Abstract
All nucleated cells make phosphatidylcholine via the CDP-choline pathway. Liver has an alternative pathway in which phosphatidylcholine is made by methylation of phosphatidylethanolamine catalyzed by phosphatidylethanolamine N-methyltransferase (PEMT). We investigated the function of PEMT and its role in animal physiology by targeted disruption of its gene, Pempt2. A targeting vector that interrupts exon 2 was constructed and introduced into mice yielding three genotypes: normal (+/+), heterozygotes (+/−), and homozygotes (−/−) for the disrupted PEMT gene. Only a trace of PE methylation activity remained in Pempt2(−/−) mice. Antibody to one form of the enzyme, PEMT2, indicated complete loss of this protein from Pempt2(−/−) mice and a decrease in Pempt2(+/−) mice, compared with Pempt2(+/+) mice. The levels of hepatic phosphatidylethanolamine and phosphatidylcholine were minimally affected. The active form of CTP:phosphocholine cytidylyltransferase, the regulated enzyme in the CDP-choline pathway, was increased 60% in the PEMT-deficient mice. Injection of [l-methyl-3H]methionine demonstrated that the in vivo PEMT activity was eliminated in the Pempt2(−/−) mice and markedly decreased in the Pempt2(+/−) mice. This experiment also demonstrated that the choline moiety derived from PEMT in the liver can be distributed via the plasma throughout the mouse where it is found as phosphatidylcholine, lysophosphatidylcholine, and sphingomyelin. Mice homozygous for the disrupted Pempt2 gene displayed no abnormal phenotype, normal hepatocyte morphology, normal plasma lipid levels and no differences in bile composition. This is the first application of the “knockout mouse” technique to a gene for phospholipid biosynthesis.
Phosphatidylcholine (PC), the most abundant mammalian phospholipid, is synthesized in all nucleated cells via a three step process: phosphorylation of intracellular choline, conversion of phosphocholine to CDP-choline, which reacts with diacylglycerol to form PC (1). A second pathway for the biosynthesis of PC is the methylation of phosphatidylethanolamine (PE). This pathway, first elucidated in 1961, occurs in mammals to a significant degree only in the liver (2, 3). S-adenosylmethionine serves as the methyl group donor for the three reactions that convert the ethanolamine head group of PE to choline via two lipid intermediates: phosphatidylmonomethylethanolamine (PMME) and phosphatidyldimethylethanolamine (PDME). Unlike yeast, which use two distinct enzymes to convert PE to PC, only one enzyme in mammals, phosphatidylethanolamine N-methyltransferase (PEMT), is necessary for the complete conversion of PE to PC (4–6).
PEMT was first purified from rat liver in 1987, and shown to be single enzyme of 19 kDa (5). Using oligonucleotides based on the N-terminal amino acid sequence of the purified protein, a cDNA encoding PEMT was cloned and expressed (6). Interestingly, the cDNA encoded a protein that, when detected by an anti-C-terminal peptide antibody, differed in its subcellular localization from that of the majority of PEMT activity. This result led to the hypothesis that two isoforms of PEMT exist: PEMT1, localized to the endoplasmic reticulum and generating the majority of PEMT activity, and PEMT2, corresponding to the cloned cDNA, which resides on mitochondria-associated membranes (6, 7). Both enzymes are capable of catalyzing all three methylation steps. The difference between these two isoforms is currently being investigated.
The structure and chromosomal localization of the mouse PEMT2 gene has been determined (8). The gene consists of seven exons between 75- and 260-bp long, spread over at least 35 kb, with the translation start site encoded by the second exon. Interspecific backcross mapping placed the gene, named Pempt2, on mouse chromosome 11, ≈31 cM from the centromere.
The physiological role of PE methylation in the liver is unclear. The majority of PC in the liver is synthesized via the CDP-choline pathway (also commonly called the Kennedy pathway), with PEMT generating 20–40% of liver PC (9). One obvious function for PEMT would be the endogenous generation of choline, which could be used for PC biosynthesis via the CDP-choline pathway, for the biosynthesis of the neurotransmitter acetylcholine, and as a source of betaine, which supplies methyl groups to the 1-carbon pool. As well, PEMT-derived PC may be specifically targeted for secretion, either with bile or with lipoproteins. Initial studies suggest that PEMT-derived PC is secreted with lipoproteins, but PEMT activity is not strictly required (10). However, inhibition of PE methylation with fibrates in primary rat hepatocytes alters lipidation of apoB48-containing lipoproteins (11). More recently, a role for PEMT in the regulation of liver growth has been suggested. When hepatocytes undergo non-neoplastic division after injection of lead nitrate, during embryonic development, and following partial hepatectomy, PEMT expression is decreased (12–14).
