Abstract
Although Bmal1 is a key component of the mammalian clock system, little is understood about the actual mechanism of circadian Bmal1 gene transcription, particularly at the chromatin level. Here we discovered a unique chromatin structure within the Bmal1 promoter. The RORE region, which is a critical cis element for the circadian regulation of the Bmal1 gene, is comprised of GC-rich open chromatin. The 3′-flanking region of the promoter inhibited rhythmic transcription in the reporter gene assay in vitro even in the presence of RORα and REV-ERBα. We also found that the nuclear matrix protein SAF-A binds to the 3′-flanking region with circadian timing, which was correlated with Bmal1 expression by footprinting in vivo. These results suggest that the unique chromatin structure containing SAF-A is required for the circadian transcriptional regulation of the Bmal1 gene in cells.
Circadian rhythms in behavior and physiology have an adaptive significance for living organisms from bacteria to humans and reflect the existence of an underlying intrinsic circadian oscillator or biological clock (10). The master clock that generates circadian rhythms in mammals is located in the suprachiasmatic nucleus (SCN) of the hypothalamus. In turn, peripheral clocks directly regulate many local rhythms that probably feed back to the SCN through hypothalamic integration (5). The molecular mechanism of the circadian oscillator consists of autoregulatory transcriptional and translational feedback loops that have both positive and negative elements (10). The key transcription factors, CLOCK and BMAL1, form heterodimers that bind to E-box enhancer sequences and activate the transcription of the three Period genes (Per1, Per2, and Per3) and two Cryptochrome genes (Cry1 and Cry2) (46). The PER and CRY proteins subsequently repress transcription at their own promoters through negative feedback caused by acting on the CLOCK-BMAL1 complex (35). This feedback loop system controls the central clock in the SCN and the peripheral clocks in most peripheral tissues (18).
Originally, BMAL1 (also known as MOP3) was characterized by high expression levels in brain and muscle cells (16). As the activity of Bmal1−/− mice immediately becomes completely arrhythmic in constant darkness, BMAL1 is apparently an essential and nonredundant component of the mammalian clock (6). The level of Bmal1 transcripts robustly oscillates in the SCN and in peripheral clock cells (25), and the circadian regulation of Bmal1 transcription contributes to the formation of interconnected feedback loops (35). The Bmal1 promoter contains two recognition motifs for ROR and REV-ERB orphan nuclear receptors (ROREs). Preitner et al. reported that REV-ERBα, which represses Bmal1 expression, is the major regulator of cyclic Bmal1 transcription (31), and Akashi and Takumi described that RORα acts to promote Bmal1 transcription (1). The opposing activities of the orphan nuclear receptors RORα and REV-ERBα are important in the maintenance of circadian clock function (34). It was also suggested that all members of the REV-ERB (α and β) and ROR (α, β, and γ) families are crucial components of the molecular circadian clock, with functional differences existing between various peripheral tissues (14). In addition, the transcriptional coactivator PGC-1α (22) and the transcriptional corepressor, the N-CoR/HDAC3 complex (44), integrate circadian Bmal1 expression.
Eukaryotic DNA is assembled into chromatin, the basic architecture of which comprises nucleosomes. The translational positions of nucleosomes depend on local variations in DNA curvature (30), helical periodicity, and/or nucleosome boundary effects (36). Nucleosomes in the transcriptional regulatory regions can act as a barrier against transcriptional initiation in a gene-specific manner (43). This repressive effect of nucleosomes is changed by rapid remodeling of the chromatin structure during gene activation, and the effect is spread throughout the entire locus. During the remodeling, histones are modified and unique regulatory events are established, which directly influence transcription in a process known as the histone code (39). Chromatin remodeling in clock genes also includes the rhythmic modification of histones. The acetylation of histone H3 (at Lys9 and Lys14) at Per1, Per2, and Cry1 and of H4 at Per1 during the transcriptional activation phase have been identified (8, 12). The di- and trimethylation of H3 (at Lys27) also proceeds at Per1 and Per2 during the repressive phase (13). The enzymes that catalyze these events are the ubiquitous histone acetyltransferase p300 (12), Sin3B (23), the histone deacetylases HDAC1 and HDAC2 (23), and the polycomb group protein EZH2 (13). In addition, CLOCK itself has histone acetyltransferase activity (9), and the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH is required for clock function in Neurospora spp. (3), suggesting clockwork-specific regulation. The chromatin structure undergoes rhythmic alterations at the Cry1 (12), Per2 (45), and frq (3) promoters. Therefore, the configurations of chromatin, in particular, those around cis-acting elements, are crucial for clock gene circadian expression. Although the transcriptional regulation of the Bmal1 gene, in particular, regulation by cis elements and transcription factors, appears to be apparent during circadian oscillations, the actual mechanism of Bmal1 gene expression at the chromatin level remains unclear, despite its functional importance.
We characterized the chromatin structure of the Bmal1 promoter and discovered that it participates in circadian transcriptional regulation.
MATERIALS AND METHODS
Cell culture.
NIH 3T3 cells were supplied by the Human Science Research Resources Bank (Osaka, Japan), and the stable lines were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and a mixture of penicillin and streptomycin in a humidified incubator at 37°C under a 5% CO2 atmosphere. To establish stable cell lines containing the luciferase reporter gene driven by the Bmal1 promoter, NIH 3T3 cells (5 × 106) were transfected with 10 μg of the luciferase reporter gene construct A (nucleotides −497 to +473), shown in Fig. 1A, and with 1 μg of pTracer-CMV (Invitrogen). Cells were selected in phleomycin D1 (Zeocin) (10 μg/ml), and clones were established as previously described (28).
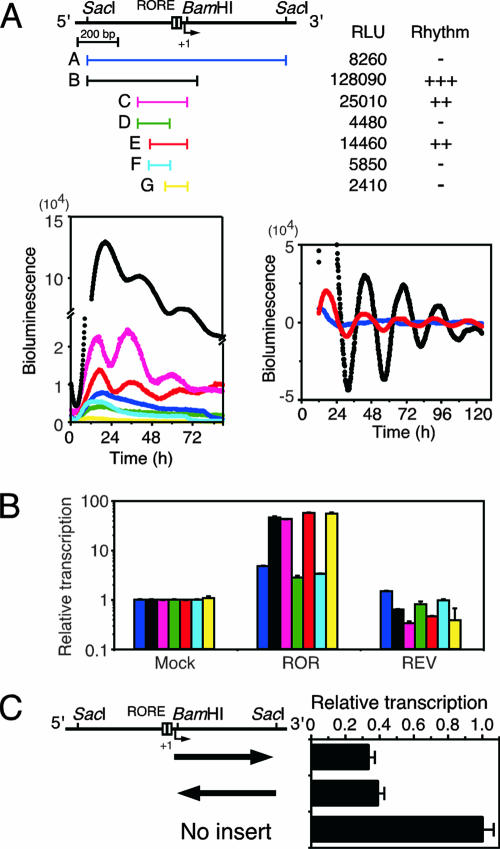
FIG. 1.
