, and 5). Replacing the strong 5′ss C2 with the weaker C3 in the presence of endogenous U1 snRNA resulted in only approximately 50% skipping, indicating that 5′ss C3 is not recognized by the endogenous U1 snRNA. However, complementation with U1C3 (Fig. 4C) completely restored the high inclusion level (Fig. 4D, compare lanes 6 and 7). The same results were obtained by using the weaker C4 site (Fig. 4D, compare lanes 8 and 9). Thus, two lines of evidence indicate that the enhancing function of site C is mediated through U1 snRNA. First, mutations that enhanced binding of endogenous U1 snRNA to site C enhanced site B selection (as evidenced by the increased inclusion with C2 with respect to that with C3). Second, complementation assays optimizing the base pairing between site C and U1 snRNA led to constitutive recognition of site B.
To further demonstrate the enhancing role of site C on the selection of site B, we created a construct with an extremely weak splice site, B4, followed by a weak splice site, C5 (Fig. 4B). This combination led to full skipping (Fig. 4D, lane 10). When this minigene was cotransfected with a mutated U1 cDNA complementary to C5, both sites B and C were activated, with similar inclusion levels of both sites (Fig. 4D, lane 11). This emphasizes that the mechanism of U1-mediated enhancement is finely tuned. When U1 bound site C with an affinity beneath a certain threshold, it led to enhancement of the selection of the adjacent site B; once this threshold was exceeded, binding of U1 to site C enhanced the selection of both 5′ss.
Two ESRs enhance selection of site A.
Whereas the ADAR minigene showed a predisposition for the selection of site B, PGT showed such a predisposition for site A. To understand the molecular basis for this predisposition, we compared the PGT Alu sequence to the ∼177,000 intronic Alu elements in the intronic data set. This analysis detected two positions located upstream of site A that differed considerably between the two groups: positions −22 and −17 (relative to 5′ss A) are both adenosines in PGT and cytosines in the intronic Alu elements (Fig. 5A). As previously shown (Fig. 3D, lane 3), activation of exonization in PGT, with site A selected, can be achieved by the strengthening of the 5′ss at site A from wt site A1 to site A2 (see Fig. 5B for a diagram depicting 5′ss strengths, and compare lanes 1 and 2 in Fig. 5C). An A→C point mutation at position −17 considerably decreased the inclusion level of the Alu exon (Fig. 5C, lane 3), and this effect was further enhanced when this mutation was combined with an additional one at position −22 (Fig. 5C, lane 4). Thus, these findings validate our bioinformatic prediction and further demonstrate the existence of a fine-tuned mechanism controlling splice site selection mediated by specific ESRs. Of the 13 instances in the exonic data set in which site A was selected, none were characterized by these two mutations, emphasizing, once again, the idiosyncratic nature of different Alu exonization events.
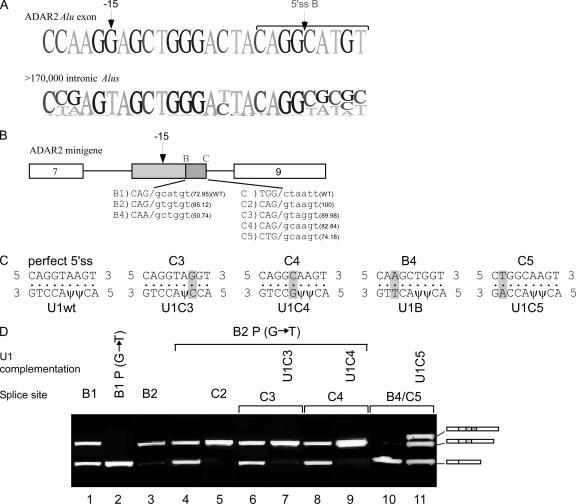
FIG. 5.