Targeted disruption of gene(s) encoding PEMT in mice seemed a particularly suitable approach to providing fundamental insight into the function of PEMT, since PEMT is not expressed in the liver until birth (13), hence embryonic lethality would not be expected. As well, the liver retains the CDP-choline pathway, which we hypothesized could compensate for the loss of PEMT by generating sufficient PC for hepatocyte viability. Because PEMT is not detectable in other tissues, serious nonhepatic consequences would not be expected.
We now report the construction of mice in which Pempt2 has been disrupted. No obvious pathophysiological phenotype developed in these mice up to 10 months old. Mice lacking Pempt2 expression breed normally. Thus it is apparent that this gene is not required for normal growth and reproduction. This is the first report of the disruption of a phospholipid biosynthetic gene in any animal species.
MATERIALS AND METHODS
Generation of PEMT-Deficient Mice.
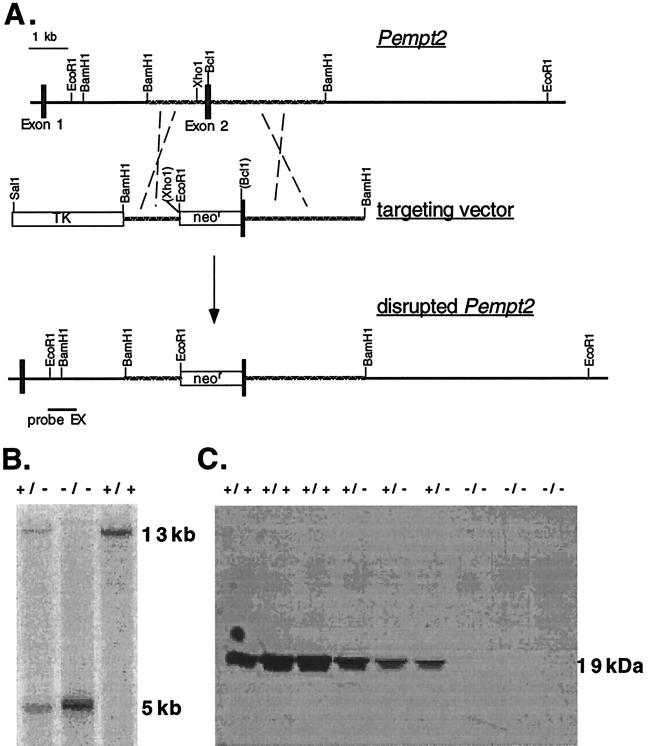
A sequence-replacement type gene targeting vector was constructed from a 6-kb BamHI subclone of λ clone 3 containing exon 2 of the mouse Pempt2 gene (8). A cassette containing a neomycin-resistance gene (neo) driven by the pgk promoter was inserted at unique XhoI and BclI sites in the subclone, replacing the majority of exon 2, including the translation start site, and the extreme 5′ end of intron 1 (Fig. 1). Therefore, the long arm of the vector consisted of the 3′ end of exon 2 and the 5′ end of intron 2. The short arm consisted of the remaining 3′ end of intron 1. A thymidine kinase (tk) gene was ligated to the 5′ end of the targeting vector. The vector was electroporated into E14 embryonic stem cells (15). Homologous recombinants were identified by Southern blot analysis of EcoRI-cleaved genomic DNA, using a 1-kb EcoRI-XhoI fragment from intron 1, 5′ to the targeting vector, as a probe. Identifications were confirmed by probing with an 800-bp HindIII-KpnI fragment 3′ to the targeting vector. One Pempt2 gene-disrupted clone was used to generate PEMT2-deficient mice using standard techniques (16).
Figure 1.