Effect of 3′-flanking region on Bmal1 transcription. (A) Oscillatory transcription is disturbed by the 3′-flanking region of the Bmal1 promoter. NIH 3T3 cells were transfected with the indicated Bmal1 promoter constructs and stimulated with dexamethasone, and then bioluminescence was measured. The analyzed regions were as follows: A, nucleotides −497 to +473 (blue); B, −497 to +74 (black); C, −275 to +27 (magenta); D, −275 to −74 (green); E, −197 to +27 (red); F, −197 to −74 (cyan); and G, −101 to +27 (yellow). Relative luciferase units (RLU) and rhythmicity results are summarized. Raw traces of all clones (bottom left) and detrended results for clones A, B, and E (bottom right) are representative of three independent experiments that generated similar results. (B) The effects of ROREs are diminished by the 3′-flanking region. Transcriptional assays were performed by using constructs containing regions A (blue), B (black), C (magenta), D (green), E (red), F (cyan), and G (yellow). RORα (ROR) and REV-ERBα (REV) expression plasmids were also introduced into NIH 3T3 cells. Normalized expression levels were calculated relative to the luciferase activities of mock transfectants. Values are means ± standard error of the mean (SEM) from triplicate assays. (C) Transcription of Bmal1 suppressed by the 3′-flanking region. The reporter plasmid pGL3- promoter, containing the 3′-flanking region (+27 to +473) and oriented as indicated by arrows, was transfected into NIH 3T3 cells. Normalized expression levels were calculated relative to the luciferase activity of pGL3-promoter. Values are means ± SEM from triplicate assays.
Real-time reporter gene assay.
The real-time reporter gene assay was performed as described previously (24). Fragments of DNA containing the Bmal1 promoter and its derivatives were cloned into pGL3-dLuc as reporter plasmids. NIH 3T3 cells transfected with these plasmids using PolyFect (Qiagen) were stimulated with 100 nM dexamethasone for 2 h and then incubated with Dulbecco's modified Eagle's medium containing 0.1 mM luciferin (Promega), 25 mM HEPES (pH 7.2), and 10% fetal bovine serum. Bioluminescence was measured and integrated for 1 min at intervals of 10 min using a Kronos AB-2500 (Atto).
Transient reporter gene assay.
Luciferase reporter gene plasmids and the internal control plasmid phRG-TK (Promega) were transfected into NIH 3T3 cells. The luciferase assay was performed using a dual-luciferase reporter assay system (Promega) as previously described (30). Transcriptional activities were normalized relative to Renilla luciferase activities.
Primer extension.
The transcription start site was determined by primer extension analysis as previously described (26). A 32P-labeled 20-mer antisense oligonucleotide (5′-ACACTCACCGTGGCTCGCTG-3′) corresponding to nucleotides +55 to +74 was synthesized, annealed to 5 μg of mouse poly(A)+ RNA, and extended with 20 units of avian myeloblastosis virus reverse transcriptase. The primer-extended DNA was resolved adjacent to a sequencing ladder primed with the same 32P-labeled 20-mer oligonucleotide by electrophoresis on 8% polyacrylamide-urea sequencing gel. Extended products were quantified using Image Gauge (Fujifilm).
Analysis of nucleosome positions using in vitro-reconstituted chromatin.
Nucleosomes were reconstituted in vitro as previously described (28). Briefly, nucleosome core particles were prepared from chicken erythrocytes as previously described (30). A 1,260-bp DNA fragment containing both the SacI fragment of the Bmal1 promoter and 290 bp of a luciferase gene fragment were prepared by PCR and were mixed with nucleosome core particles. The mixtures were dialyzed against 1 liter of TEP (10 mM Tris-Cl [pH 7.5], 0.1 mM EDTA, 0.1 mM phenylmethylsulfonyl fluoride [PMSF]) containing 0.4 M NaCl for 3 h and then against 1 liter of TEP with 16 mM NaCl overnight. The reconstituted nucleosomes were digested with micrococcal nuclease (MNase) at a final concentration of 15 units/ml at 25°C for 2, 5, and 10 min. DNA was purified and analyzed by Southern blotting.
Indirect end labeling.
Nucleosomal positions in vivo were determined by indirect end labeling as described by Bryan et al. (4), with a slight modification. Nuclei were isolated from NIH 3T3 cells and resuspended at a DNA concentration of 1 mg/ml in 15 mM Tris (pH 7.5), 15 mM NaCl, 60 mM KCl, 1 mM CaCl2, 0.34 M sucrose, 15 mM β-mercaptoethanol, and 0.5 mM spermidine. MNase (30 units/ml) was added, and the reaction was allowed to proceed for 10 min at 25°C. Genomic DNA was isolated and analyzed by Southern blotting.
Southern blotting.
After electrophoretic resolution on 2% agarose gels was performed, DNA was transferred onto a membrane, hybridized, and detected as previously described (28).
ChIP assay.
The chromatin immunoprecipitation (ChIP) assay was performed as previously described (29). Briefly, NIH 3T3 cells were incubated with 1% formaldehyde for 10 min at room temperature to cross-link proteins and DNA. The cells were lysed, and chromatin was fragmented and immunoprecipitated using antihistone antibody (Santa Cruz Biotechnology), anti-Myc antibody (Roche Diagnostics), anti-Flag antibody (Sigma), anti-hnRNP-U antibody (Sigma), and protein A/G-Sepharose (Santa Cruz Biotechnology). Immunocomplexes were eluted with 1% sodium dodecyl sulfate (SDS) and 0.1 M NaHCO3, and then cross-links were reversed by heating the DNA at 65°C for 4 h. The cross-links of input DNA samples were similarly reversed. DNA from the samples was purified and amplified by PCR. The primer sequences were as follows: for the nuclease-sensitive region, 5′-GAACGCGAATTGGTTTGGGTTGTCCG-3′ and 5′-ACACTCACCGTGGCTCGCTGCGAGC-3′; for the nuclease-resistant region, 5′-ACGGAGGTGCCTGTTTACCC-3′ and 5′-TTTAAGGGGCGCAGCCTC-3′; for G3PDH, 5′-ACCACAGTCCATTCCTACACAG-3′ and 5′-CTGGTCCTCTGTGTAAGCAAGGATGC-3′; and for luciferase, 5′-TCCGGTACTGTTGGTAAAGCCACCATG-3′ and 5′-ATATCGTTTCATAGCTTCTGCCAACCG-3′.
Nuclear “halo” assay.
“Halo” nuclei prepared by extraction with 2 M NaCl as previously described (37) were suspended in 10 mM Tris-Cl (pH 7.5), 2 mM MgCl2, and 0.5 mM PMSF and then digested with 100 units/ml of DNase I (Takara) for 30 min, 1 h, and 2 h at 37°C. After precipitation by centrifugation at 10,000 × g for 20 min, the remaining DNA was purified by proteinase K digestion and phenol extraction and analyzed by PCR. The primer sequences are as follows: for Bmal1, 5′-ACGGAGGTGCCTGTTTACCC-3′ and 5′-TCCAGAAGCTCCCGGGGATGTATC-3′, and for G3PDH, 5′-ACCACAGTCCATTCCTACACAG-3′ and 5′-CTGGTCCTCTGTGTAAGCAAGGATGC-3′.
CpG methylation analysis.
CpG islands were identified in the Bmal1 promoter using the algorithm at MethPrimer (www.urogene.org/methprimer) (21). Methylation analysis was performed by bisulfite modification using EpiTect bisulfite (Qiagen), followed by PCR cloning and sequence analysis.
Western blotting.
SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting were performed as previously described (27). Proteins were resolved by 7% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Amersham). Nonspecific binding was blocked with 3% dry milk in phosphate-buffered saline. Proteins were probed with anti-SAF-A antibody (Sigma) or anti-lamin A/C antibody (Santa Cruz Biotechnology) and then incubated with horseradish peroxidase-conjugated anti-mouse immunoglobulin G (Upstate). Immunoreactive proteins were visualized using ECL (Amersham) according to the manufacturer's instructions.
EMSA.