Selection of 5′ss A in ADAR2 Alu exon. (A) The upper sequence shows the last 26 nt and first 6 nt of the exon-intron junction of 5′ss A of the PGT Alu exon. The lower sequence is the sequence in the corresponding region of the alignment of ∼177,000 introns in the intronic data set. Positions −22 and −17 and a putative ESR are indicated above the exon sequence. (B) Diagram of PGT minigene. 5′ss A, selected for this experiment, is marked above the box. wt and mutated 5′ss are shown in the lower part, marked A1 and A2; Senapathy scores are given in parentheses. Position −17A upstream of 5′ss A is marked above the box. (C) Analysis of in vivo splicing assay as described in the legends of previous figures. Lane 1, splicing products of wt PGT gene; lanes 2 to 4, splicing products of exon 10 mutants containing an A2 site with the indicated mutations. (D) Diagram of ADAR2 minigene. The two alternative 5′ss positions (A and B) selected in this experiment are marked above the box. wt and mutated 5′ss, marked as A to A2, B0, and B1, are shown in the lower part; Senapathy scores are given in parentheses. (E) In vivo splicing assay as described for panel C. Lane 1, splicing products of wt ADAR2 gene; lanes 2 to 8, splicing products of exon 8 after mutation to eliminate 5′ss B (indicated as 5′ss B0), together with the indicated mutations; lanes 9 to 12, splicing products of exon 8 mutants containing 5′ss B1, together with the indicated mutations.
To further validate these findings, we created a 5′ss at site A of the ADAR2 Alu exon (Fig. 5D, A1 or A2) and eliminated the functional 5′ss at site B (by mutation from 5′ss B1 to B0). Creation of sites A1 and A2 was not sufficient for exonization (Fig. 5E, lanes 2 and 3), and neither was a single C→A mutation at position −22 (Fig. 5E, lane 4). However, a single C→A mutation at position −17 did result in a low level of alternative selection of site A in the ADAR minigene (Fig. 5E, lane 5). The alternative selection of site A was greatly increased when the mutations at positions −17 and −22 were combined (Fig. 5E, lane 7). This validates our findings regarding the ability of these two positions to enhance the selection of site A.
Having achieved alternative exonization of site A in the ADAR minigene, we next sought to determine how this site interacts with the wt, alternatively selected site B. The presence of a 5′ss at site A had little effect on 5′ss selection at site B (Fig. 5E, lanes 9 and 10, respectively). Interestingly, C→A mutations at positions −17 and −22 (as in the PGT Alu exon) resulted in constitutive selection of 5′ss B1, regardless of the presence of 5′ss A1 (Fig. 5E, compare lanes 11 and 12). This suggests that these two positions have a general silencing role that is not restricted to site A only but can also act upon site B, although to a lesser extent.
Environmental effect on exonization.
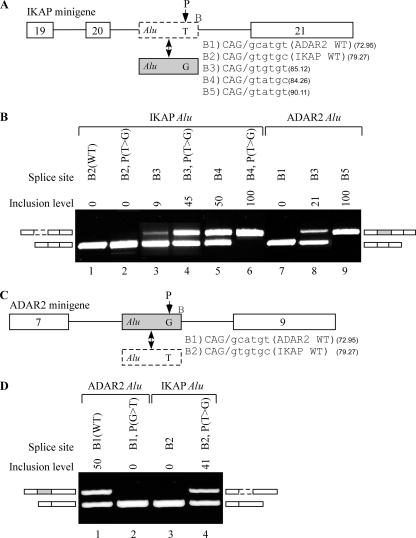
We next sought to further validate the importance of position −15 (upstream of 5′ss B) and the region surrounding it. In order to do so, we cloned another minigene generated from the human genomic sequence of the IKAP gene (IKBKAP). The IKAP minigene contains three constitutively spliced exons, exons 19 to 21, with an Alu sequence in intron 20 (Fig. 6A; the intronic Alu element is marked as an empty box with dashed borders). A mutation from T to C at position 6 in intron 20 of the IKAP gene leads to a tissue-specific skipping of exon 20, resulting in familial dysautonomia disease (1, 38).
FIG. 6.