(A) The normal Pempt2 gene, the gene targeting vector, and the disrupted Pempt2 allele. The disrupted allele was identified by Southern blot analysis of EcoRI-digested DNA, using the 5′ probe EX. (B) Genotyping of mice by Southern blot analysis. The normal Pempt2 allele yielded a 13-kb EcoRI fragment detected by probe EX, while the disrupted allele yielded a 5-kb EcoRI fragment. (C) Absence of PEMT2 protein in Pempt2 knockout mice. Immunoblot analysis of liver homogenates demonstrated that PEMT2 protein is present only in mice carrying at least one normal Pempt2 allele. Each lane contained 50 μg protein. PEMT2 has an apparent molecular mass of 19 kDa.
Tissue Collection.
Mice were sacrificed following an overnight fast, and liver and other organs were removed. Tissues were cut into small pieces with scissors, then homogenized with a glass-Teflon homogenizer in five volumes of 50 mM Tris⋅HCl (pH 7.4), 150 mM NaCl, 1.0 mM phenylmethylsulfonyl fluoride, 1.0 mM EDTA, 2.0 mM DTT, 0.025% sodium azide. Plasma was isolated by centrifugation following blood collection by cardiac puncture, with EDTA as an anticoagulant.
Determination of Enzyme Mass and Activity.
Total protein mass in homogenates was measured using the BCA (bicinchoninic acid) protein assay (Pierce), with BSA as standard. To determine enzyme mass, homogenates were boiled for 5 min in Laemmli buffer (17), then separated electrophoretically on a 12% polyacrylamide gel containing 0.1% SDS (50 μg protein per lane). Proteins were transferred to Immobilon-P membranes (Millipore), and probed with an anti-PEMT2-specific antibody (6). Bands were visualized using an enhanced chemiluminescence system following manufacturer’s instructions (Amersham).
Homogenates were centrifuged at 600 × g for 5 min to remove unbroken cells, and the supernatant was assayed for PEMT activity (18). Methylated, chloroform-soluble products were separated by thin-layer chromatography on silica gel G60 plates with chloroform/propionic acid/n-propanol/water (30/20/60/10) as solvent. Bands were visualized by iodine vapor, and those corresponding to methylated PE products (PMME, PDME, and PC) were scraped and radioactivity measured.
For CTP:phosphocholine cytidylyltransferase (CT) activity measurements, soluble and membrane fractions of the 600 × g supernatant were separated by centrifugation at 350,000 × g for 15 min. Fractions were assayed in the presence of PC/oleate vesicles, as described (19), with modifications (20).
Phospholipid Analyses.
Phospholipids were isolated from total homogenates by the method of Bligh and Dyer (21), then separated by thin-layer chromatography with chloroform/methanol/acetic acid/water (50/30/8/4) as the solvent. Bands were visualized with iodine vapor, then scraped. The phosphorous content was then measured (22).
In Vivo Metabolism of [3H]Methionine.
[l-methyl-3H]-labeled methionine (62.5 μCi; 1 Ci = 37 GBq) was diluted to a final volume of 200 μl with PBS, then injected into mice through the tail vein. After 5 min, 1 h, and 24 h, mice were sacrificed, organs and plasma were harvested as described above, and choline-containing phospholipids (PC, lyso-PC, and sphingomyelin) isolated by thin-layer chromatography. Phospholipids were analyzed for mass and radiolabel incorporation.
RESULTS
Generation of Pempt2-Deficient Mice.
A gene targeting vector was designed and constructed to interrupt exon 2 of the Pempt2 gene, including the translation start site (Fig. 1A). The vector was electroporated into embryonic stem cells, and seven targeted embryonic stem cell clones out of 131 neomycin-resistant clones were identified by Southern blot analysis. One of these clones was used to generate chimeric mice (strains 129/J and C57BL/6) capable of germline transmission of the altered Pempt2 allele. The chimeric mice were bred with C57BL/6 females to generate mice heterozygous for the disrupted Pempt2 gene. These mice were interbred to generate all three genotypes: Pempt2(+/+) carrying two copies of the normal allele, Pempt2(+/−) carrying one normal and one disrupted allele, and Pempt2(−/−), carrying two copies of the disrupted allele (Fig. 1B). From 110 offspring, 32 Pempt2(+/+), 54 Pempt2(+/−), and 24 Pempt2(−/−) were identified. This result is close to the expected Mendelian ratio of 1:2:1, suggesting that disruption of Pempt2 does not affect gamete viability.