We performed the electrophoretic mobility shift assay (EMSA) as previously described (30). Briefly, DNA probes were end labeled with [γ-32P]ATP using T4 polynucleotide kinase (New England Biolabs). Portions (10 μl) of the DNA probe were suspended in 20 μl of 16 mM HEPES (pH 7.5), 150 mM KCl, 16% (vol/vol) glycerol, 1.6 mM MgCl2, 0.8 mM dithiothreitol, 0.4 mM PMSF, 1 mM EDTA, 0.8 mg/ml of bovine serum albumin, 0.06 mg/ml of poly(dI-dC), and 0.01% NP-40 and were incubated with proteins. Nuclear extracts were prepared from NIH 3T3 cells with or without exogenously overexpressed His6-tagged SAF-A protein as previously described (30). Protein complexes containing His6-tagged SAF-A were purified using Ni-nitrilotriacetic acid spin columns (Qiagen). Purified SAF-A/hnRNP-U protein was purchased from Vaxron (Rockaway, NJ). The anti-SAF-A antibody was added for supershift assays. These mixtures were resolved by electrophoresis on 4% polyacrylamide gels in 40 mM Tris-acetate, 1 mM EDTA, and 5% glycerol.
RT-PCR.
We performed reverse transcriptase PCR (RT-PCR) as previously described (28) using the primer sets as follows: for Bmal1, 5′-GGCCGAATGATTGCTGAGGAAATCATGG-3′ and 5′-TTACAGCGGCCATGGCAAGTCACTAAAG-3′, and for G3PDH, 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′. The PCR fragments were resolved by electrophoresis on 2% agarose gels and visualized by ethidium bromide staining.
Real-time quantitative PCR.
Real-time quantitative PCR was performed using LightCycler (Roche) with a LightCycler FastStart DNA master SYBR green I kit (Roche), and the primer sets were the same as those used in the ordinary PCR. PCR products cloned into the pGEM-T Easy vector (Promega) were used as an authentic template. Relative expression levels were evaluated using LightCycler software, version 3.5.
LM-PCR.
Ligation-mediated PCR (LM-PCR) was performed as previously described (29). Nuclei were isolated and resuspended at a DNA concentration of 1 mg/ml in 15 mM Tris (pH 7.5), 15 mM NaCl, 60 mM KCl, 1 mM CaCl2, 0.34 M sucrose, 15 mM β-mercaptoethanol, and 0.5 mM spermidine. MNase (30 units/ml) was added, and the reaction was allowed to proceed for 10 min at 25°C. Genomic DNA was purified and used for first-strand synthesis extending up to the cleaved sites, using first-synthesis primers (nuclease-sensitive region, 5′-ACACTCACCGTGGCTCGCTGCGAGC-3′; nuclease-resistant region, 5′-CCGGCGGGGAGCGGATTGGTCG-3′). The synthesized DNA was ligated to a double-stranded linker (sense, 5′-GAATTCAGATC-3′; antisense, 5′-GCGGTGACCCGGGAGATCTGAATTC-3′) and amplified by PCR using the linker antisense oligonucleotide and forward primers (nuclease-sensitive region, 5′-GCACCCGCACTCGGATCCCGCGG-3′; nuclease-resistant region, 5′-ACGGAGGTGCCTGTTTACC-3′). The amplified fragments were linearly amplified using a 32P-labeled extension primer (nuclease-sensitive region, 5′-AAGTCCGGCGCGGGTAAACAG-3′; upstream nuclease-resistant region, 5′-TGTTTACCCGCGCCGGACTTG-3′; downstream nuclease-resistant region, 5′-ACGGTGAGTGTCGGCATGGC-3′), and the amplified fragments were resolved on 8% denaturing polyacrylamide gels.
RESULTS
The 3′-flanking region of the Bmal1 promoter inhibits oscillatory transcription.
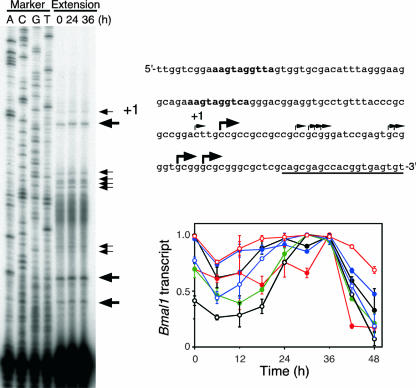
We determined the minimal promoter region required for rhythmic expression of the Bmal1 gene using real-time reporter gene assays. Analysis of transcriptional regulation using various constructs containing the Bmal1 promoter region showed that construct E (nucleotides −197 to +27) displayed a weak but clear oscillation and thus comprised the minimal required region (Fig. 1A). In addition, the period of the transcriptional rhythm was shorter for construct E than for construct B (−497 to +74) (for construct E, 24.28 ± 0.77 h; for construct B, 26.17 ± 1.04 h; P < 0.05, t test). Construct G (−101 to +27) had weak transcriptional activity without rhythmicity, although it included ROREs. Preitner et al. have reported that Bmal1 transcription initiates at various sites in mouse liver (31), and we also observed multiple transcriptional initiations in NIH 3T3 cells stimulated with dexamethasone (Fig. 2). Expression profiles of each transcript were scattered, whereas all transcripts were expressed at the highest level 36 h after dexamethasone stimulation (Fig. 2). These results suggest that several regulatory elements are involved in oscillatory Bmal1 transcription. The transcriptional activity of construct A (−497 to +473), which contained an additional 3′-flanking region (+75 to +473), was surprisingly weak and not at all rhythmic compared with the activity of construct B (−497 to +74) (Fig. 1A). This suggests that the 3′-flanking region plays a critical role in rhythmic Bmal1 transcription. We examined transcriptional activity in NIH 3T3 cells coexpressed with RORα or REV-ERBα to evaluate the relationship between the effect of the 3′-flanking region and the effect of the ROREs. Whereas the exogenous expression of RORα and that of REV-ERBα caused the predicted approximately 47-fold increase and 50% decrease, respectively, in the transcriptional activity of construct B, RORα and REV-ERBα were less effective in construct A, exerting a fivefold increase and no decrease, respectively (Fig. 1B). These data suggest that the 3′-flanking region plays an important role in RORE-dependent circadian expression. We analyzed reporter constructs with the simian virus 40 promoter connected with the 3′-flanking region (nucleotides +27 to +473) and found that the promoter activities of these constructs decreased to ∼40% of the simian virus 40 promoter activity independently of the direction in which the 3′-flanking region was oriented (Fig. 1C). These results suggest that the 3′-flanking region inhibits the promoter activity but also damps both the positive and the negative effects upon the ROREs independently of orientation.
FIG. 2.
Analysis of transcription initiation sites. After stimulation with 100 nM dexamethasone for 2 h, NIH 3T3 cells were incubated for the indicated times, and mRNA was prepared for primer extension analysis (left). Arrows show the positions of the extended products. Primers for extension and ROREs are underlined and in bold, respectively (top right). The transcription initiation site of the longest product is designated nucleotide +1. The band intensity was measured, and the relative amounts of products are shown (bottom right).
Unique chromatin structure of the Bmal1 promoter.