Exonization of intronic Alu exon. (A) Diagram of the IKAP minigene, containing exons 19 to 21 (empty boxes) and an intronic Alu exon, shown by an empty dashed box in intron 20. The gray box represents the ADAR2 Alu exon. The five different B sites are listed to the right of the Alu box. The nucleotide at position −15 of 5′ss B is shown. (B) In vivo splicing assay as described in the legend to Fig. 2. Lane 1, splicing products of wt IKAP containing 5′ss B2; lanes 2 to 6, splicing products of IKAP minigene with the indicated mutations; lanes 7 to 9, splicing products of IKAP minigene in which the IKAP Alu intron was replaced with the ADAR2 Alu exon containing the wt 5′ss (B1), mutated B3, and B5, respectively. (C) Diagram of the ADAR2 minigene, containing exons 7 to 9. The Alu intron of the IKAP minigene is shown as a dashed box in the lower part of the panel. Two different 5′ss sequences are listed to the right of the Alu box. (D) In vivo splicing assay as described for panel B. Lane 1, splicing products of wt ADAR2 minigene, in which exon 8 contains 5′ss B1; lane 2, splicing products of ADAR2 minigene in which the Alu intron contains a −15 G→T mutation; lanes 3 and 4, splicing products of ADAR2 minigene in which the ADAR2 Alu exon was replaced with the potential IKAP Alu exon containing the indicated 5′ss and the position −15 G→T mutation.
The IKAP Alu element contains a 3′ss identical to the ADAR2 Alu exon 3′ss and a stronger 5′ss at site B (Fig. 6A). Yet the IKAP Alu element does not undergo exonization either under endogenous conditions or in the minigene (Fig. 6B, lane 1). We attempted to enhance the recognition of site B by introducing a T→G mutation at position −15, but this was not sufficient to achieve exonization (Fig. 6B, lane 2). A slight amount of exonization was achieved upon strengthening of the 5′ss at site B from B2 to B3 (Fig. 6B, lane 3), and we were able to substantially increase the selection of site B (from 9% to 45%) after introducing a T→G mutation at position −15 (Fig. 6B, lane 4). Additional strengthening of the 5′ss B site from B3 to B4 increased the inclusion level, and a combination of this strengthened 5′ss with the T→G mutation resulted in constitutive splicing of the Alu exon in the IKAP minigene (Fig. 6B, lanes 5 and 6, respectively). These results demonstrate two phenomena. First, position −15 relative to site B has a strong effect on selection of this site, and mutations at this position can result in exonization of an otherwise silent, intronic Alu element. Second, the fact that splice site B was not selected in the IKAP gene, even following a mutation at position −15, whereas its weaker counterpart in the ADAR gene was selected, suggests that factors other than site B and position −15 are involved in determining exonization.
To confirm this conclusion, we replaced the Alu exon residing within the IKAP minigene with its counterpart from the ADAR2 minigene (including its weak wt 5′ss B1). This resulted in full skipping of the ADAR2 Alu exon in the IKAP minigene, in stark contrast with its alternatively spliced pattern in the wt environment (Fig. 6B, lane 7). As in the case of the IKAP minigene, gradual strengthening of the ADAR2 Alu exon 5′ss (from 5′ss B1 to B3 and B5) resulted in gradual increases in its selection (from 0% to 21% to 100%) (Fig. 6B, lanes 7 to 9). To complement our findings, we replaced the Alu exon of the ADAR2 gene with the corresponding IKAP Alu sequence, including its putative wt 5′ss B2 (Fig. 6C). This exon was skipped (Fig. 6D, lane 3). However, exon selection was achieved via a T→ G mutation at position −15 (Fig. 6D, lane 4). This strongly contrasts with the complete skipping observed in the wt environment of the IKAP minigene. These two analyses demonstrate that while 5′ss strength, strongly modulated by position −15, has an important effect on inclusion levels, further factors not residing within the Alu exon, but presumably in sequences adjacent to it, determine the profile of the final transcript.
DISCUSSION
With over 1 million copies, Alu elements are the most abundant repetitive element in the human genome, comprising more than 10% of the human genome (22, 36). Alu elements enrich and diversify the primate transcriptome (2, 9) through exonization events (16, 25, 36, 42). Such events occur as a consequence of mutations accumulating within intronic Alu elements that lead the splicing machinery to select them as alternatively spliced, internal exons (36, 40). Close regulation of these exonization events is vital. Very low inclusion levels of the Alu exon will not serve to enrich the transcriptome, whereas very high inclusion levels will be deleterious since they will compromise the original transcriptomic repertoire.