PEMT2 Is Absent from Pempt2(−/−) Mice.
We investigated whether or not disruption of the Pempt2 gene eliminated expression of PEMT2 protein. Immunoblot analysis was performed on liver homogenates from mice, using an antibody against the C terminus of rat PEMT2 (6). The 19-kDa protein was present in Pempt2(+/+) and (+/−) mice, but completely absent from Pempt2(−/−) mice (Fig. 1C). The band intensity appeared reduced in heterozygotes compared with normal mice, suggesting a gene dosage effect.
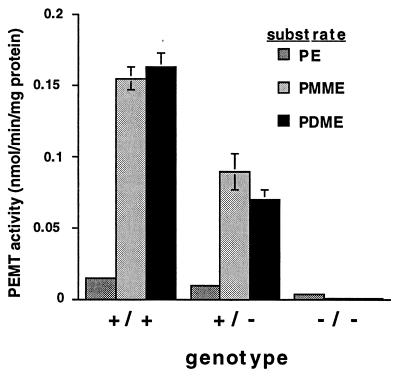
PEMT activity was measured in liver homogenates from mice of all three genotypes. With PMME and PDME as substrates, PEMT activity in the homozygous knockout mice was <0.3% of that seen in normal mice (Fig. 2). Activity in heterozygotes was approximately half that of wild-type mice, suggesting a gene dosage effect on activity. This result demonstrates that both PEMT1 and PEMT2 are encoded by Pempt2. With PE as substrate, PEMT activity of the normal mice was ≈10% of that against PMME and PDME. It has been reported (5) that PE is a relatively poor substrate for PEMT compared with PMME and PDME. However, homozygous knockout mice retained ≈20% of the PE methylation activity seen in their normal littermates. The importance of this residual activity remains to be determined.
Figure 2.
Elimination of methylation activity of PE, PMME, and PDME in Pempt2-disrupted mice. Total liver homogenates from mice of all three genotypes were assayed for PEMT activity. For all three genotypes, n = 4. Bars = SD.
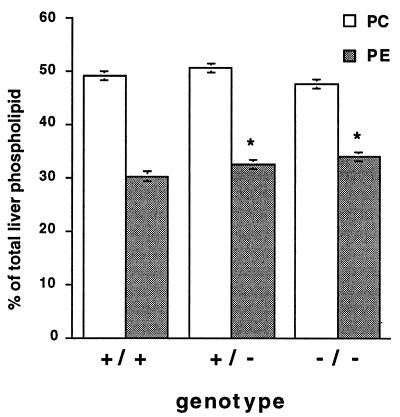
PC and PE Levels in Liver Are Minimally Affected by Elimination of Pempt2.
Lipids were extracted from liver homogenates, separated by thin-layer chromatography, and quantitated. The PC content was not significantly altered, and PE levels were raised slightly, from 30.2% of total phospholipids in Pempt2(+/+) mouse livers, to 32.4% in Pempt2(+/−) mouse livers, to 33.9% in Pempt2(−/−) mouse livers (Fig. 3). The level of total liver phospholipids did not change significantly according to genotype, varying from 83.8 ± 1.8 nmol/mg protein in Pempt2(+/+) mice, to 82.7 ± 4.2 nmol/mg protein Pempt2(+/−) mice to 86.3 ± 3.9 nmol/mg protein in Pempt2(−/−) mice. As well, levels of other phospholipid species were unaffected.
Figure 3.
Phospholipid composition of livers from mice of all three genotypes. Levels of PC and PE in liver homogenates were measured and expressed relative to total phospholipid. In all cases, n = 4. Bars = SD. *P < 0.05 compared with (+/+).
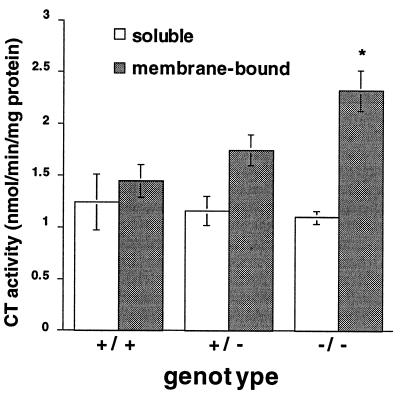
CT Activity Is Increased in Livers of Pempt2-Disrupted Mice.