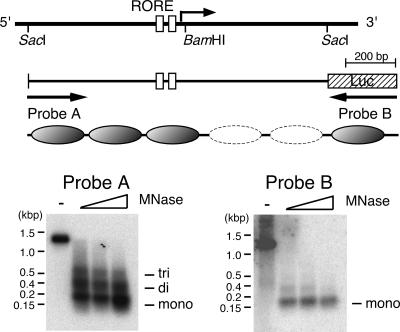
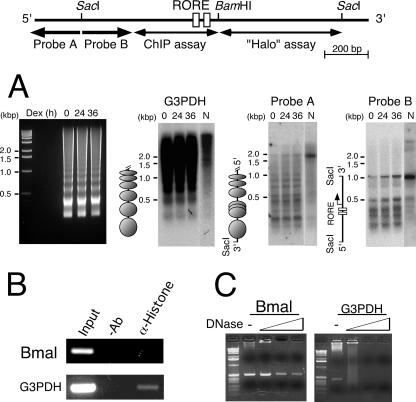
The Bmal1 gene is transcribed in NIH 3T3 cells with circadian oscillation even when the gene contains the 3′-flanking region (see Fig. 8A), suggesting that the chromatin structure around this region is important. In addition, the nucleotide sequence of the Bmal1 promoter region is GC rich (66%) without TATA boxes, implying a unique chromatin structure. GC-rich chromatin, such as CpG islands, differs in three aspects from bulk chromatin: histone H1 is present in very small amounts, histones H3 and H4 are highly acetylated, and nucleosome-free regions are present (41). Chromatin reconstituted in vitro by using the Bmal1 promoter region (nucleotides −497 to +473) generated tri-nucleosomes from the 5′ SacI site (Fig. 3, probe A) but no nucleosomes at the 3′-flanking region (Fig. 3, probe B), suggesting that the 3′-flanking region has the intrinsic and unique task of disrupting nucleosome formation. We analyzed the chromatin structure of this region in NIH 3T3 cells and found that the 200-bp ladder that is characteristic of the G3PDH gene was located in the region upstream from the 5′ SacI site of the Bmal1 promoter, indicating a normal nucleosome structure (Fig. 4A, probe A). In probe B in Fig. 4A, the promoter region around the ROREs was hypersensitive to MNase, whereas the 3′-flanking region was completely protected from nuclease digestion. These results suggested that the region around the ROREs is an open chromatin structure and that the 3′-flanking region is wrapped in some proteins.
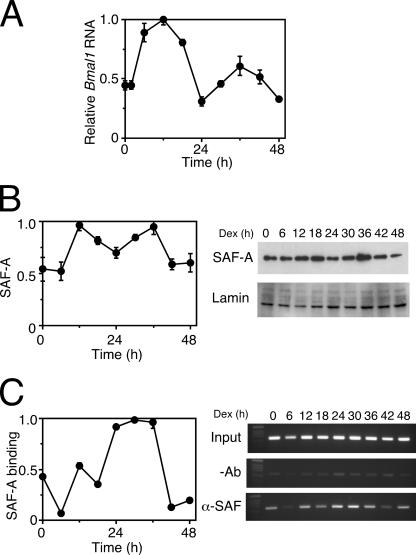
FIG. 8.
The binding of SAF-A correlates with Bmal1 transcription. (A) Oscillatory transcription of the Bmal1 gene. NIH 3T3 cells were stimulated with 100 nM dexamethasone, and then transcripts were analyzed by a real-time quantitative RT-PCR. The levels of RNA were normalized to the level of G3PDH expression, and the peak value was set at 1. Values are means ± SEM from triplicate assays. (B) SAF-A protein in the NIH 3T3 cell nuclei. NIH 3T3 cells were stimulated with 100 nM dexamethasone (Dex), and then nuclei were isolated and analyzed by Western blotting. The levels of SAF-A protein were normalized to lamin protein, and the peak value was set to 1. Values are means ± SEM from triplicate assays. SAF-A, with anti-SAF-A antibody; Lamin, with anti-lamin A/C antibody. (C) Oscillatory binding of SAF-A to the 3′-1 region. After stimulation with dexamethasone, NIH 3T3 cells were analyzed by ChIP assays of the 3′-1 region, followed by a real-time quantitative PCR (left). PCR products were also analyzed on a 2% agarose gel (right). Binding values were normalized to input DNAs, and the peak value was set at 1. Values are means ± SEM from triplicate assays. −Ab, without antibody; α-SAF-A, with anti-SAF-A antibody.
FIG. 3.
The 3′-flanking region of the Bmal1 promoter prevents nucleosome formation in vitro. Nucleosomes were reconstituted in vitro using 1,260-bp DNA fragments; digested with MNase at a final concentration of 15 units/ml at 25°C for 2, 5, and 10 min; and analyzed by Southern blotting using probe A (nucleotides −497 to −236). Fragments on the same membrane were rehybridized with probe B (290-bp luciferase [Luc] gene fragment). Deduced nucleosome positions in reconstituted chromatin are shown as ovals. The dashed ovals show the region that has little nucleosome structure. tri, di, and mono, tri-, di-, and mononucleosome respectively.
FIG. 4.
Unique chromatin structure of the Bmal1 promoter in vivo. (A) Nucleosome positions in the Bmal1 promoter. NIH 3T3 cells were cultured for the indicated periods after stimulation with dexamethasone (Dex), and then isolated nuclei and naked genomes were incubated with MNase. DNA was purified, digested with SacI, and blotted onto the membrane that also hybridized with probes A (nucleotides −691 to −497), B (−497 to −236), and G3PDH. The image of the agarose gel which was used for the Southern blotting is shown in the left panel. Deduced nucleosome positions in vivo are shown as ovals. (B) Nucleosomes are not featured in the 5′-flanking region around ROREs. ChIP assays using NIH 3T3 cells were performed without antibody (−Ab) and with antihistone antibody (α-Histone). (C) The intranuclear 3′-flanking region of the Bmal1 promoter is protected from DNase I digestion. “Halo” nuclei were prepared from NIH 3T3 cells and digested with DNase I, and purified residual DNA was used as a template for PCR. The control was G3PDH.
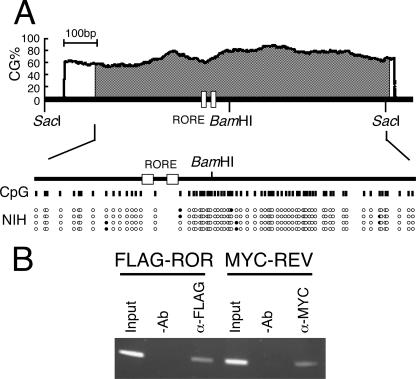
To confirm this notion, we analyzed the promoter region by ChIP assays using an antihistone antibody. The G3PDH gene was amplified, whereas the Bmal1 promoter region was not, suggesting that the Bmal1 promoter region around the ROREs has few nucleosomes (Fig. 4B). It has been shown that the CpG island is undermethylated when the gene is active and extensively methylated when the gene is inactive (42). In fact, bisulfite sequencing showed a hypomethylated Bmal1 promoter region with a CpG island in NIH 3T3 cells (Fig. 5A). ChIP assays using NIH 3T3 cells expressing FLAG-RORα or MYC-REV-ERBα showed that both of these transcription factors interacted with the ROREs (Fig. 5B). Taken together, these results suggest that the ROREs in the Bmal1 promoter of NIH 3T3 cells are open structures, which allow access by transcription factors such as RORα and REV-ERBα.
FIG. 5.
Tandem ROREs are accessible for transcriptional regulators in vivo. (A) Tandem ROREs in the hypomethylated CpG island. The examination of the Bmal1 promoter sequence revealed a prominent CpG island situated at nucleotides −351 to +494 (shaded area) using the algorithm at MethPrimer (www.urogene.org/methprimer). The genomic sequence of NIH 3T3 cells was analyzed after modification with bisulfite. Vertical lines indicate CpG sites in the Bmal1 promoter region. Filled and open circles indicate methylated and unmethylated CpG sites, respectively. (B) Binding of transcriptional regulators to tandem ROREs in NIH 3T3 cells. Flag-RORα or Myc-REV-ERBα was expressed in NIH 3T3 cells and analyzed by ChIP assays 24 h later, using anti-Flag and anti-Myc antibodies. −Ab, without antibody; α-Flag, with anti-Flag antibody; α-Myc, with anti-Myc antibody.