Here we have combined bioinformatic and molecular methodologies to understand the requirements for Alu exonization events. Specifically, we have concentrated on the factors governing 5′ss selection, and we have found four main levels controlling 5′ss selection. The first, presumably most dominant, is 5′ss strength. The importance of 5′ss strength was underscored in our initial bioinformatic analysis, which showed that the strongest splice site tended to be selected. In the course of our molecular analysis, we consistently enhanced 5′ss selection by strengthening the 5′ss, in agreement with many previous studies (e.g. see references 24, 35, and 42). The second level is a U1-mediated interplay between adjacent splice sites. Such interplay was observed between sites B and C, with the latter enhancing the former, and also between sites A and B, with the latter repressing the former. U1 complementation assays demonstrated that these effects were mediated by the strength of base pairing between U1 and the 5′ss. The third level is specific ESRs. By means of a simple but efficient bioinformatic approach, we were able to identify several exonic positions that strongly affect splice site selection. The fact that the ESRs we identified in a particular Alu exon usually do not tend to appear among other, similarly characterized Alu exons suggests that different ESRs have evolved independently among different Alu exons. Finally, we have shown that within different environments, otherwise identical exons differ in their susceptibility to exonization.
The major discovery of this study is the U1-mediated interplay between the different splice sites. Scanning the literature, we found one previous study by Hwang and Cohen that implicated U1 snRNA in such a mechanism (17). In this study, the authors used complementation assays to demonstrate that U1 binding to a sequence downstream of the 5′ss enhances the selection of the 5′ss. However, in their study, they used an artificial exogenous U1 snRNA devoid of a GT/GC-binding catalytic site, and therefore the applicability of these findings to the choice of two adjacent splice sites under endogenous conditions was unclear. Our results strongly support and complement the model proposed by Hwang and Cohen by indicating that binding of U1 snRNA with an intact catalytic site to a potential splice site enhanced selection of a more upstream one. It appears that U1 snRNA may act to enhance the selection of an upstream 5′ss by binding a more downstream one, but once the strength of the downstream site has exceeded a certain threshold, U1 snRNA may also lead to selection—albeit not necessarily exclusive—of the more downstream site.
Thus, the impact of these findings may well exceed the relatively limited scope of Alu exonization. An analysis of >60,000 constitutive exons and >3,000 alternative exons in the human genome revealed that 5.6% of constitutive and 5% of alternative exons contain a 5′ss with a Senapathy strength of >70 within 20 nt of the selected 5′ss (the distance between 5′ss B and C is 20 nt). In these cases, as in Alu exons, binding of U1 snRNA to the downstream 5′ss may well play a part in determining 5′ss selection. Moreover, in a recent study searching for putative intronic splicing regulators based on sequence conservation across seven mammals in the 100 nt downstream of introns, a cluster of motifs highly resembling the 5′ss was found and was associated with alternative splicing (47). Based on these bioinformatic findings, it was suggested that U1 snRNP may be involved in the recognition of this motif, in the context of regulation of alternative splicing. Our analysis is in line with this prediction and validates it in the case of Alu exons.
Our work emphasizes the interplay between the above-mentioned factors determining exonization. We have demonstrated the interaction between strengths of splice sites and the presence of adjacent splice sites. Beyond a certain threshold, splice sites are selected regardless of the existence of a further splice site downstream. Beneath a certain threshold, splice sites are not selected regardless of the existence of a further splice site downstream. Between these two thresholds, an interplay exists, with the existence of one splice site affecting the selection of the other. We also demonstrated the complex interaction between strength, adjacent splice sites, and ESRs. ESRs can independently lead to alternative exonization, as can an interaction between adjacent splice sites, but when these two factors work in tandem they can lead to constitutive exonization, which can also be achieved by manipulation of splice site strength alone. Finally, we have shown how splice site strengths, interplay between adjacent splice sites, and ESRs lead to different results when superimposed upon different pre-mRNA environments. Within different environments, the thresholds of the various factors governing exonization may differ (e.g., the threshold governing exonization of a given splice site was shown to vary in two different minigenes). The ultimate profile of the transcript is determined by a combination of these four factors.