To determine whether the CDP-choline pathway for PC biosynthesis is stimulated to maintain normal PC levels in PEMT-deficient mice, the activity of CT, which catalyzes the rate limiting step in this pathway (23–25), was measured. The enzyme exists in two forms: a soluble, inactive form and a membrane-bound, active form. In livers of Pempt2(−/−) mice, membranous CT activity was increased by 60% (P < 0.05), from 1.45 nmol/min/mg protein in normal mice to 2.32 nmol/min/mg protein in homozygous knockouts (Fig. 4). Heterozygotes showed intermediate values. Thus, it appears that mice compensate for the loss of PEMT activity by stimulation of the CDP-choline pathway to maintain liver PC levels.
Figure 4.
Membrane-bound CT activity was increased in Pempt2 knockout mice. CT activity was measured in soluble and membrane-bound fractions of liver homogenates. In all cases n = 4. Bars = SD. *P < 0.05 compared with (+/+).
Incorporation of [3H]Methionine into Choline-Containing Phospholipids.
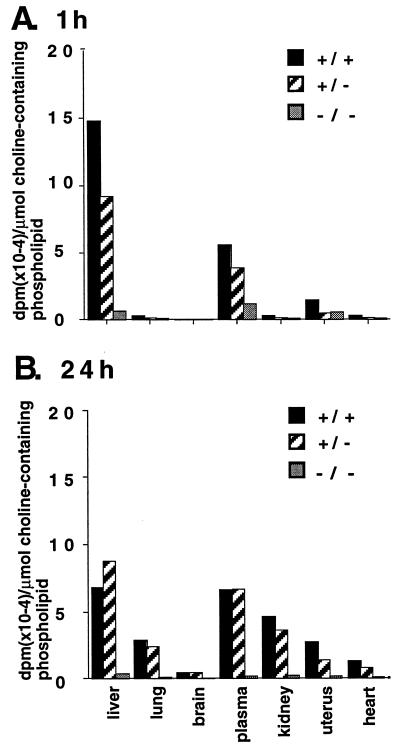
We investigated the extent to which the conversion of PE to PC in vivo was impaired in Pempt2(+/−) and (−/−) mice. Mice of each genotype were injected with [l-methyl-3H]methionine via the tail vein. Organs and plasma were harvested either 5 min, 1 h, or 24 h after injection, and radiolabeling of PC, lysoPC, and sphingomyelin was measured. Labeled PC (90,000 dpm/μmol choline-containing phospholipid) was recovered from the liver, but not plasma of the Pempt2(+/+) mouse 5 min. after injection. Less labeled PC (30929 dpm/μmol choline-containing phospholipid) was recovered from the liver of the Pempt2(+/−) mouse, and the Pempt2(−/−) mouse contained <1% (738 dpm/mmol choline-containing phospholipid) of label associated with PC in its liver compared with the Pempt2(+/+) mouse. After 1 h, the level of radiolabeled choline-containing phospholipids in liver was highest in the Pempt2(+/+) mouse (Fig. 5A), less in the Pempt2(+/−) mouse (62.0% of +/+), and much less in the Pempt2(−/−) mouse (4.4% of +/+). This result correlates with the pattern of PEMT activity in liver homogenates (Fig. 2). In all genotypes, >85% of radioactivity was in PC, with minor amounts in lysoPC and sphingomyelin. After 1 h, the plasma also contained significant levels of radiolabeled choline-containing phospholipids, primarily PC (79.9%) and lysoPC (20.1%), suggesting that PC and lysoPC (with a choline moiety derived from PE methylation) are secreted from the liver into the bloodstream. The level of radiolabeled choline-containing phospholipids in other tissues was minimal, consistent with PEMT being a liver-specific enzyme.
Figure 5.
Incorporation of radiolabeled methionine into choline-containing phospholipids. [l-methyl-3H]methionine was injected into the bloodstream of mice of each of the three genotypes. Plasma and organs were harvested after 1 h (A) or 24 h (B). Phospholipids were extracted and separated by thin-layer chromatography. Bands corresponding to choline-containing phospholipids (PC, lyso PC, and sphingomyelin) were scraped, and radiolabel incorporation measured, as well as total lipid phosphorous. Results are expressed as dpm per μmol choline-containing phospholipid. The mouse homozygous for the disrupted Pempt2 allele had only 4.4% the level of radiolabeled methionine incorporation into choline-containing phospholipid seen in the normal mouse after 1 h, and 4.9% the level of incorporation after 24 h.