A long-range region beyond 500 bp protected from nuclease attack generally suggests the existence of the nuclear matrix. We therefore prepared a nuclear “halo” using NIH 3T3 cells digested with DNase I. We analyzed the remaining DNA in which the matrix-associated region was concentrated from the viewpoint of the presence or absence of a 3′-flanking region. The 3′-flanking region was retained even under the most stringent digestion conditions (2 h at 37°C) with DNase I (Fig. 4C). On the other hand, the G3PDH gene, which generates nucleosomes (Fig. 4A), disappeared under the stringent conditions as expected (Fig. 4C). Multiple barriers against nuclease digestion and resistance to dissociation by high concentrations of salt suggest that some proteins are tightly bound to the 3′-flanking region, which is a unique chromatin structure similar to the nuclear matrix but not to the nucleosomes of bulk chromatin observed in the G3PDH gene. In summary, the Bmal1 promoter region comprises mainly three parts: a general nucleosome structure upstream from the 5′ SacI site, an open chromatin structure around the ROREs, and a nuclear matrix-like structure at the 3′-flanking region.
The nuclear matrix protein SAF-A binds directly to the 3′-flanking region.
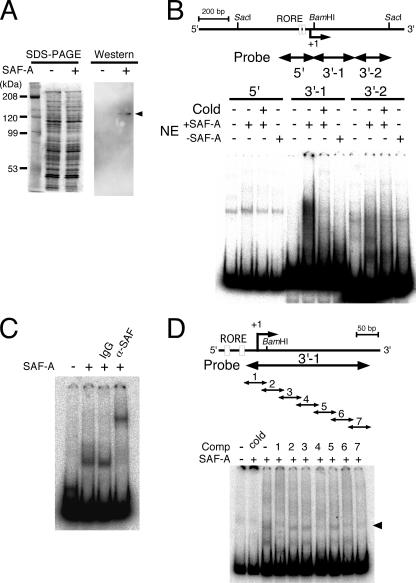
To identify the protein that binds to the 3′-flanking region, we investigated the activities of SAF-A (also known as hnRNP-U), which is a nuclear matrix protein (32) with circadian expression in the central pacemaker of circadian clocks in the brain, namely, the SCN (40). We prepared protein complexes from NIH 3T3 cells with or without the His6-tagged SAF-A expression plasmid using a Ni-nitrilotriacetic acid column (Fig. 6A). EMSAs using these complexes indicated that the SAF-A protein complex specifically bound to the 3′-1 probe (nucleotides −27 to +266) but not to the 3′-2 probe (nucleotides +262 to +473) in the 3′-flanking region (Fig. 6B). The broad, shifted band with a higher molecular weight than that shown in Fig. 6C might reflect variety among SAF-A protein complexes (lane 6 in Fig. 6B). When purified SAF-A protein was analyzed by EMSA using the 3′-1 probe, a band clearly supershifted after incubation with the anti-SAF-A antibody (Fig. 6C). To minimize the binding sequence in the 3′-1 region, the EMSA was performed using serial 50-bp competitors of various regions of the 3′-1 probe sequence. When competitors 1 (−27 to +23), 4 (+94 to +143), 6 (+174 to +223), and 7 (+214 to +263) were added to the reaction mixture, the sifted band disappeared (Fig. 6D), indicating that these regions (−27 to +23, +94 to +143, +174 to +223, and +214 to +263) are important for SAF-A binding. Taken together, these results indicate that SAF-A protein directly binds to the 3′-1 region (−27 to +266).
FIG. 6.
SAF-A binds to the region protected from DNase I digestion. (A) Nuclear complex containing SAF-A. Nuclear complexes prepared from NIH 3T3 cells with (+) or without (−) His6-tagged SAF-A expression plasmids were resolved by 7% SDS-PAGE and Western blotting against anti-SAF-A antibody. Arrowhead, SAF-A. (B) SAF-A complexes bound to the 3′-1 region. The EMSA was performed using SAF-A-containing nuclear complex. The positions of probes 5′ (nucleotides −197 to +39), 3′-1 (−27 to +266), and 3′-2 (+262 to +473) are shown as arrows. Cold, 200-fold molar excess of unlabeled probe; NE, nuclear complex with (+SAF-A) or without (−SAF-A) SAF-A. (C) SAF-A bound directly to the 3′-1 region. The EMSA was performed using the 3′-1 probe and purified SAF-A protein. IgG, control immunoglobulin G; α-SAF-A, anti-SAF-A antibody. (D) SAF-A binding sites in the 3′-1 region were determined by EMSA using purified SAF-A protein and a 200-fold molar excess of competitors (Comp). Arrows show the positions of the probe and competitors (1 to 7). Competitors were as follows: cold, nucleotides −27 to +266; 1, −27 to +23; 2, +14 to +63; 3, +54 to +103; 4, +94 to +143; 5, +134 to +183; 6, +174 to +223; and 7, +214 to +263. Arrowhead, shifted band.
SAF-A is required for circadian expression.
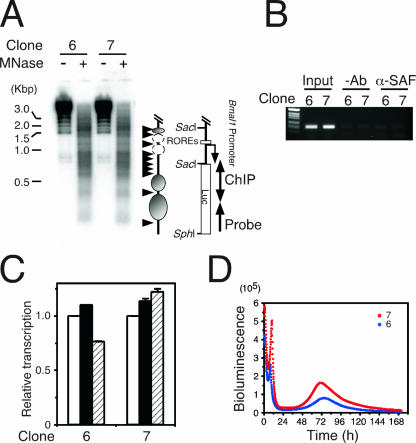
To confirm the role of SAF-A in the circadian regulation of Bmal1 expression, we established stable clones of NIH 3T3 cells transfected with reporter construct A (nucleotides −497 to +473) (Fig. 1A). Six of 10 clones had promoter activity, and 2 with high levels of luciferase activity (clones 6 and 7) were further analyzed. Indirect end labeling with MNase digestion showed that clones 6 and 7 displayed similar images and that the 3′-flanking region observed as a protected region in parental NIH 3T3 cells (probe B in Fig. 4A) was hypersensitive to MNase in the integrated gene (Fig. 7A). The ChIP assay indicated that SAF-A is not located in the 3′-flanking region of the integrated gene in the stable cell lines (Fig. 7B). Reporter gene assays indicated that RORE functions were incomplete in both clones (Fig. 7C). These results strengthened the notion that SAF-A and the ROREs function cooperatively through the chromatin structure. Although endogenous Bmal1 transcription was circadian in both clones (data not shown), real-time reporter gene assays showed that the oscillation of the reporter gene was not circadian in either clone (Fig. 7D), suggesting that SAF-A is required for circadian regulation. Notably, the reporter gene did not oscillate in a circadian manner in any clones.
FIG. 7.
SAF-A is involved in oscillatory Bmal1 transcription. (A) Analysis of the chromatin structure of integrated genes. Indirect end labeling was performed using stable clones 6 and 7. Arrow, probe for Southern blotting; arrowheads, MNase-sensitive sites; ovals, nucleosomes; dashed ovals, nucleosomes in small amounts. Luc, luciferase. (B) SAF-A does not bind to the 3′-end-flanking region of the integrated gene. Stable clones 6 and 7 were analyzed using ChIP assays with anti-SAF-A antibody, and integrated gene-specific or luciferase (A) regions were analyzed by PCR. −Ab, without antibody; α-SAF-A, with anti-SAF-A antibody. (C) The functions of ROREs are inhibited in integrated genes. RORα (filled bars) and REV-ERBα (hatched bars) expression plasmids were introduced into stable clones 6 and 7 and analyzed by luciferase gene assays. Normalized expression levels were calculated relative to the luciferase activities of mock transfectants (open bars). Values are means ± SEM from triplicate assays. (D) Transcription from the promoter in integrated genes without circadian oscillation. Stable clones 6 (blue) and 7 (red) were analyzed by real-time reporter gene assays. Actual bioluminescence is shown.
The binding of SAF-A correlates with Bmal1 gene expression.