Combinatorial control of splicing, involving various levels of regulation, has been observed in other studies. Modafferi and Black examined how individual elements from the src gene combine to regulate splicing of a reporting exon and thus to determine its tissue specificity (30). Lee et al. characterized the splicing of NMDAR1 exon 21 and found that two RNA elements within this exon coregulated splicing repression of this exon in response to a neuronal stimulus (23). Similar results were found by Han et al.; they defined the roles of hnRNP proteins and the association of exon silencing with the UAGG and GGGG motifs, concluding that the multicomponent silencing code may play an important role in the tissue-specific regulation of the CI cassette exon (15). The interaction between weak splice sites and ESRs has also been the basis for computational prediction of exonic splicing enhancers (11).
The examples described above, as well as our findings in this study, indicate that splicing of every exon is a result of a complex interaction between multiple factors. This network can usually not be inferred using bioinformatic means, since sequences governing exonization events may differ from one exon to another. Alu exonization events provide a unique opportunity for understanding the minimal steps required for intronic Alu elements to become exonic sequences and therefore serve as an ideal platform with which to analyze multifactorial elements that govern exon selection. We have successfully taken advantage of this platform to bioinformatically identify specific ESRs leading to specific exonization events. Such techniques may be of particular importance for understanding the pathogenesis of Alu exonizations leading to diseases such as Alport syndrome, SLY syndrome, and OAT deficiency. Better understanding of the factors leading to exonization in these diseases may be of both diagnostic and, ultimately, therapeutic value.
Supplementary Material
Acknowledgments
We thank Iris Schlachter for her assistance in the experimental work, and we thank Hadas Keren and Eddo Kim for their critical readings of the manuscript.
This work was supported by grants from the Israel Science Foundation (40/05), MOP Germany-Israel, GIF, and DIP. O.R. is supported by EURASNET. S.S. is a fellow of the Edmond J. Safra bioinformatic program at Tel Aviv University.
Footnotes
Published ahead of print on 10 March 2008.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Anderson, S. L., R. Coli, I. W. Daly, E. A. Kichula, M. J. Rork, S. A. Volpi, J. Ekstein, and B. Y. Rubin. 2001. Familial dysautonomia is caused by mutations of the IKAP gene. Am. J. Hum. Genet. 68753-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ast, G. 2004. How did alternative splicing evolve? Nat. Rev. Genet. 5773-782. [DOI] [PubMed] [Google Scholar]
- 3.Black, D. L. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem. 72291-336. [DOI] [PubMed] [Google Scholar]
- 4.Black, D. L. 2000. Protein diversity from alternative splicing: a challenge for bioinformatics and post-genome biology. Cell 103367-370. [DOI] [PubMed] [Google Scholar]
- 5.Caceres, J. F., and A. R. Kornblihtt. 2002. Alternative splicing: multiple control mechanisms and involvement in human disease. Trends Genet. 18186-193. [DOI] [PubMed] [Google Scholar]
- 6.Caceres, J. F., S. Stamm, D. M. Helfman, and A. R. Krainer. 1994. Regulation of alternative splicing in vivo by overexpression of antagonistic splicing factors. Science 2651706-1709. [DOI] [PubMed] [Google Scholar]
- 7.Carmel, I., S. Tal, I. Vig, and G. Ast. 2004. Comparative analysis detects dependencies among the 5′ splice-site positions. RNA 10828-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chabot, B., C. LeBel, S. Hutchison, F. H. Nasim, and M. J. Simard. 2003. Heterogeneous nuclear ribonucleoprotein particle A/B proteins and the control of alternative splicing of the mammalian heterogeneous nuclear ribonucleoprotein particle A1 pre-mRNA. Prog. Mol. Subcell. Biol. 31 59-88. [DOI] [PubMed] [Google Scholar]
- 9.Cordaux, R., D. J. Hedges, S. W. Herke, and M. A. Batzer. 2006. Estimating the retrotransposition rate of human Alu elements. Gene 373134-137. [DOI] [PubMed] [Google Scholar]