Between 1 and 24 h after injection of [l-methyl-3H]methionine, the level of labeled PC, lysoPC, and sphingomyelin in the liver of the Pempt2(+/+) mouse decreased, falling slightly below that in the Pempt2(+/−) (Fig. 5B). Concurrently, the level of radiolabeled choline-containing phospholipids in other organs examined increased significantly, in both normal and heterozygous mice. At all times, incorporation of label into brain phospholipids was minimal. This result suggests that choline derived from PE methylation in the liver is distributed throughout the body following secretion, where it is found in PC, lysoPC, and sphingomyelin. Again, the level of radiolabeled choline-containing phospholipids was low in all tissues of the Pempt2(−/−) mouse.
Our ability to trace the incorporation of [3H]methyl groups into aqueous choline is compromised by the promiscuity of methionine as a precursor for other water-soluble compounds. This fact renders the isolation of pure labeled choline and phosphocholine by thin-layer chromatography impossible.
Mice Homozygous for the Disruption of Pempt2 Display No Obvious Physiological Defects.
Mice homozygous for the disrupted Pempt2 allele displayed no obvious pathophysiological or behavioral defects, nor did they appear to have a reduced life span or altered body weight. Breeding ability of Pempt2(−/−) was indistinguishable from Pempt2(+/+) and (+/−) mice. Histological examination of liver sections from adult mice revealed no obvious changes in hepatocyte morphology. In animals maintained under normal laboratory conditions, there were no significant changes in lipoprotein profile, nor in plasma triacylglycerol, cholesterol or phospholipid levels. Finally, there were no significant differences in the bile acid or phospholipid concentrations in the bile, nor in the volume of bile collected from the gallbladder.
DISCUSSION
Using well-established gene targeting techniques, we generated mice carrying a disrupted allele of the Pempt2 gene. This represents the first application of the “knockout” mouse technique to a gene for phospholipid biosynthesis. Mice carrying two copies of the disrupted allele lacked not only PEMT2, but virtually all methylation activity against the lipid substrate intermediates PMME and PDME. This result indicates that we have also eliminated PEMT1, the endoplasmic reticulum form of PEMT. Therefore, PEMT1 and PEMT2 are encoded by the same gene, Pempt2. The difference between these two forms of PEMT, which results in their divergent subcellular localizations, must be generated posttranscriptionally. Possible differences in posttranscriptional and posttranslational modifications between PEMT1 and PEMT2, including RNA editing, alternative splicing, phosphorylation, glycosylation, and fatty acylation, will be the focus of future study. Normal mice incorporated [l-methyl-3H]methionine into PC within 5 min of tail vein injection. However, mice homozygous for the null allele had a greatly reduced ability (<1% of wild type) to incorporate [l-methyl-3H]methionine into choline-containing phospholipids, further confirming the elimination of PEMT activity. Mice heterozygous for the null allele had ≈50% of the PEMT activity of normal mice, suggesting a gene dosage effect, without increased expression of the remaining functional copy of the Pempt2 gene as compensation.
Mice homozygous for the disruption of Pempt2 retained a low level of methylation activity against the initial lipid substrate PE. Therefore, it appears that a second PE methylation activity might exist in mice, with lower activity against PMME and PDME. We are investigating whether this activity is responsible for the radiolabel associated with PC generated in Pempt2(−/−) mice following [l-methyl-3H]methionine injection.
Mice deficient in PEMT activity showed very little change in their liver phospholipid composition. Despite the fact that PE methylation has been hypothesized to generate between 20–40% of liver PC (9), the Pempt2(−/−) mice showed no significant change in either PC (nmol/μg protein) or PC contribution to total liver phospholipid, compared with normal mice. PE, the initial substrate for methylation, was slightly elevated in the livers of mice lacking PEMT activity. Thus, either the PEMT contribution to total liver PC levels has been overestimated, or the CDP-choline pathway is stimulated to compensate for the loss of PEMT activity and to maintain hepatic PC levels. This latter hypothesis was tested by measurement of the activity of CT, the rate limiting step in the CDP-choline pathway. Homozygous knockout mice had 60% higher CT activity bound to membranes compared with normal mice. Since the membrane-bound form of CT is the active form, we hypothesize that CT is stimulated to compensate for the loss of PEMT activity. Previous studies also demonstrate a reciprocal regulation of PEMT and CT activities (12–14). For example, induction of hepatocyte proliferation increases CT activity while decreasing PEMT activity.