SAF-A protein and Bmal1 show similar phase-angle and oscillation patterns (40). We therefore postulated that SAF-A plays an important role in Bmal1 oscillation in the cell-autonomous core clock. To test this hypothesis, we analyzed the temporal expression profile of the SAF-A protein in NIH 3T3 cells after stimulation with 100 nM dexamethasone. Figure 8A shows that the Bmal1 gene was rhythmically expressed, with a peak at 36 h and a trough at 24 h in NIH 3T3 cells after stimulation with dexamethasone. Western blots using the anti-SAF-A antibody also yielded similar results for SAF-A protein expression in the nuclei of NIH 3T3 cells (Fig. 8B). We then examined the temporal binding of endogenous SAF-A on the 3′-1 region by ChIP assays, and the binding profile of SAF-A to the 3′-1 region was similar to those of both the SAF-A protein levels and Bmal1 gene expression (Fig. 8C). The correlation among Bmal1 gene expression, nuclear SAF-A protein levels, and SAF-A binding to the 3′-1 region suggests that SAF-A plays an important role in augmenting the rhythmic expression of the Bmal1 gene.
Rhythmic alteration of the chromatin structure.
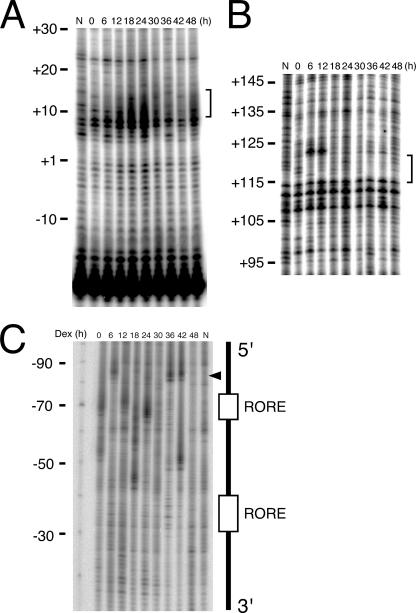
The rhythmic binding of the SAF-A protein to the 3′-1 region means rhythmic changes in structural component(s), including the chromatin structure. We evaluated such changes by using LM-PCR of NIH 3T3 cells stimulated with dexamethasone. The bracketed regions of competitors 1 (nucleotides −27 to +23) and 4 (nucleotides +94 to +143) in Fig. 6 showed a clear rhythmic pattern, with a peak of footprints at 24 h and troughs at 6 h and at 36 to 42 h, which were consistent with the timing of SAF-A binding to the 3′-1 region (Fig. 9A and B). The same regions of competitors 6 (+174 to +223) and 7 (+214 to +263) were not analyzed because we do not have appropriate sequences for the LM-PCR primer. Considering that the 3′-1 region inhibited the ROREs (Fig. 1), we also analyzed the RORE regions to understand the effect of the chromatin alterations at the 3′-1 region upon the ROREs. The digested bands adjacent to the upstream RORE showed a circadian cycle, indicating that the circadian changes of the chromatin structure had also occurred around the ROREs (Fig. 9C). The correlation between SAF-A binding and the altered chromatin structure suggests that SAF-A participates in the regulation of circadian Bmal1 expression through chromatin alteration in the cell-autonomous core clock.
FIG. 9.
Rhythmic alterations of chromatin structure around the Bmal1 promoter. Footprinting in vivo in the region corresponding to competitors 1 (A) and 4 (B). Brackets indicate regions showing rhythmic footprinting alterations. (C) Footprinting in vivo around the ROREs. The arrowhead indicates bands with rhythmic changes of intensity. Numbers and “N” above the gels indicate time after stimulation and naked DNA, respectively. Results are representative of three independent experiments that generated similar results. Dex, dexamethasone.
DISCUSSION
Unique chromatin in the Bmal1 promoter participates in Bmal1 oscillation.
Chromatin is a fundamental architecture in the nucleus that is thought to be involved in the regulation of gene transcription. We discovered using the β-globin gene that nucleosomes, the most basic units of chromatin, modulate enhancer activity (28) and chromatin remodeling (29). Many recent reports have noted that histone tail modifications are involved in clock gene regulation, implicating the importance of the chromatin structure in clock gene regulation (2). The ROREs of the Bmal1 promoter are surrounded by a unique open chromatin structure (Fig. 4A and B), and this might be related to the intrinsic characteristics of the DNA sequence of the Bmal1 promoter, such as the high G/C content (66%) and the CpG island (Fig. 3 and 5A). Tazi and Bird reported that CpG islands consist of a 100- to 200-bp region that is free of nucleosomes and thus hypersensitive to nuclease, and the flanking nucleosomes with the highly acetylated histones H3 and H4 and less histone H1 (41) indicated a transcriptionally active state. This is consistent with our observation of the chromatin structure around the ROREs, and transcriptional regulators such as RORα and REV-ERBα can access their cognate motifs in the hypomethylated region (Fig. 5B) and exert their functions (Fig. 1A and B). The Bmal1 promoter also has multiple transcription initiation sites (Fig. 2) without a TATA sequence, suggesting similarity with housekeeping genes (42). Other clock genes, such as Per1, Per2, and Per3, also have high G/C contents (63%, 60.5%, and 75%, respectively) at the promoter region. Furthermore, the CpG island in the Per1 promoter is hypomethylated (15), and the E2 enhancer at the Per2 promoter is hypersensitive to DNase I (45). These data indicate that chromatin structures are common to all clock gene promoters.
Functional cis elements in the Bmal1 promoter.
Real-time reporter gene assays showed that construct E (nucleotides −197 to +27) (Fig. 1A) was the minimal region required for circadian Bmal1 transcription. However, its circadian period was shorter than that of construct B (−497 to +74) (Fig. 1A), suggesting the presence of a cis element(s) other than ROREs within the region of nucleotides −497 to −198 (Fig. 1A). Yu et al. found several cis elements within this region and reported that Bmal1 transcription is activated by CRY proteins and PER2 and is repressed by the BMAL1-CLOCK heterodimer (47). We also confirmed the same effects in this region (data not shown), but the target element(s) remains unknown. On the other hand, as the 3′-flanking region of the Bmal1 promoter (+27 to +473) inhibited both the positive and the negative effects of the ROREs (Fig. 1B) and basal transcription (Fig. 1C), the exclusion of such inhibitory effects is important for oscillatory Bmal1 transcription by the 3′-flanking region. The structure of the 3′-flanking region was rigid and similar to that of the nuclear matrix (Fig. 4A and C), the function of which is to neutralize the inhibitory effects of the 3′-flanking region on the transcriptional circadian regulation of Bmal1.
SAF-A is involved in the circadian regulation of the Bmal1 gene.
We showed that SAF-A directly binds to the 3′-flanking region in the Bmal1 promoter (Fig. 6). Nevertheless, the rigid structure at the 3′-flanking region might not be the nuclear matrix because the region is composed of GC-rich sequences, whereas the DNA regions that anchor chromosomal DNA to the nuclear matrix are usually AT rich (7).