- 10.Eperon, I. C., O. V. Makarova, A. Mayeda, S. H. Munroe, J. F. Caceres, D. G. Hayward, and A. R. Krainer. 2000. Selection of alternative 5′ splice sites: role of U1 snRNP and models for the antagonistic effects of SF2/ASF and hnRNP A1. Mol. Cell. Biol. 208303-8318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fairbrother, W. G., and L. A. Chasin. 2000. Human genomic sequences that inhibit splicing. Mol. Cell. Biol. 206816-6825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gabut, M., M. Mine, C. Marsac, M. Brivet, J. Tazi, and J. Soret. 2005. The SR protein SC35 is responsible for aberrant splicing of the E1α pyruvate dehydrogenase mRNA in a case of mental retardation with lactic acidosis. Mol. Cell. Biol. 253286-3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goren, A., O. Ram, M. Amit, H. Keren, G. Lev-Maor, I. Vig, T. Pupko, and G. Ast. 2006. Comparative analysis identifies exonic splicing regulatory sequences—the complex definition of enhancers and silencers. Mol. Cell 22769-781. [DOI] [PubMed] [Google Scholar]
- 14.Graveley, B. R. 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 17100-107. [DOI] [PubMed] [Google Scholar]
- 15.Han, K., G. Yeo, P. An, C. B. Burge, and P. J. Grabowski. 2005. A combinatorial code for splicing silencing: UAGG and GGGG motifs. PLoS Biol. 3e158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hasler, J., and K. Strub. 2006. Alu elements as regulators of gene expression. Nucleic Acids Res. 345491-5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hwang, D. Y., and J. B. Cohen. 1996. Base pairing at the 5′ splice site with U1 small nuclear RNA promotes splicing of the upstream intron but may be dispensable for slicing of the downstream intron. Mol. Cell. Biol. 163012-3022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Johnson, J. M., J. Castle, P. Garrett-Engele, Z. Kan, P. M. Loerch, C. D. Armour, R. Santos, E. E. Schadt, R. Stoughton, and D. D. Shoemaker. 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 3022141-2144. [DOI] [PubMed] [Google Scholar]
- 19.Jurka, J., V. V. Kapitonov, A. Pavlicek, P. Klonowski, O. Kohany, and J. Walichiewicz. 2005. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 110462-467. [DOI] [PubMed] [Google Scholar]
- 20.Kim, E., A. Magen, and G. Ast. 2007. Different levels of alternative splicing among eukaryotes. Nucleic Acids Res. 35125-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kruskal, J. B. 1983. An overview of sequence comparison, p. 1-44. In D. Sankoff and J. B. Kruskal (ed.), Time warps, string edits and macromolecules: the theory and practice of sequence comparison. Addison-Wesley Publishing Co., Reading, MA.
- 22.Lander, E. S., et al. 2001. Initial sequencing and analysis of the human genome. Nature 409860-921. [DOI] [PubMed] [Google Scholar]
- 23.Lee, J. A., Y. Xing, D. Nguyen, J. Xie, C. J. Lee, and D. L. Black. 2007. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 5e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lei, H., I. N. Day, and I. Vorechovsky. 2005. Exonization of AluYa5 in the human ACE gene requires mutations in both 3′ and 5′ splice sites and is facilitated by a conserved splicing enhancer. Nucleic Acids Res. 333897-3906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lev-Maor, G., R. Sorek, N. Shomron, and G. Ast. 2003. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 3001288-1291. [DOI] [PubMed] [Google Scholar]
- 26.Levy, A., N. Sela, and G. Ast. 2007. TranspoGene and microTranspoGene: transposed elements influence on the transcriptome of seven vertebrates and invertebrates. Nucleic Acids Res. 36D47-D52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim, L. P., and C. B. Burge. 2001. A computational analysis of sequence features involved in recognition of short introns. Proc. Natl. Acad. Sci. USA 9811193-11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Matlin, A. J., F. Clark, and C. W. Smith. 2005. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell. Biol. 6386-398. [DOI] [PubMed] [Google Scholar]
- 29.Mitchell, G. A., D. Labuda, G. Fontaine, J. M. Saudubray, J. P. Bonnefont, S. Lyonnet, L. C. Brody, G. Steel, C. Obie, and D. Valle. 1991. Splice-mediated insertion of an Alu sequence inactivates ornithine delta-aminotransferase: a role for Alu elements in human mutation. Proc. Natl. Acad. Sci. USA 88815-819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Modafferi, E. F., and D. L. Black. 1999. Combinatorial control of a neuron-specific exon. RNA 5687-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Modrek, B., and C. Lee. 2002. A genomic view of alternative splicing. Nat. Genet. 3013-19. [DOI] [PubMed] [Google Scholar]