Radiolabeled methionine injected into the bloodstream was incorporated into PC in the liver of a normal mouse within as short a time as 5 min. After 24 h, the radiolabeled choline generated by PE methylation was distributed throughout the body of a normal mouse, showing that PEMT pathway can serve as a source of PC, not just for the liver, but for other tissues as well. However, it remains to be determined whether this PC is carried to other tissues as PC on the surface of lipoproteins, or as lysoPC, or is distributed to other tissues as choline itself, and subsequently incorporated into PC via the CDP-choline pathway. PC could be secreted from the liver directly following its synthesis by PEMT, or generated from the catabolism of PE-derived PC to choline or phosphocholine, which is used for PC synthesis via the CDP-choline pathway.
Mice lacking PEMT that were fed a normal chow diet did not display any abnormal phenotype. Therefore, it appears that the lack of PEMT can be compensated. We have demonstrated that CT activity was increased in the homozygous knockout mice, suggesting that the CDP-choline pathway substitutes for PEMT, and that the PC generated by this pathway is functionally equivalent to PC generated by PEMT. In other words, if PEMT-derived PC were the source of PC in normal mice for a particular cellular function, for example lipoprotein and bile secretion, PC from the CDP-choline pathway appears to be able to fulfill the same roles in Pempt2(−/−) mice. The question of whether PC generated by the two pathways is functionally equivalent has been studied (6, 26–28). Rodents fed a diet deficient in choline survive, apparently because PC and choline are generated by PEMT, whose expression in the liver is increased (26). As well, hepatoma cell lines that completely lack PEMT expression, and primary hepatocytes exposed to PEMT inhibitors, survive with PC generated via the CDP-choline pathway (6, 27). Conversely, Chinese hamster ovary cells carrying a lethal temperature-sensitive mutation in the CT gene cannot be rescued by overexpression of PEMT2 cDNA, despite the restoration of PC levels (28). Therefore, although CT can compensate for the loss of PEMT, the converse does not appear to be true: PEMT cannot compensate for the loss of CT.
The availability of these mice will allow us to address a number of other issues with respect to PEMT and its metabolic function. (i) What would the phenotype be if the mice were fed a choline-deficient diet? (ii) Can dimethylethanolamine or monomethylethanolamine substitute for choline in PEMT-deficient mice? (iii) Is the conversion of choline to betaine or acetylcholine altered in PEMT-deficient mice?
Acknowledgments
The authors thank Charlie Lerner (The Jackson Laboratory) for the culture, transfection, and selection of the embryonic stem cells; Brenda Roszell and Michelle Schrader (University of Alberta) for maintenance of the mouse colony; Sandra Ungarian, Ross Waite, and Liqing Yu (University of Alberta) for expert technical assistance. This work was supported by grants from the Medical Research Council of Canada. C.J.W. was supported by studentships from the Medical Research Council of Canada and the Alberta Heritage Foundation for Medical Research. D.E.V. is a Medical Scientist of the Alberta Heritage Foundation for Medical Research. L.B.A. is a Scholar of the Medical Research Council of Canada and the Alberta Heritage Foundation for Medical Research.
ABBREVIATIONS
- PEMT
phosphatidylethanolamine N-methyltransferase
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PMME
phosphatidylmonomethylethanolamine
- PDME
phosphatidyldimethylethanolamine
- CT
CTP:phosphocholine cytidylyltransferase
Note Added in Proof
The gene name Pempt2 has been changed to Pempt, reflecting the fact that this gene encodes both PEMT1 and PEMT2 isoforms. This change has been registered with the Mouse Genome Database (The Jackson Laboratory, accession no. MGD-MRK-25966).