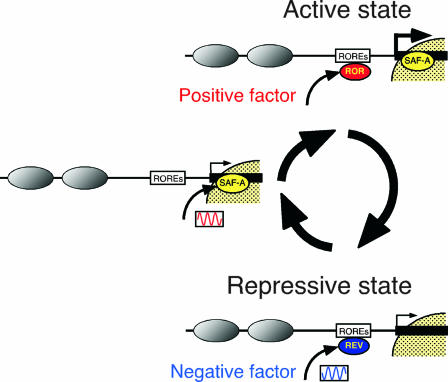
SAF-A/hnRNP-U has several possible roles in circadian gene expression. The first role is the regulation of mRNA turnover (48), since, for example, members of the hnRNP family, but not SAF-A, regulate the circadian oscillation of the mRNA for serotonin N-acetyltransferase (20). SAF-A binds to and modifies the function of the glucocorticoid receptor (GR) (11), for which the ligand or glucocorticoid is the major cue for circadian oscillation. We confirmed that SAF-A binds to the genomic DNA (Fig. 8C), but our ChIP assays did not uncover GR binding to the Bmal1 promoter (data not shown). However, the regulation of mRNA turnover and the modulation of GR function by SAF-A should be elucidated in the future. Lastly, we discuss the function of SAF-A as a component of the chromatin structure. The oscillatory profile of SAF-A expression and binding was similar to that of Bmal1 transcription in NIH 3T3 cells (Fig. 8), which is consistent with the findings for the SCN (40), suggesting that SAF-A plays an important role in the cell-autonomous clocks. This implies that SAF-A binding to the 3′-flanking region affects RORα's binding to the ROREs (Fig. 10). The importance of SAF-A in Bmal1 circadian transcription was also confirmed, using the stable cell lines with an integrated Bmal1 reporter gene (Fig. 7). In our preliminary experiments, the SAF-A homologue knockdown flies had a longer-period rhythm (not shown), suggesting the importance of SAF-A to circadian rhythms.
FIG. 10.
Model of transcriptional regulation of the Bmal1 gene. The Bmal1 promoter region has a unique chromatin structure, with open chromatin around ROREs and a nuclear matrix-like structure at the 3′-flanking region. First, SAF-A protein binds to the 3′-flanking region with circadian rhythm and alters the local chromatin structure. Thereafter, RORα (ROR) binds to ROREs and activates transcription. In contrast, SAF-A protein deviates from the 3′-flanking region under a repressive state and gains changes in the chromatin structure. Thereafter, the negative transcription factor REV-ERBα (REV) binds to ROREs with the reverse rhythmic phase of SAF-A to repress transcription. Finally, the Bmal1 gene is transcribed with accurate circadian rhythm. Gray ovals, nucleosomes; yellow region, the nuclear matrix-like structure.
Oscillatory regulation of Bmal1 transcription at the chromatin level.
We discovered that the chromatin structure undergoes rhythmic alterations in vivo at the region around the ROREs and at the 3′-flanking region in response to SAF-A binding (Fig. 9). These data indicate cooperative alteration of the chromatin structure between the 3′-flanking region and the ROREs. Despite the critical function of tandem ROREs for Bmal1 oscillation, how individual ROREs play key roles at the chromatin level remains obscure, although Yin and Lazar indicated from the results of transient reporter gene assays that both ROREs are required (44). Here, we demonstrated rhythmic chromatin alterations immediately upstream of the distal RORE in vivo, suggesting that at least the distal RORE is required for Bmal1's oscillatory transcription in vivo. Figure 10 summarizes a model of the regulation of Bmal1 oscillation.
Physiological circadian rhythms are features of organisms ranging from bacteria to humans and represent robustness in response to environmental changes. To adapt circadian rhythms to various environments, lower organisms regulate gene expression by arranging chromosomal compaction (38) and modifying DNA topology (17, 33). In mammals, Bmal1 is a key player in the circadian clock system (6), and chromatin dynamics are involved in physiological circadian rhythms. The regulation of Bmal1 transcription through altering the chromatin structure observed herein might represent evolutionary progress of the architectural regulation of gene expression.
Acknowledgments
We thank G. Dreyfuss (University of Pennsylvania) for providing the anti-hnRNP-U antibody (19). We are grateful to K. Tsutsui (Okayama University) for helpful discussions.
This work was supported by operational subsidies from AIST and by grants from the Ministry of Education, Culture, Sports, Science and Technology and from the Mitsubishi Chemical Corporation.
Footnotes
Published ahead of print on 10 March 2008.
REFERENCES
- 1.Akashi, M., and T. Takumi. 2005. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 12441-448. [DOI] [PubMed] [Google Scholar]
- 2.Belden, W. J., J. J. Loros, and J. C. Dunlap. 2006. CLOCK leaves its mark on histones. Trends Biochem. Sci. 31610-613. [DOI] [PubMed] [Google Scholar]
- 3.Belden, W. J., J. J. Loros, and J. C. Dunlap. 2007. Execution of the circadian negative feedback loop in Neurospora requires the ATP-dependent chromatin-remodeling enzyme CLOCKSWITCH. Mol. Cell 25587-600. [DOI] [PubMed] [Google Scholar]
- 4.Bryan, P. N., H. Hofstetter, and M. L. Birnstiel. 1981. Nucleosome arrangement on tRNA genes of Xenopus laevis. Cell 27459-466. [DOI] [PubMed] [Google Scholar]
- 5.Buijs, R. M., and A. Kalsbeek. 2001. Hypothalamic integration of central and peripheral clocks. Nat. Rev. Neurosci. 2521-526. [DOI] [PubMed] [Google Scholar]
- 6.Bunger, M. K., L. D. Wilbacher, S. M. Moran, C. Clendenin, L. A. Radcliffe, J. B. Hogenesch, M. C. Simon, J. S. Takahashi, and C. A. Bradfield. 2000. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell 1031009-1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cockerill, P. N., and W. T. Garrard. 1986. Chromosomal loop anchorage of the kappa immunoglobulin gene occurs next to the enhancer in a region containing topoisomerase II sites. Cell 44273-282. [DOI] [PubMed] [Google Scholar]
- 8.Curtis, A. M., S. B. Seo, E. J. Westgate, R. D. Rudic, E. M. Smyth, D. Chakravarti, G. A. FitzGerald, and P. McNamara. 2004. Histone acetyltransferase-dependent chromatin remodeling and the vascular clock. J. Biol. Chem. 2797091-7097. [DOI] [PubMed] [Google Scholar]
- 9.Doi, M., J. Hirayama, and P. Sassone-Corsi. 2006. Circadian regulator CLOCK is a histone acetyltransferase. Cell 125497-508. [DOI] [PubMed] [Google Scholar]
- 10.Dunlap, J. C. 1999. Molecular bases for circadian clocks. Cell 96271-290. [DOI] [PubMed] [Google Scholar]
- 11.Eggert, M., J. Michel, S. Schneider, H. Bornfleth, A. Baniahmad, F. O. Fackelmayer, S. Schmidt, and R. Renkawitz. 1997. The glucocorticoid receptor is associated with the RNA-binding nuclear matrix protein hnRNP U. J. Biol. Chem. 27228471-28478. [DOI] [PubMed] [Google Scholar]
- 12.Etchegaray, J.-P., C. Lee, P. A. Wade, and S. M. Reppert. 2003. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature 421177-182. [DOI] [PubMed] [Google Scholar]
- 13.Etchegaray, J.-P., X. Yang, J. P. DeBruyne, A. H. F. M. Peters, D. R. Weaver, T. Jenuwein, and S. M. Reppert. 2006. The polycomb group protein EZH2 is required for mammalian circadian clock function. J. Biol. Chem. 28121209-21215. [DOI] [PubMed] [Google Scholar]
- 14.Guillaumond, F., H. Dardente, V. Giguere, and N. Cermakian. 2005. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J. Biol. Rhythms 20391-403. [DOI] [PubMed] [Google Scholar]