- 32.Needleman, S. B., and C. D. Wunsch. 1970. J. Mol. Biol. 48443-453. [DOI] [PubMed] [Google Scholar]
- 33.Netzer, K. O., O. Pullig, U. Frei, J. Zhou, K. Tryggvason, and M. Weber. 1993. COL4A5 splice site mutation and alpha 5(IV) collagen mRNA in Alport syndrome. Kidney Int. 43486-492. [DOI] [PubMed] [Google Scholar]
- 34.Roca, X., R. Sachidanandam, and A. R. Krainer. 2005. Determinants of the inherent strength of human 5′ splice sites. RNA 11683-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roca, X., R. Sachidanandam, and A. R. Krainer. 2003. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 316321-6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sela, N., B. Mersch, N. Gal-Mark, G. Lev-Maor, A. Hotz-Wagenblatt, and G. Ast. 2007. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 8R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shapiro, M. B., and P. Senapathy. 1987. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 157155-7174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Slaugenhaupt, S. A., A. Blumenfeld, S. P. Gill, M. Leyne, J. Mull, M. P. Cuajungco, C. B. Liebert, B. Chadwick, M. Idelson, L. Reznik, C. Robbins, I. Makalowska, M. Brownstein, D. Krappmann, C. Scheidereit, C. Maayan, F. B. Axelrod, and J. F. Gusella. 2001. Tissue-specific expression of a splicing mutation in the IKBKAP gene causes familial dysautonomia. Am. J. Hum. Genet. 68598-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith, C. W., and J. Valcarcel. 2000. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends Biochem. Sci. 25381-388. [DOI] [PubMed] [Google Scholar]
- 40.Sorek, R. 2007. The birth of new exons: mechanisms and evolutionary consequences. RNA 131603-1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sorek, R., G. Ast, and D. Graur. 2002. Alu-containing exons are alternatively spliced. Genome Res. 121060-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorek, R., G. Lev-Maor, M. Reznik, T. Dagan, F. Belinky, D. Graur, and G. Ast. 2004. Minimal conditions for exonization of intronic sequences: 5′ splice site formation in Alu exons. Mol. Cell 14221-231. [DOI] [PubMed] [Google Scholar]
- 43.Spena, S., M. L. Tenchini, and E. Buratti. 2006. Cryptic splice site usage in exon 7 of the human fibrinogen Bbeta-chain gene is regulated by a naturally silent SF2/ASF binding site within this exon. RNA 12948-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stickeler, E., F. Kittrell, D. Medina, and S. M. Berget. 1999. Stage-specific changes in SR splicing factors and alternative splicing in mammary tumorigenesis. Oncogene 183574-3582. [DOI] [PubMed] [Google Scholar]
- 45.Sugnet, C. W., K. Srinivasan, T. A. Clark, G. O'Brien, M. S. Cline, H. Wang, A. Williams, D. Kulp, J. E. Blume, D. Haussler, and M. Ares. 2006. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput. Biol. 2e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sun, H., and L. A. Chasin. 2000. Multiple splicing defects in an intronic false exon. Mol. Cell. Biol. 206414-6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Voelker, R. B., and J. A. Berglund. 2007. A comprehensive computational characterization of conserved mammalian intronic sequences reveals conserved motifs associated with constitutive and alternative splicing. Genome Res. 171023-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamada, S., S. Tomatsu, W. S. Sly, R. Islam, D. A. Wenger, S. Fukuda, K. Sukegawa, and T. Orii. 1995. Four novel mutations in mucopolysaccharidosis type VII including a unique base substitution in exon 10 of the beta-glucuronidase gene that creates a novel 5′-splice site. Hum. Mol. Genet. 4651-655. [DOI] [PubMed] [Google Scholar]
- 49.Yang, X., M. R. Bani, S. J. Lu, S. Rowan, Y. Ben-David, and B. Chabot. 1994. The A1 and A1B proteins of heterogeneous nuclear ribonucleoparticles modulate 5′ splice site selection in vivo. Proc. Natl. Acad. Sci. USA 916924-6928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zavolan, M., and E. van Nimwegen. 2006. The types and prevalence of alternative splice forms. Curr. Opin. Struct. Biol. 16362-367. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.