References
- 1.Kennedy E P. In: Phosphatidylcholine Metabolism. Vance D E, editor. Boca Raton, FL: CRC; 1989. pp. 1–8. [Google Scholar]
- 2.Bremer J, Greenberg D M. Biochim Biophys Acta. 1961;46:205–216. [Google Scholar]
- 3.Ridgway N D. In: Phosphatidylcholine Metabolism. Vance D E, editor. Boca Raton, FL: CRC; 1989. pp. 103–120. [Google Scholar]
- 4.Kodaki T, Yamashita S. J Biol Chem. 1987;262:15428–15435. [PubMed] [Google Scholar]
- 5.Ridgway N D, Vance D E. J Biol Chem. 1987;262:17231–17239. [PubMed] [Google Scholar]
- 6.Cui Z, Vance J E, Chen M H, Voelker D R, Vance D E. J Biol Chem. 1993;268:16655–16663. [PubMed] [Google Scholar]
- 7.Vance J E. J Biol Chem. 1990;265:7248–7256. [PubMed] [Google Scholar]
- 8.Walkey C J, Cui Z, Agellon L B, Vance D E. J Lipid Res. 1996;37:2341–2350. [PubMed] [Google Scholar]
- 9.Sundler R, Åkesson B. J Biol Chem. 1975;250:3359–3367. [PubMed] [Google Scholar]
- 10.Vance J E, Vance D E. J Biol Chem. 1986;261:4486–4491. [PubMed] [Google Scholar]
- 11.Nishimaki-Mogami T, Suzuki K, Takahashi A. Biochim Biophys Acta. 1996;1304:21–31. doi: 10.1016/s0005-2760(96)00100-2. [DOI] [PubMed] [Google Scholar]
- 12.Houweling M, Cui Z, Tessitore L, Vance D E. Biochim Biophys Acta. 1997;1346:1–9. doi: 10.1016/s0005-2760(97)00011-8. [DOI] [PubMed] [Google Scholar]
- 13.Cui Z, Shen Y-J, Vance D E. Biochim Biophys Acta. 1997;1346:10–16. doi: 10.1016/s0005-2760(97)00012-x. [DOI] [PubMed] [Google Scholar]
- 14.Tessitore L, Cui Z, Vance D E. Biochem J. 1997;322:151–154. doi: 10.1042/bj3220151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doetschman T C, Eistetter H, Katz M, Kemler R. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 16.Hogan B, Beddington R, Costantini F, Lacy E. Manipulating the Mouse Embryo: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1994. [Google Scholar]
- 17.Laemmli U K. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 18.Ridgway N D, Vance D E. Methods Enzymol. 1992;209:366–374. doi: 10.1016/0076-6879(92)09045-5. [DOI] [PubMed] [Google Scholar]
- 19.Weinhold P A, Rounsifer M E, Feldman D A. J Biol Chem. 1986;261:5104–5110. [PubMed] [Google Scholar]
- 20.Yao Z, Jamil H, Vance D E. J Biol Chem. 1990;265:4326–4331. [PubMed] [Google Scholar]
- 21.Bligh E G, Dyer W J. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 22.Bartlett G R. J Biol Chem. 1959;234:466–468. [PubMed] [Google Scholar]
- 23.Tijburg L B M, Geelen M J H, van Golde L M G. Biochim Biophys Acta. 1989;1004:1–19. doi: 10.1016/0005-2760(89)90206-3. [DOI] [PubMed] [Google Scholar]
- 24.Vance D E. Biochem Cell Biol. 1990;68:1151–1165. doi: 10.1139/o90-172. [DOI] [PubMed] [Google Scholar]
- 25.Kent C. Prog Lipid Res. 1991;29:87–105. doi: 10.1016/0163-7827(90)90010-i. [DOI] [PubMed] [Google Scholar]
- 26.Cui Z, Vance D E. J Biol Chem. 1996;271:2839–2843. doi: 10.1074/jbc.271.5.2839. [DOI] [PubMed] [Google Scholar]
- 27.Nishimaki-Nogami T, Suzuki K, Okochi E, Takahashi A. Biochim Biophys Acta. 1996;1304:11–20. doi: 10.1016/s0005-2760(96)00101-4. [DOI] [PubMed] [Google Scholar]
- 28.Houweling M, Cui Z, Vance D E. J Biol Chem. 1995;270:16277–16282. doi: 10.1074/jbc.270.27.16277. [DOI] [PubMed] [Google Scholar]