- 15.Hsu, M.-C., C.-C. Huang, K.-B. Choo, and C.-J. Huang. 2007. Uncoupling of promoter methylation and expression of Period1 in cervical cancer cells. Biochem. Biophys. Res. Commun. 360257-262. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda, M., and M. Nomura. 1997. cDNA cloning and tissue-specific expression of a novel basic helix-loop-helix/PAS protein (BMAL1) and identification of alternative spliced variants with alternative translation initiation site usage. Biochem. Biophys. Res. Commun. 233258-264. [DOI] [PubMed] [Google Scholar]
- 17.Ishida, N. 2007. Circadian clock, cancer and lipid metabolism. Neurosci. Res. 57483-490. [DOI] [PubMed] [Google Scholar]
- 18.Ishida, N., M. Kaneko, and R. Allada. 1999. Biological clocks. Proc. Natl. Acad. Sci. USA 968819-8820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kiledjian, M., and G. Dreyfuss. 1992. Primary structure and binding activity of the hnRNP U protein: binding RNA through RGG box. EMBO J. 112655-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, T.-D., J.-S. Kim, J. H. Kim, J. Myung, H.-D. Chae, K.-C. Woo, S. K. Jang, D.-S. Koh, and K.-T. Kim. 2005. Rhythmic serotonin N-acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol. Cell. Biol. 253232-3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li, L. C., and R. Dahiya. 2002. MethPrimer: designing primers for methylation PCRs. Bioinformatics 181427-1431. [DOI] [PubMed] [Google Scholar]
- 22.Liu, C., S. Li, T. Liu, J. Borjigin, and J. D. Lin. 2007. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nature 447477-482. [DOI] [PubMed] [Google Scholar]
- 23.Naruse, Y., K. Oh-hashi, N. Iijima, M. Naruse, H. Yoshioka, and M. Tanaka. 2004. Circadian and light-induced transcription of clock gene Per1 depends on histone acetylation and deacetylation. Mol. Cell. Biol. 246278-6287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohno, T., Y. Onishi, and N. Ishida. 2007. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res. 35648-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oishi, K., H. Fukui, and N. Ishida. 2000. Rhythmic expression of BMAL1 mRNA is altered in Clock mutant mice: differential regulation in the suprachiasmatic nucleus and peripheral tissues. Biochem. Biophys. Res. Commun. 268164-171. [DOI] [PubMed] [Google Scholar]
- 26.Onishi, Y., S. Hashimoto, and H. Kizaki. 1998. Cloning of the TIS gene suppressed by topoisomerase inhibitors. Gene 215453-459. [DOI] [PubMed] [Google Scholar]
- 27.Onishi, Y., M. Kato, and Y. Hanyu. 2004. Preparation and characterization of an anti-DNA monoclonal antibody showing size selectivity toward DNA fragments. Hybrid. Hybridomics 23311-317. [DOI] [PubMed] [Google Scholar]
- 28.Onishi, Y., and R. Kiyama. 2001. Enhancer activity of HS2 of the human β-LCR is modulated by distance from the key nucleosome. Nucleic Acids Res. 293448-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Onishi, Y., and R. Kiyama. 2003. Interaction of NF-E2 in the human β-globin locus control region before chromatin remodeling. J. Biol. Chem. 2788163-8171. [DOI] [PubMed] [Google Scholar]
- 30.Onishi, Y., Y. Wada-Kiyama, and R. Kiyama. 1998. Expression-dependent perturbation of nucleosomal phases at HS2 of the human β-LCR: possible correlation with periodic bent DNA. J. Mol. Biol. 284989-1004. [DOI] [PubMed] [Google Scholar]
- 31.Preitner, N., F. Damiola, L. Lopez-Molina, J. Zakany, D. Duboule, U. Albrecht, and U. Schibler. 2002. The orphan nuclear receptor REV-ERBα controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell 110251-260. [DOI] [PubMed] [Google Scholar]
- 32.Romig, H., F. O. Fackelmayer, A. Renz, U. Ramsperger, and A. Richter. 1992. Characterization of SAF-A, a novel nuclear DNA binding protein from HeLa cells with high affinity for nuclear matrix/scaffold attachment DNA elements. EMBO J. 113431-3440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Salvador, M. L., U. Klein, and L. Bogorad. 1998. Endogenous fluctuations of DNA topology in the chloroplast of Chlamydomonas reinhardtii. Mol. Cell. Biol. 187235-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sato, T., S. Panda, L. Miraglia, T. Reyes, R. Rudic, P. McNamara, K. Naik, G. FitzGerald, S. Kay, and J. Hogenesch. 2004. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron 43527-537. [DOI] [PubMed] [Google Scholar]
- 35.Shearman, L. P., S. Sriram, D. R. Weaver, E. S. Maywood, I. Chaves, B. Zhen, K. Kume, C. C. Lee, T. J. van der Horst, M. H. Hastings, and S. M. Reppert. 2000. Interacting molecular loops in the mammalian circadian clock. Science 2881013-1019. [DOI] [PubMed] [Google Scholar]
- 36.Sivolob, A. V., and S. N. Khrapunov. 1995. Translational positioning of nucleosomes on DNA: the role of sequence-dependent isotropic DNA bending stiffness. J. Mol. Biol. 247918-931. [DOI] [PubMed] [Google Scholar]
- 37.Small, D., B. Nelkin, and B. Vogelstein. 1985. The association of transcribed genes with the nuclear matrix of Drosophila cells during heat shock. Nucleic Acids Res. 132413-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith, R. M., and S. Williams. 2006. Circadian rhythms in gene transcription imparted by chromosome compaction in the cyanobacterium Synechococcus elongatus. Proc. Natl. Acad. Sci. USA 1038564-8569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strahl, B. D., and C. D. Allis. 2000. The language of covalent histone modifications. Nature 40341-45. [DOI] [PubMed] [Google Scholar]
- 40.Tamaru, T., Y. Isojima, K. Nagai, and K. Takamatsu. 2003. Circadian expression of hnRNP U, a nuclear multi-potent regulatory protein, in the suprachiasmatic nucleus. Neurosci. Lett. 341111-114. [DOI] [PubMed] [Google Scholar]
- 41.Tazi, J., and A. Bird. 1990. Alternative chromatin structure at CpG islands. Cell 60909-920. [DOI] [PubMed] [Google Scholar]
- 42.Wolf, S. F., and B. R. Migeon. 1985. Clusters of CpG dinucleotides implicated by nuclease hypersensitivity as control elements of housekeeping genes. Nature 314467-469. [DOI] [PubMed] [Google Scholar]
- 43.Wyrick, J. J., F. C. Holstege, E. G. Jennings, H. C. Causton, D. Shore, M. Grunstein, E. S. Lander, and R. A. Young. 1999. Chromosomal landscape of nucleosome-dependent gene expression and silencing in yeast. Nature 402418-421. [DOI] [PubMed] [Google Scholar]
- 44.Yin, L., and M. A. Lazar. 2005. The orphan nuclear receptor Rev-erbα recruits the N-CoR/histone deacetylase 3 corepressor to regulate the circadian Bma1 gene. Mol. Endocrinol. 191452-1459. [DOI] [PubMed] [Google Scholar]
- 45.Yoo, S.-H., C. H. Ko, P. L. Lowrey, E. D. Buhr, E.-J. Song, S. Chang, O. J. Yoo, S. Yamazaki, C. Lee, and J. S. Takahashi. 2005. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc. Natl. Acad. Sci. USA 1022608-2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young, M. W., and S. A. Kay. 2001. Time zones: a comparative genetics of circadian clocks. Nat. Rev. Genet. 2702-715. [DOI] [PubMed] [Google Scholar]
- 47.Yu, W., M. Nomura, and M. Ikeda. 2002. Interactivating feedback loops within the mammalian clock: BMAL1 is negatively autoregulated and upregulated by CRY1, CRY2, and PER2. Biochem. Biophys. Res. Commun. 290933-941. [DOI] [PubMed] [Google Scholar]
- 48.Yugami, M., Y. Kabe, Y. Yamaguchi, T. Wada, and H. Handa. 2007. hnRNP-U enhances the expression of specific genes by stabilizing mRNA. FEBS Lett. 5811-7. [DOI] [PMC free article] [PubMed] [Google Scholar]